Overlap Graphs
To prepare libraries for this pipeline:
poretools fasta --type 2D <path/to/base/called/reads/> > <name.fasta>bwa mem -x on2d <indexed_reference.fasta> <name.fasta> | samtools view -bS - | samtools sort -o <name.sorted.bam> -samtools depth <name.sorted.bam> > <name.coverage>head <name.coverage># This finds the name of the ‘chromosome’; there may be >1.
awk '$1 == "<chromosomename>" {print $0}' <name.coverage> > chr1.coverage- Repeat for paired library
- Fill in
and into pool1 and pool2 in `depth_coverage.R` - Stats acquired using
poretools stats <path/to/base/called/reads
Reported p20 and p40 values represent percentage of the Zika genome which has at coverage of at least 20/40 reads, respectively.
Overlap graphs and stats on pass libraries: USVI Library 1
NB01-NB07 Overlap
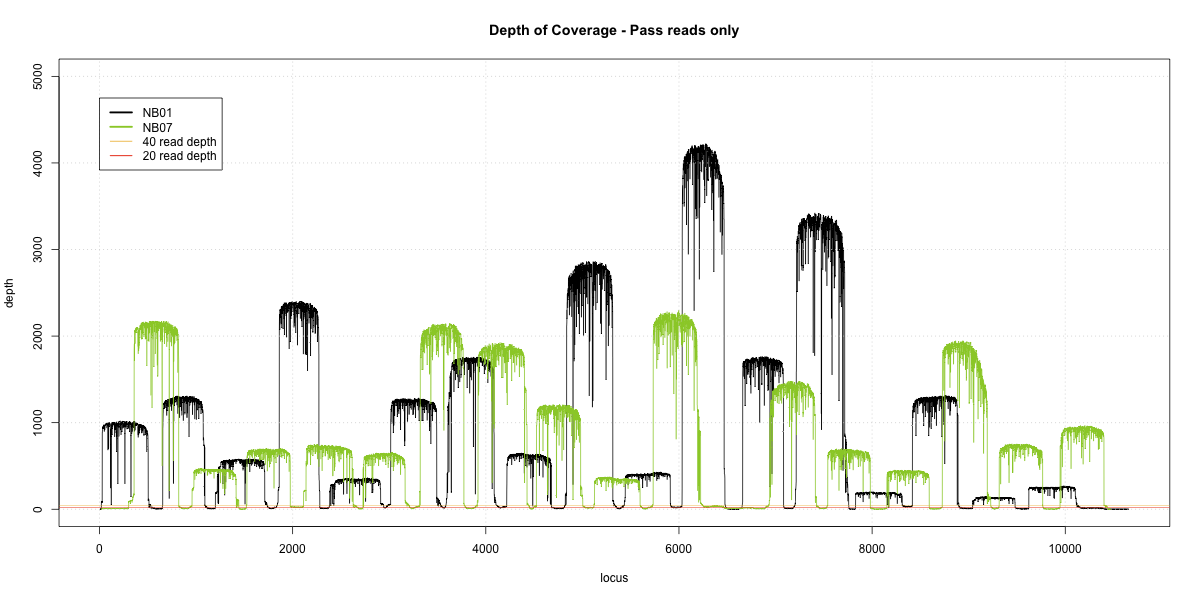
Complete library
- p20: 0.9815 (+1%)
- p40: 0.9758 (+1%)
