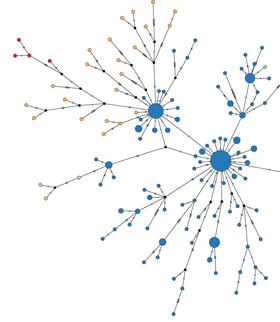
In collaboration with a large consortium from Harvard, the Broad Institute, the University of Edinburgh, Tulane University, the CDC, Kenema Government Hospital and others, we’ve just published a paper in Cell detailing the transmission dynamics of Ebola virus circulating in Sierra Leone between May and December last year. Here, we present 232 new Ebola genome sequences and combine these sequences with 86 previously characterized genomes to give a broad picture of the evolution of the virus during the Sierra Leone epidemic. We find evidence for a sustained epidemic within Sierra Leone driven by endemic transmission rather than by repeated introductions from Guinea or Liberia. We also investigate evidence for evolutionary adaptation to the human host, finding that genome-wide selection is largely purifying, aiming to maintain the Ebola genome in a fixed state rather than selecting for new mutations. However, we do see some signal in the glycoprotein gene for faster rates of evolution in regions that are putative antibody epitopes. We attribute this signal to within-host selection for immune escape during the course of an infection.
This was a fantastic collaboration to be a part of. Being able to make timely contributions to our understanding of the evolution and epidemiology of an outbreak will have important implications for intervention strategies. Going forward, immediate public release of outbreak sequence data will be needed to make actionable inferences. In the case of an outbreak, each sample is only truly valuable when put in context with samples gathered in other regions or other points in time. Because of this, data sharing is crucial to make proper conclusions.