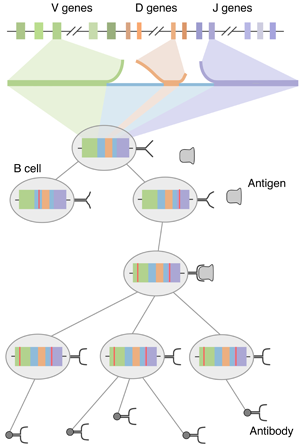
Another paper posted to the arXiv… this one on a new and exciting research direction for me, looking at sequence changes within an individual’s B cell repertoire as an evolutionary process, amenable to modern phylogenetic inference methods. Basically, in order to promote immunity that protects against a wide range of possible pathogens, the adaptive immune response creates antibodies that target specific antigens. Antibodies are encoded in the immunoglobulin genes of B cells. Rather than attempting to encode all possible antibodies within the human genome (which would be impossible), B cell immunoglobulin sequences are seeded by a process of random rearrangement of germline (inherited) sequences, termed VDJ recombination. After this initial rearrangement, B cells that successfully (if partially) bind to antigen are stimulated to reproduce, during which time additional mutations are induced through somatic hypermutation. Mutated B cells that bind better will outcompete poorer binders via Darwinian selection resulting in B cell affinity maturation towards circulating antigens. A schematic of this whole process can be seen here.
It’s been exciting starting to look at B cell affinity maturation from an evolutionary perspective. Erick Matsen has really inspired this interest and it’s been great helping him, Connor McCoy and Vladimir Minin develop this project. In this paper, we fit substitution models and analyzed selective pressures in a short-read deep-sequencing data set of B cell receptor genes from three healthy individuals (courtesy of Harlan Robins). We found strikingly similar patterns across individuals in terms of exactly which sites evolved and which nucleotides replacements occurred. In this case, we did a pairwise analysis comparing observed read to matched germline sequence. Future methodological development will focus on identifying B cells that descend from a common rearrangement event and reconstructing their phylogenies.