Tracking novel coronavirus epidemic spread through genome sequencing
Trevor Bedford (@trvrb)
14 Feb 2020
AAAS Annual Meeting
Seattle, WA
Slides at: bedford.io/talks
Traditionally, we learn about an epidemic by tracking cases
Genetic sequencing to reconstruct pathogen spread
Epidemic process
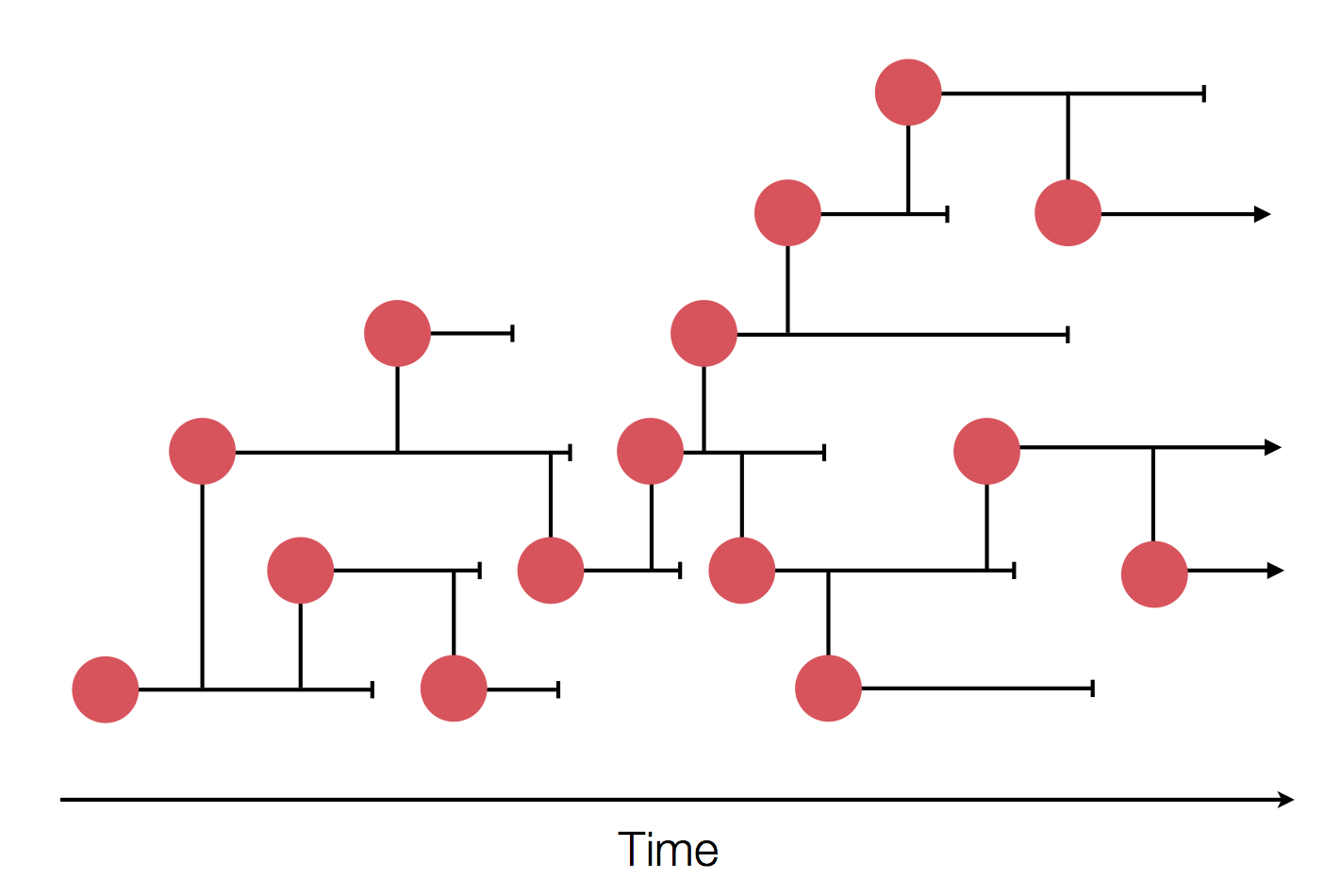
Sample some individuals
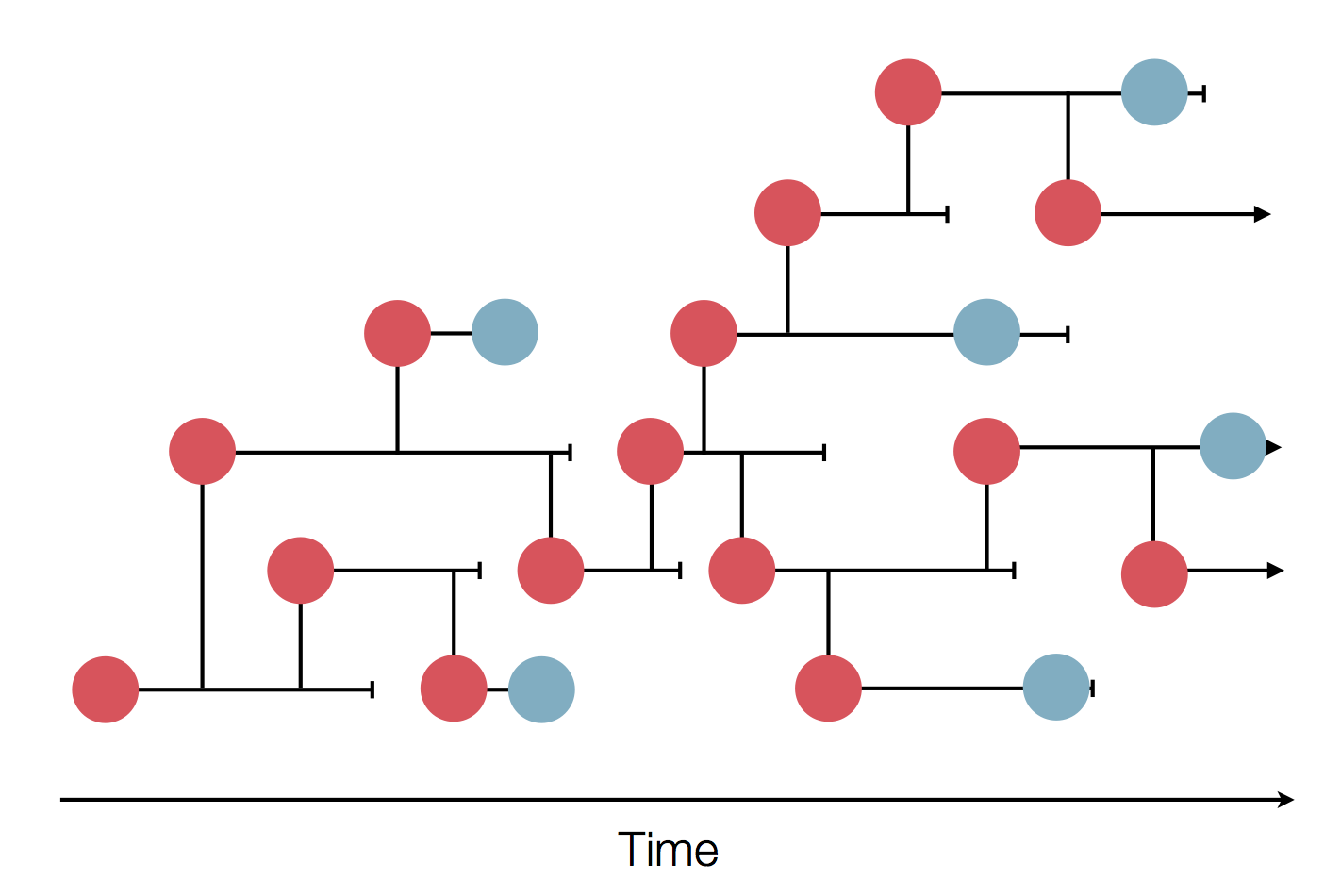
Sequence and determine phylogeny
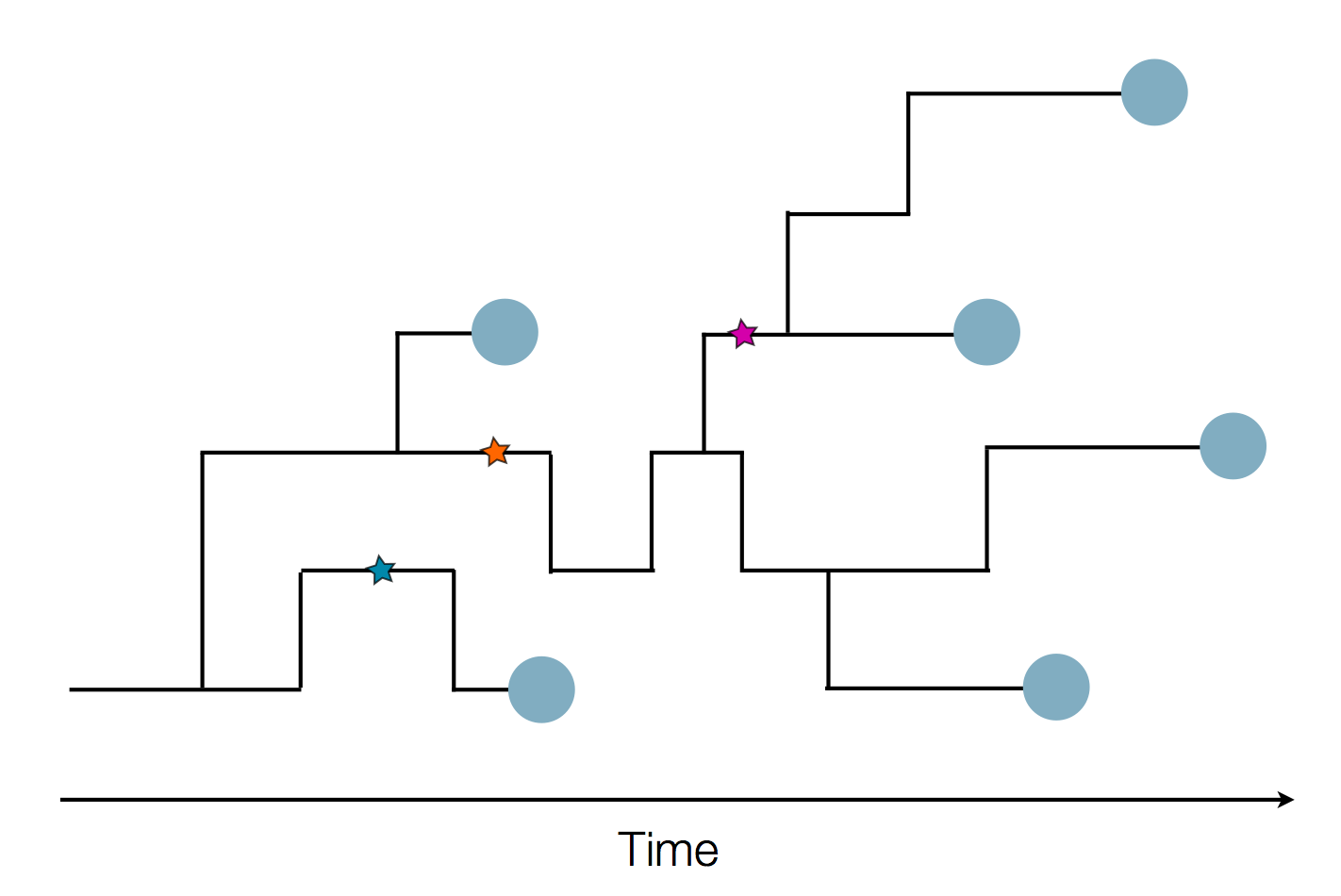
Sequence and determine phylogeny
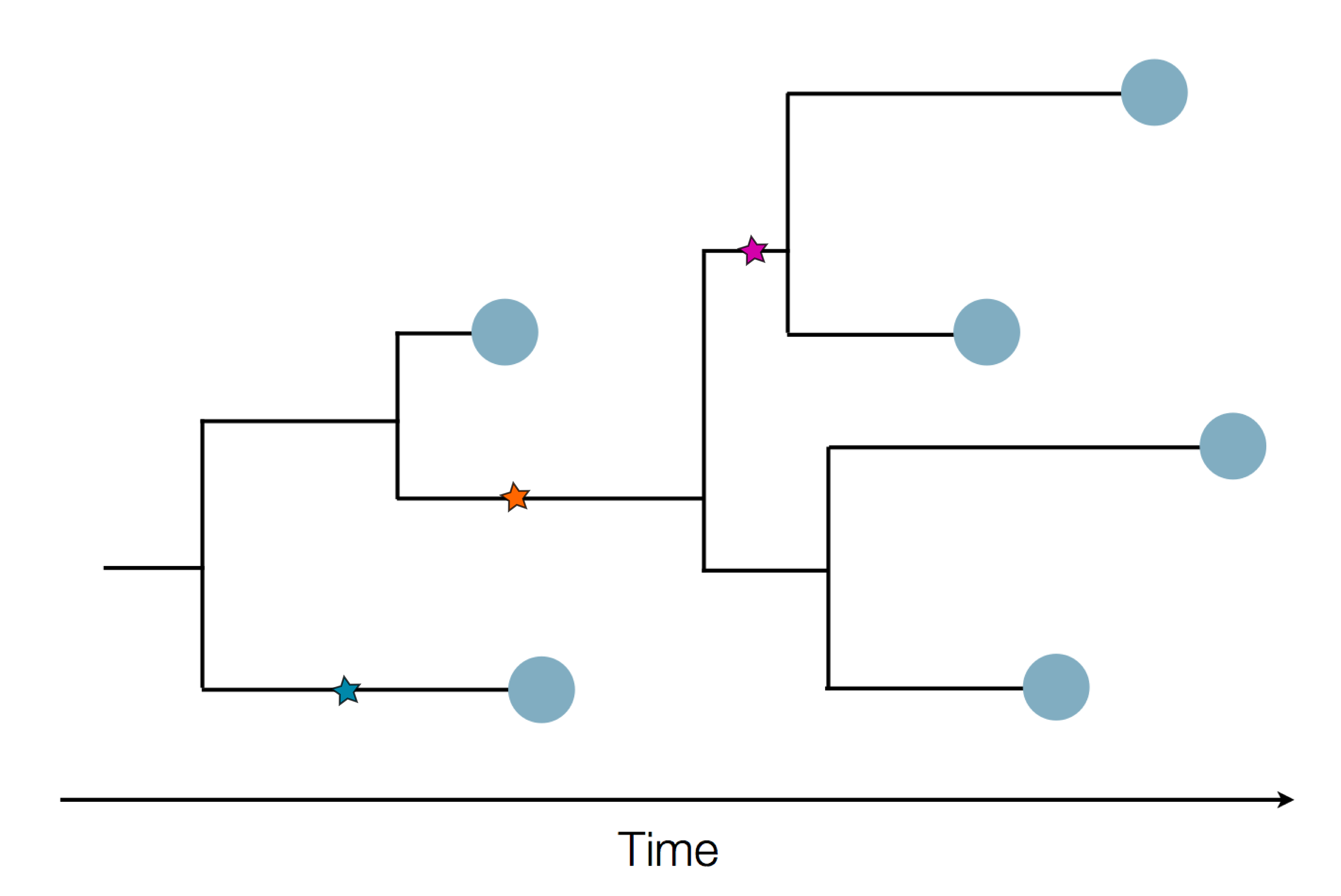
Revolutionary tool for understanding outbreaks
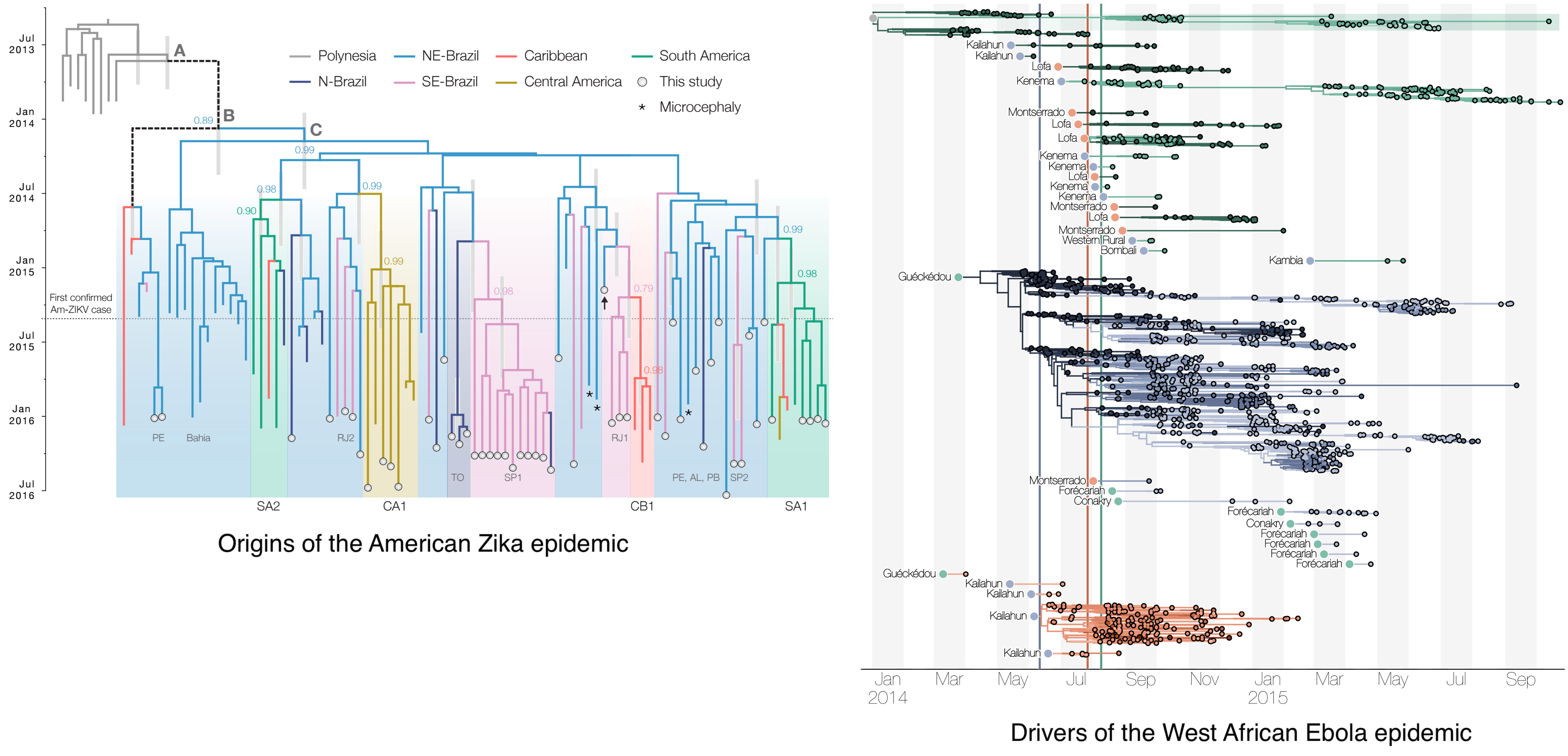
Genomic analyses were mostly done in a retrospective manner
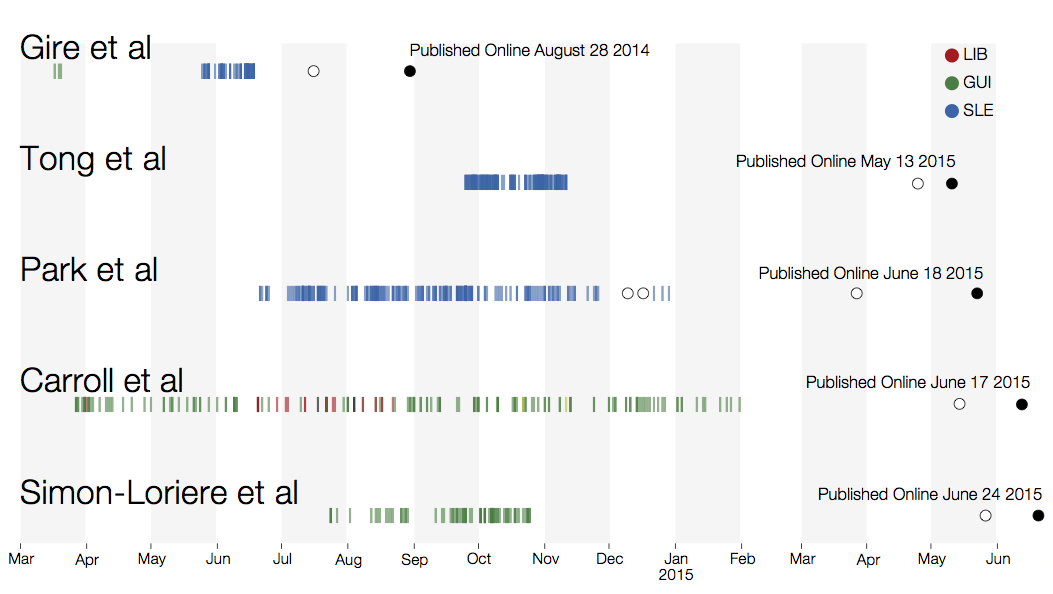
Key challenges to making genomic epidemiology actionable
- Timely analysis and sharing of results critical
