Continuing SARS-CoV-2 evolution under population immune pressure
Trevor Bedford (@trvrb)
Fred Hutchinson Cancer Center / Howard Hughes Medical Institute
6 Apr 2022
Vaccines and Related Biological Products Advisory Committee Meeting
FDA
Slides at: bedford.io/talks
Disclosures
- I receive grant support from the National Institutes of Health and the Howard Hughes Medical Institute to research methods for evolutionary forecasting of SARS-CoV-2.
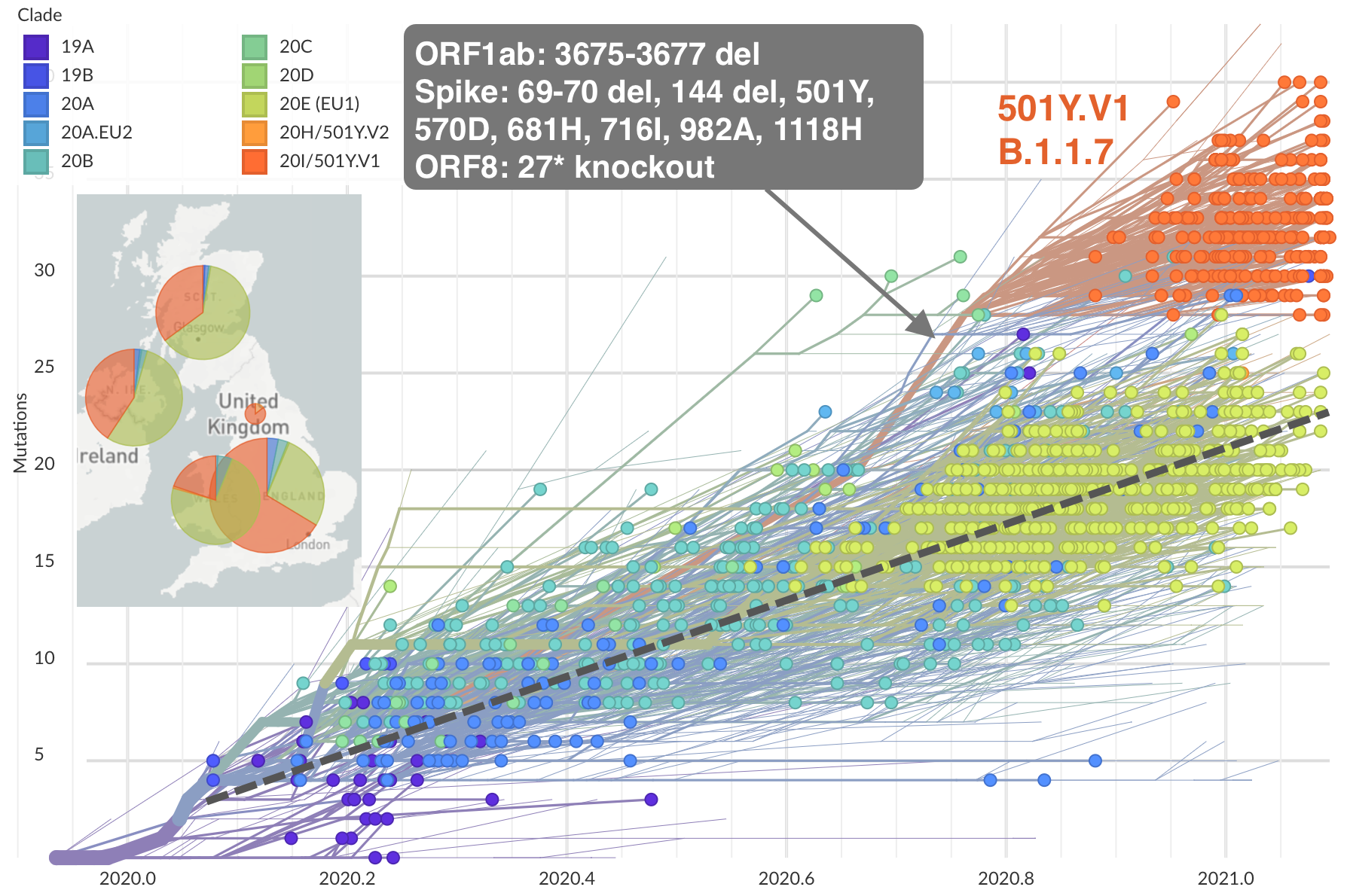
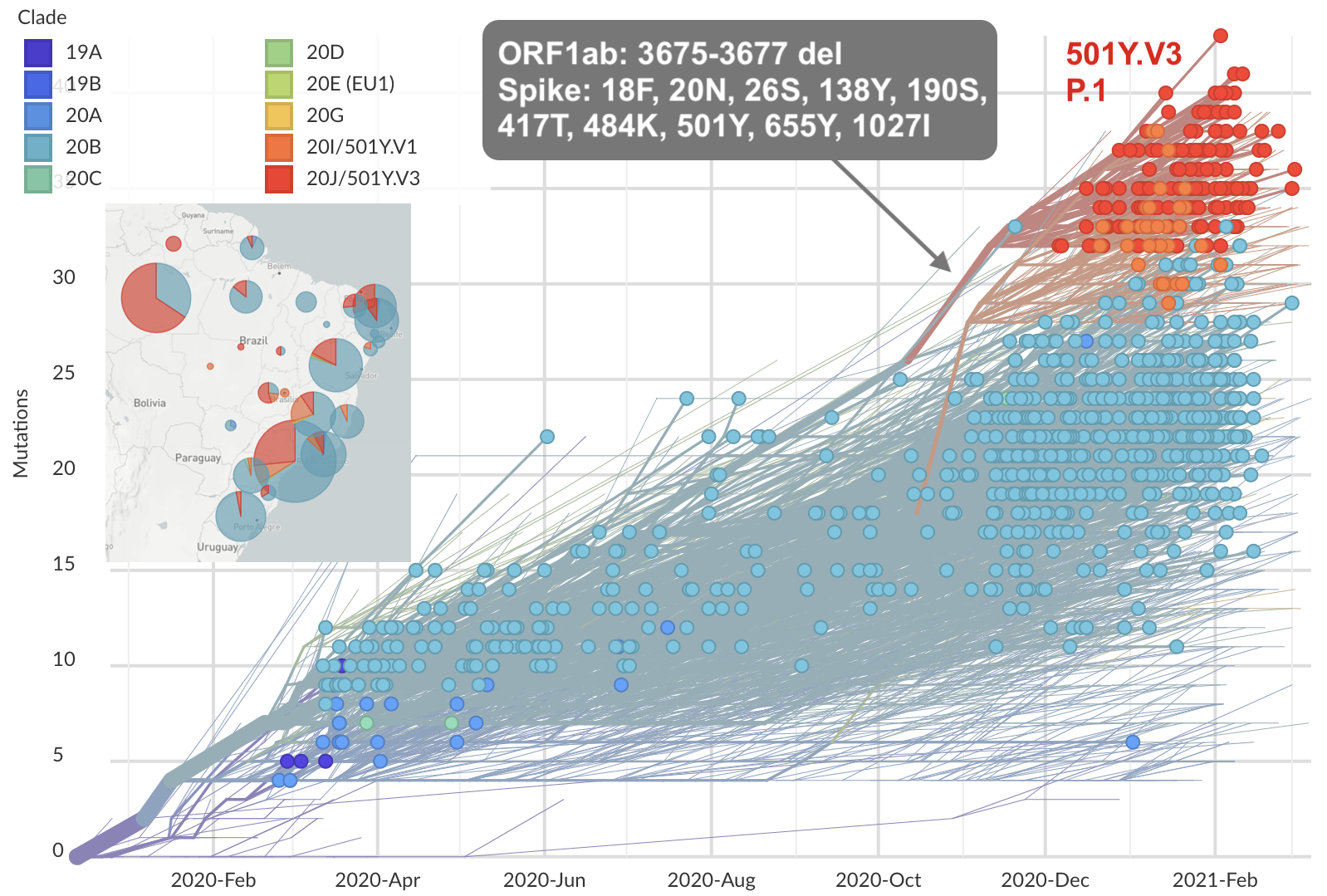
Repeated emergence of variant of concern viruses
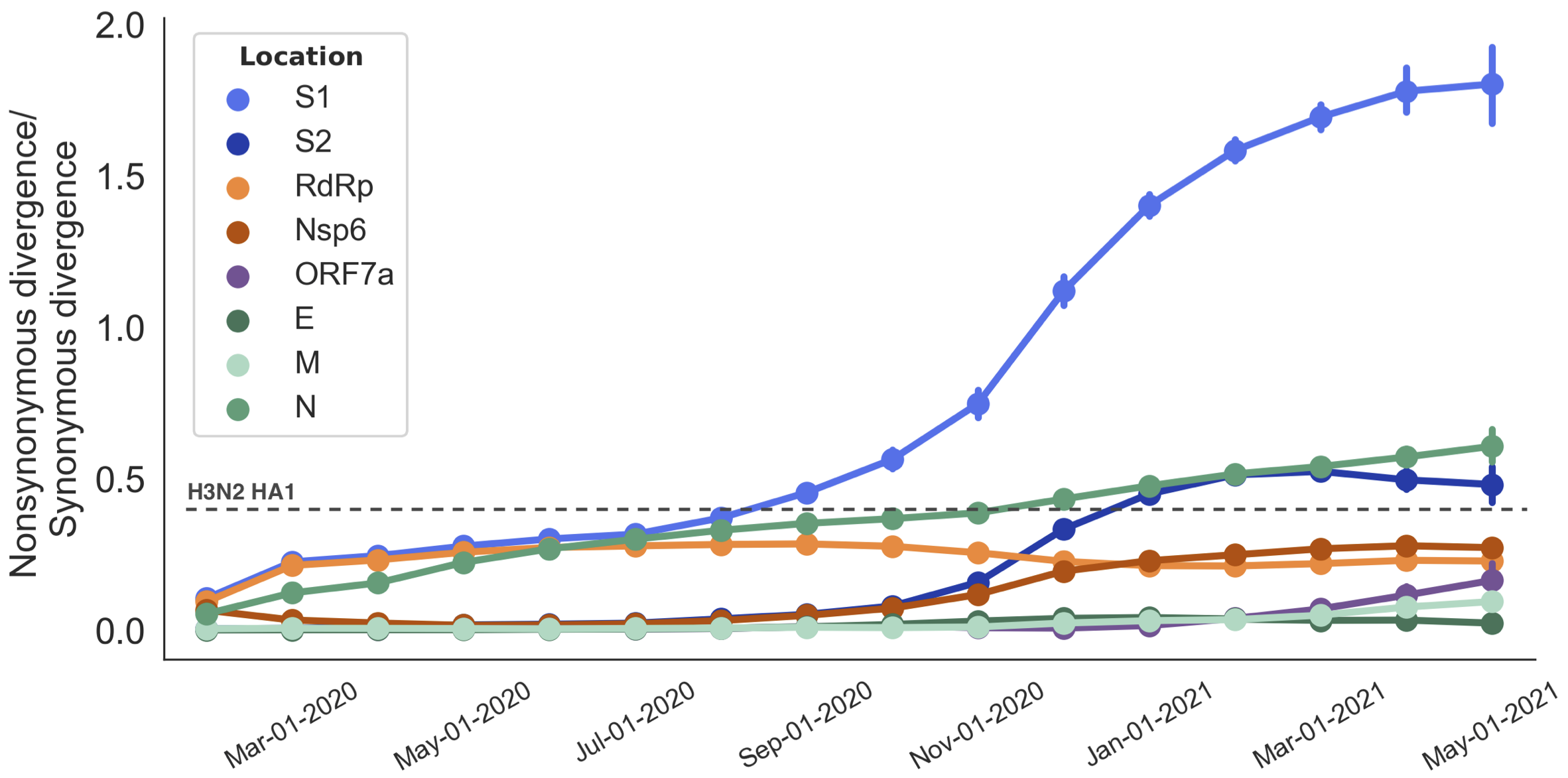
Rapid adaptive evolution focused on S1 subunit of spike protein
Genetic relationships of globally sampled SARS-CoV-2 to present
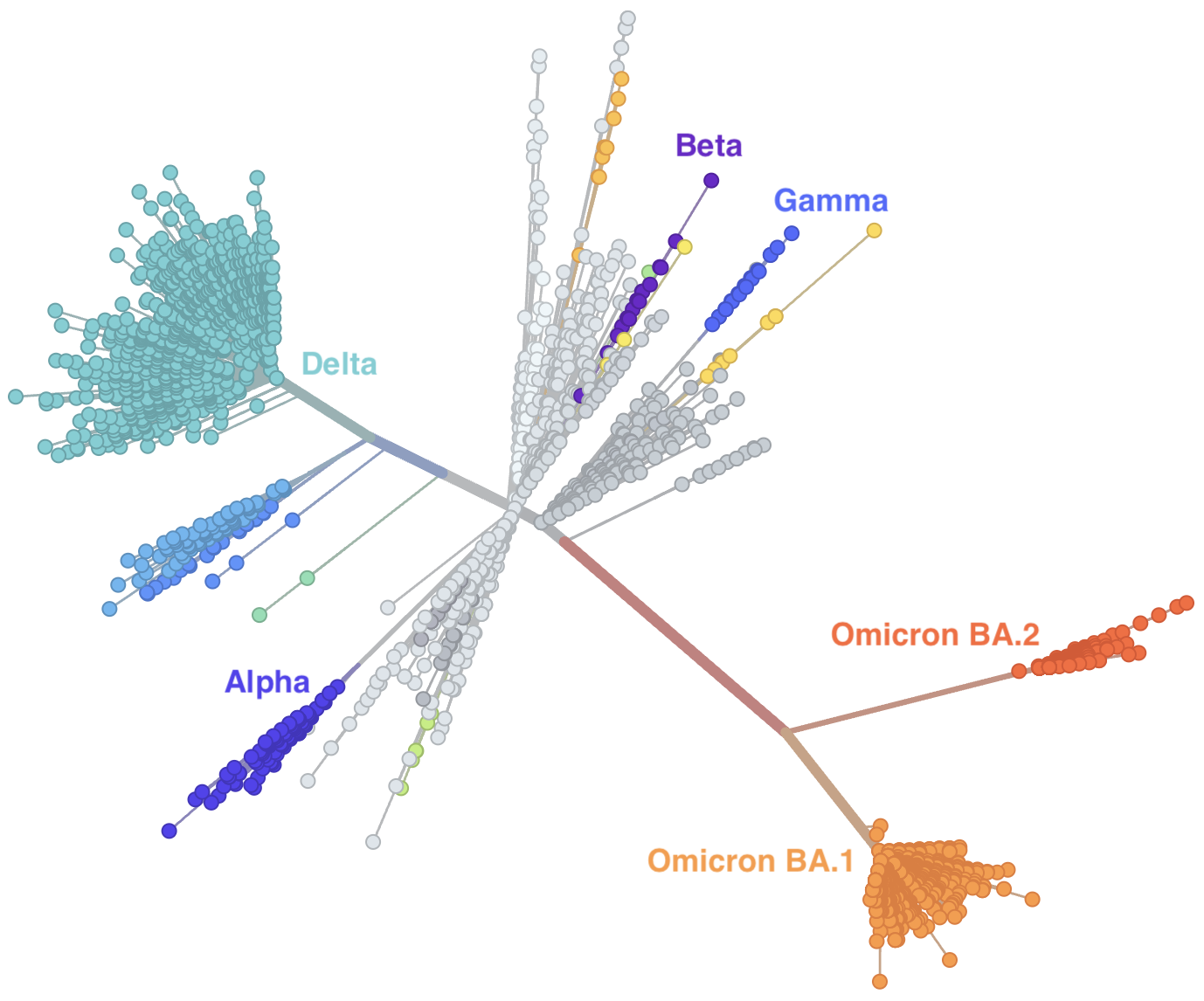
