If you are reading this blog, you are probably already onboard with releasing data openly as it’s generated. You may even have led the charge in getting other researchers and/or journals to be more open with protocols, data, and analyses. You probably don’t need a reminder of why open data sharing is awesome, but sometimes it’s nice to take stock and remember that what we’re pushing for has real tangible value.
Louise has been investigating the genomic epidemiology of the mumps outbreak in Washington, and I’ve been helping out a bit too. If you want some big picture details about the project, you can read more about it here and here. As part of this project, Louise been managing Nextstrain Mumps. Recently, Patrick Stapleton from Public Health Ontario shared his sequences with us (thank you!!). Louise rebuilt Nextstrain with them, and we were totally struck by how much context the Ontario viruses provided for one of our “one-off” Washingtonian sequences.
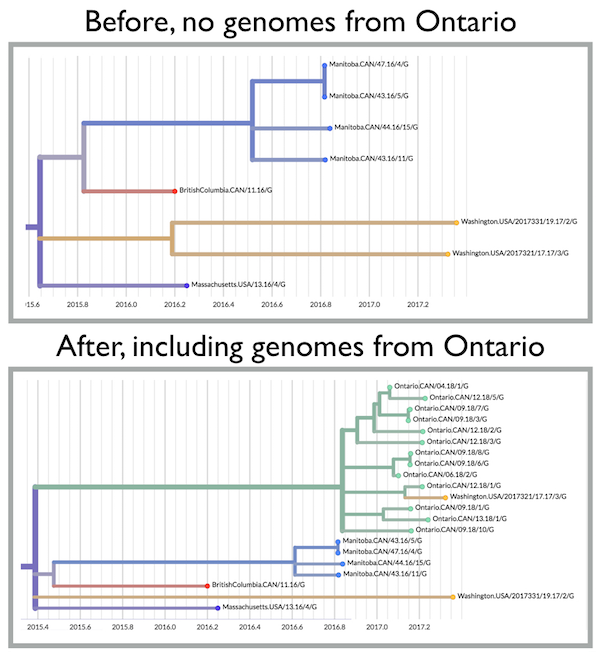
Take a look at the image with the before and after. Before, we see that we have a couple of viruses that don’t nest within the primary Washington outbreak clade. They are most closely related to Canadian viruses (Manitoba, BC), but those branches are pretty long, an indicator that there’s unsampled transmission occurring. This crops up not infrequently in sparse datasets, but I still always find myself wanting to know where this transmission chain was circulating before it pops up on our radar. Importantly, this isn’t just about my curiosity; knowing where importations come from, and their frequency, is important for tailoring surveillance efforts and designing or evaluating infection control measures.
Here we get lucky on two counts: 1) other people are sequencing mumps, and 2) they like sharing data! With the Ontario viruses included in the tree, we see that Washington.USA/2017321 is a very clear introduction of mumps from Ontario into Washington. Given the high genetic similarity between this Washington strain and the viruses from Ontario, it seems pretty likely that this was a direct introduction or perhaps a very recent introduction followed by a short transmission chain.
This may seem trivial, but you can play through some different scenarios to show that it’s not. Before, we don’t really have any idea what is going on with Washington.USA/2017321. We might ask, does this strain represent a tiny chunk of a lot of transmission that is going unobserved? If so, do we need to ramp up surveillance in a particular population? With just a bit of context we realize that no, we don’t need to pour a whole bunch of resources into figuring out what’s going on here. We have a travel-associated introduction, and while it’s good to follow up on close contacts, we probably don’t need to take significant resources away from another cluster to look into this one.
This is such a clear example of how much more value we can get out of genomic surveillance when we pool our data. Other people’s sequences provide context for our own. With this project, we’ve been incredibly fortunate to have lots of people sharing sequences with us. Many thanks to Jenn Gardy and Jeff Joy in British Columbia, Shirlee Wohl in Massachusetts, and Patrick Stapleton in Ontario. And of course thank you to all the authors who have put sequences up openly on GenBank. The mumps phylogeny would look ridiculous without you.