Run the phylogeographic analysis
Open BEAST and select the file pandemic_geo.xml.
This analysis (because of the function to infer ancestral states) requires that BEAGLE be loaded.
Select ‘Use BEAGLE library if available’.
Generally, I turn on SSE as this should give a decent speed increase and set the ‘Rescaling scheme’ to ‘Dynamic’.
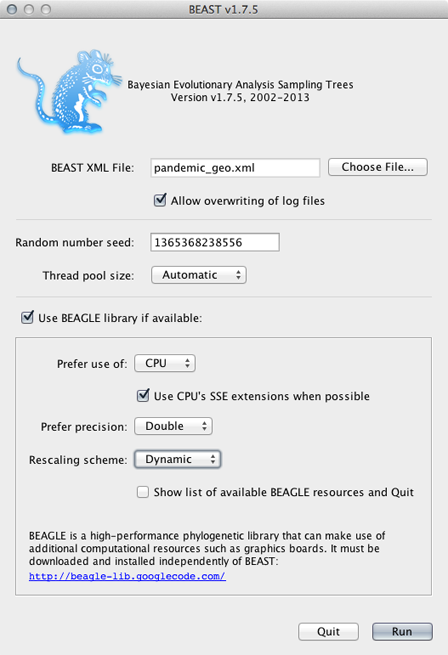
Click on ‘Run’.
This will produce the output files pandemic_geo.log, pandemic_geo.rates, pandemic_geo.root and pandemic_geo.trees.
This analysis took ~24 hours to complete on a single cluster node.
I’ve included these files with the practical in the output/ directory.