- Dissemination must be scalable
- Integrate many data sources
- Results must be easily interpretable and queryable
Nextstrain
Project to conduct real-time molecular epidemiology and evolutionary analysis of emerging epidemics
with
![]() Richard Neher,
Richard Neher,
![]() James Hadfield,
James Hadfield,
![]() Emma Hodcroft,
Emma Hodcroft,
![]() Thomas Sibley,
Thomas Sibley,
![]() John Huddleston,
John Huddleston,
![]() Louise Moncla,
Louise Moncla,
![]() Misja Ilcisin,
Misja Ilcisin,
![]() Kairsten Fay,
Kairsten Fay,
![]() Jover Lee,
Jover Lee,
![]() Allison Black,
Allison Black,
![]() Colin Megill,
Colin Megill,
![]() Sidney Bell,
Sidney Bell,
![]() Barney Potter,
Barney Potter,
![]() Charlton Callender
Charlton Callender
Jan 10: nCoV is a SARS-like betacoronavirus
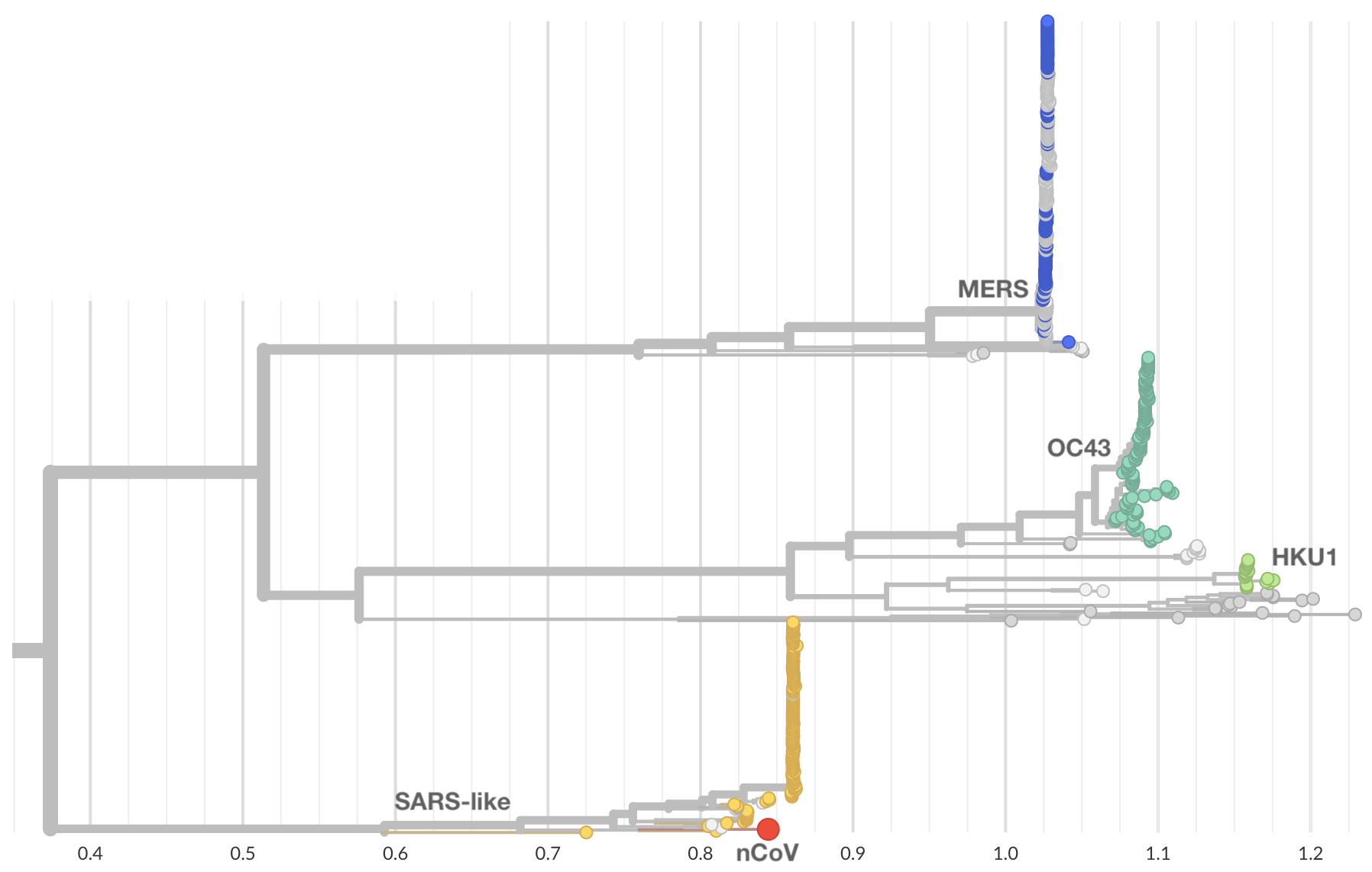