Rapid displacement of existing diversity by emerging variants
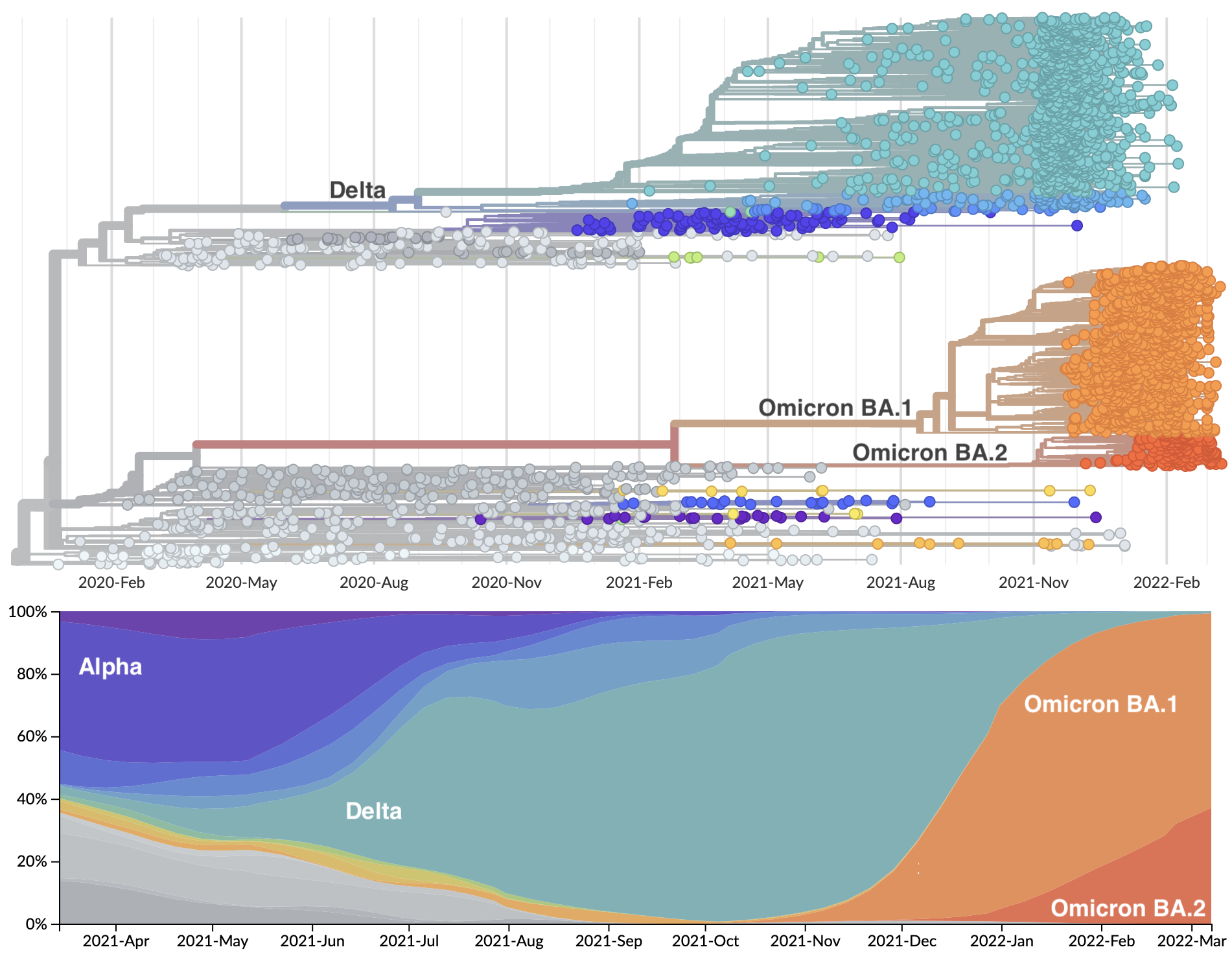
Omicron shows particular excess of mutations at S1
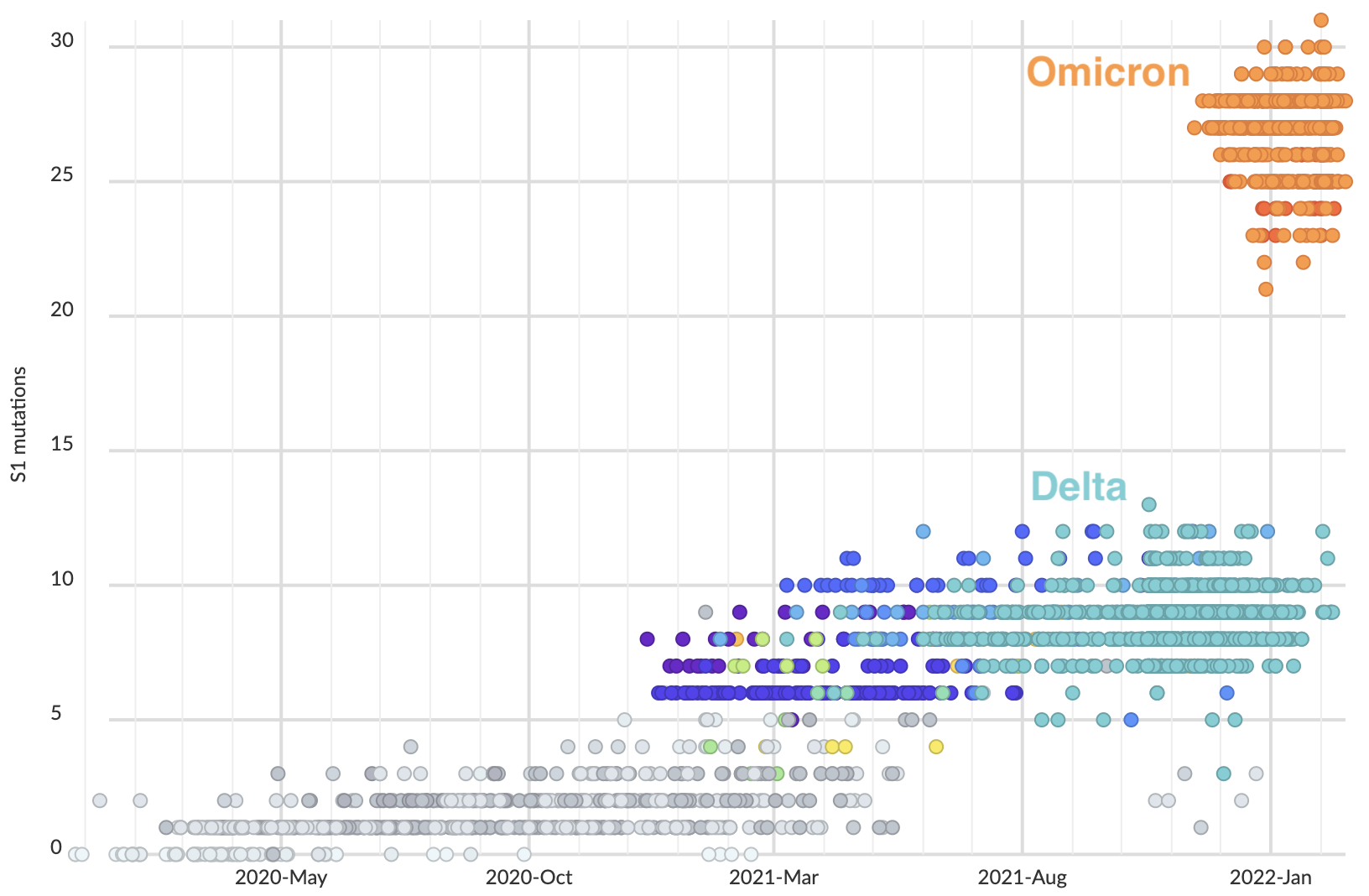
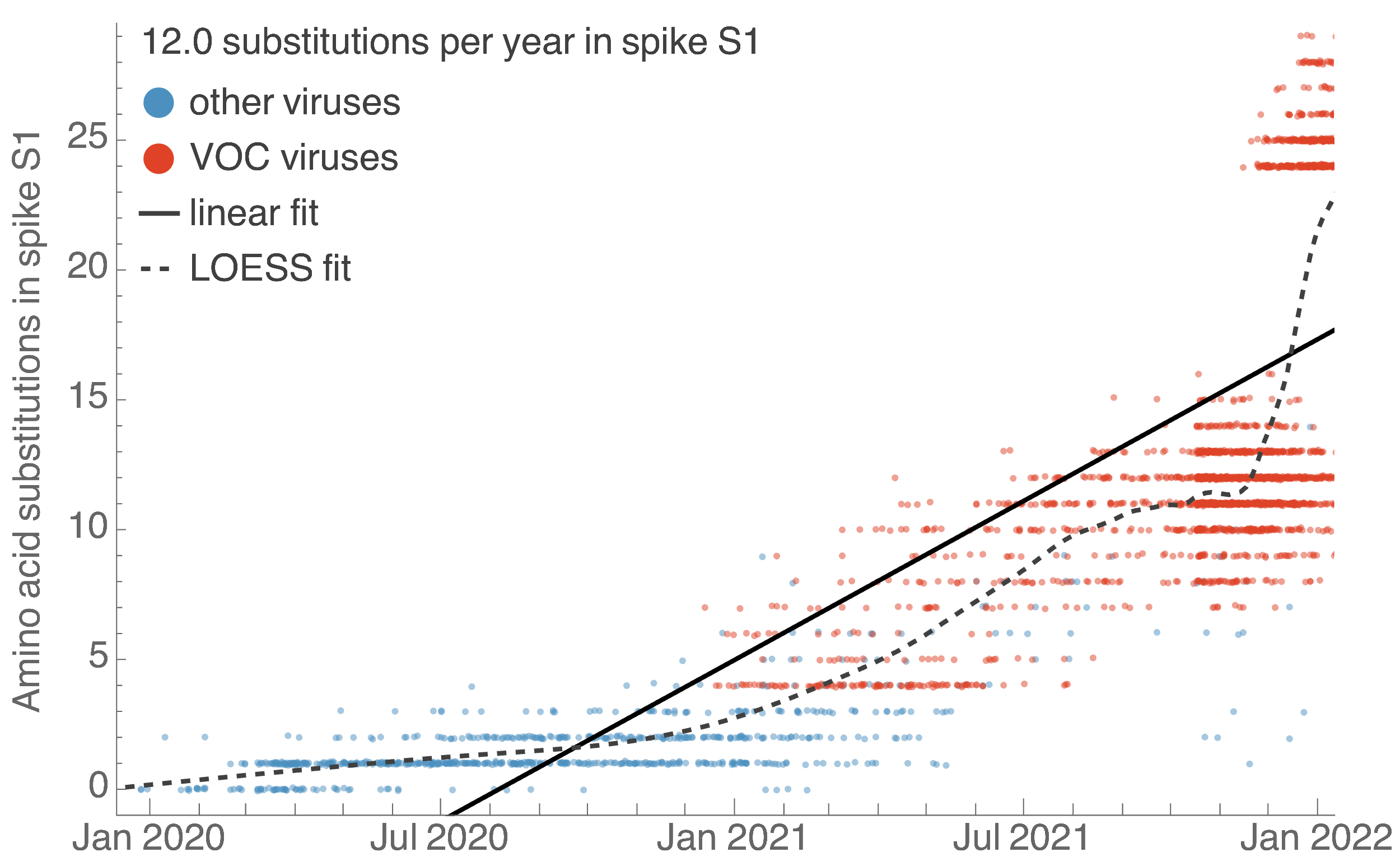
S1 evolved at a rate of 12 amino acid changes per year in 2021
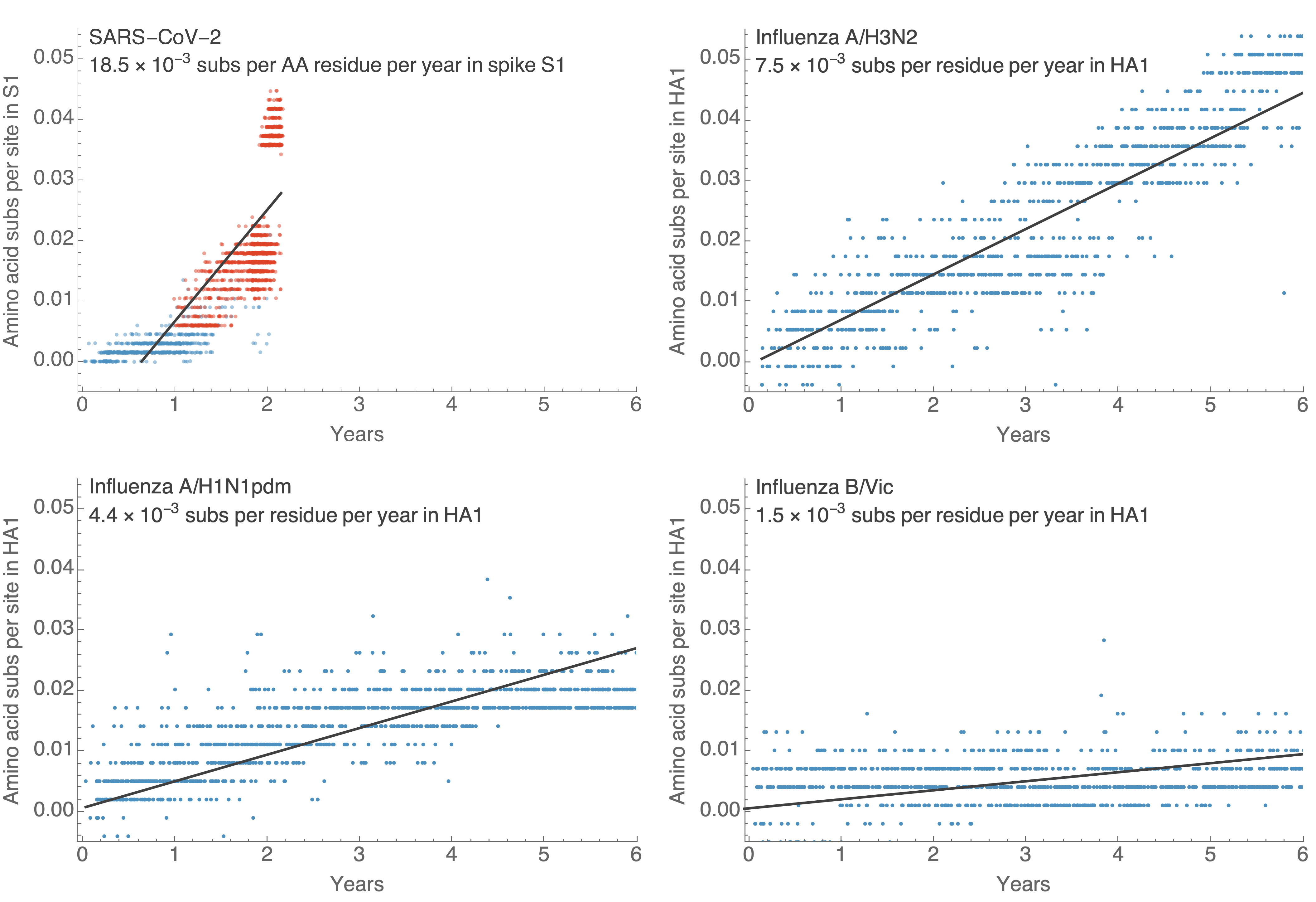
This is remarkably fast relative to seasonal influenza