- Difference: 1%
Incomplete library
- p20: 0.9702
- p40: 0.9671
NB02-NB08 Overlap
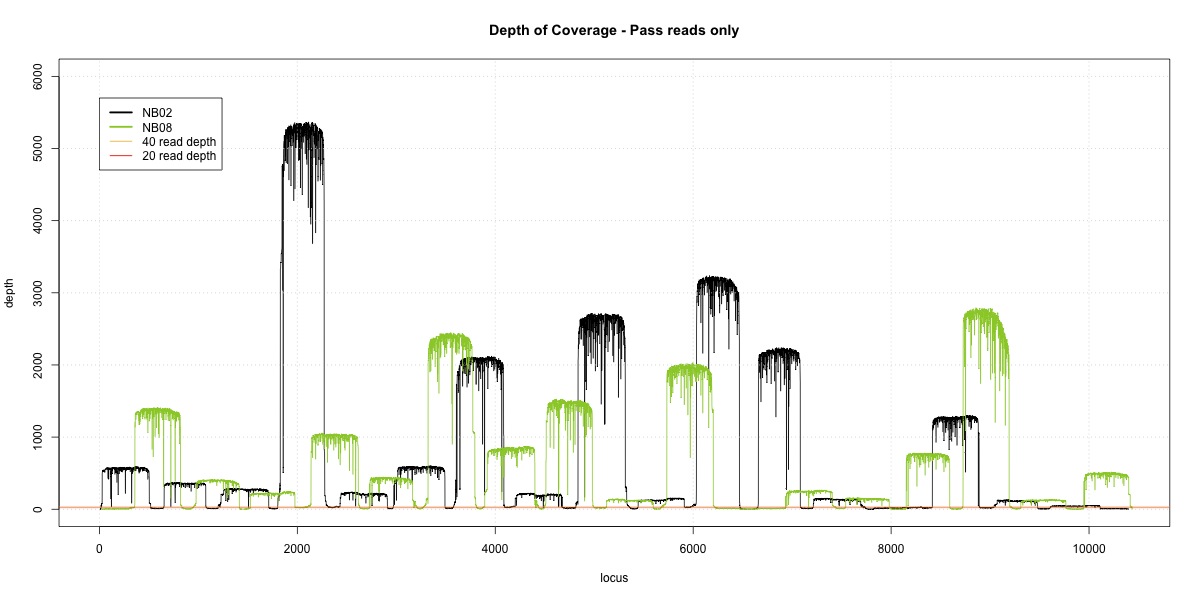
Complete library
- p20: 0.9732 (+13%)
- p40: 0.9631 (+25%)
- Difference: 1%
Incomplete library
- p20: 0.8412
- p40: 0.7145
NB03-NB09 Overlap
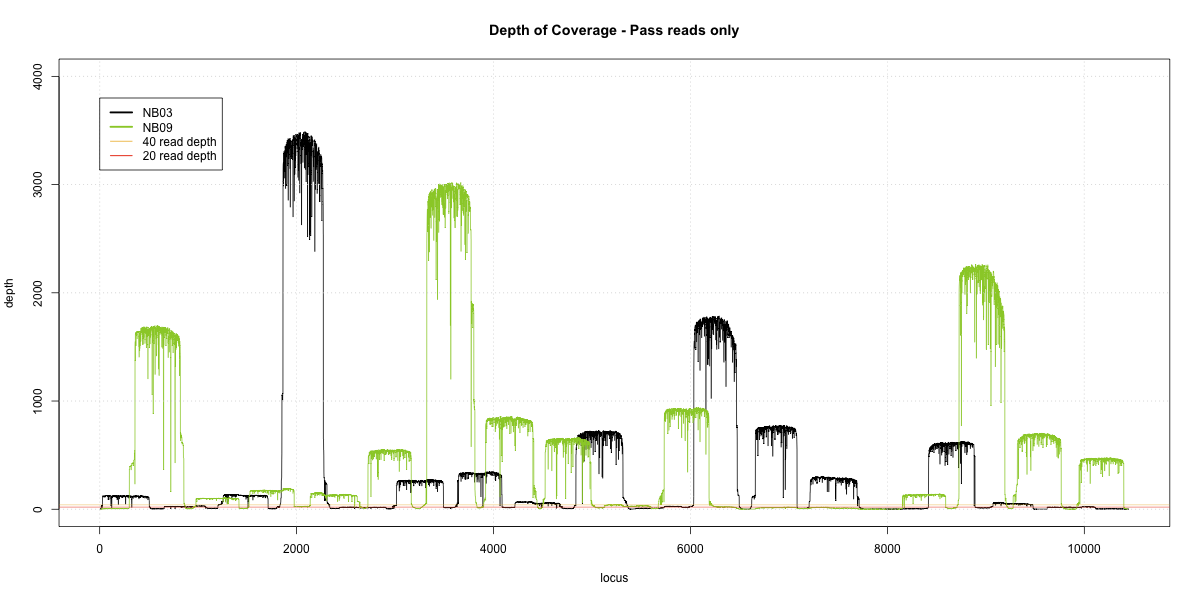
