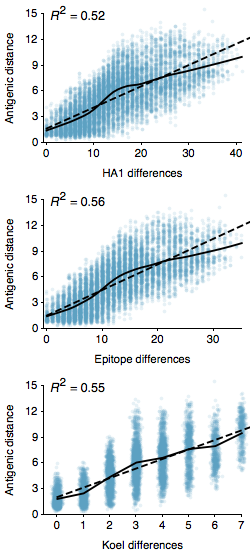
This is another follow-up to the “Integrating influenza antigenic dynamics…” paper and is in part a response to the impressive new paper by Koel et al., where the authors use reverse genetics to identify 7 sites that appear to be responsible for the majority of antigenic evolution seen in influenza A/H3N2 from 1968 to 2003. In the “Integrating influenza antigenic dynamics…” paper I calculated the correlation between amino acid changes in the 330 sites of the HA1 region of the hemagglutinin protein. The HA1 region of the hemagglutinin protein lies on the surface of the virus particle, making it a target for the humoral immune response. Here, we see that mutations to the HA1 region correlate with antigenic distance between pairs of A/H3N2 viruses separated by at most 10 years, with a coefficient of determination of R^2 = 0.52 and an absolute predictive error of 1.61 antigenic units.
Early work by Wiley et al. identified sites within HA1 that are the main targets for specific antibodies. Muñoz and Deem provide an easy to incorporate list of these sites, yielding 129 sites for analysis. Here, I looked at the correlation between substitutions at these sites and antigenic distance, finding a slightly stronger correlation than with just HA1, with R^2 = 0.56 and a predictive error of 1.50 antigenic units. So we see that these 129 sites provide a stronger predictor than HA1 as whole. These sites have previously been shown to be undergo more rapid adaptive evolution than other sites on HA1 (Wolf et al.).
The new paper by Koel et al. suggests that just 7 sites contribute to the majority of antigenic change in A/H3N2. Here, I show the correlation between substitutions at just these 7 sites and antigenic distance, yielding approximately the same strength of correlation with R^2 = 0.55 and an absolute predictive error of 1.51 antigenic units. Each substitution at one of these sites results, on average, on 1.11 units of antigenic change. There are almost certainly antigenic differences attributable to sites beyond these 7, but it’s still impressive how well these few sites work to predict antigenic distance relative to the full complement of HA1 sites.