Pathogen phylogenetics for decision making
Trevor Bedford (@trvrb)
Fred Hutchinson Cancer Center / Howard Hughes Medical Institute
14 Jul 2023
SURP Seminar
Fred Hutch
Slides at: bedford.io/talks
We work at the interface of virology, evolution and epidemiology
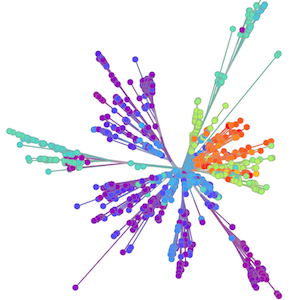
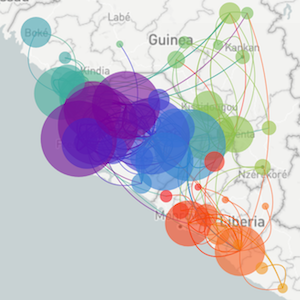

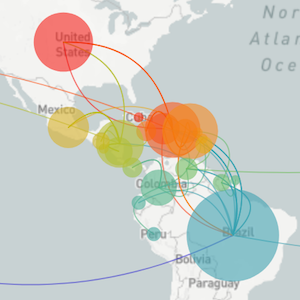
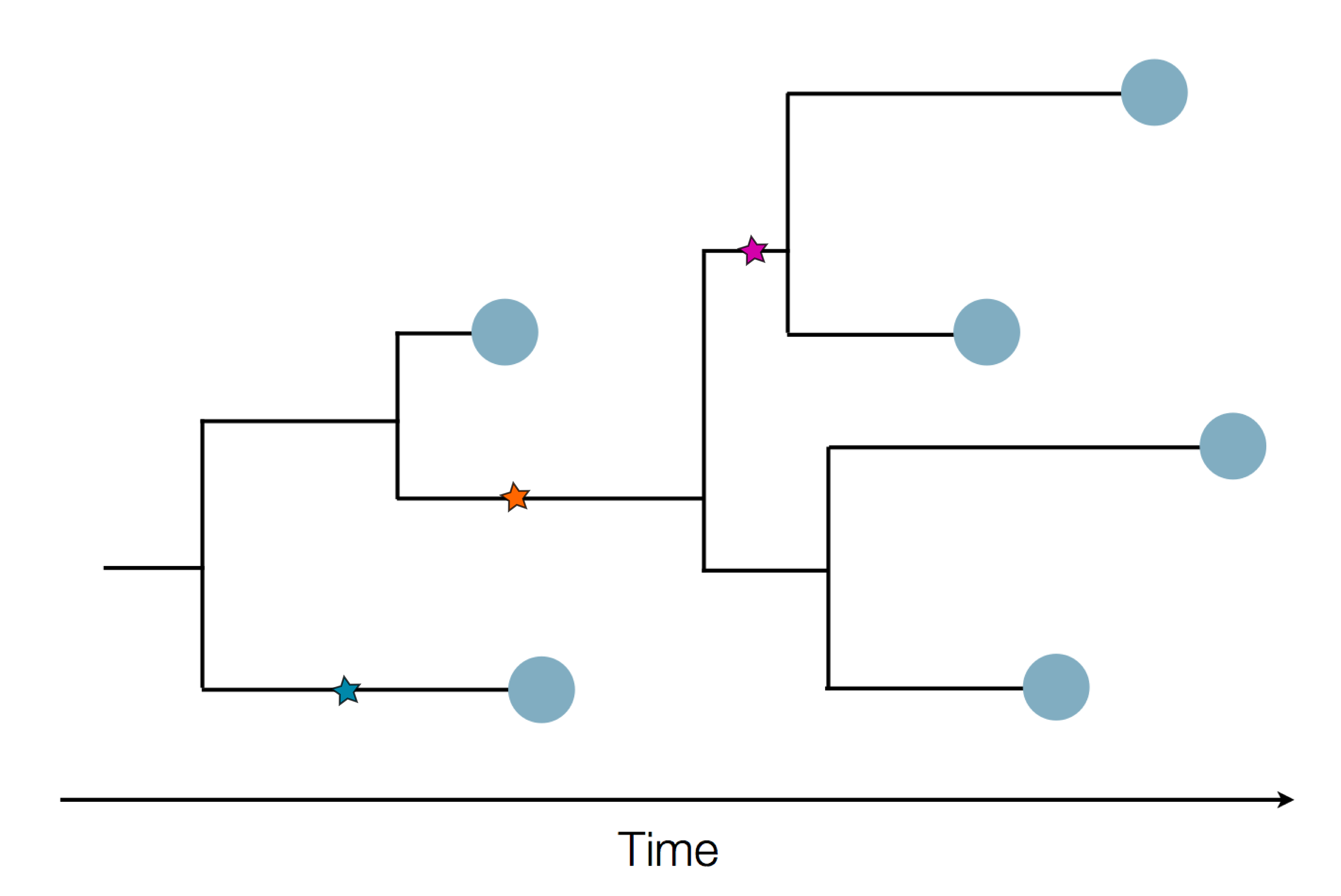
Sequencing to reconstruct pathogen spread
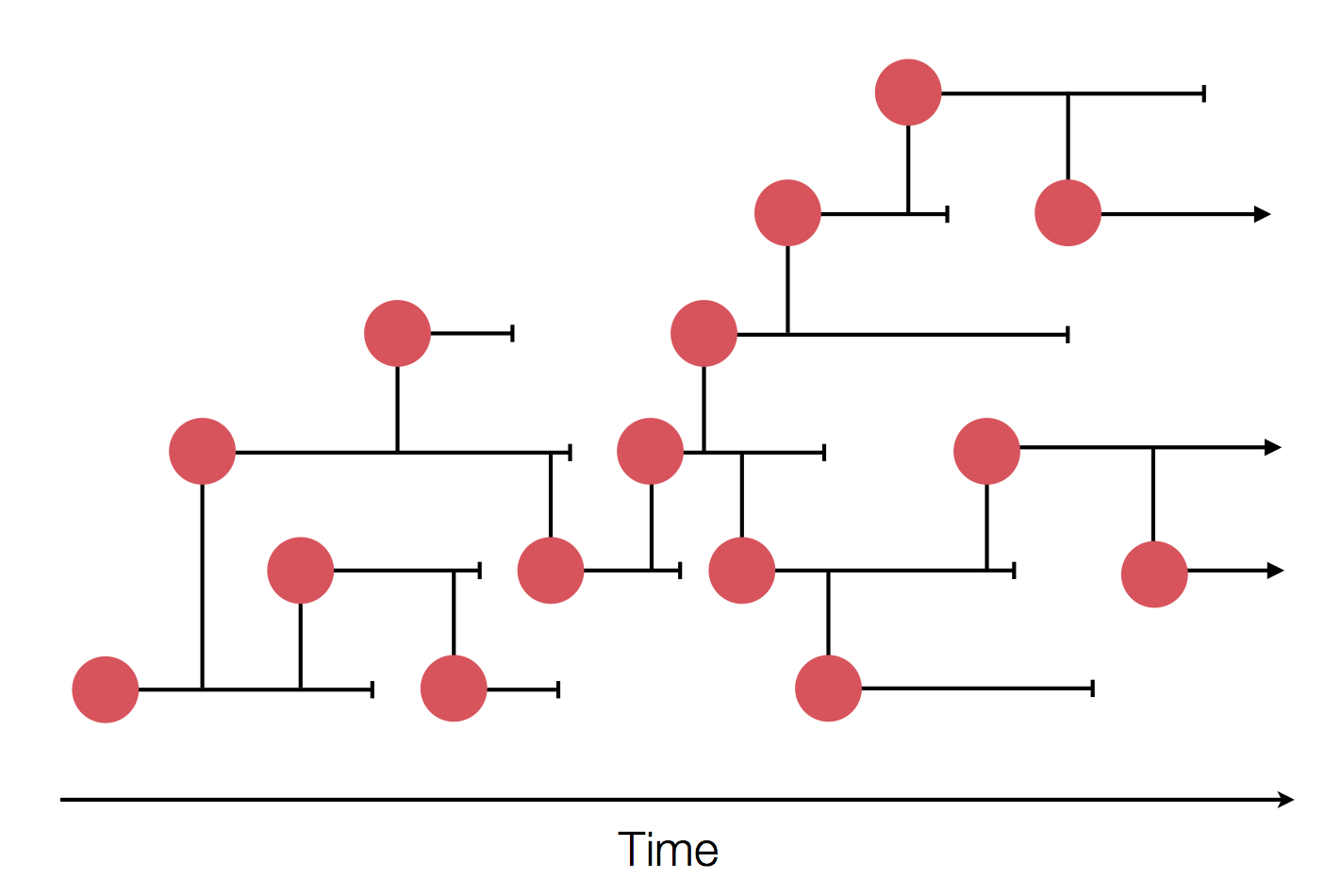
Epidemic process
Sample some individuals
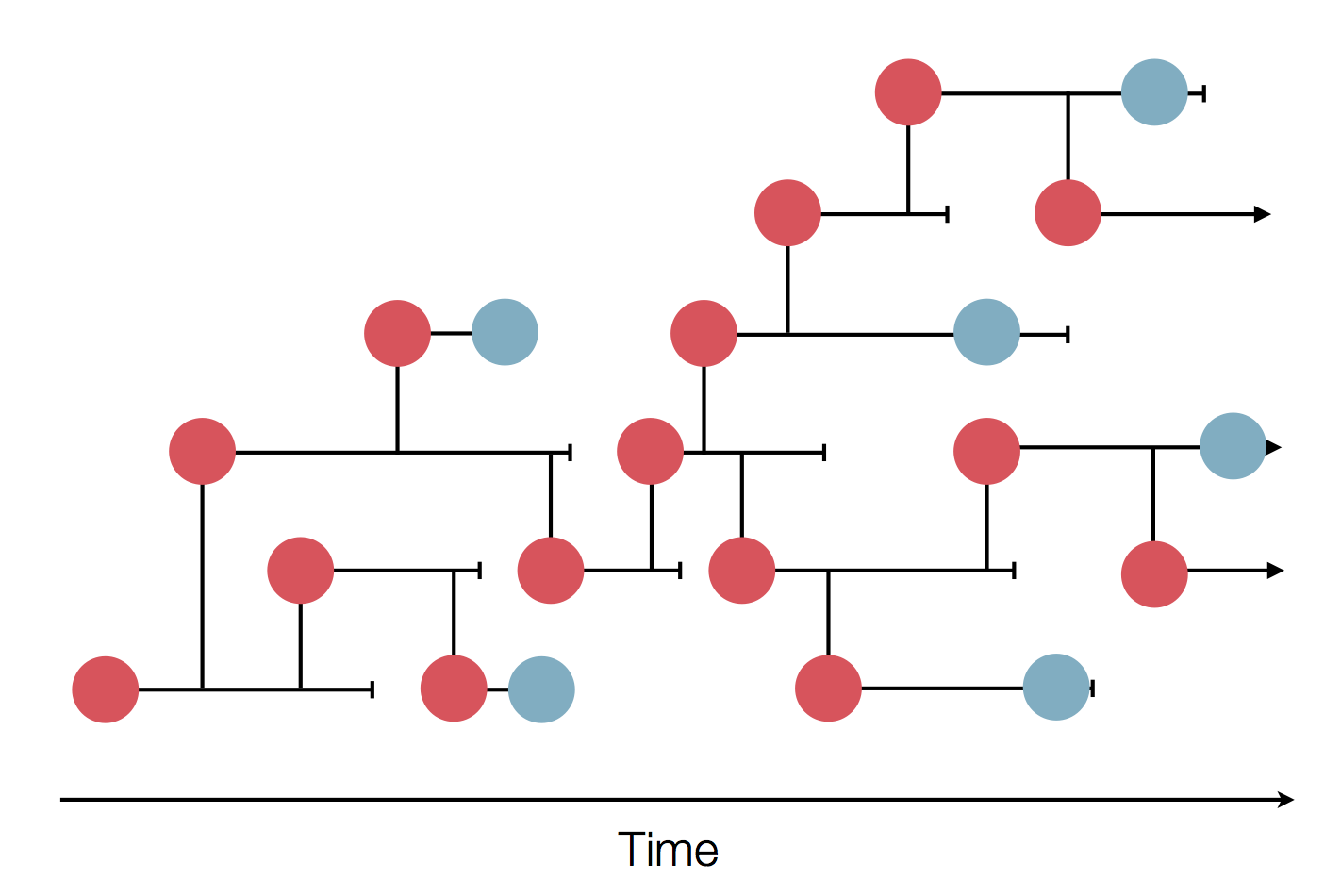
Sequence and determine phylogeny
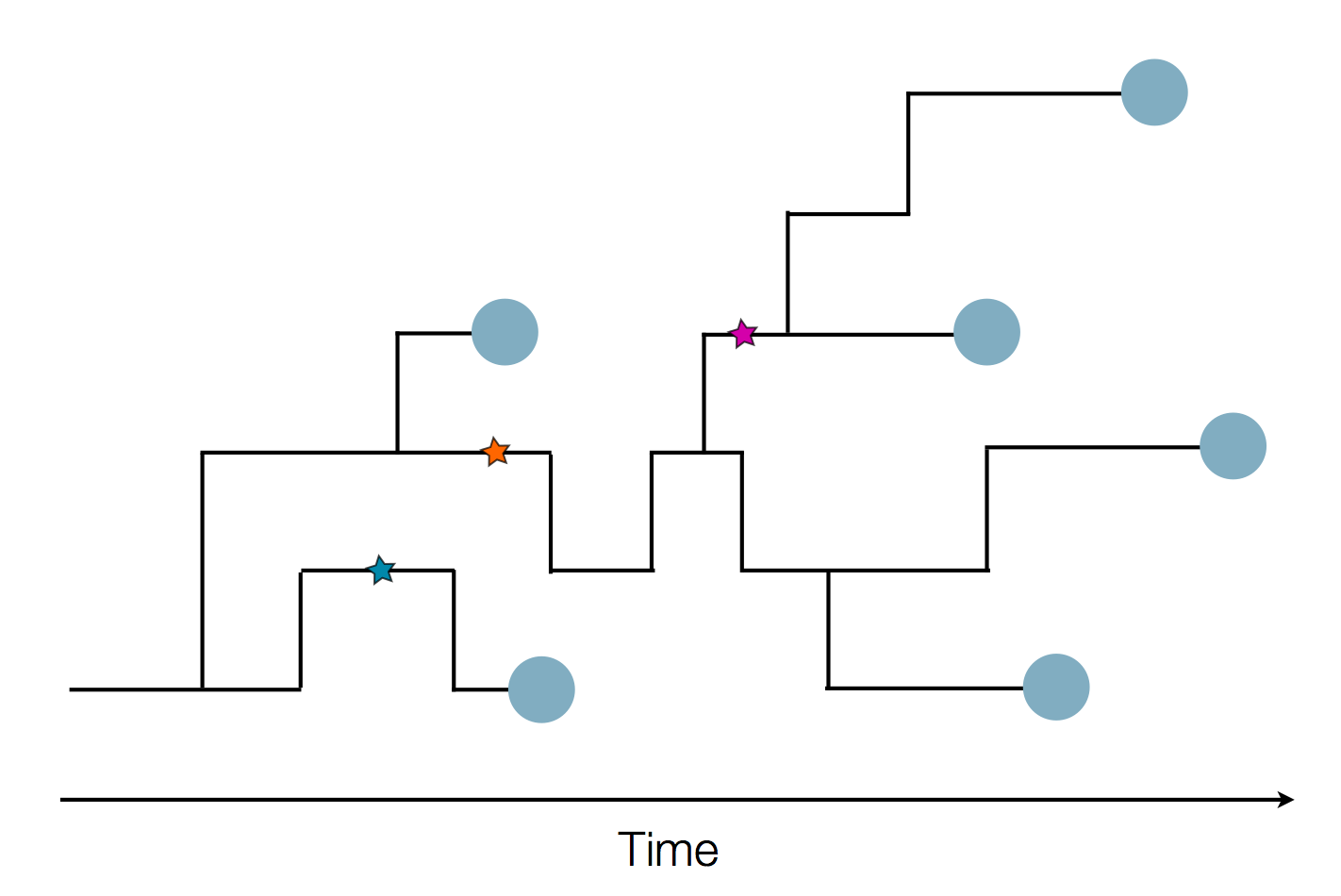
Sequence and determine phylogeny
Genomic epidemiology during the COVID-19 pandemic