Jan 11: These viruses have a natural reservoir in bats
Jan 11: Initial 5 nCoV genomes from Wuhan showed highly restricted genetic diversity
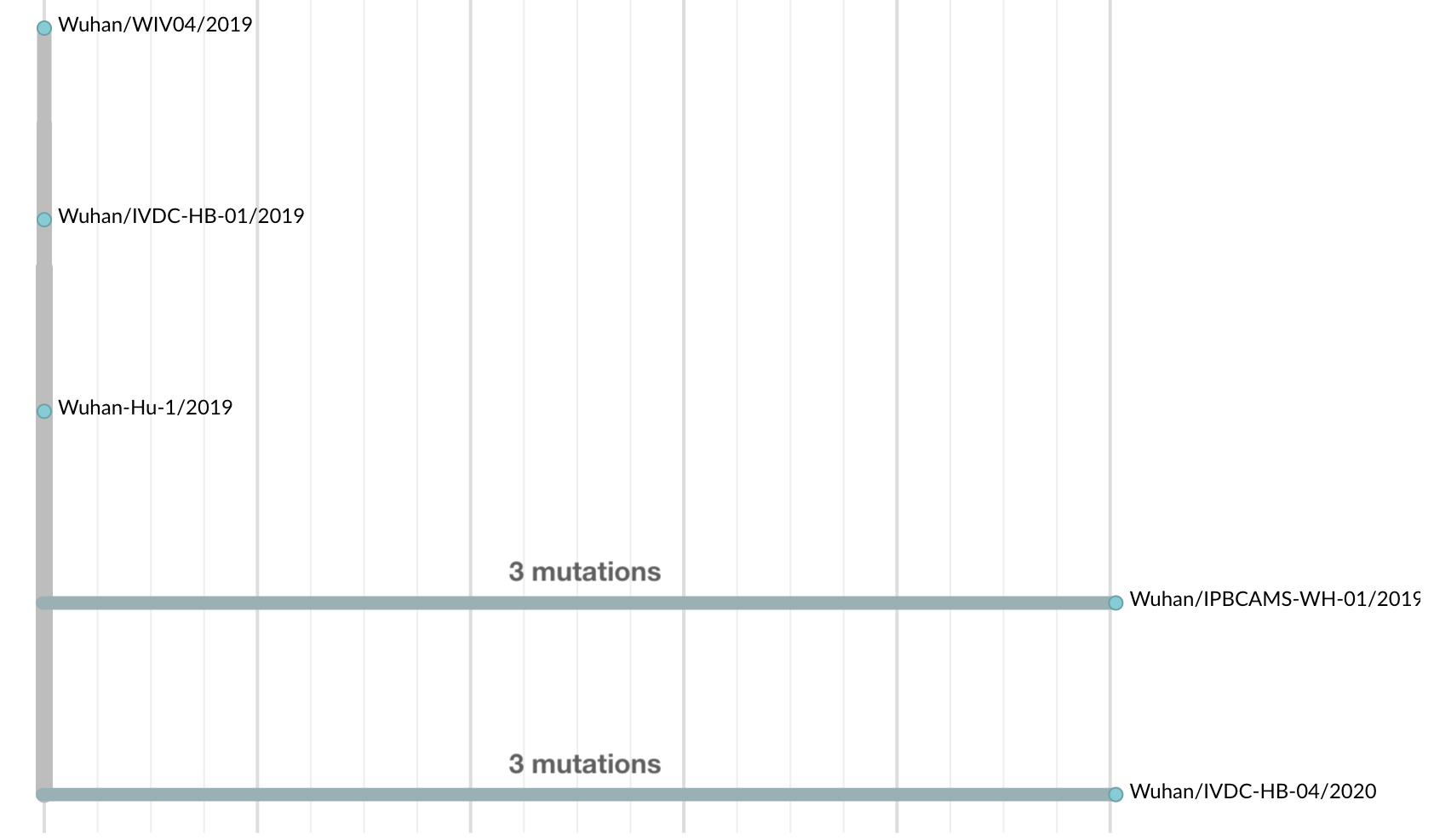
Initially thought clustering due to epi investigation of linked cases at Huanan seafood market
Jan 17: Additional 2 nCoV genomes from Thailand travel cases also lacked diversity
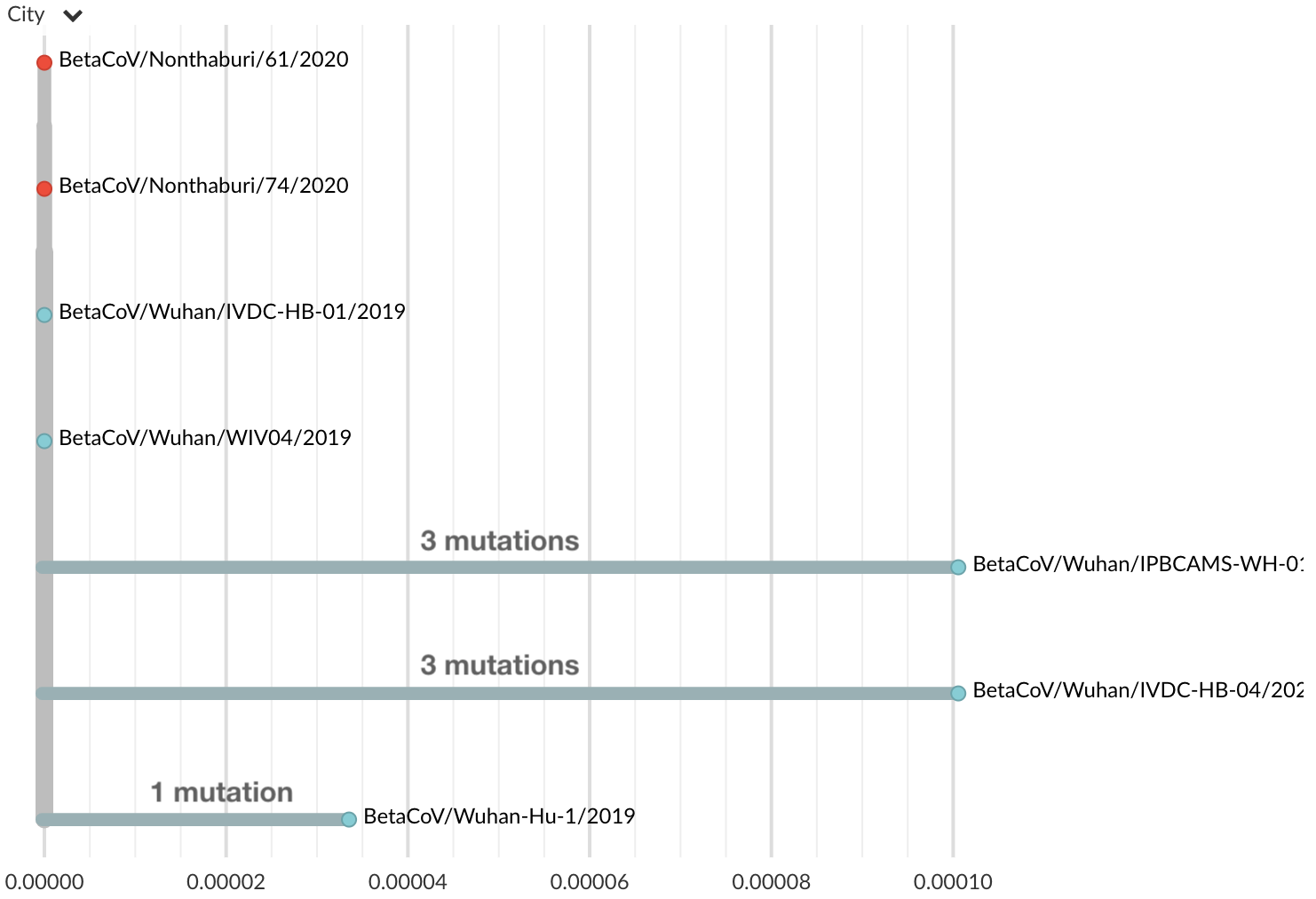
Jan 19: Additional 5 nCoV genomes from Wuhan still lacked diversity
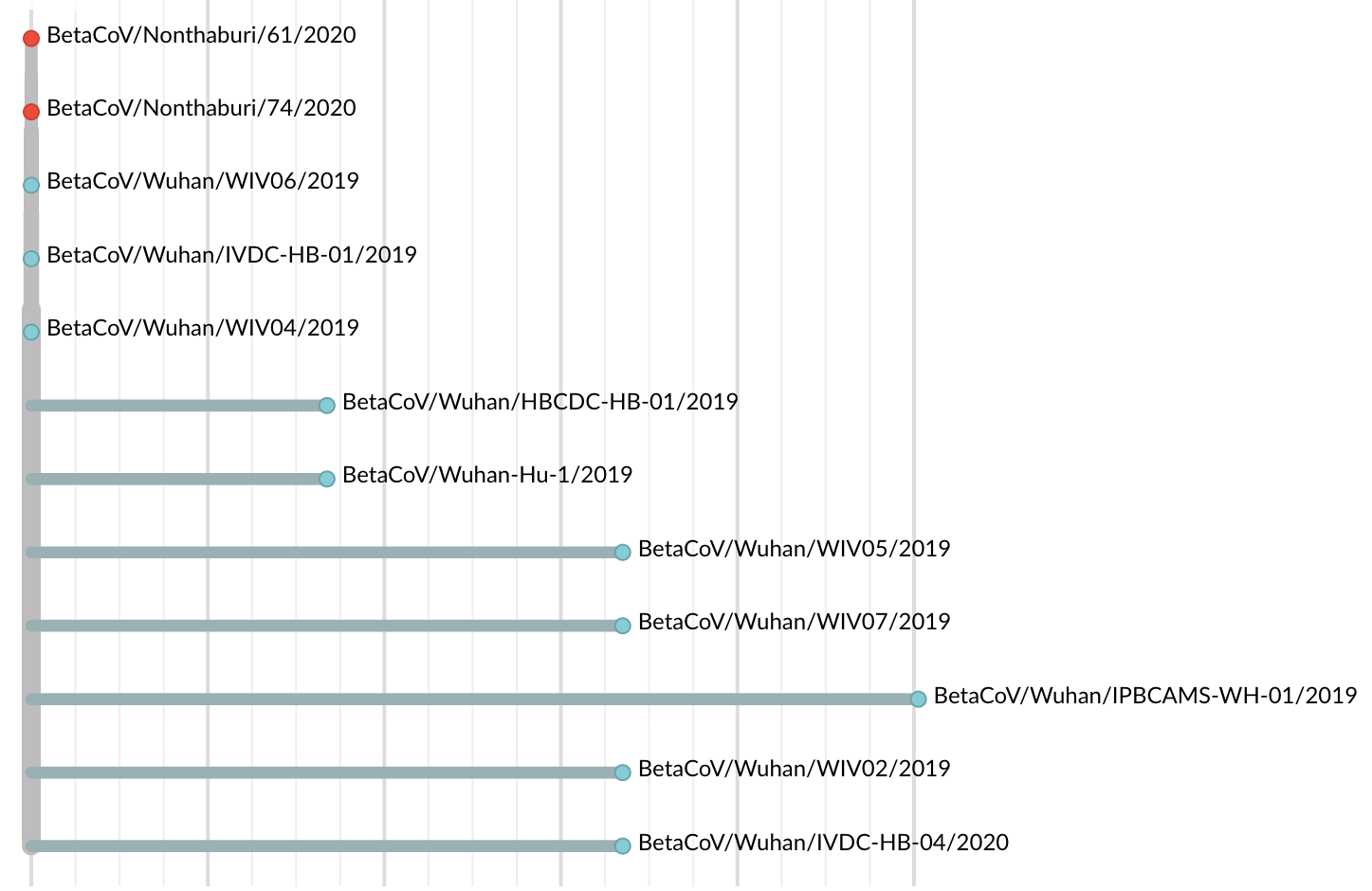
😧
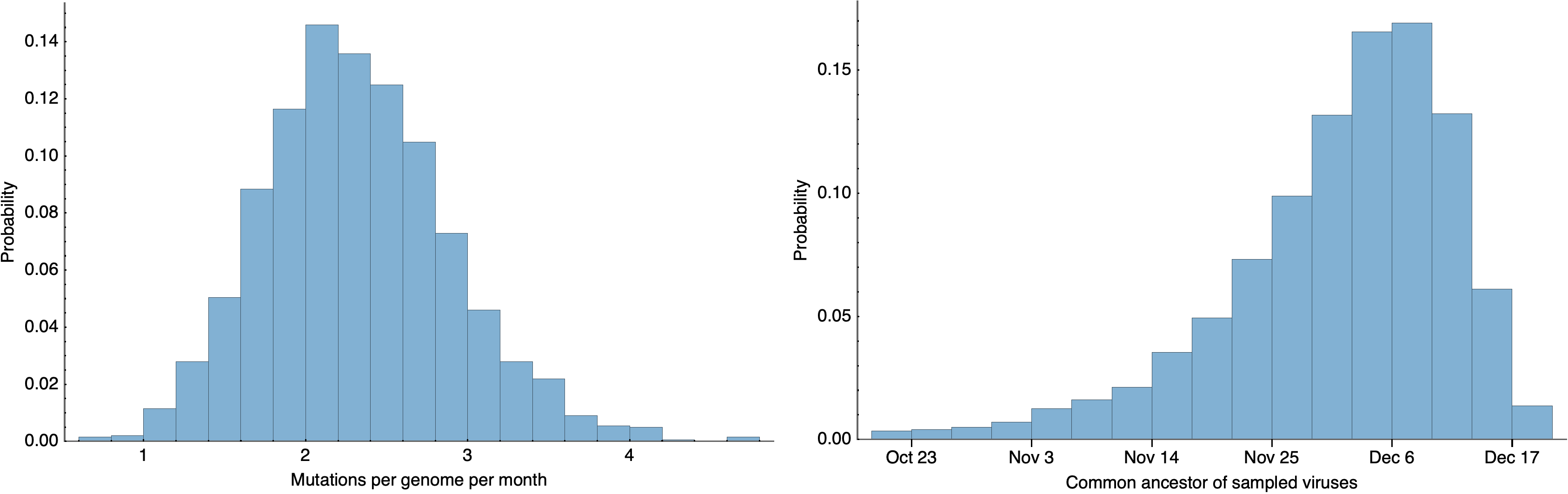
Single introduction into the human population between Nov 15 and Dec 15 and human-to-human epidemic spread from this point forward
Spent the week of Jan 20 alerting public health officials, and since then have aimed to keep nextstrain.org/ncov updated within ~1hr of new sequences being deposited