Complete library
- p20: 0.9191 (+25%)
- p40: 0.8723 (+37%)
- Difference: 4%
Incomplete library
- p20: 0.6571
- p40: 0.5022
NB04-NB010 Overlap
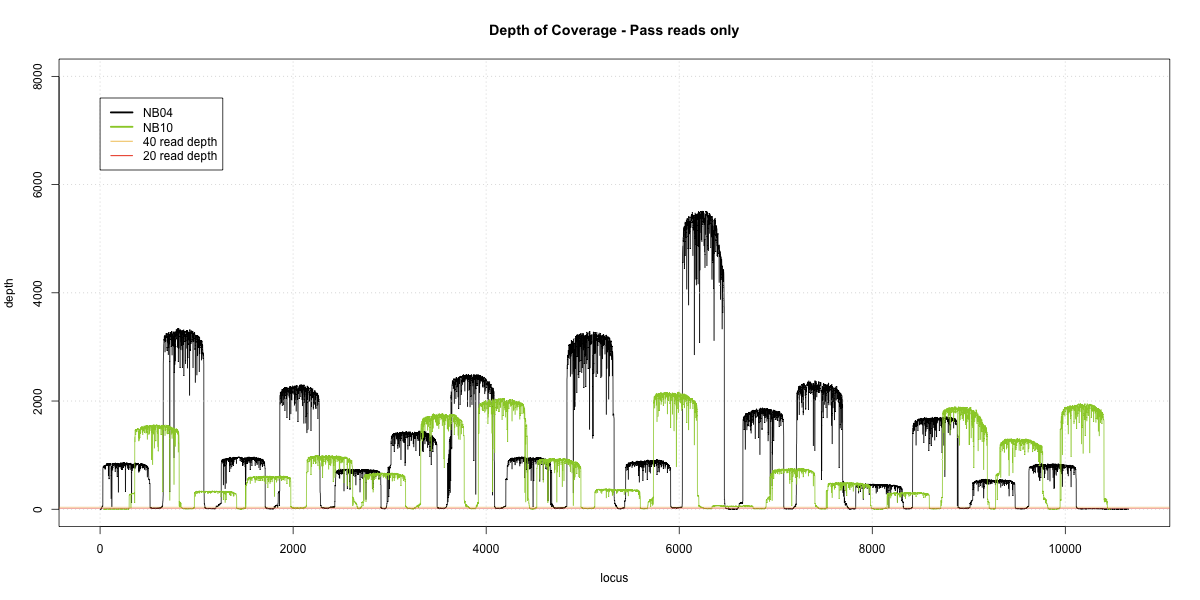
Complete library
- p20: 0.9932 (+6%)
- p40: 0.9925 (+9%)
- Difference: 0.1%
Incomplete library
- p20: 0.9384