Genomic epidemiology during the COVID-19 pandemic
(Or my experiences as a very public scientist during the COVID-19 pandemic)
Wuhan emergence and human-to-human spread
Jan 11: First five genomes from Wuhan showed a novel SARS-like coronavirus
Initially thought clustering due to epi investigation of linked cases at Huanan seafood market
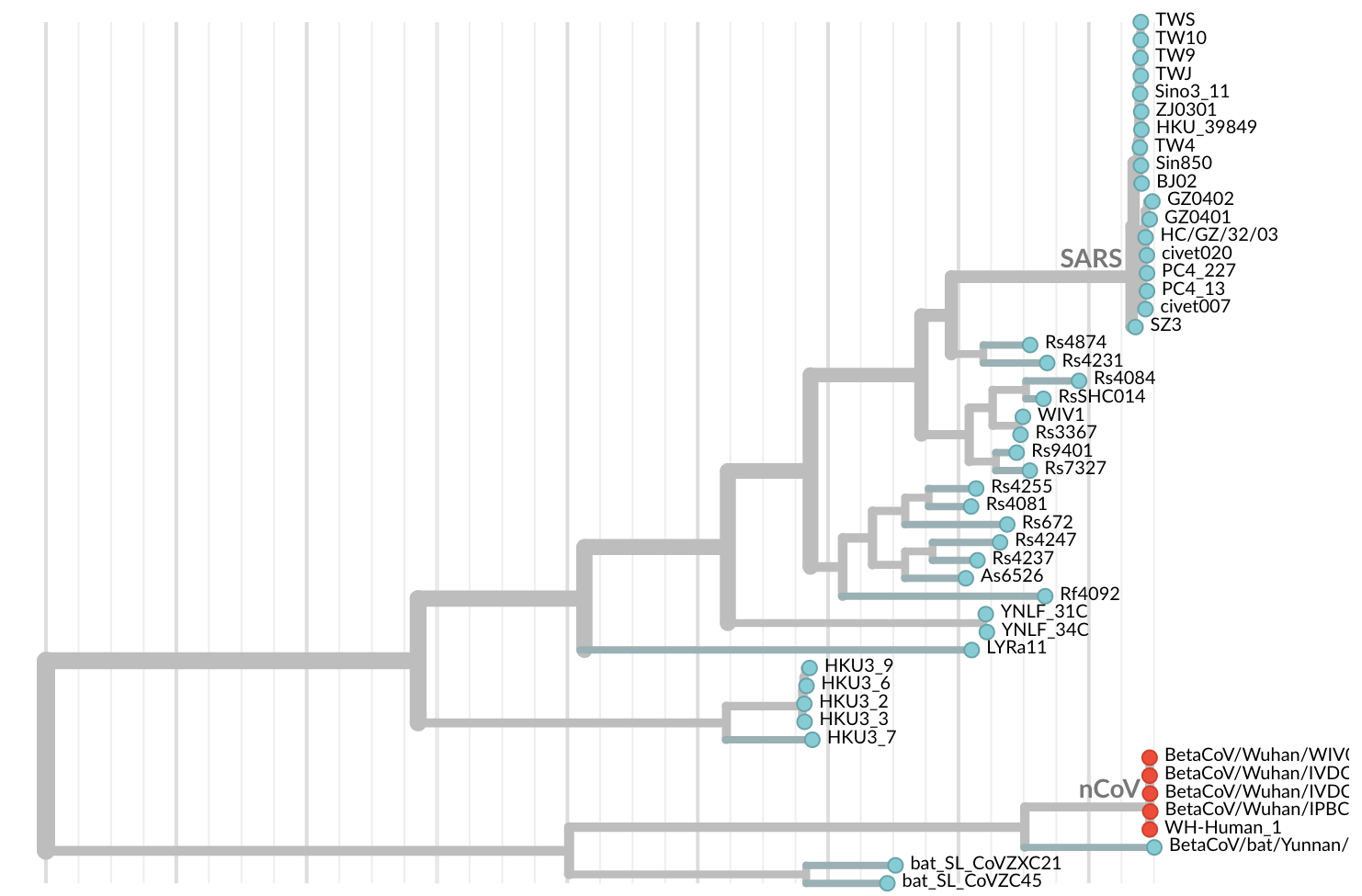
Jan 19: First 12 genomes from Wuhan (blue) and Bangkok (red) showed lack of genetic diversity
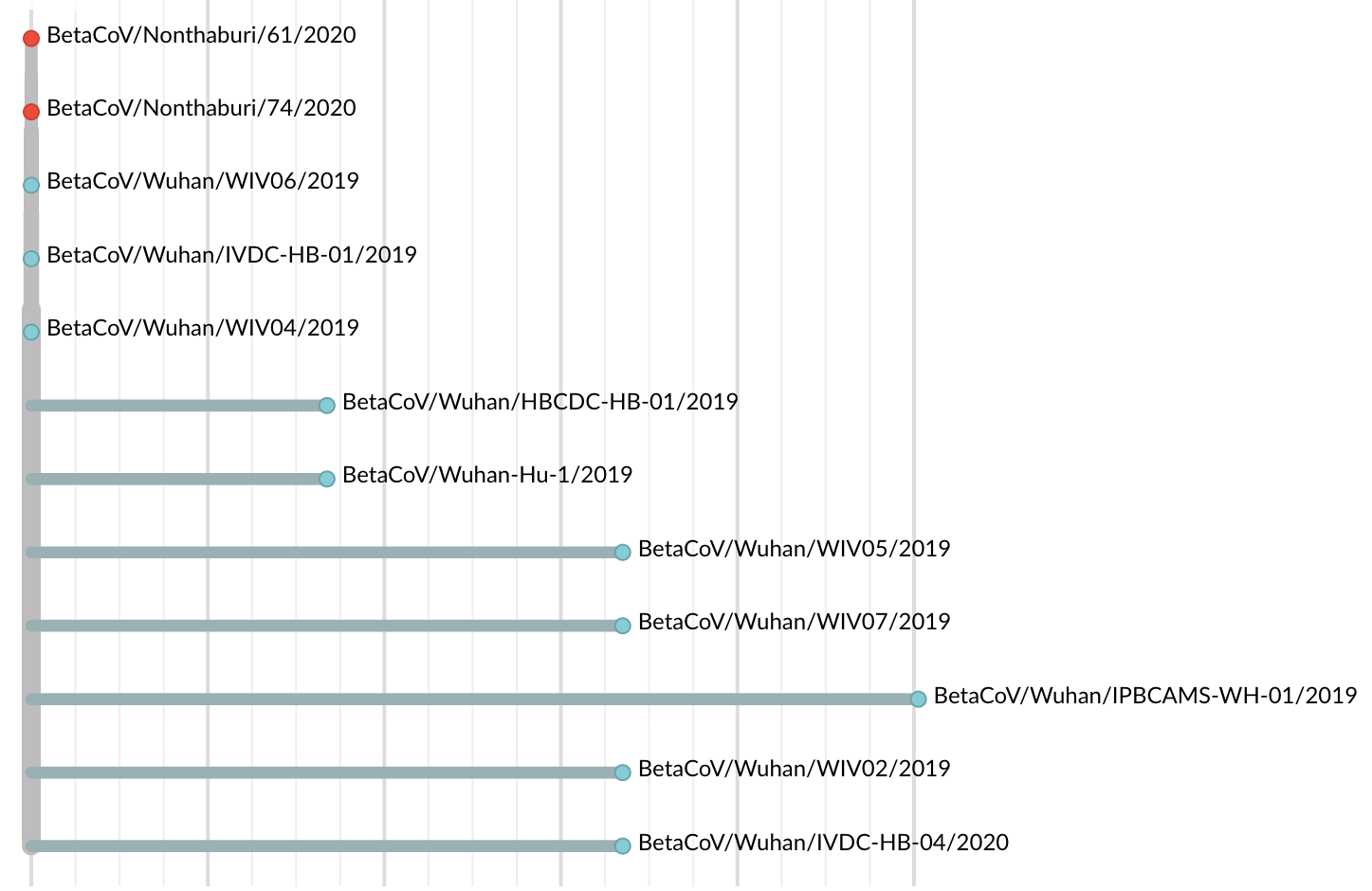
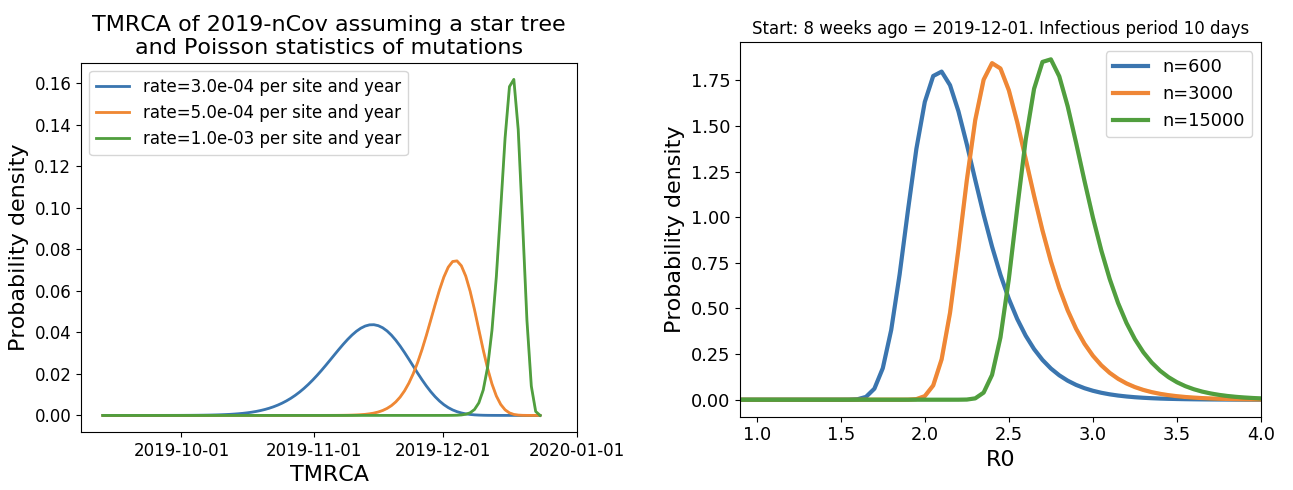
Jan 23: Introduction into the human population between Nov 15 and Dec 15 and subsequent rapid human-to-human spread
Jan 23: Email blast to colleagues at PHSKC, WA DOH, CDC, BMGF, NIH, UW, Fred Hutch

