I’ve been remiss for the past year about posting our biannual flu report publicly. We’ve now however posted our Sep 2019 flu report to bioXriv where it details recent seasonal influenza evolution during 2019 and projections for spread over the next 12 months to Sep 2020. Our timing with this report is designed to correspond to the timing of the World Health Organization’s Vaccine Composition Meeting being held this week in Geneva. Richard Neher has lead much of this analysis, with John Huddleston providing fitness model projections and Barney Potter contributing to data curation.
With each of the reports, we generally end up focusing on a handful of emerging clades within each influenza lineage and tracking their rate of global spread and viral characteristics. In one current example, H3N2 viruses have diversified into a large number of competing lineages, however, over the course of 2019 we’ve seen the emergence and spread of A1b/197R viruses as well as A1b/137F viruses. Over the course of the past ~9 months these clades have grown from nearly 0% global frequency to a combined >50% global frequency. Previously, Richard and colleagues had identified local branching index (LBI) as a strong predictor of future strain success. The idea is basically that clades that are currently outcompeting their relatives are estimated to be higher fitness and so are predicted to continue to increase in frequency into the future. In previous reports, we’ve used LBI to project which clades will come in to dominate.
More recently, John has sought to build a fitness model that makes quantitative predictions of clade frequencies based on LBI as well as viral characteristics. There is some description of this model in the September report. We’re hoping to have a preprint and source code shared shortly. However, we’ve now elected to start including live model predictions for H3N2 at nextstrain.org/flu. The bottom panel shows frequencies of different clades up to present as well as a forecast over the following 12 months:
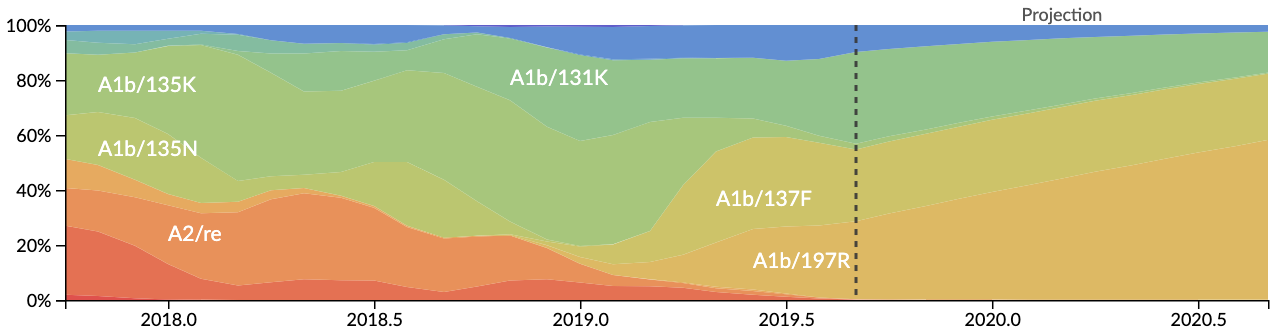
Here, it’s clear that the model follows LBI in predicting the further growth of 197R viruses. Additionally, in the “color by” dropdown menu you can now select “fitness” to show fitness estimates for each virus and also select “distance to future population” to show amino acid match of sampled viruses to the predicted future population.
These forecasts will now be made automatically alongside our weekly site updates.