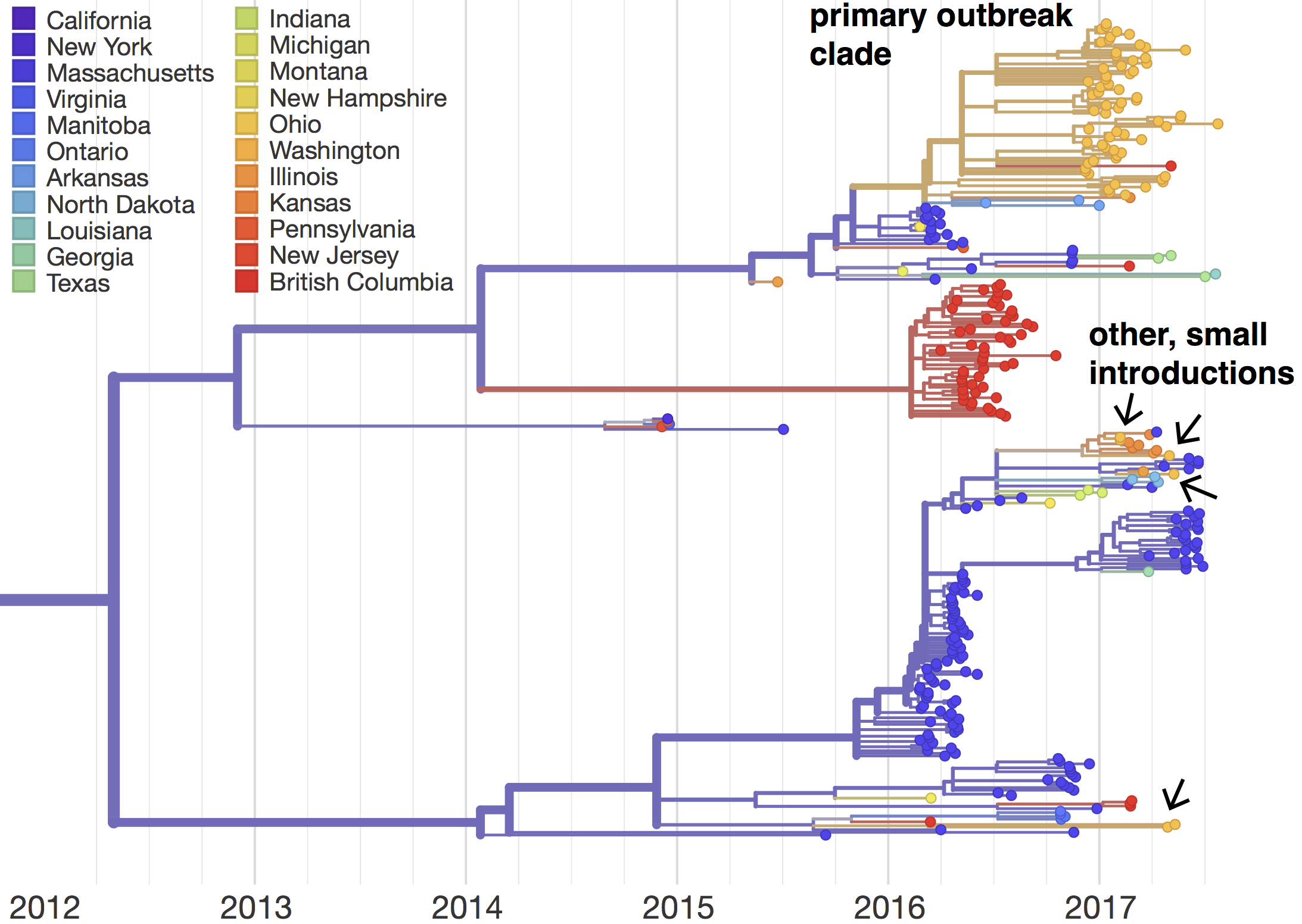
In 2016 and 2017, there were mumps virus outbreaks reported across the United States, with 5,629 total cases reported by the CDC. Washington state experienced one of the highest incidence rates nationwide, reporting 891 confirmed cases between October 2016 and September 2017. To characterize the timing and number of mumps introductions into Washington, and to describe how the virus spread within the state, we have been collaborating with the Washington State Department of Health to sequence mumps viruses from buccal swabs collected throughout the outbreak. You can find all of our protocols for sample processing and sequencing on our mumps-seq GitHub page, and a downloadable fasta file with all consensus sequences here. We have sequenced 72 near-complete mumps genomes from Washington so far, and will probably take a break from sequencing for awhile to see what we can learn from these genomes. All of our genomes have been added to nextstrain.org/mumps, where they can be viewed in the context of other mumps viruses sequenced from 2016/2017 outbreaks in the US and Canada.
So far, preliminary analysis of our 72 genomes shows that the Washington mumps outbreak consists of at least 5 separate introductions of mumps into Washington. The majority of sequences (66/72) cluster within a single clade that we will call the primary outbreak clade. Viruses in the primary outbreak clade are nested within the diversity of viruses from the Massachusetts outbreak, and are closely related to viruses from Arkansas and Kansas, suggesting that outbreaks across the United States are related. The vast majority of viruses from British Columbia form their own distinct clade, suggesting that viruses from Washington are more related to those from Massachusetts than they are to those from British Columbia. However, there is a single British Columbia sequence nested within the primary outbreak clade, suggesting some, limited transmission between Washington and British Columbia.
The phylogeny also shows 4 additional introductions of mumps into Washington, although these introductions lead to a small number of cases. These smaller introduction events are also linked to Massachusetts and Kansas viruses, suggesting that there may be epidemiologic links among these states. Together, these data suggest that there were multiple introductions of mumps viruses into Washington state, but that most of these introductions did not lead to sustained transmission that we were able to sample. A single introduction event, occurring around March 2016 (95% CI: Feb 2016 – April 2016), likely seeded the majority of transmission during the outbreak. Our phylogenetic tree suggests that this introduction likely stemmed from Massachusetts; however, many US states that reported mumps cases have not been sampled, so we cannot rule out that mumps passed through another intermediate location.
We will be continuing to analyze these data to more firmly estimate the timing of introduction events, and to estimate transmission rates among different subsets of the Washington population. We would like to give a huge shoutout to our collaborators at the Washington State Department of Health, especially Chas DeBolt, Ailyn Perez-Osorio, Misty Lang, and Nhan Le, for all of their work characterizing the outbreak and collecting and preparing samples. We would also like to send a huge thank you to Jen Gardy, Jeff Joy, Pardis Sabeti, and Shirlee Wohl for sharing their sequence data with us on nextstrain.org. None of this work would make sense without the ability to contextualize our outbreak, so thank you all so much for making this possible!