Jan 26: Media reporting based on technical report and interviews

Ongoing tracking of genomic data via Nextstrain
- Phylogenetic analysis at nextstrain.org/ncov up since Jan 19, 2020
- During this time, watching GISAID and attempting immediate updates as new data appeared
- Update process became increasing automated over the subsequent weeks and months

Feb 14: AAAS meeting and BMGF dinner
- Most media interest at the time was on "open science" angle, rather than "pandemic" angle, no one seemed to want to hear the bad news
- I gave press conference and presentation at the AAAS Annual Meeting, concluding "We have not yet had time to ascertain effects of intervention measures, but my hope for containment is slim"
- Dinner with directors of the BMGF where I presented to Mr. Gates on genomic conclusions
Following Tufte's advice, I printed out a handout


Cryptic transmission and testing in Seattle
 Seattle Flu Study
Seattle Flu Study
Project initiated in 2018-2019 season and continued into the 2019-2020 flu season
Lead investigators: Helen Chu, Michael Boeckh, Janet Englund, Michael Famulare, Barry Lutz, Deborah Nickerson, Mark Rieder, Lea Starita, Matthew Thompson, Trevor Bedford, Jay Shendure
Co-investigators: Amanda Adler, Jeris Bosua, Elisabeth Brandstetter, Kairsten Fay, Chris Frazar, Peter Han, Reena Gulati, James Hadfield, ShiChu Huang, Misja Ilcisin, Michael Jackson, Anahita Kiavand, Louise Kimball, Enos Kline, Kirsten Lacombe, Jover Lee, Jennifer Logue, Victoria Lyon, Kira Newman, Miguel Paredes, Thomas Sibley, Monica Zigman Suchsland, Cassia Wagner, Caitlin Wolf




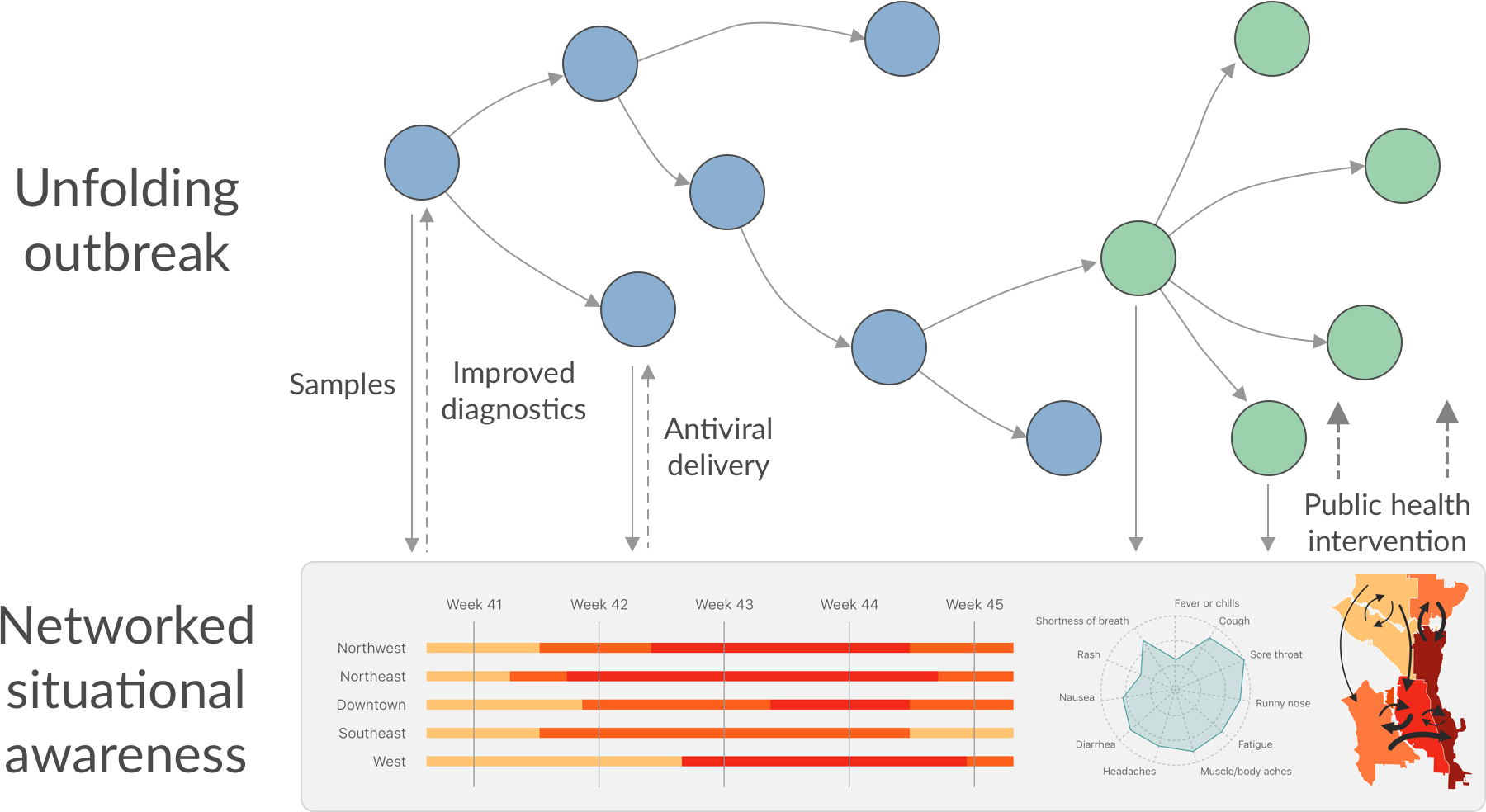
Feb 2020: Struggle to test samples that were in hand