Omicron spike mutations substantially drop VE against infection
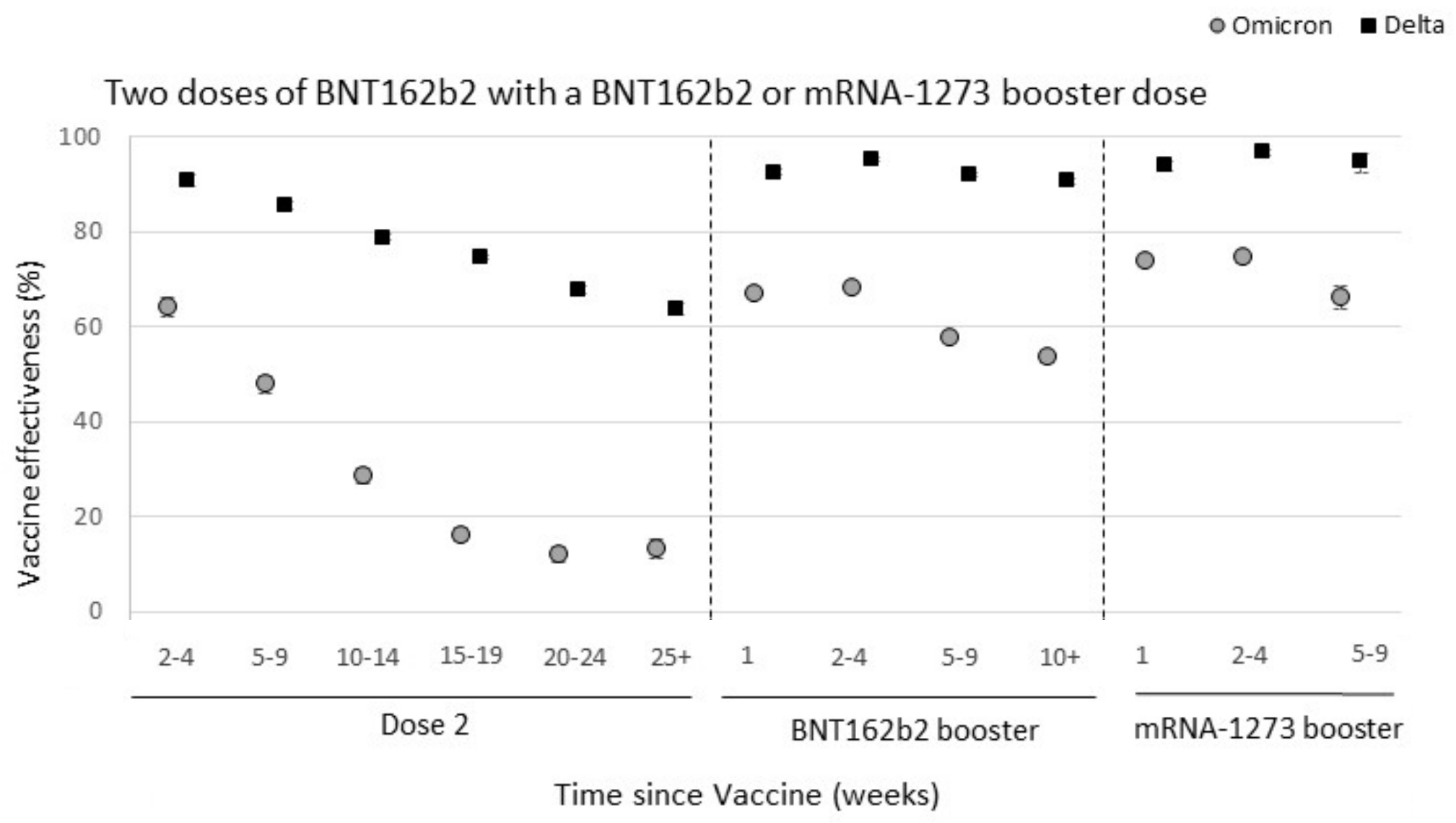
Significant immune escape drove large-scale epidemics
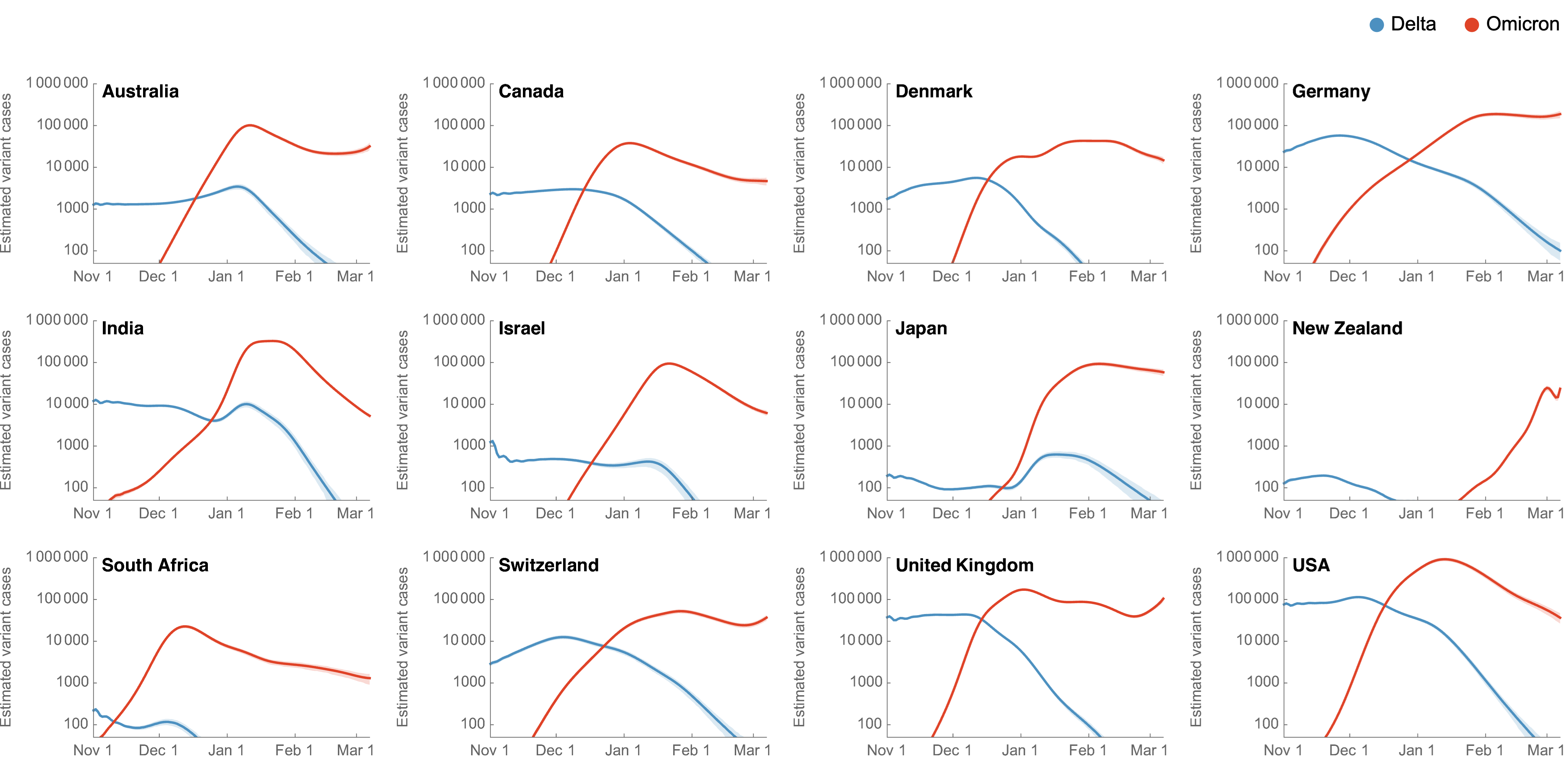
Omicron attack rate
- We estimate US has seen 9.8% of the population as confirmed cases of Omicron through Mar 1, with the large majority accumulating after Dec 15