- p40: 0.9087
NB05-NB11 Overlap
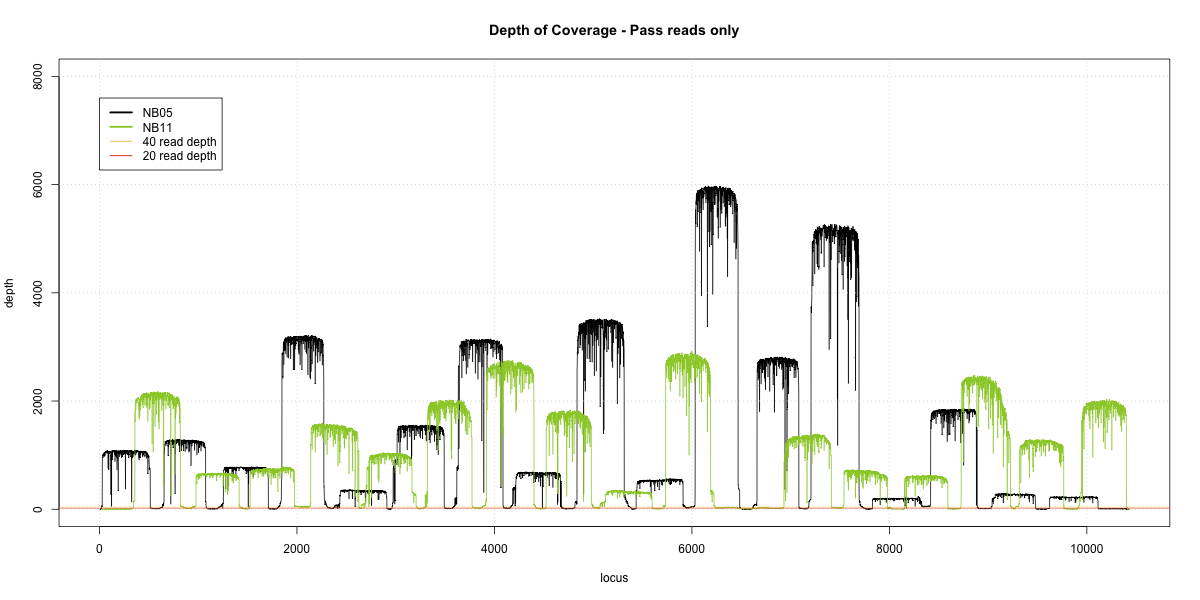
Complete library
- p20: 0.9779 (+7%)
- p40: 0.9749 (+10%)
- Difference: 0.5%
Incomplete library
- p20: 0.9066
- p40: 0.8737
NB06-NB12 Overlap
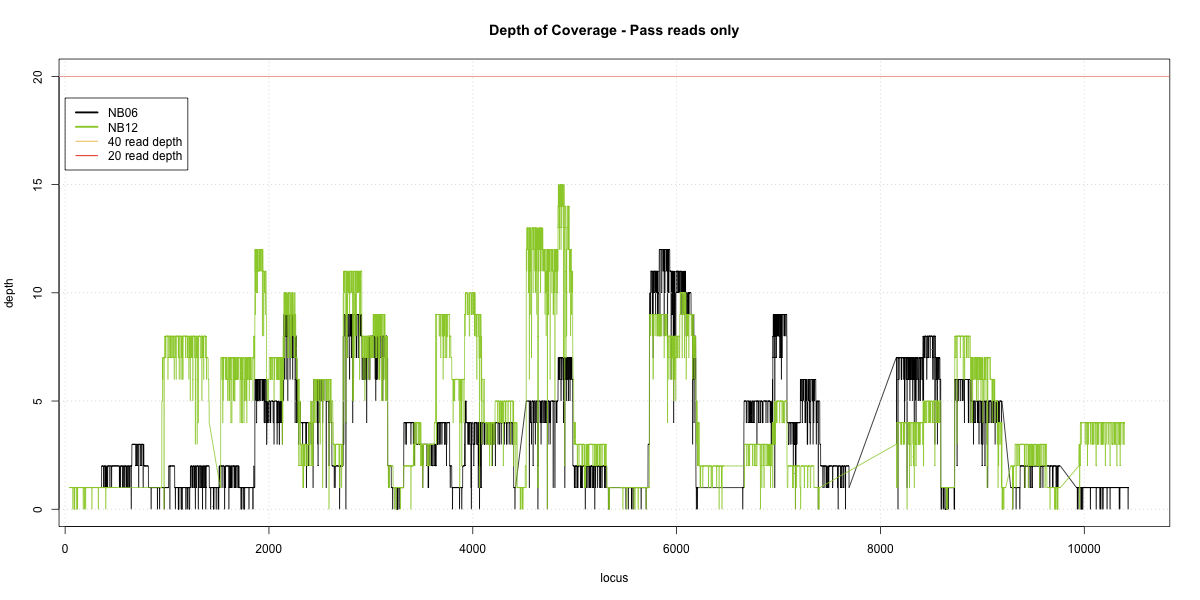
Complete library
- p20: 0.0163 (+1%)