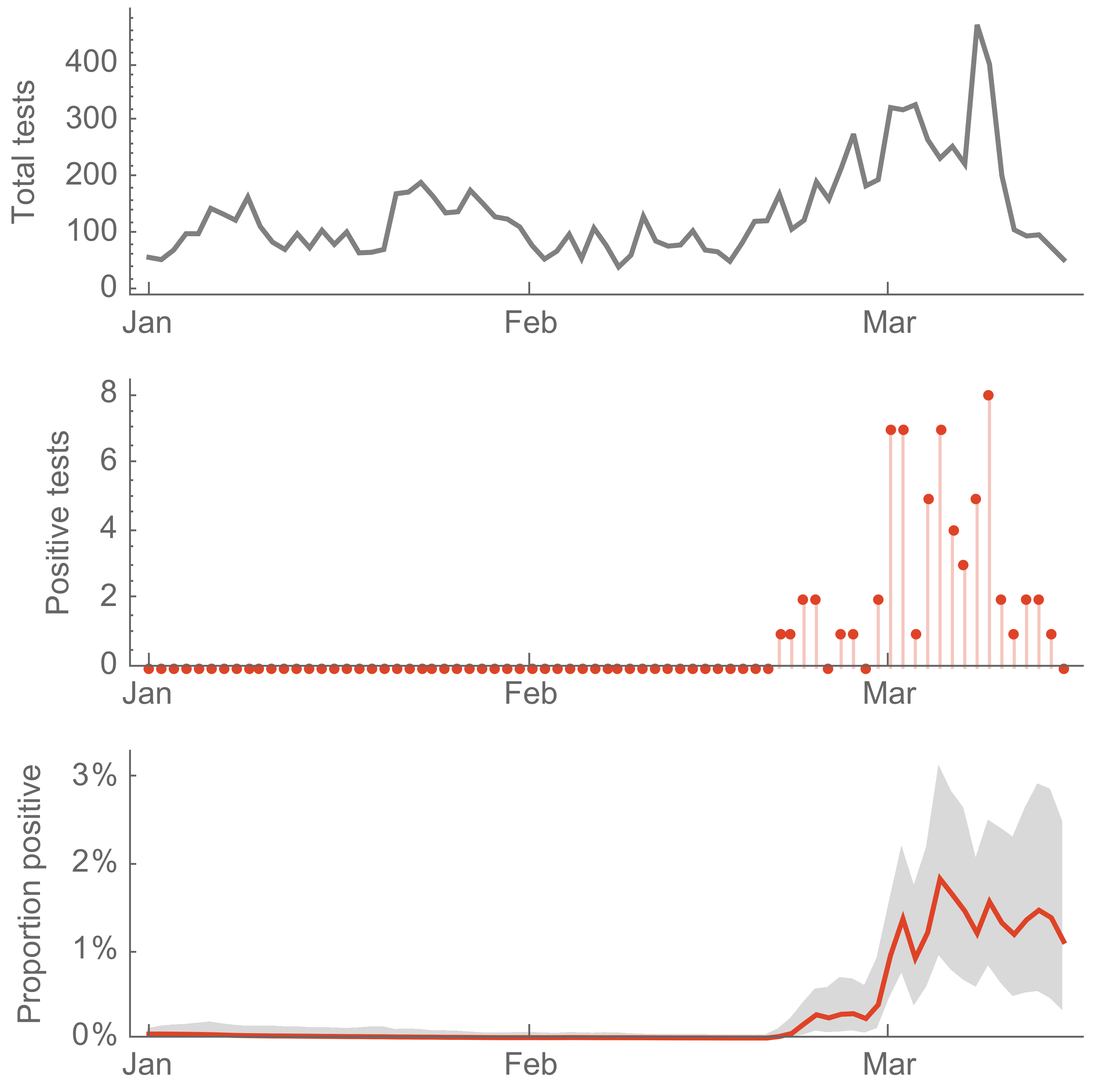
We started testing samples on Tue Feb 24 with capacity for ~400 tests a day and find the first positive on Thur Feb 27
Sequencing this positive showed surprising connection




Calls with Mayor Durkan and Governor Inslee, which focus on understanding results and modeled expectations for epidemic spread

Screening of acute respiratory infections for SARS-CoV-2
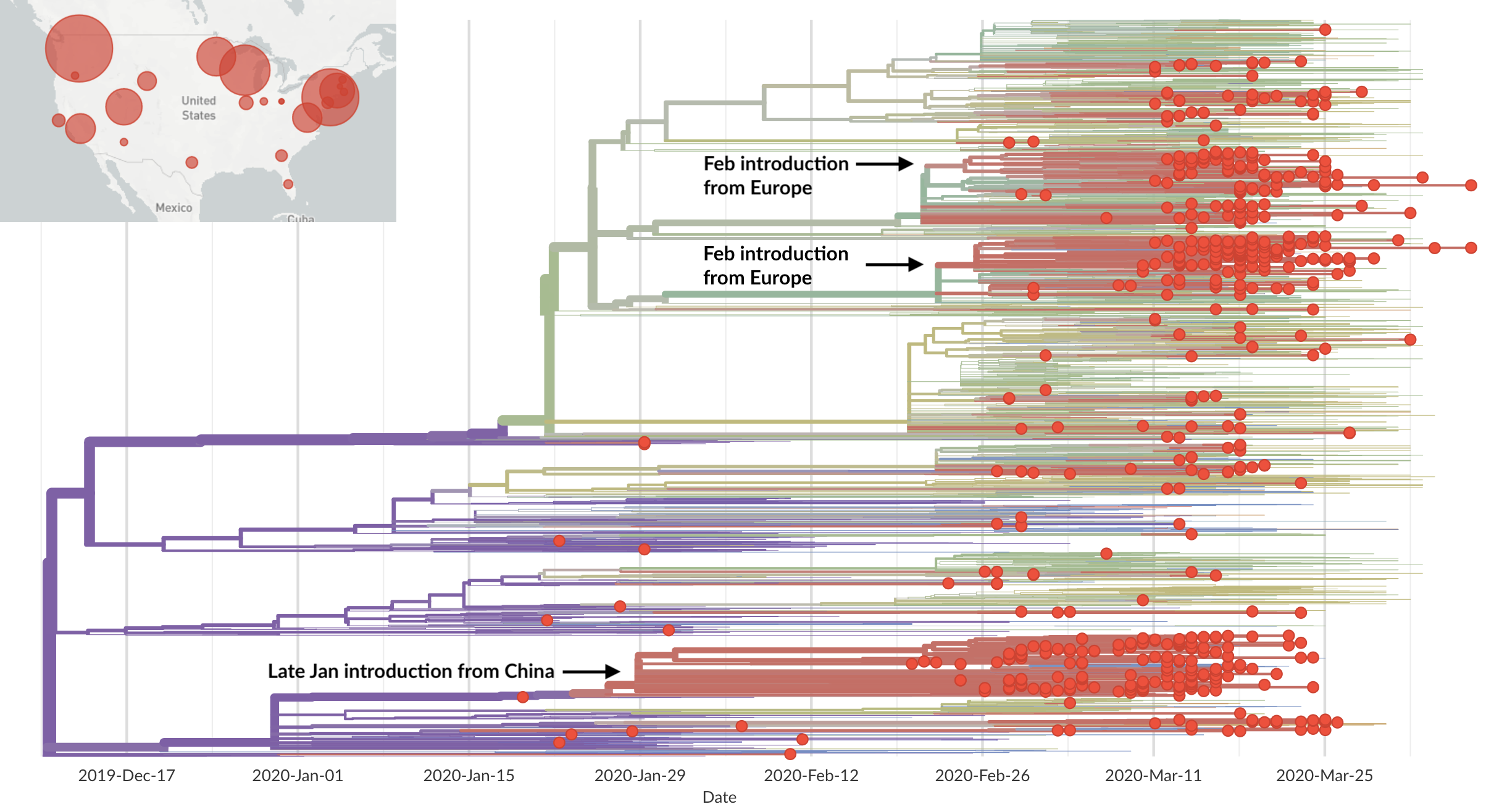
Sequencing of viruses collected prior to March 15 detects origins and rate of local spread
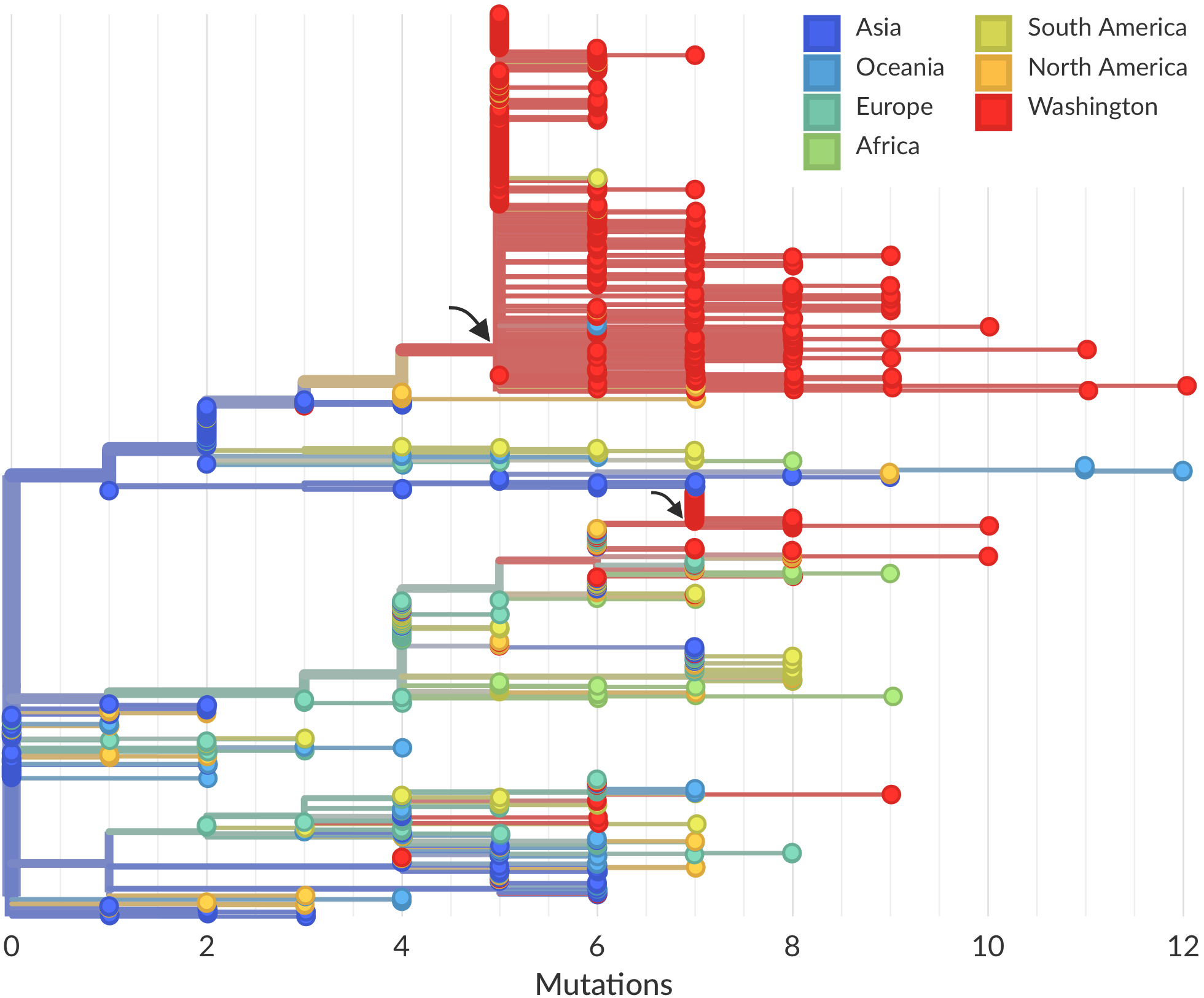
Sequencing of viruses collected prior to March 15 detects origins and rate of local spread
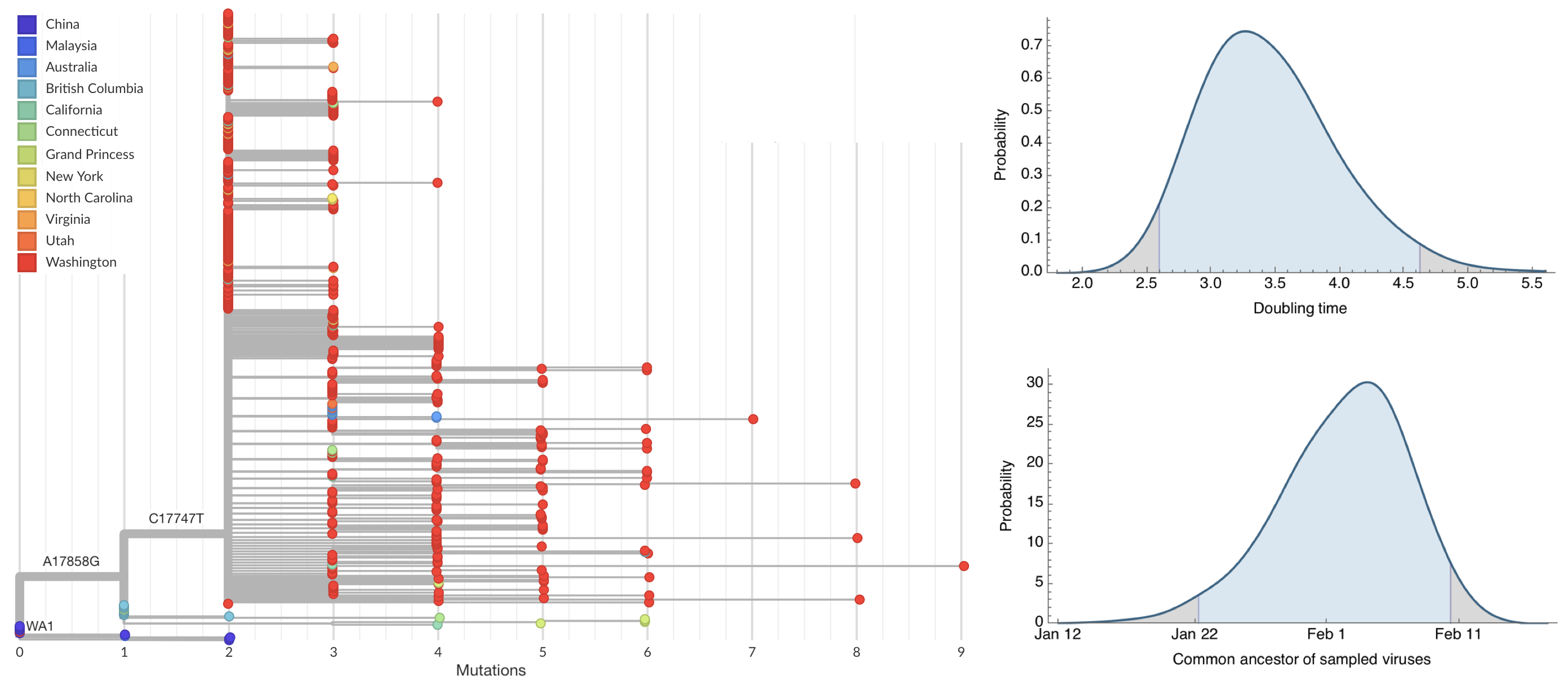
Rare introduction from China that spread widely, most US epidemic arrived via Europe
Continued public communication
Engagement with scientists, public health, policy makers and public through Twitter
- I discover a strategy in which I can dive deeply into a topic / question, present results on Twitter and then take interviews / meetings from reporters / colleagues
- Multiple reporters can pull quote from Twitter rather than having to do repeated interviews