- Assuming a case detection rate of 1 in 5 infections, we estimate almost 50% of the US infected with Omicron, most in the span of ~10 weeks
SARS-CoV-2 will continue to evolve to escape population immunity, though with multiple potential avenues

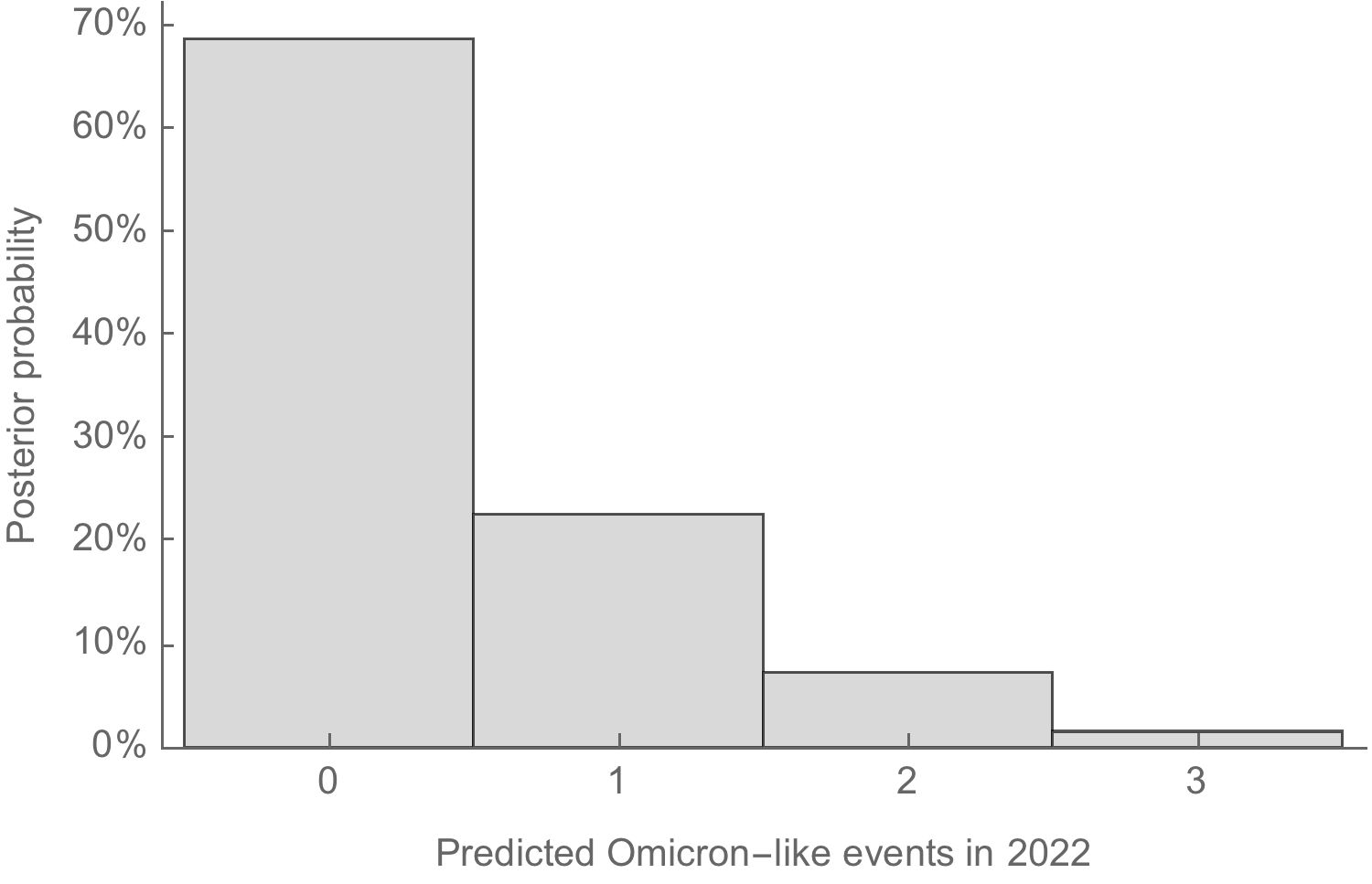
With 1 observation in 2.35 years of virus evolution, it's currently unclear how common Omicron-like events will be

This rate distribution gives a naive prediction of Omicron-like emergence events occurring in the next 12 months
Likely scenarios over the next 12 months
- (More likely) Evolution within Omicron BA.2 to further increase intrinsic transmission and to escape from Omicron-derived immunity. This scenario sees lower attack rates with 2022-2023 epidemic driven by drift + waning + seasonality.
- (Less likely) Another Omicron-like emergence event in which a chronic infection initiated in ~2021 incubates a new wildly divergent virus. This scenario sees high attack rates with epidemic driven by variant emergence.
Acknowledgements
SARS-CoV-2 genomic epi: Data producers from all over the world, GISAID and the Nextstrain team
Bedford Lab:
![]() John Huddleston,
John Huddleston,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Louise Moncla,
Louise Moncla,
![]() Maya Lewinsohn,
Maya Lewinsohn,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Cassia Wagner,
Cassia Wagner,
![]() Miguel Paredes,
Miguel Paredes,
![]() Nicola Müller,
Nicola Müller,
![]() Marlin Figgins,
Marlin Figgins,
![]() Denisse Sequeira,
Denisse Sequeira,
![]() Victor Lin,
Victor Lin,
![]() Jennifer Chang
Jennifer Chang






