Fitness flux in SARS-CoV-2
Trevor Bedford
Fred Hutchinson Cancer Center / Howard Hughes Medical Institute
4 Dec 2025
Science Meeting
HHMI
Slides at: bedford.io/talks
Genetic relationships of globally sampled SARS-CoV-2 from 2020 to present
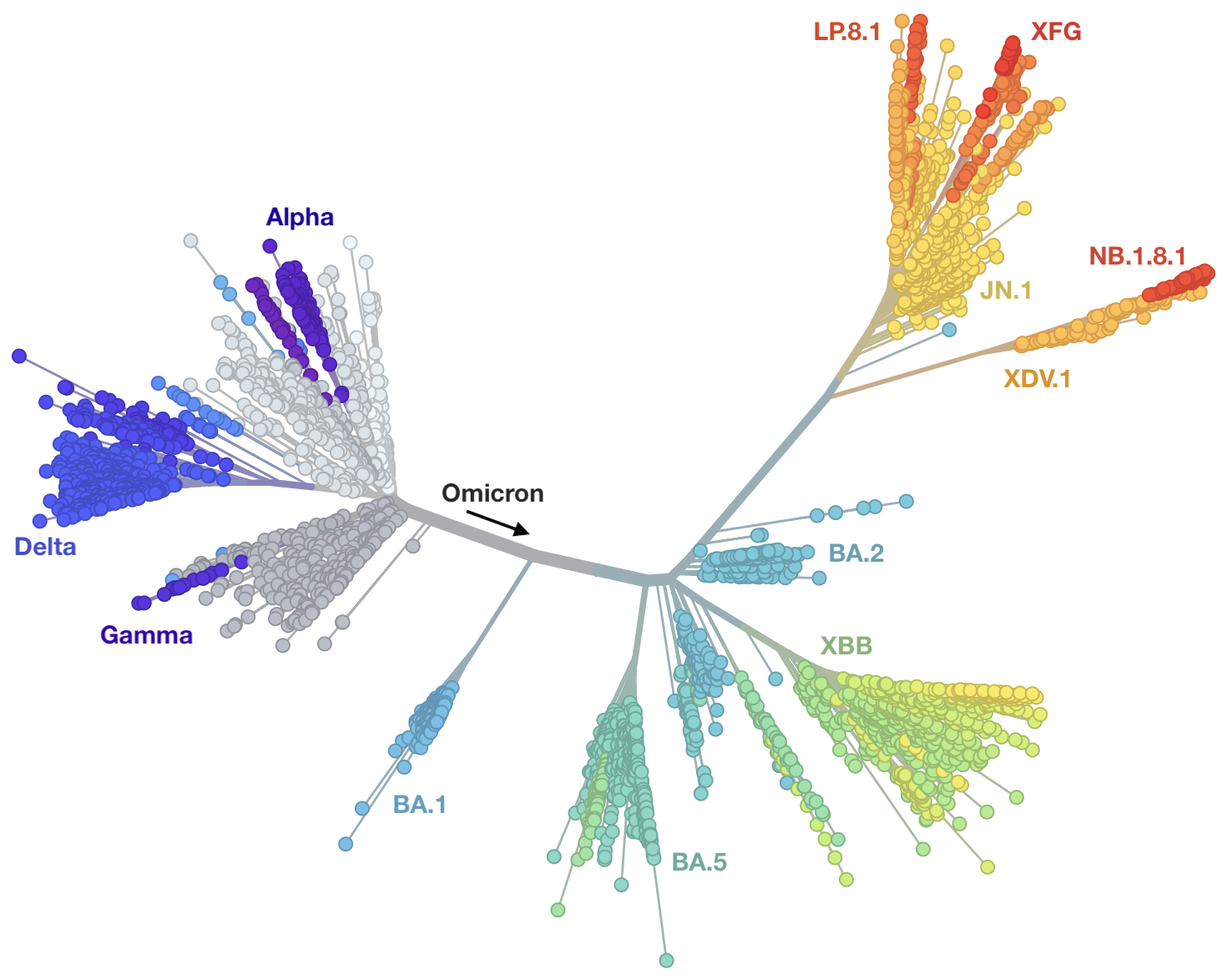 nextstrain.org
nextstrain.org
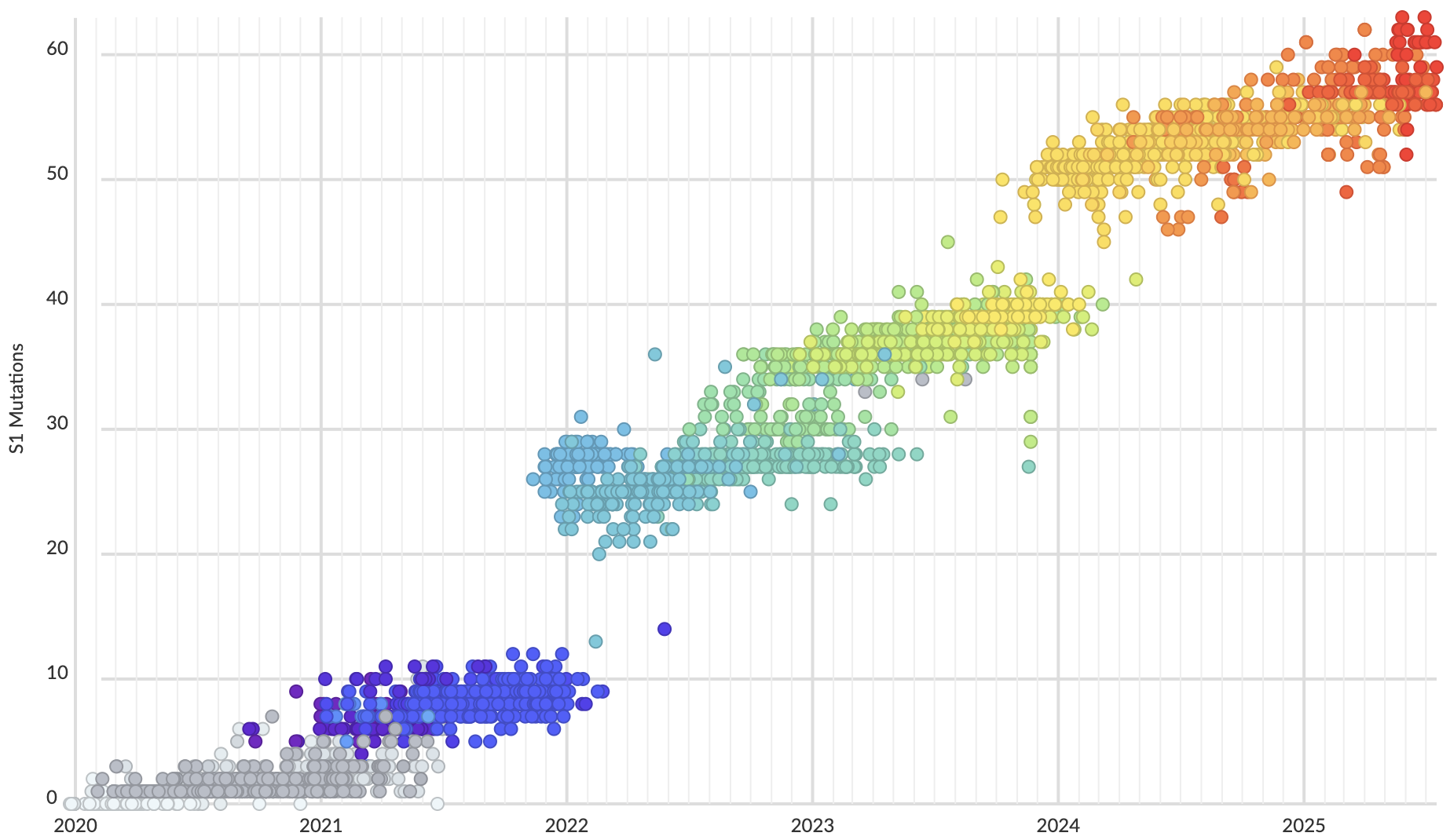
Rapid displacement of existing diversity by emerging variants
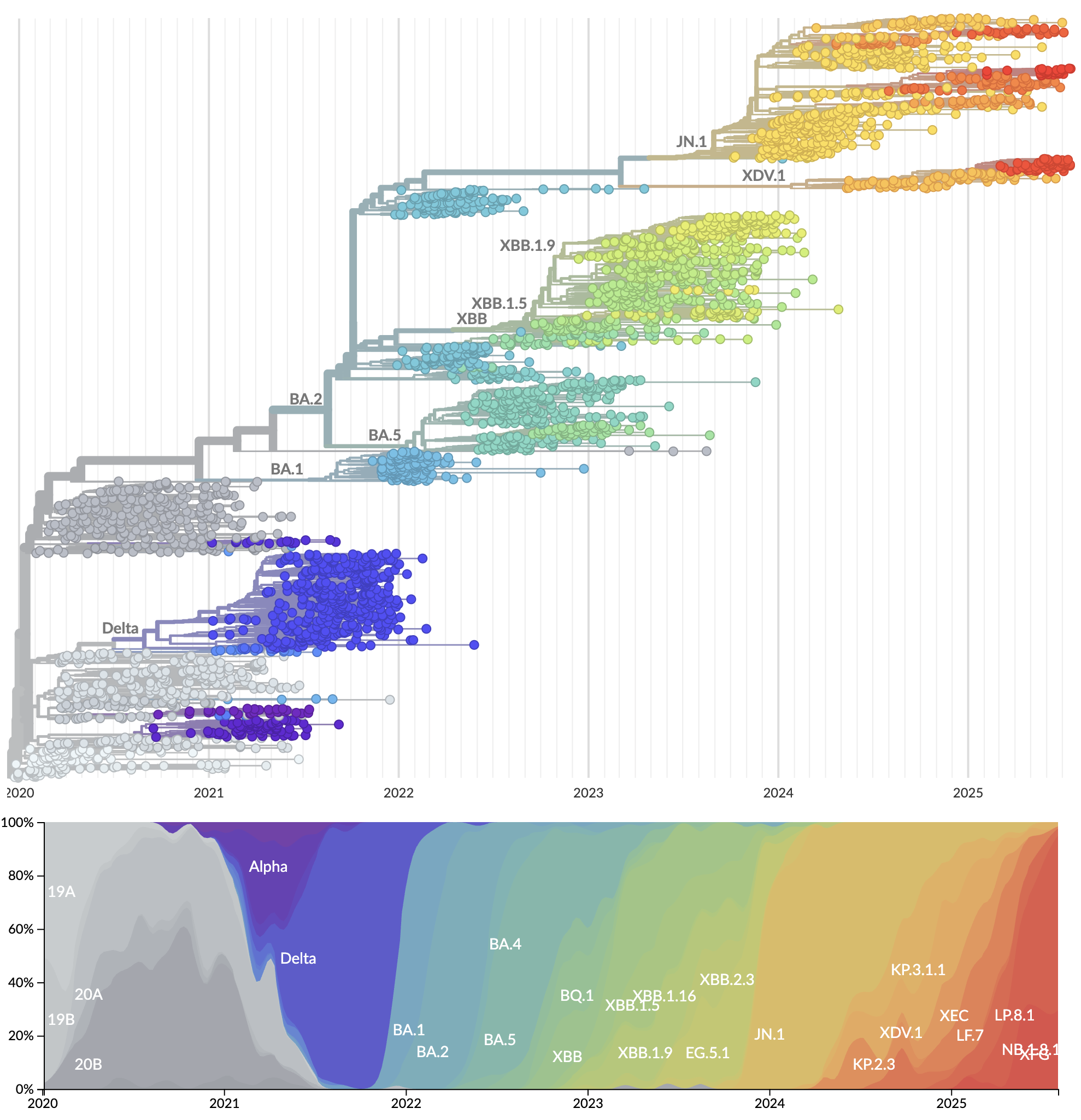
