Together with Richard Neher, we’ve just released a website, called nextflu, that is meant as a real-time view into the evolution of seasonal human influenza, focusing on influenza subtype H3N2. Influenza H3N2 is the fastest evolving of circulating influenza viruses and is responsible for the bulk of human mortality and morbidity. Because of its rate of evolution, it is also the most difficult subtype to match during vaccine strain selection. We built nextflu to try to make sense of ongoing H3N2 evolution and to provide a tool to better pick vaccine strains.
For the site, data is downloaded from GISAID and a pipeline for cleaning and processing sequences is run. The resulting files are then rendered in the browser using d3.js. Primary outputs of this analysis are an annotated phylogeny showing relatedness between circulating influenza isolates and estimated mutation frequency trajectories through time. A large degree of interactivity is provided, allowing coloring of the tree by trait or genotype and rewinding time to see what isolates were circulating in previous seasons.
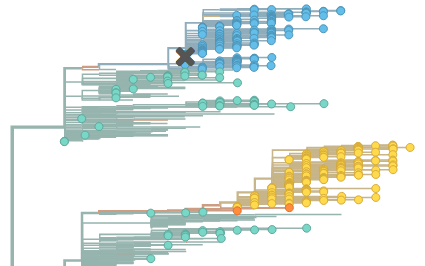
I’m really excited about this work, and I can imagine this sort of real-time tracking and automated analysis being useful for a variety of viruses, such as H5N1, norovirus or Ebola. A major goal at this point (or at least something to strive for) is build in automated forecasting of which strains are likely to succeed based on historical patterns. At this point, we are showing predictors that have previously been found to correlate with strain success, such as number of epitope mutations and local branching patterns, but the goal is definitely quantitative forecasts of strain frequencies through time.
We’ve strove to be as open as possible with this work. All source code is freely available. If you have specific suggestions for improving nextflu, please, open an issue.
