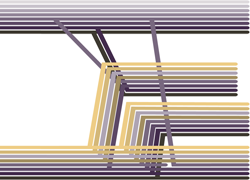
We’ve just posted a paper to the arXiv examining reassortment patterns between influenza B lineages. Influenza B viruses circulate alongside the better known influenza A and also cause seasonal epidemics within the human population. Flu B has evolves slower than influenza A and shows less antigenic drift. However, there have gradually emerged two distinct lineages of influenza B denoted as the Victoria lineage and the Yamagata lineage. These two lineages diverged in their evolution in the hemagglutinin (HA) protein, which is the primary determinant of the human immune response. Consequently, at this point, infection or vaccination with one lineage no longer protects against infection by the other lineage. Owing to this, recent vaccine formulations often include both a Victoria B virus and a Yamagata B virus.
However, although HA has diverged substantially between lineages, other influenza proteins do not show the same distinction between Victoria and Yamagata lineages. For this study, the very talented Gytis Dudas looked at reassortment patterns between Victoria and Yamagata viruses across all 8 gene segments and came to some very interesting conclusions. Although most segments have frequently reassorted and lost genetic diversity, the PB1 and PB2 segments that encode two subunits of influenza’s RNA polymerase have continually coassorted with HA. We hypothesize a previously undetected functional linkage between these proteins. These findings also have interesting parallels with speciation research in more standard organisms, i.e. not viruses, where it’s often found that a co-adapted gene complex emerges that resists recombination and promotes speciation between populations.