Mutations in S1 domain of spike protein driving displacement
 nextstrain.org
nextstrain.org
SARS-CoV-2 evolution fast relative to previous endemic viruses
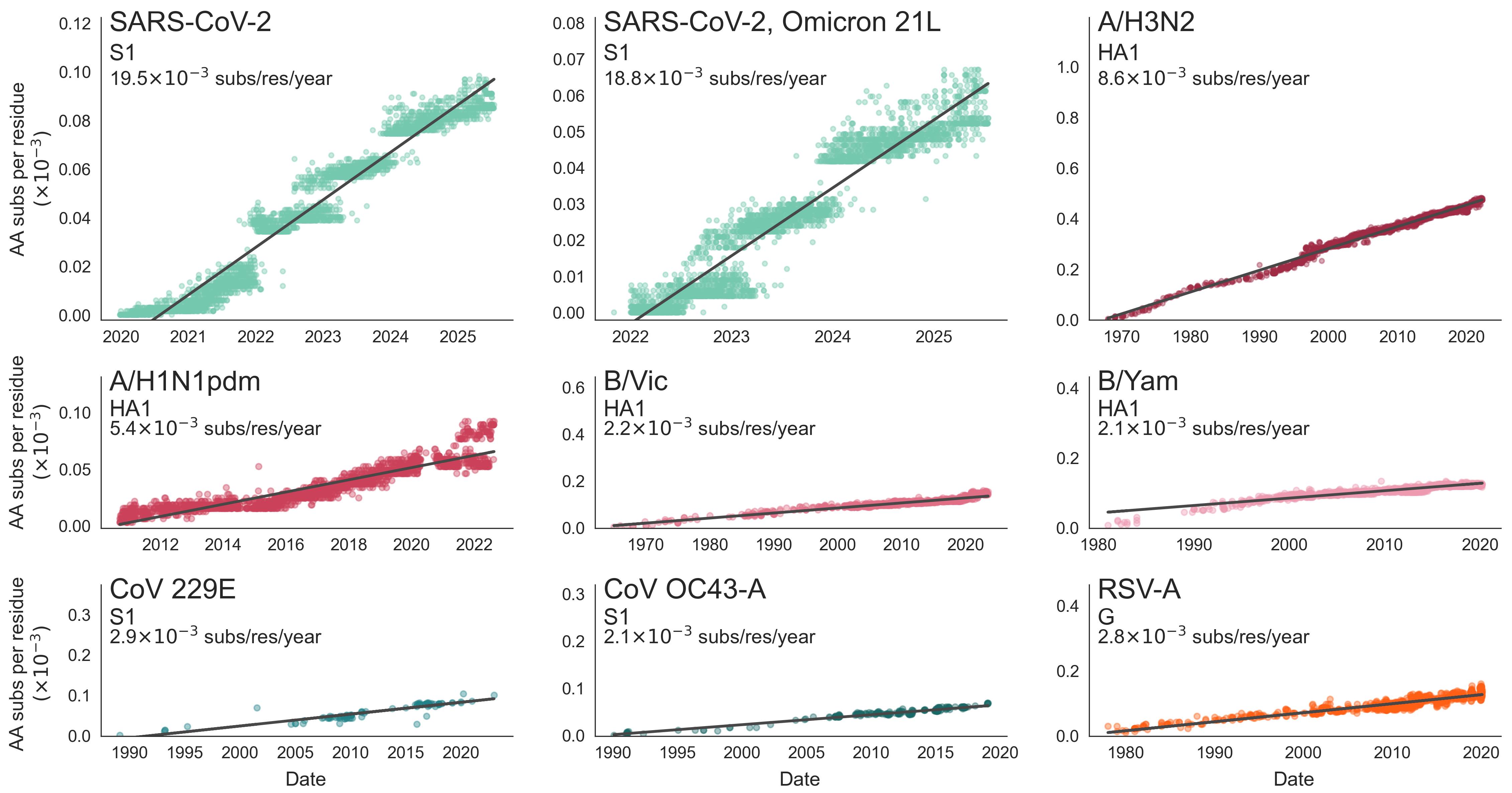
This talk
- Frequency dynamics