Feb 13: >100 sequences shared by groups all over the world
Almost real-time with many genomes shared within 3-6 days of sampling
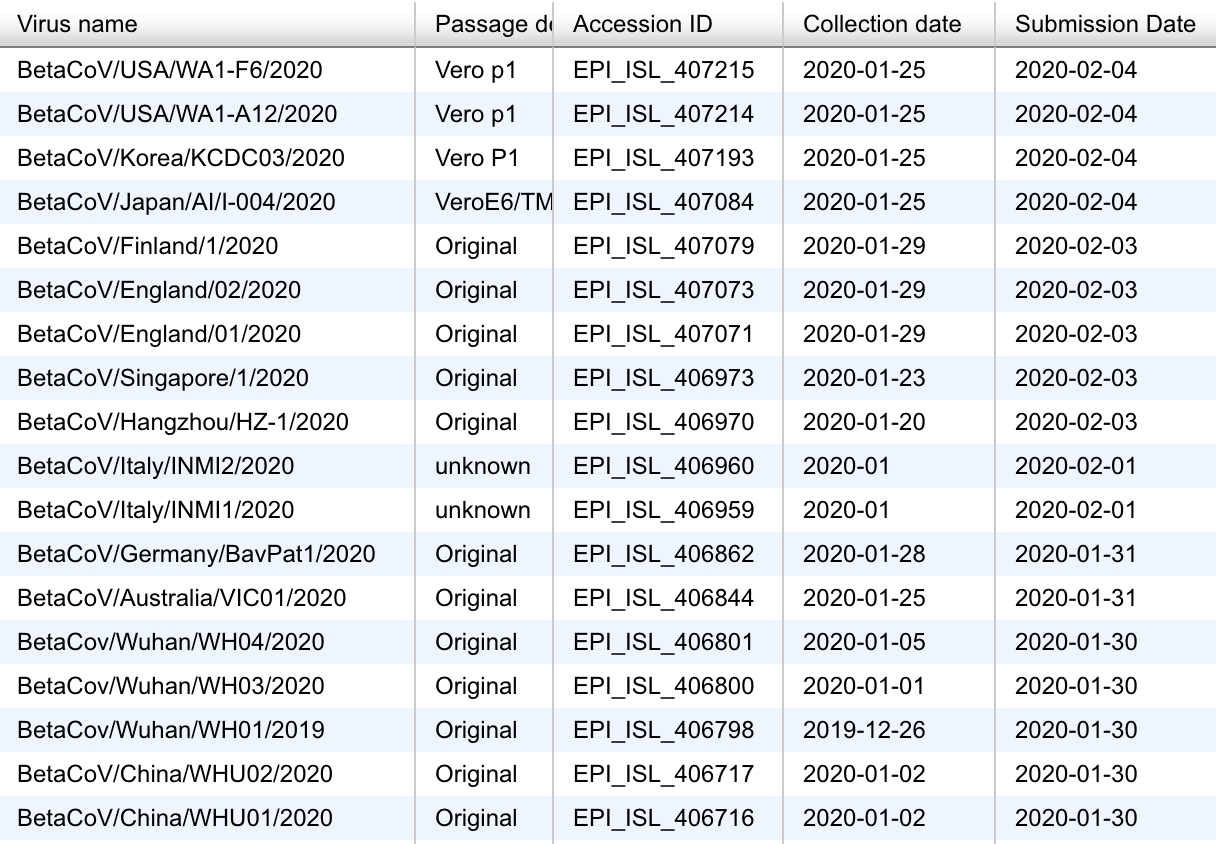
Current state of nextstrain.org/ncov with 88 genomes as of Feb 13
Phylodynamic estimates of epidemic growth
Rate of evolution and time of common ancestor
Viral genetic diversity and population size
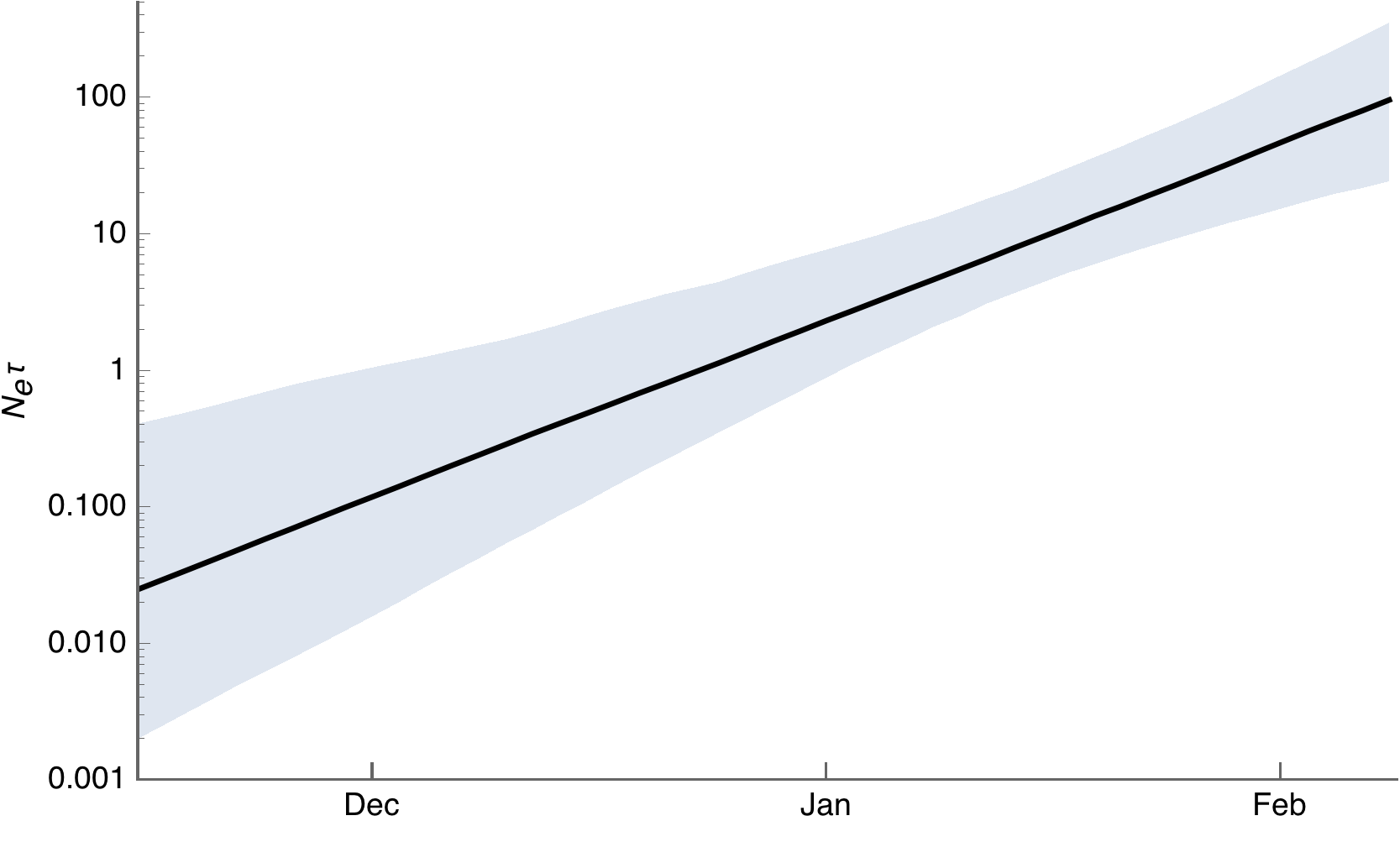
Transformed to infection prevalence
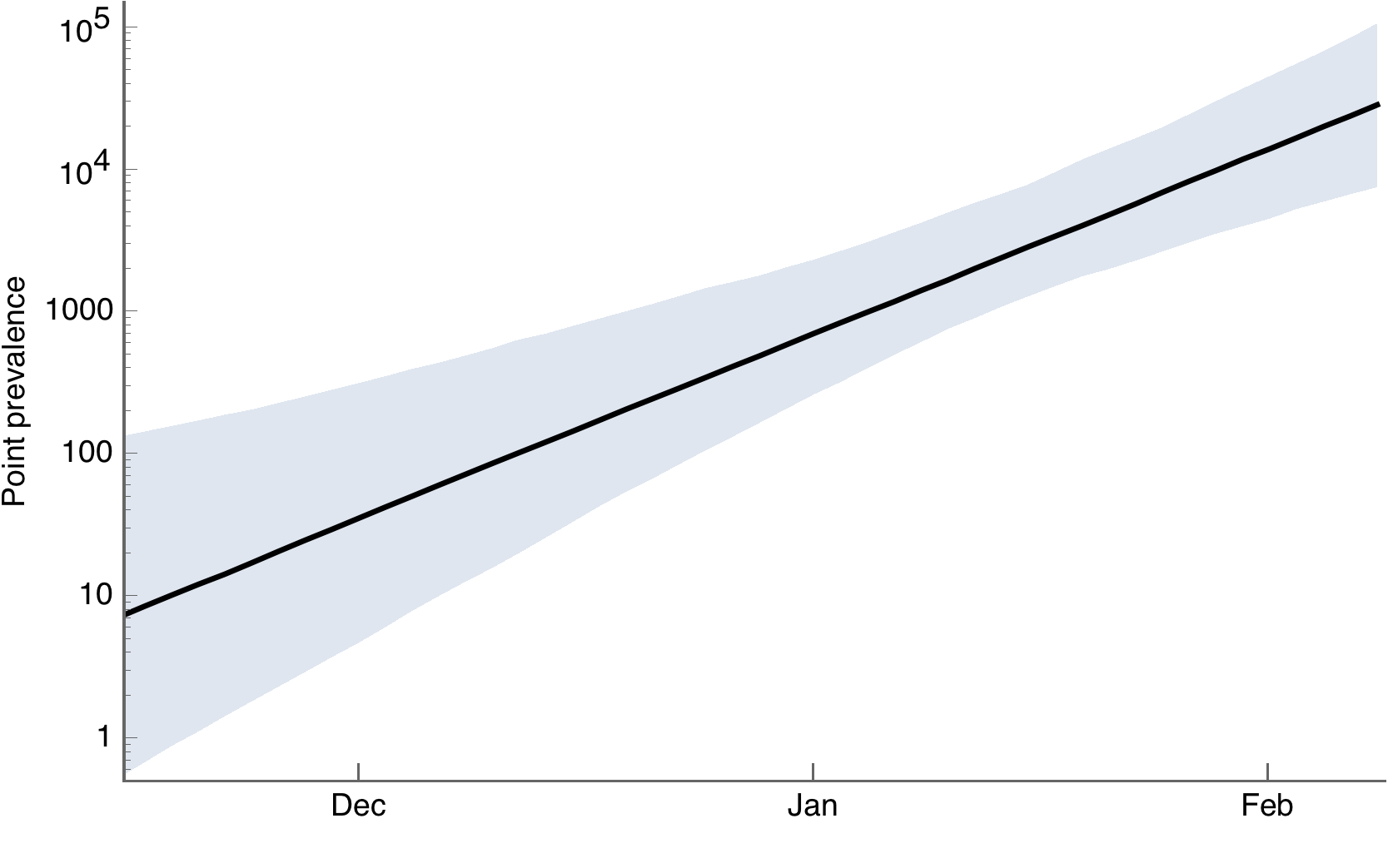
And to cumulative incidence
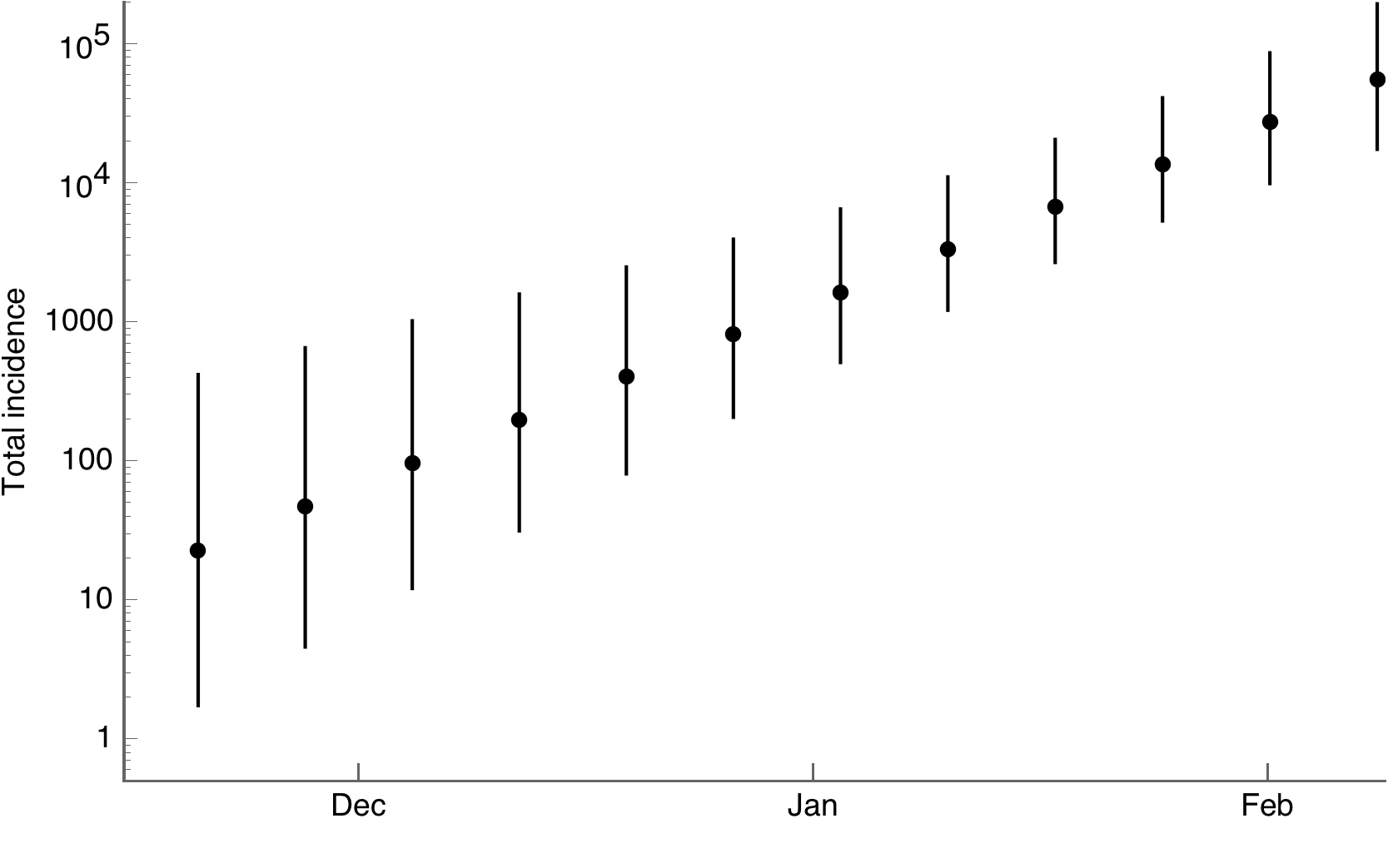
Scientific communication surrounding outbreak has completely flipped with everything posted to bioRxiv, modeling groups posting live analyses and crowd-sourced line lists 🙌🏻
This communication between academics and public health officials has spilled over with huge interest from general public