Broadly, I make it my goal to help public and policy makers understand what's happening with the pandemic. Although there are practical applications of genomic epi and modeling for specific pathogens, I think that understanding is really what these approaches offer more broadly.
I've had many conversations over the course of the pandemic, but I don't think there's been any fundamental difference between conversations with reporters, policy makers or friends and family. In each case, I'm trying to convey my understanding of the world and uncertainty of this understanding in a fashion that's comprehensible.
I believe public health messaging has been repeatedly scientifically subverted with messaging for intended behavior. This was seen with messaging over masks, airborne transmission, natural immunity, B.1.1.7 causing more severe illness, etc...

Phylogenetics or genomic epi, by itself, is just one avenue towards this sort of understanding, and should be combined with other sources to model / understand what's going on.
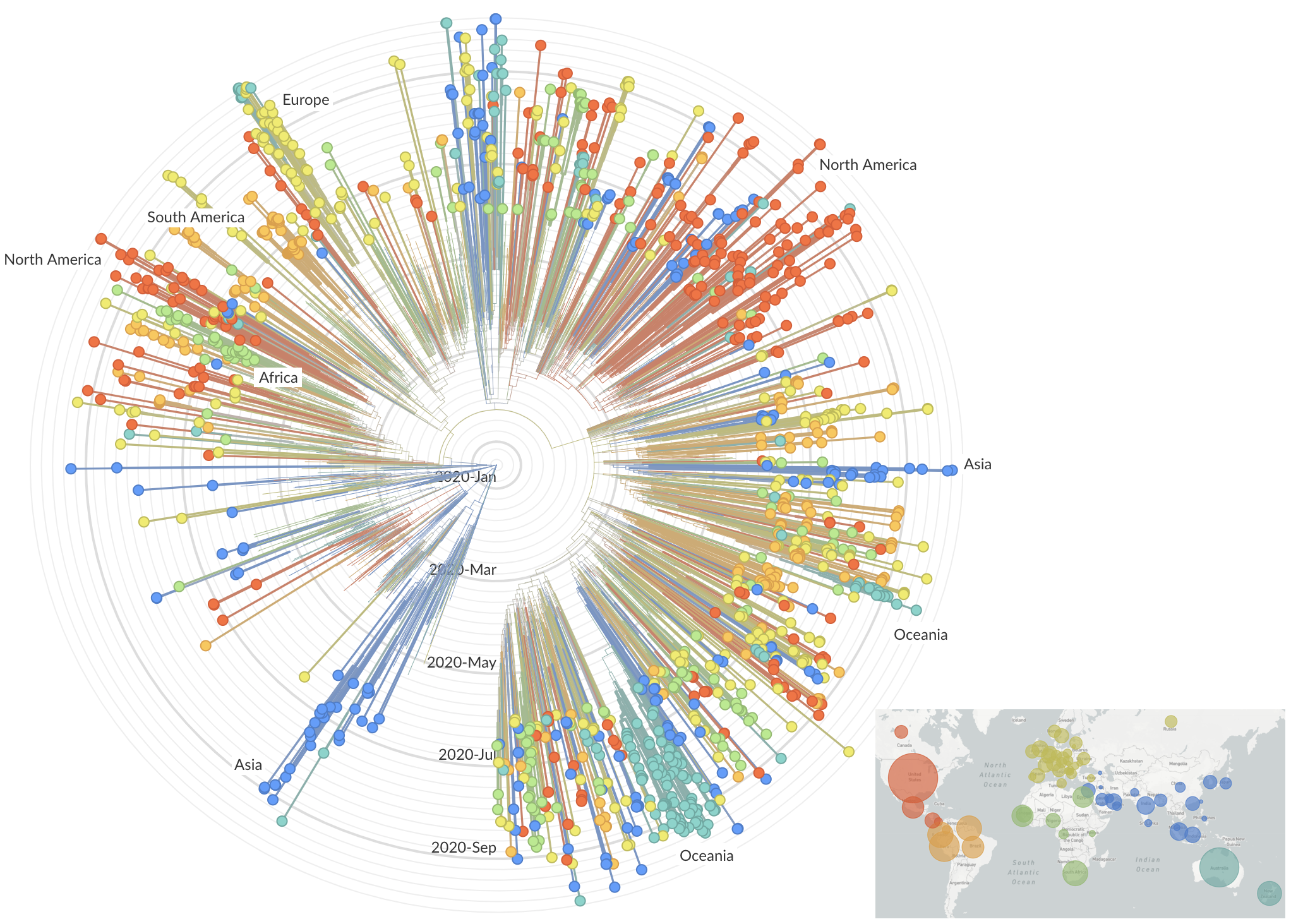
Emergence of variants of concern
After initial wave, with mitigation
efforts and decreased travel,
regional clades emerge
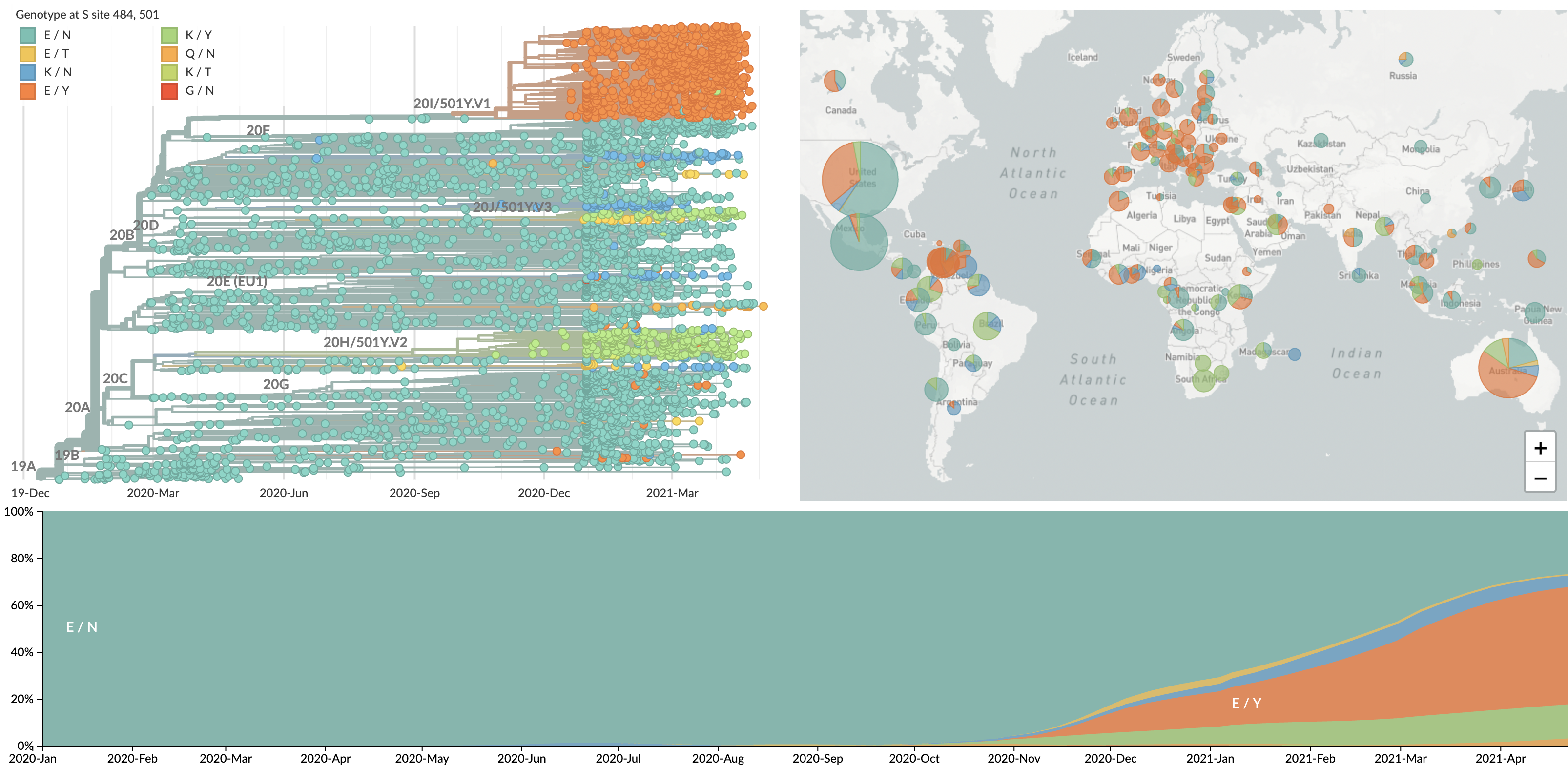
Repeated emergence of 484K and 501Y across the world
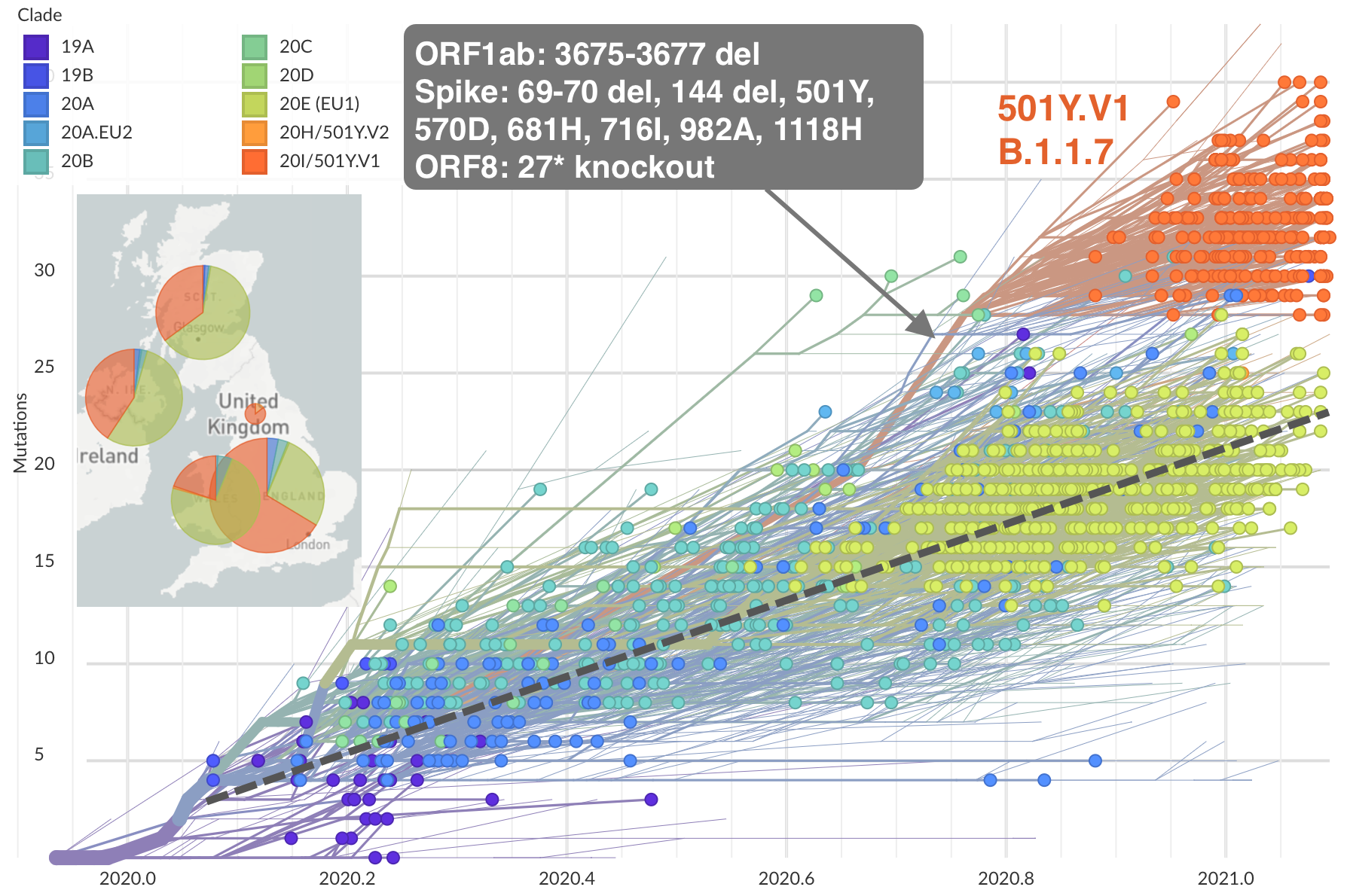
Emergence of Alpha (B.1.1.7) in the UK
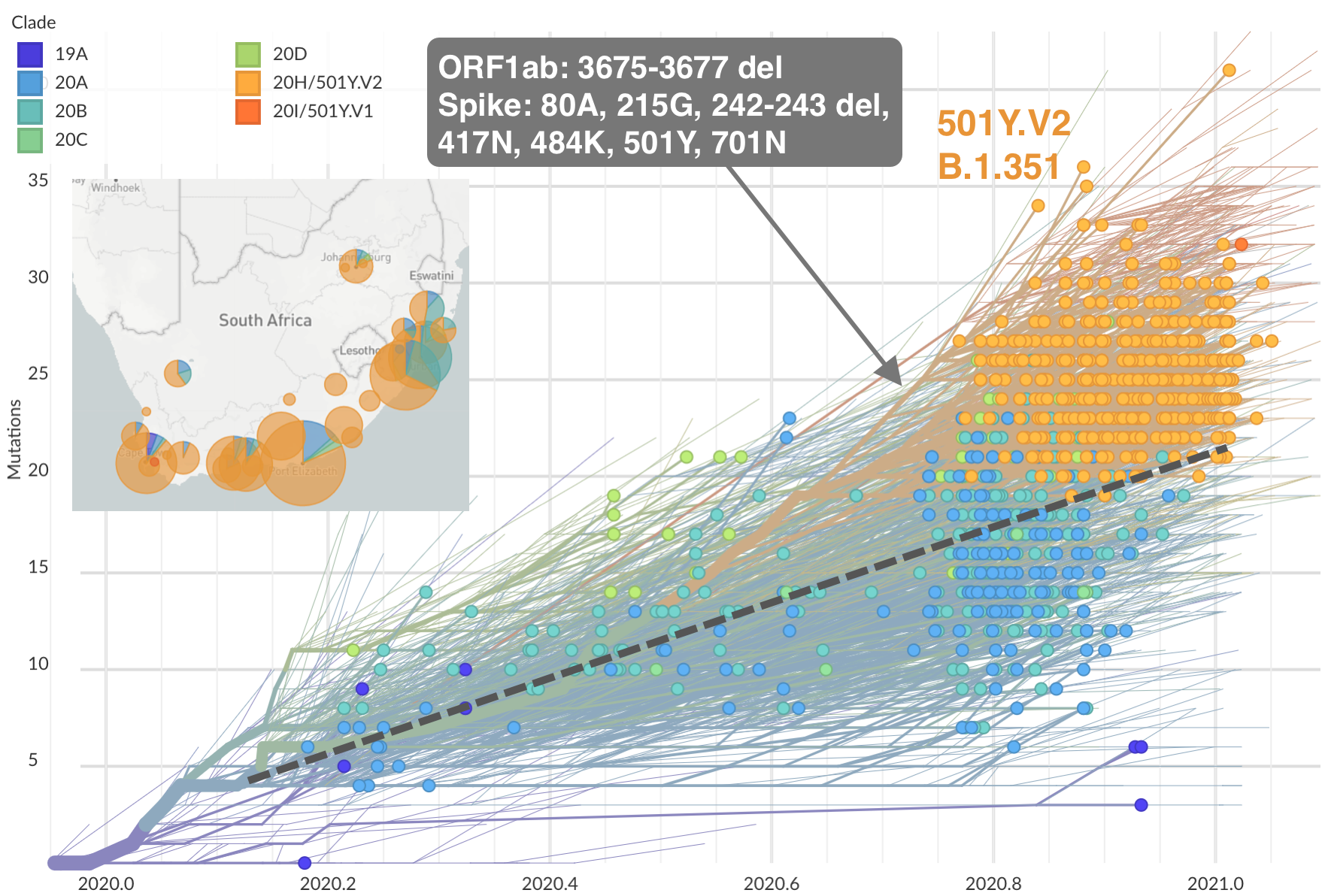
Emergence of Beta (B.1.351) in the South Africa
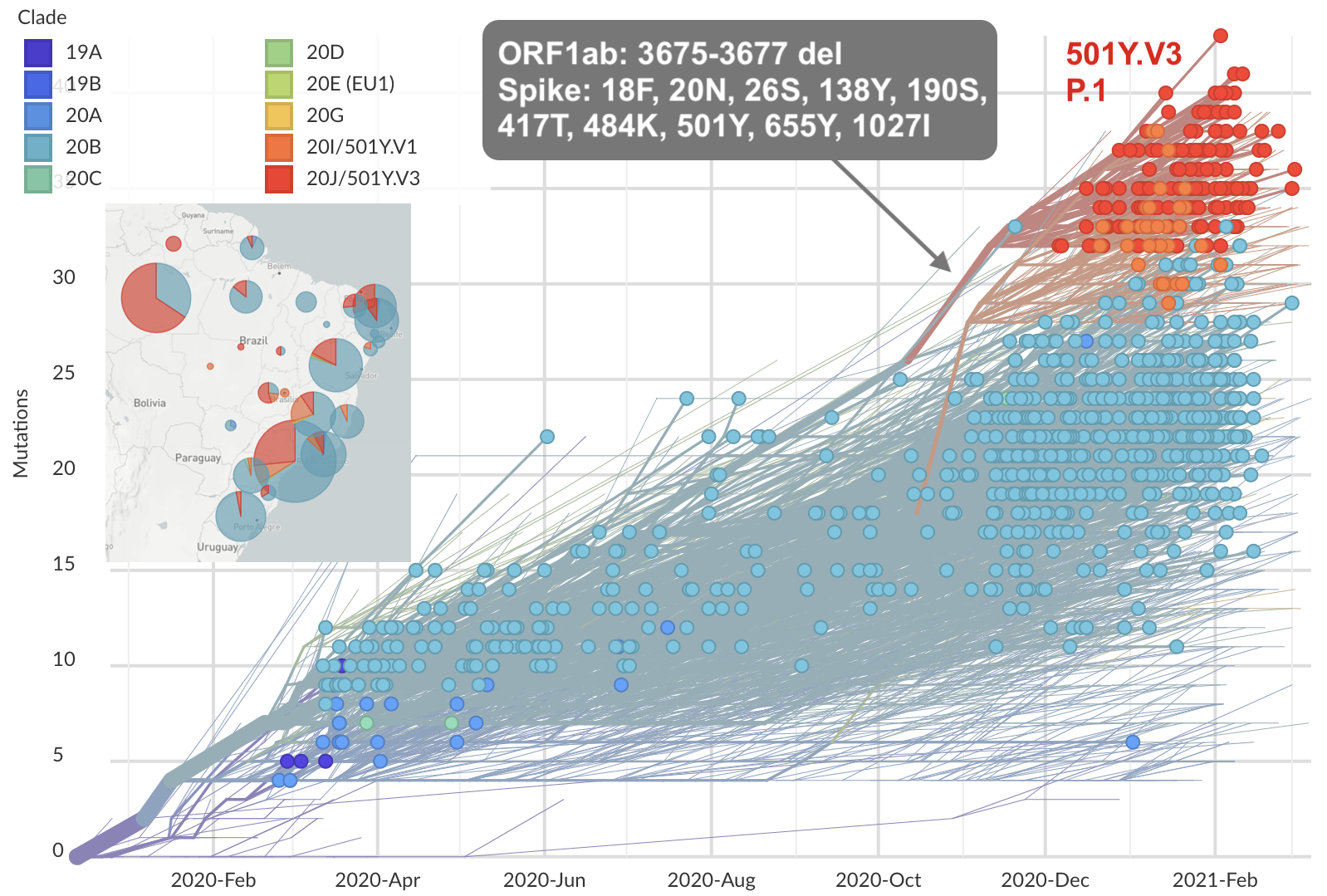
Emergence of Gamma (P.1) in the Brazil
Lobbying for improved genomic surveillance
Disappointing that focus has been on within-country sequencing rather than global surveillance