- Fitness flux
- Mutational fitness effects
Frequency dynamics
Multinomial logistic regression (MLR)
Simple haploid population genetic model is equivalent to statistical multinomial logistic regression
The frequency $x_i(t)$ of variant $i$ at time $t$ is determined by its initial frequency $p_i$ along with its fitness $f_i$ following
$$x_i(t) = \frac{p_i \, \mathrm{exp}(f_i \, t)}{\sum_j p_j \, \mathrm{exp}(f_j \, t) }$$
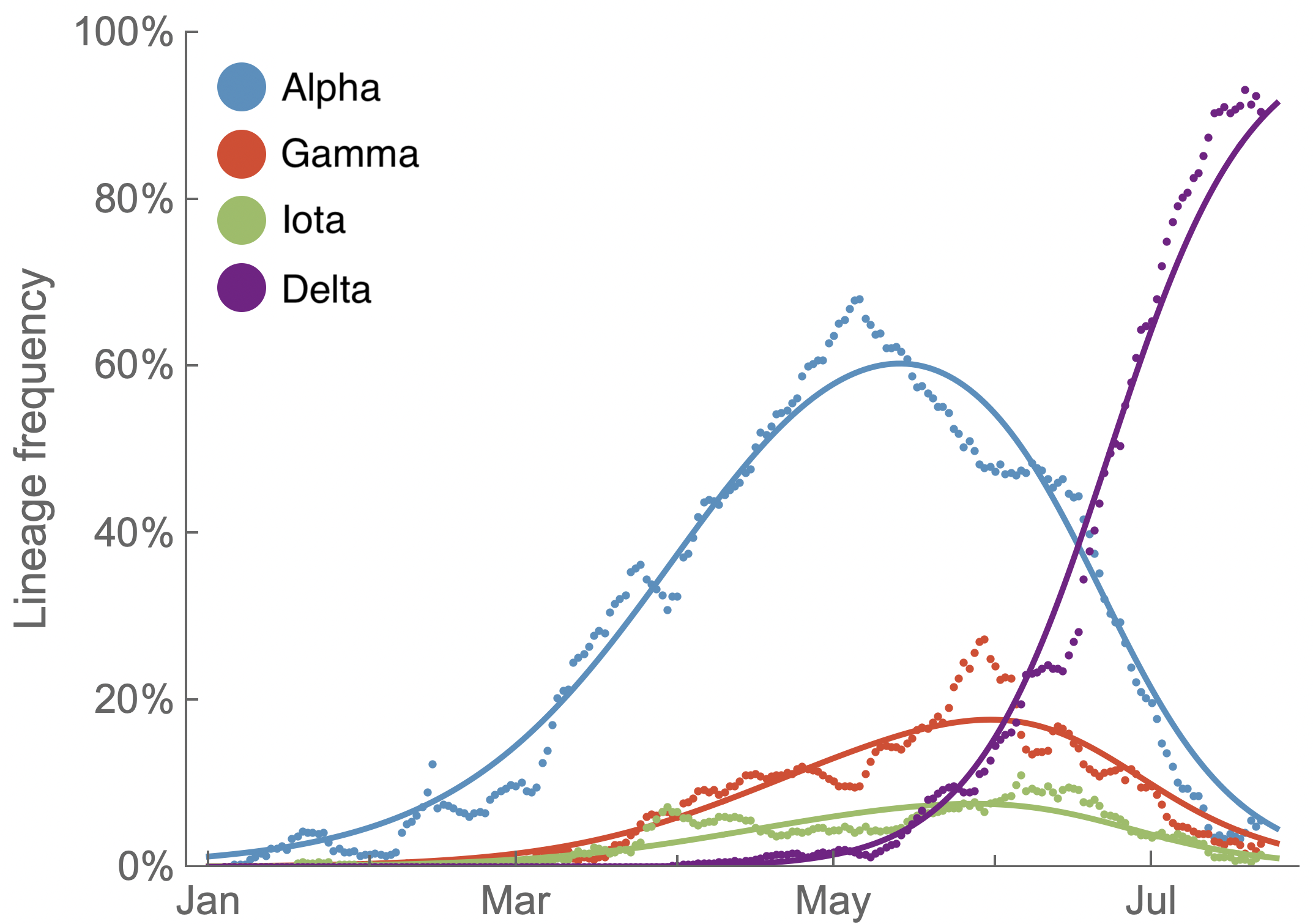