- p40: 0 (+0%)
- Difference: 1%
Incomplete library
- p20: 0
- p40: 0
Overlap graphs and stats on pass libraries: USVI Library 3
NB01-NB07 Overlap
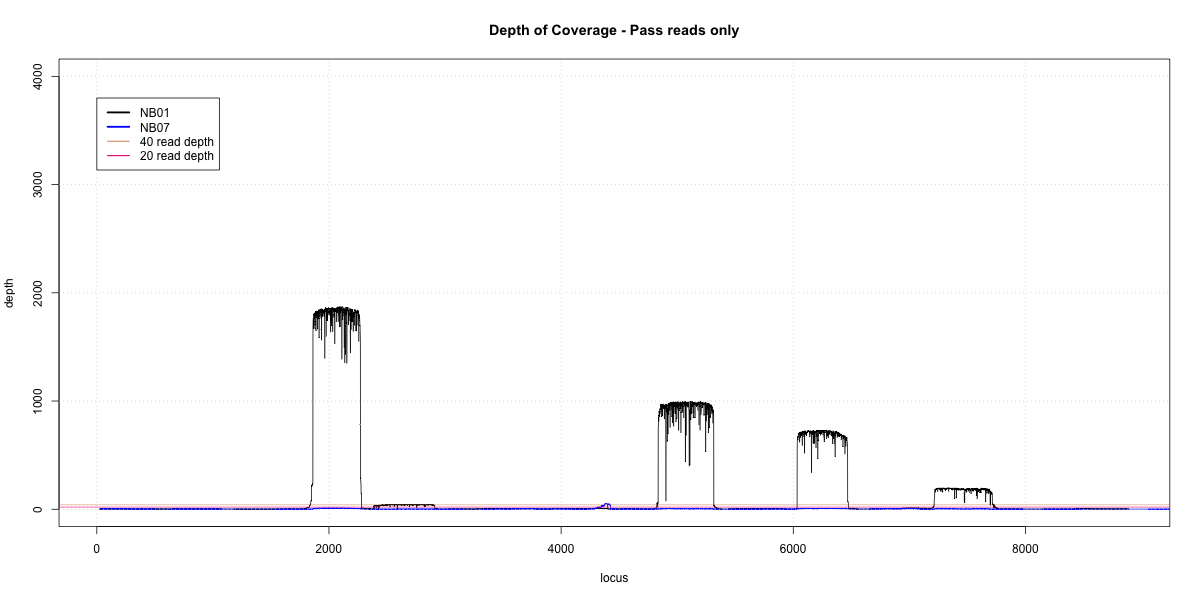
p20: 0.2896 p40: 0.2658
NB02-NB08 Overlap
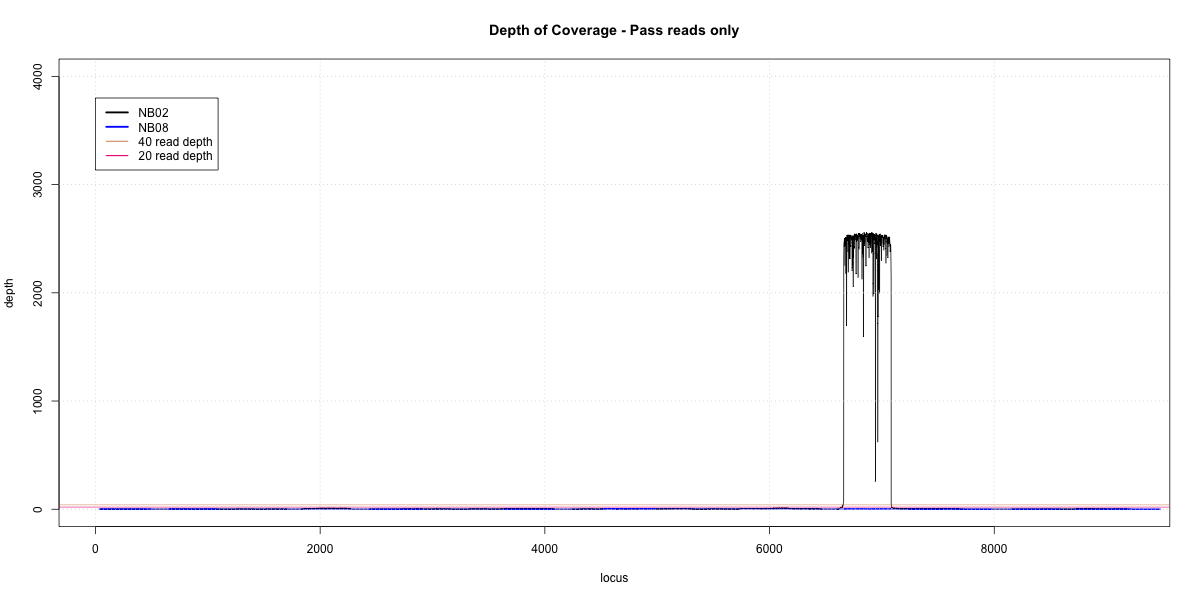
p20: 0 p40: 0
NB03-NB09 Overlap