Understanding characteristics and origins of variant viruses


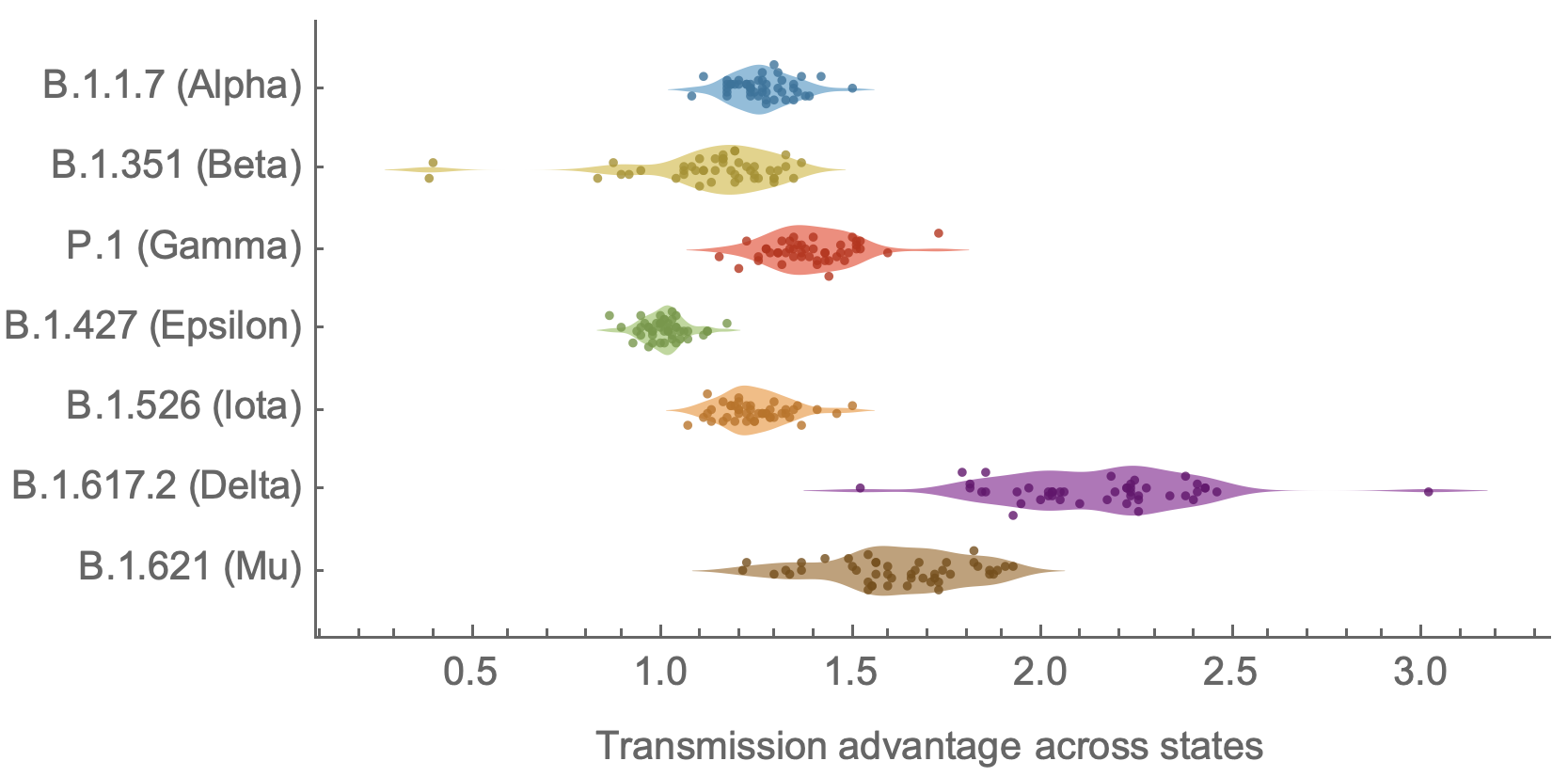
Increasingly, focus on tracking variant spread and estimating growth rates