This is having knock-on effects on science communication and spread of misinformation
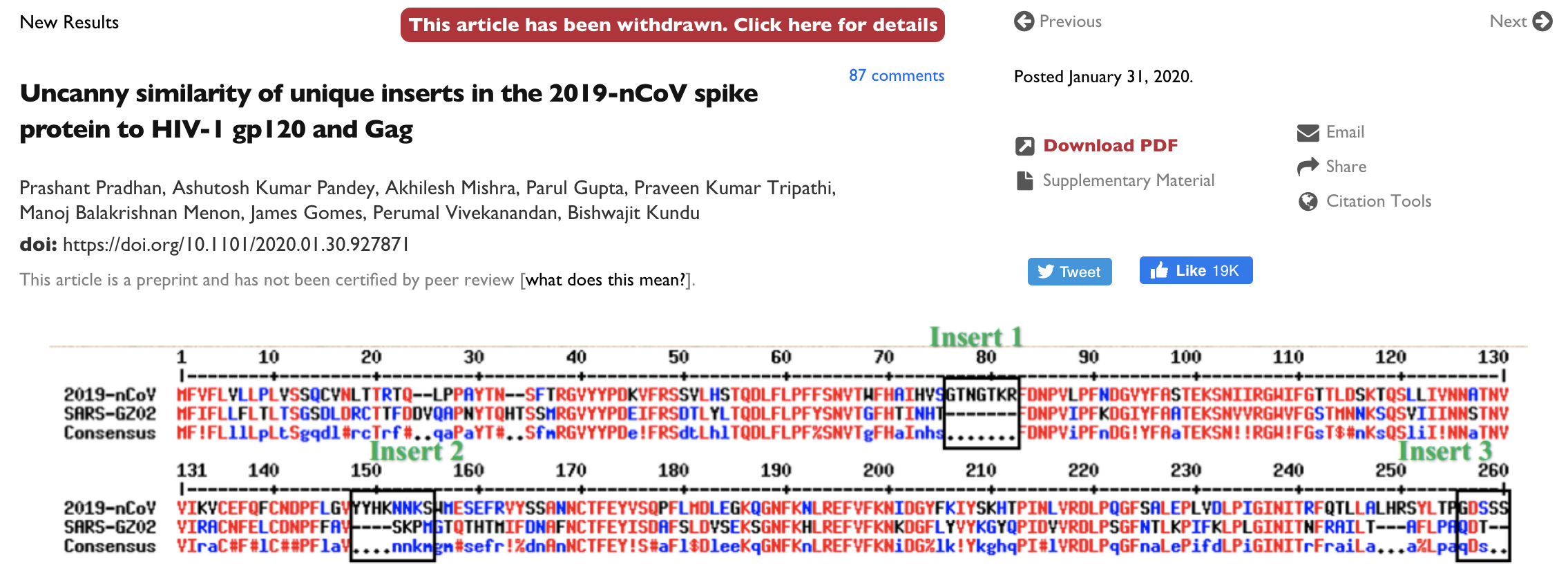
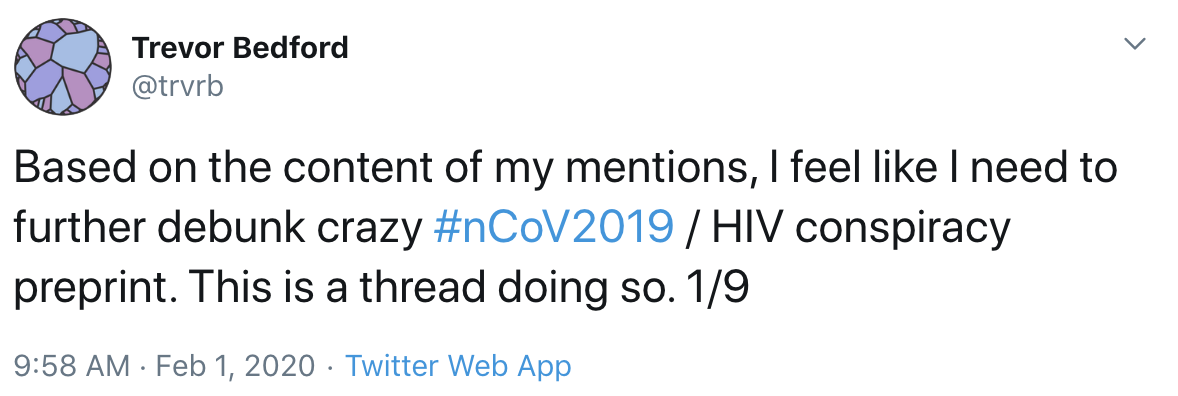
Moving forward
- Epidemic spread still appears to be sustained with estimates of R0 between ~2-3 and epidemic doubling time of ~6 days
- We have not yet had time to ascertain effects of intervention measures, but my hope for containment is slim
- Biggest question for me now surrounds severity
- I expect genomic data to be most immediately useful to help pin down emerging community transmission
Acknowledgements
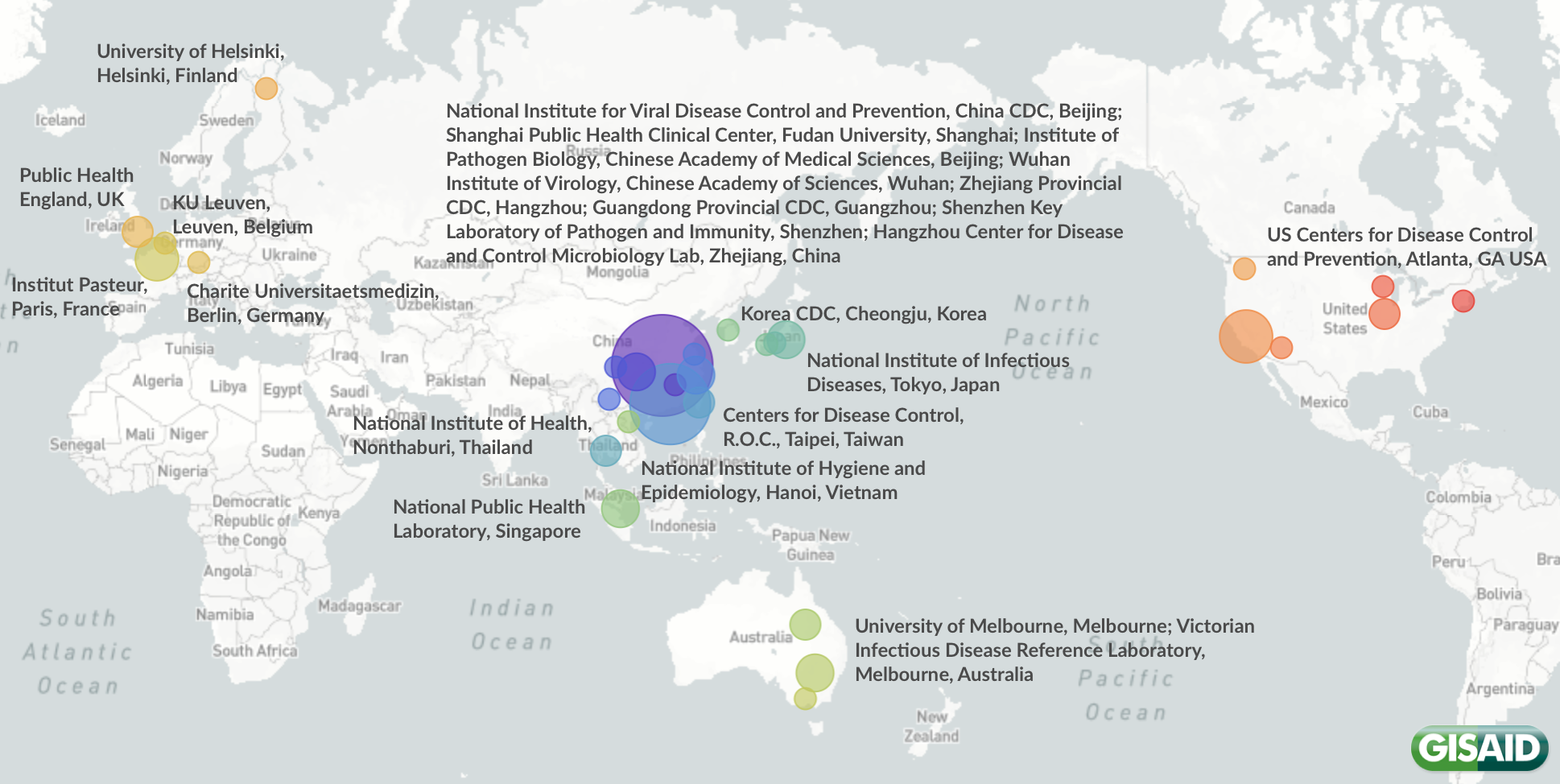
Data producers from all over the world: BGI & Institute of Microbiology, Chinese Academy of Sciences & Shandong First Medical University & Shandong