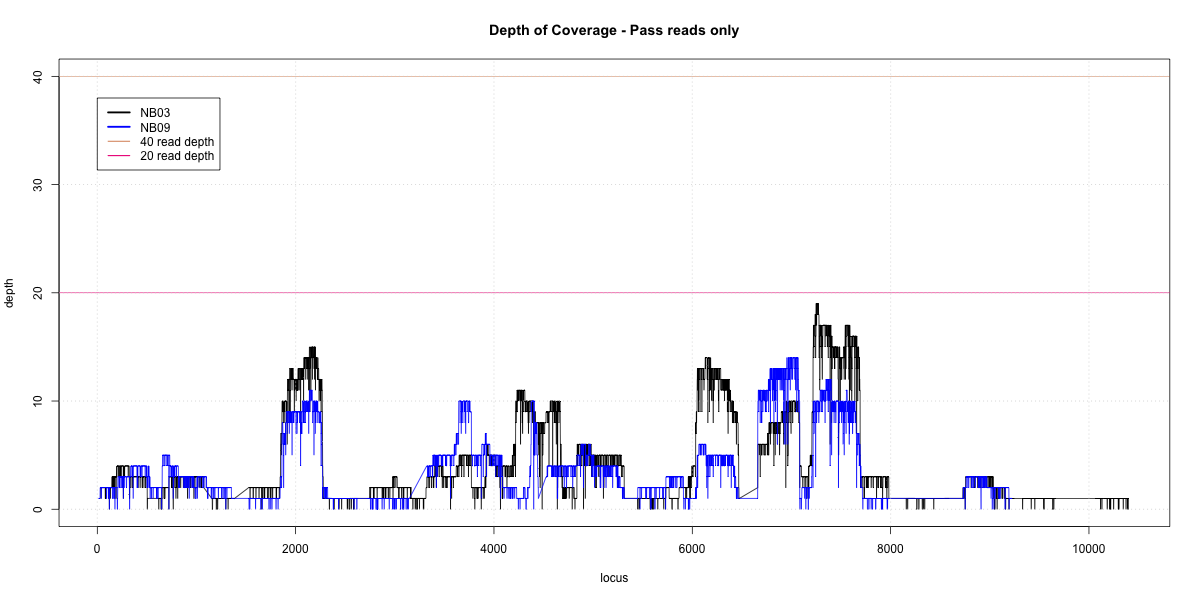
p20: 0.0925 p40: 0
NB04-NB10 Overlap
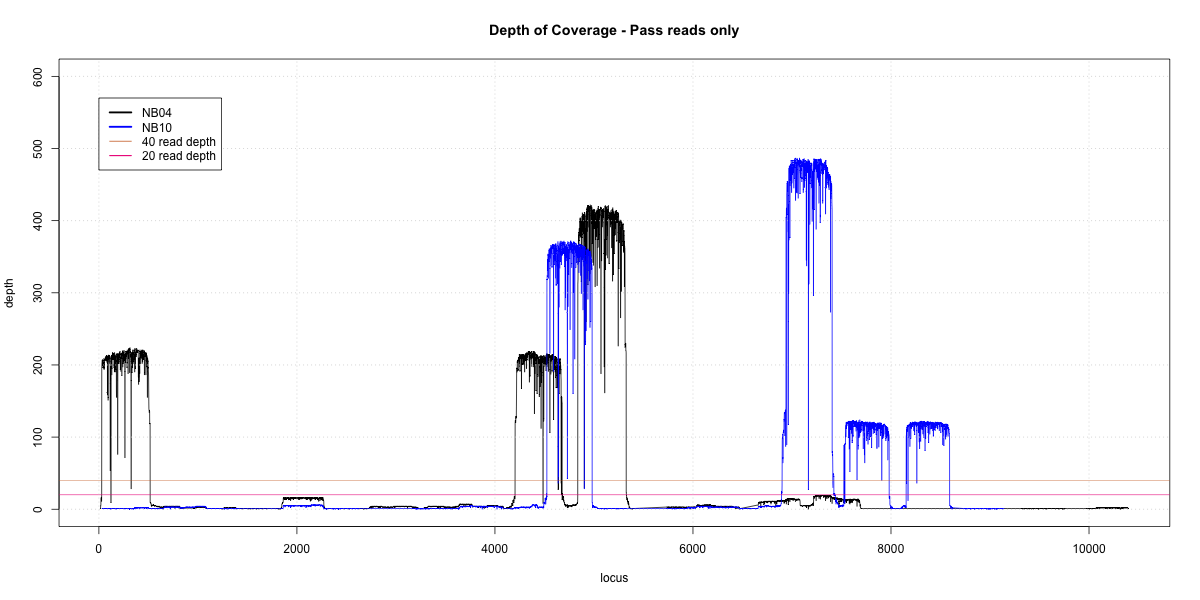
p20: 0.3825 p40: 0.3624
NB05-NB11 Overlap
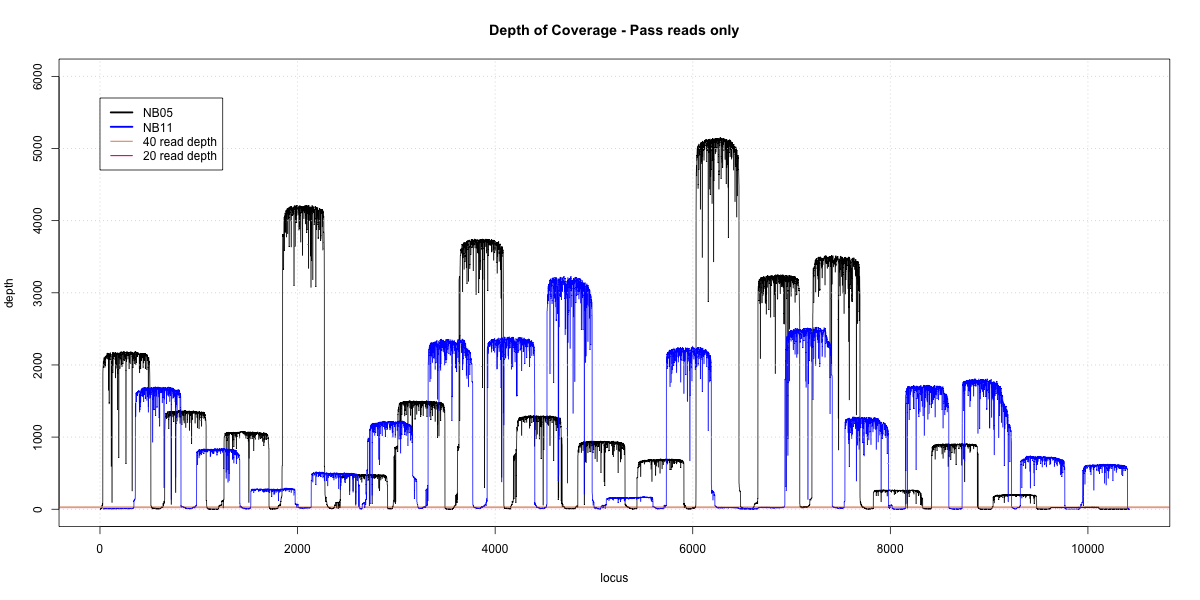
p20: 0.974 p40: 0.9675
NB06-NB12 Overlap
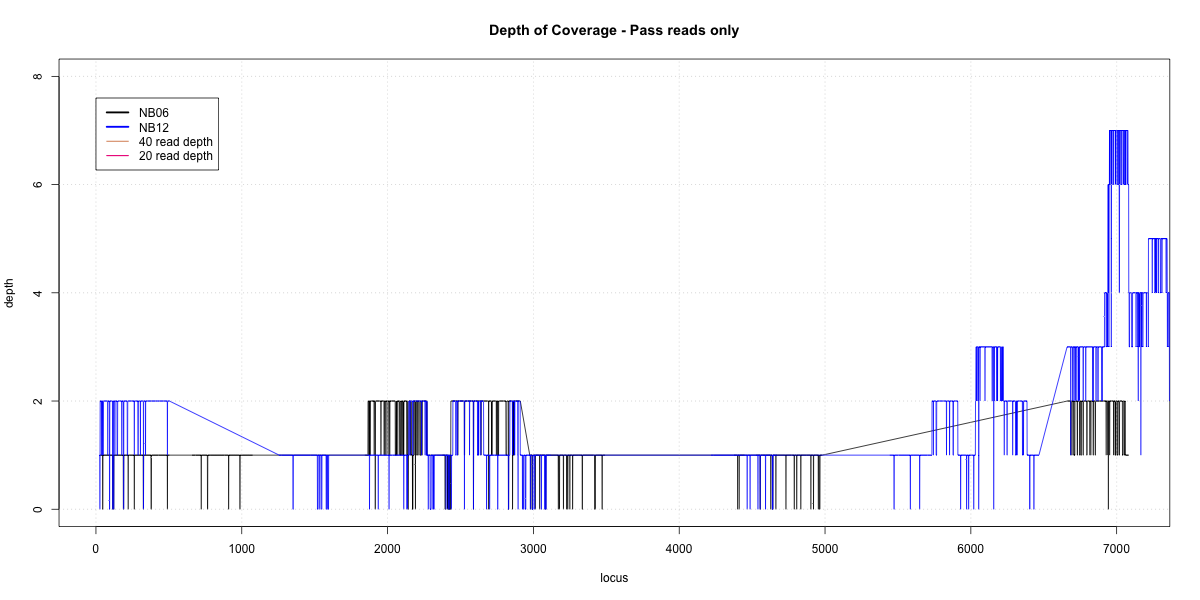
p20: 0 p40: 0