Consistent differences in variant-specific transmission rate across states
Emergence of Omicron variant
Nov 26: Lineage B.1.1.539 / clade 21K / Omicron variant emerging from basal diversity
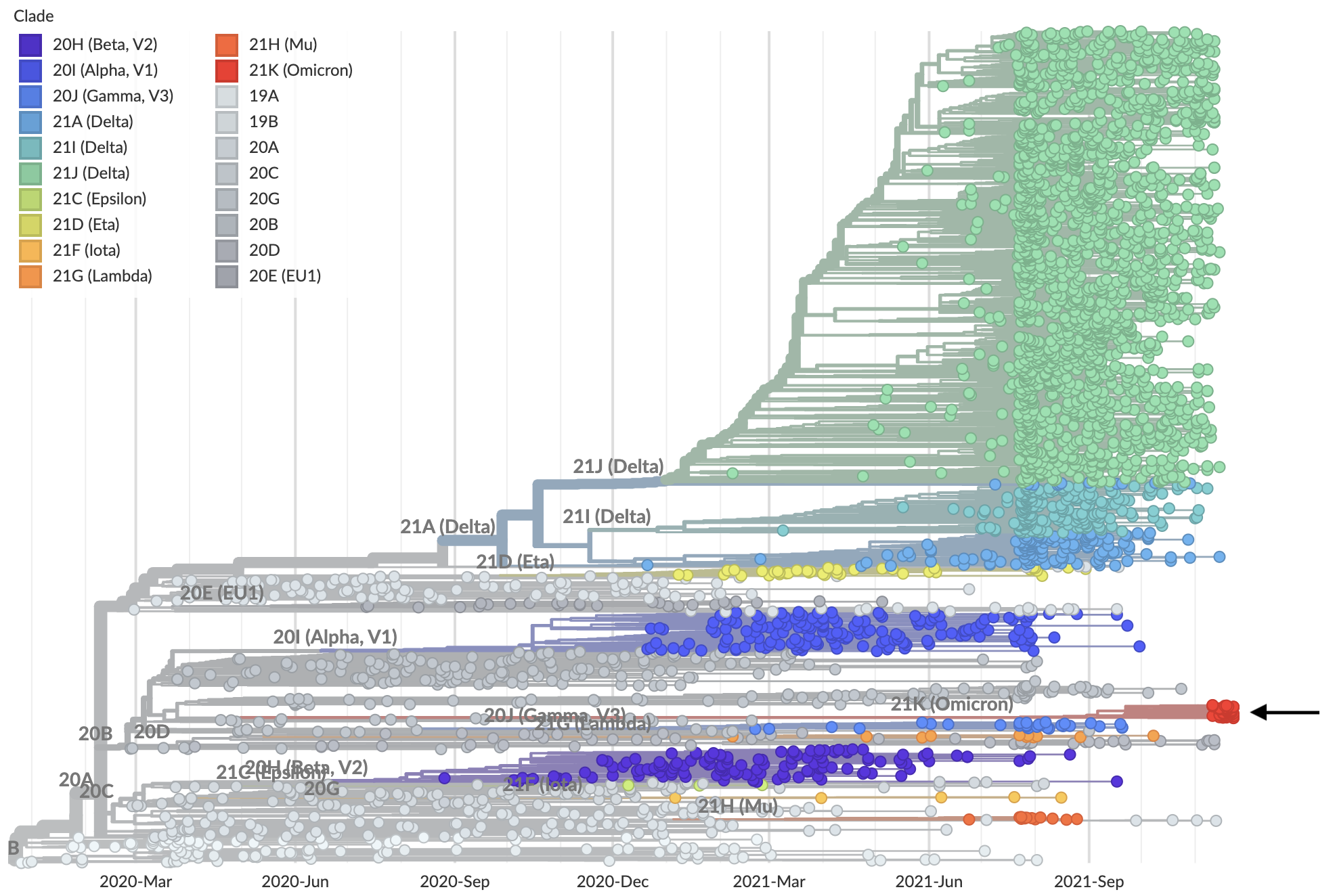
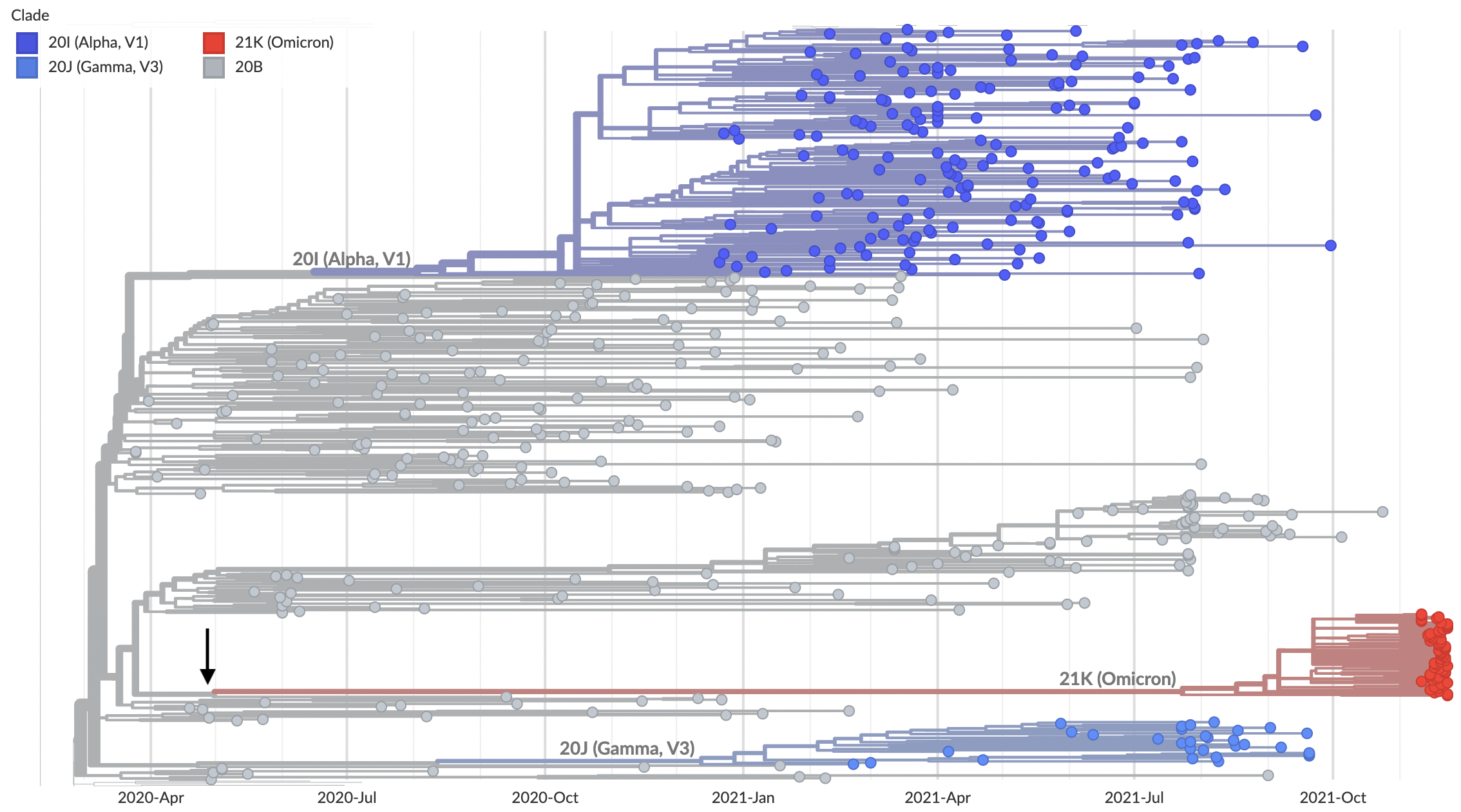
Nov 26: Long branch connecting closest sequenced viruses
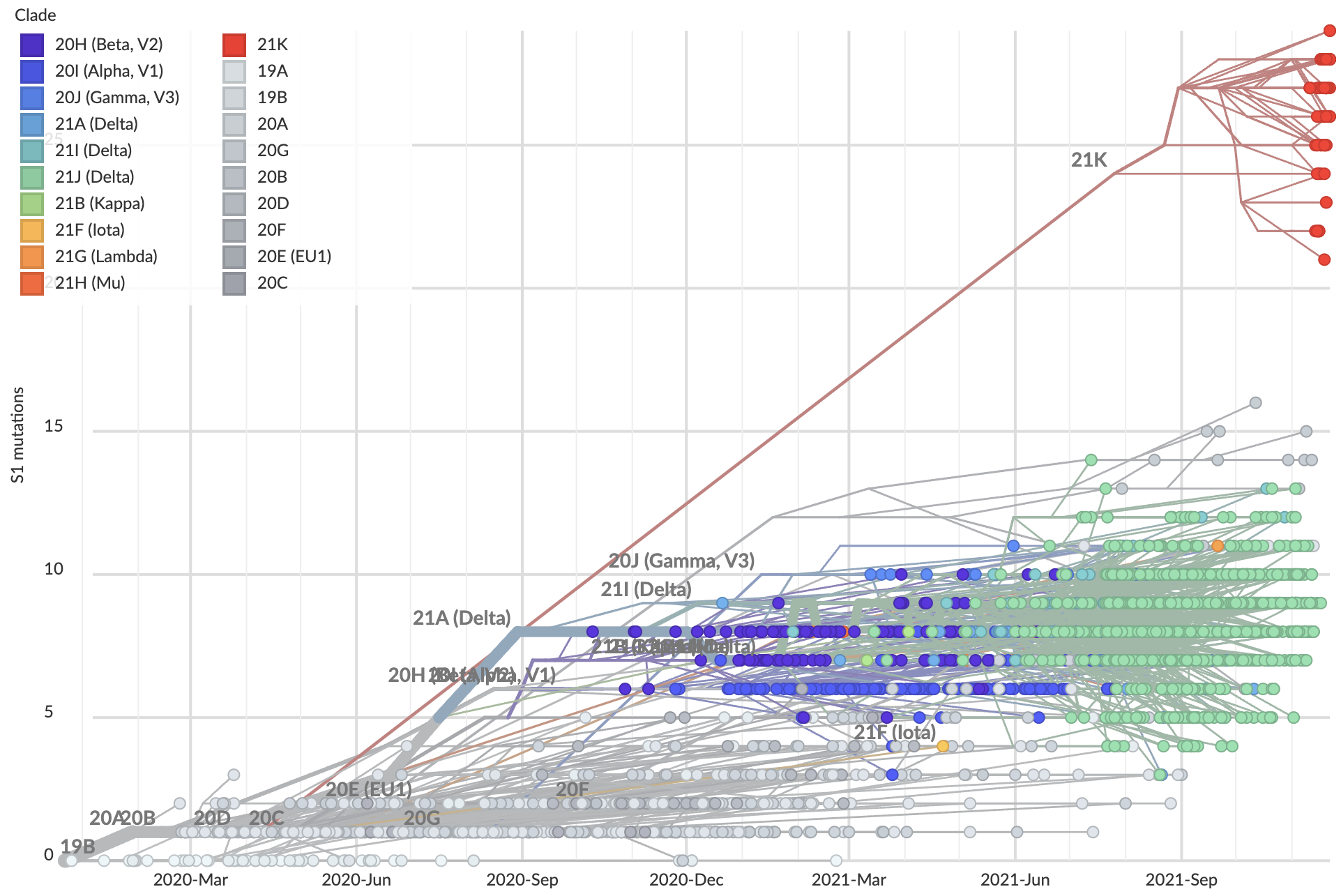
Nov 26: Omicron viruses with huge excess of mutations in S1
Dec 4: Projections from rapid epidemic spread in South Africa

Dec 4: Projections from rapid epidemic spread in South Africa


Dec 16: Warning of large incipient Omicron epidemics

Warning public health, policy makers and the public of timing and intensity of incoming Omicron wave.
Given decrease in individual-level severity, policy maker worry primarily concerned hospital capacity

Continuing evolution of SARS-CoV-2 post-Omicron
Genetic relationships of globally sampled SARS-CoV-2 to present

Rapid displacement of existing diversity by emerging variants

Mutations in S1 domain of spike protein driving displacement

S1 evolution remarkably fast relative to seasonal influenza

Continued escape from neutralization by existing population immunity necessitating vaccine updates
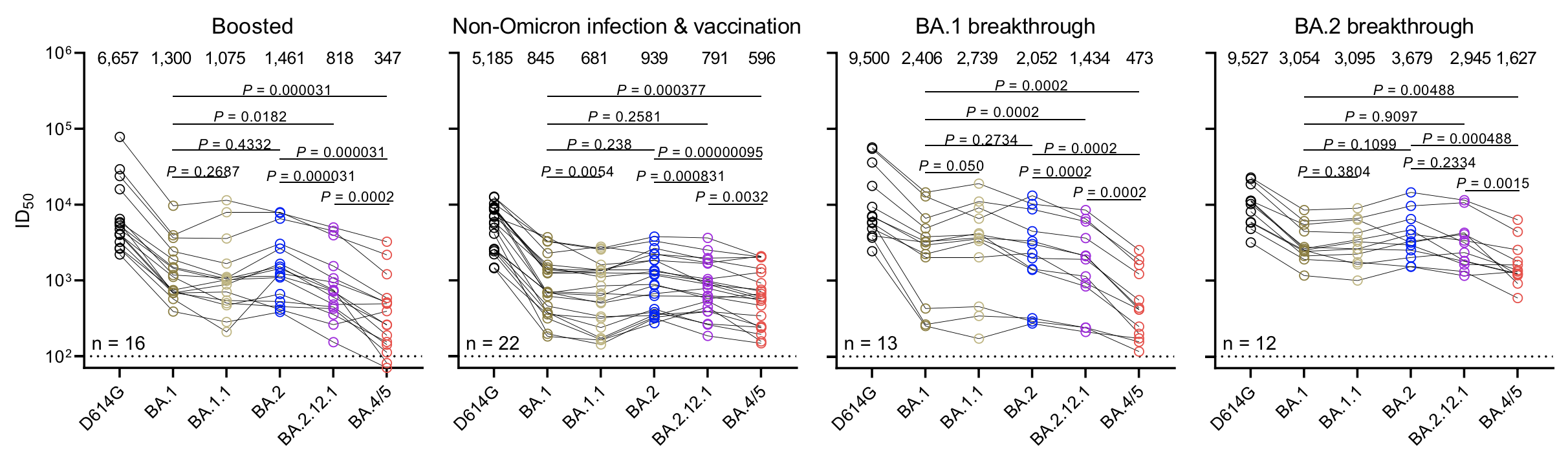
Takeaways
- Phylogenetics and genomic epidemiology most impactful early on, when case-based surveillance is poor
- Phylogenetic analysis should be combined with other sources of knowledge
- I believe in transparency and public sharing of scientific results / understanding, even if the primary audience is other scientists
- My (admittedly) ivory tower perspective highlights the critical need to preserve scientific accuracy over falling prey to well meaning propaganda

Acknowledgements
SARS-CoV-2 genomic epi: Data producers from all over the world, GISAID and the Nextstrain team
Bedford Lab:
![]() John Huddleston,
John Huddleston,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Cassia Wagner,
Cassia Wagner,
![]() Miguel Paredes,
Miguel Paredes,
![]() Nicola Müller,
Nicola Müller,
![]() Marlin Figgins,
Marlin Figgins,
![]() Victor Lin,
Victor Lin,
![]() Jennifer Chang,
Jennifer Chang,
![]() Allison Li,
Allison Li,
![]() Eslam Abousamra,
Eslam Abousamra,
![]() Donna Modrell,
Donna Modrell,
![]() Nashwa Ahmed,
Nashwa Ahmed,
![]() Cécile Tran Kiem
Cécile Tran Kiem






