We have a new preprint up on bioRxiv describing the genomic epidemiology of Zika virus in Colombia!
Via a collaboration with the Instituto Nacional de Salud de Colombia and the Universidad del Rosario, we sequenced 8 new Colombian genomes directly from clinical samples, and did the first detailed genomic analysis of Zika in Colombia.
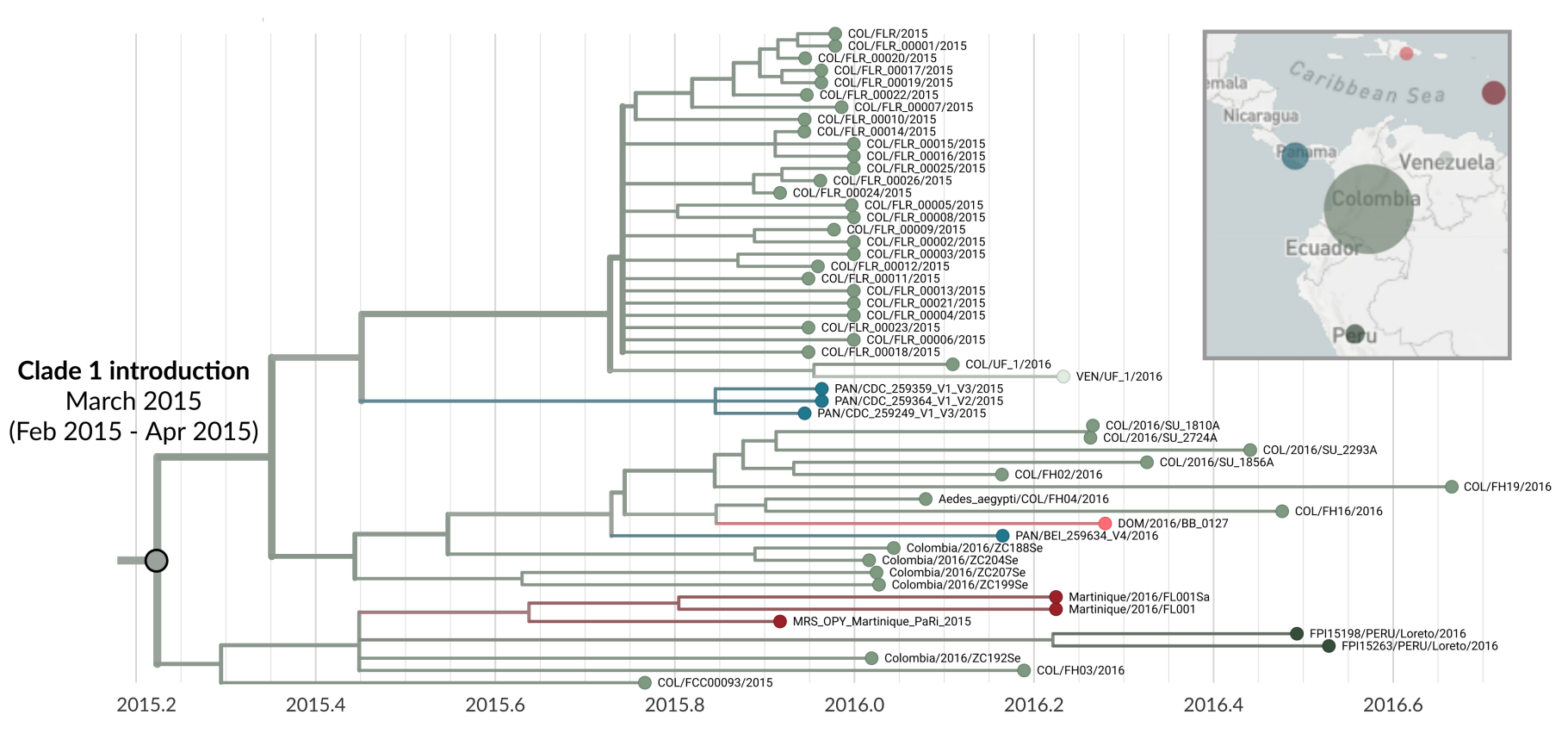
For this study we performed a phylogeographic analysis of 386 American Zika genomes, of which 46 were sampled from Colombia (38 previously available genomes and 8 from this study). We found that Colombian samples grouped into two separate clades, indicating at least two separate introductions of Zika to Colombia. Remarkably, despite evidence for multiple introductions, the majority of transmission within Colombia appears to descend from only one introduction. We infer that this large clade was introduced to Colombia around March 2015, indicating that Zika likely spread to Colombia before it was even confirmed to be circulating in Brazil. Finally, we see lots of evidence in the tree for movement of Zika from Colombia to other countries. Most of this movement is to countries sharing a border with Colombia. For instance, viruses from Panama, Venezuela, and Peru are descended from viruses in the primary Colombian clade, and the small secondary clade shows viral migration from Colombia into Ecuador.
We should add that the bulk of the lab work for this study occurred in Bogotá, Colombia. The folks there were so helpful and supportive of the work, and we are grateful for the chance to have worked with such an incredible team (not to mention the opportunity to explore Colombia!).
In the spirit of open science we’ve tried to make our data and analyses freely available and reproducible. If you’re interested in the laboratory protocols and bioinformatic pipeline we’re using for genome assembly you can find that all on github.com/blab/zika-seq. If you would like to reproduce the analysis presented in the manuscript, you can find all data files and build instructions on github.com/blab/zika-colombia. Finally, you can also interactively explore the phylogeny at nextstrain.org/community/blab/zika-colombia.
As always getting global collaborations off the ground requires the effort of many people. We would especially like to thank Juan-David Ramírez for sharing precious samples, and Diana Rojas and Betz Halloran for their work on getting the data sharing agreement with the INS in place. We are also incredibly grateful to the virology team at the INS, especially Katherine Laiton-Donato, Lissethe Pardo, Dioselina Peláez-Carvajal, and Marcela Mercado-Reyes, who worked tirelessly with us down in Bogotá to ensure this study’s success. We sincerely hope that this is just the first of such collaborations!