Academy of Medical Sciences; Beijing Genomics Institute (BGI), Shenzhen, China; Centers for Disease Control, R.O.C., Taipei, Taiwan; Charite Universitaetsmedizin Berlin, Berlin, Germany; Chongqing Municipal Center for Disease Control and Prevention, Chongqing, China; Victorian Infectious Disease Reference Laboratory, Melbourne, Australia; National Institute of Health, Nonthaburi, Thailand; Guangdong Provincial Center for Diseases Control and Prevention; Guangdong Provinical Public Health, Guangzhou, China; Department of Microbiology, Zhejiang Provincial Center for Disease Control and Prevention, Hangzhou, China; Department of Virology, University of Helsinki, Helsinki, Finland; Guangdong Provincial Center for Diseases Control and Prevention, Guangzhou, China; Hangzhou Center for Disease and Control Microbiology Lab, Zhejiang, China; Hubei Provincial Center for Disease Control and Prevention, Wuhan, China; Chinese Academy of Medical Sciences & Peking Union Medical College, Beijing, China; Institute of Viral Disease Control and Prevention, China CDC, Beijing, China; KU Leuven, Leuven, Belgium; Korea Centers for Disease Control & Prevention (KCDC), Cheongju, Korea; Microbial Genomics Core Lab, National Taiwan University, Taipei, Taiwan; NSW Health Pathology - Institute of Clinical Pathology and Medical Research; Centre for Infectious Diseases and Microbiology Laboratory Services; Westmead Hospital; University of Sydney, Westmead, Australia; University of Sydney, Westmead, Australia; National Institute of Hygiene and Epidemiology, Hanoi, Vietnam; National Institute for Communicable Disease Control and Prevention (ICDC), China CDC, Beijing, China; National Institute for Viral Disease Control and Prevention, China CDC, Beijing, China; National Public Health Laboratory, Singapore; Institut Pasteur, Paris, France; Centers for Disease Control and Prevention, Atlanta, USA; National Institute of Infectious Diseases, Tokyo, Japan; Duke-NUS Medical School, Singapore; Public Health Virology Laboratory, Brisbane, Australia; Public Health England, London, United Kingdom; National Clinical Research Center for Infectious Disease, Shenzhen, China; Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, China, made possible through GISAID
Nextstrain: Richard Neher, James Hadfield, Emma Hodcroft, Tom Sibley, John Huddleston, Louise Moncla, Misja Ilcisin, Kairsten Fay, Jover Lee, Allison Black Colin Megill, Sidney Bell, Barney Potter, Charlton Callender