Multinomial logistic regression fits variant frequencies well
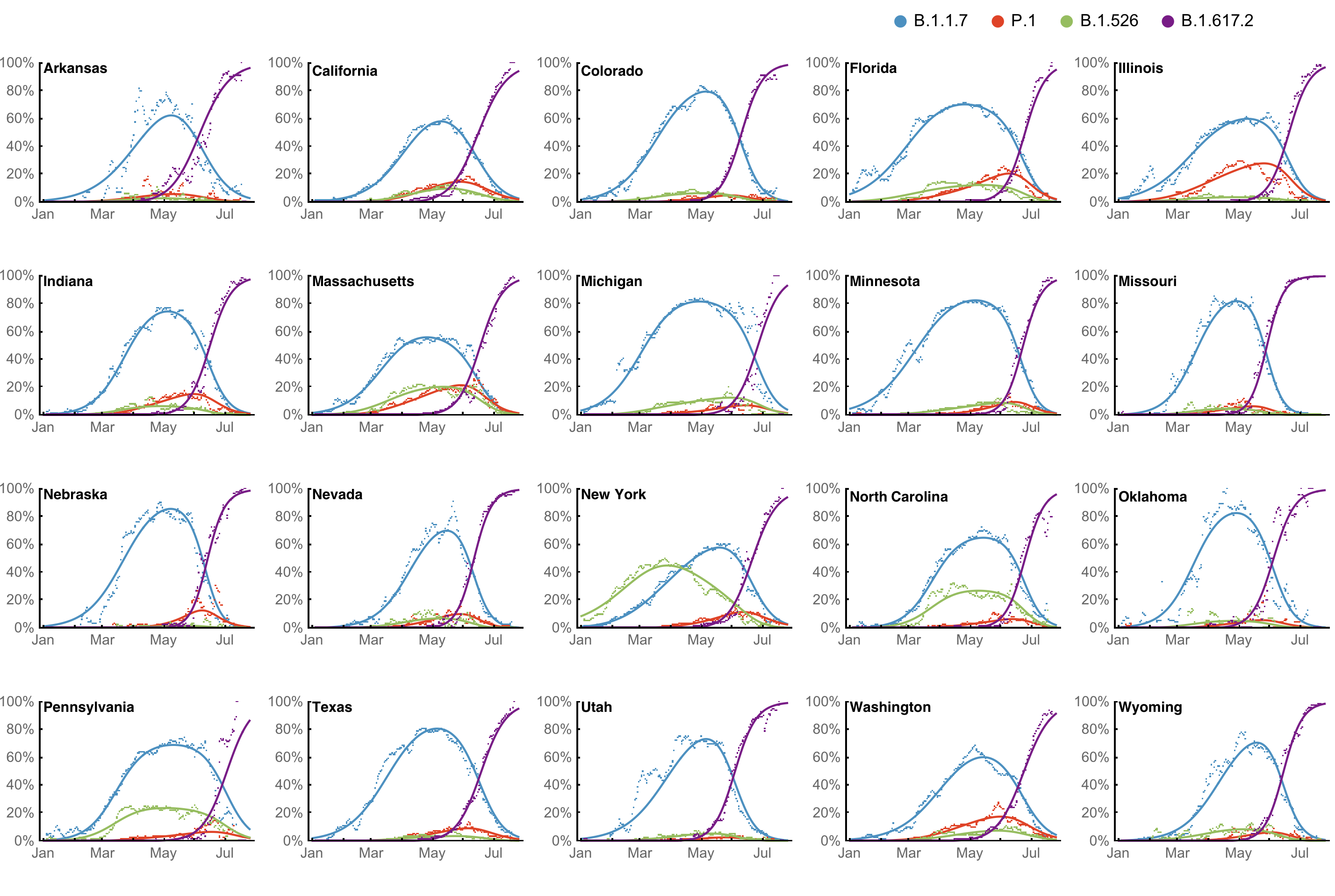
MLR models generate accurate short-term forecasts
30 days out, countries range from 5 to 15% mean absolute error
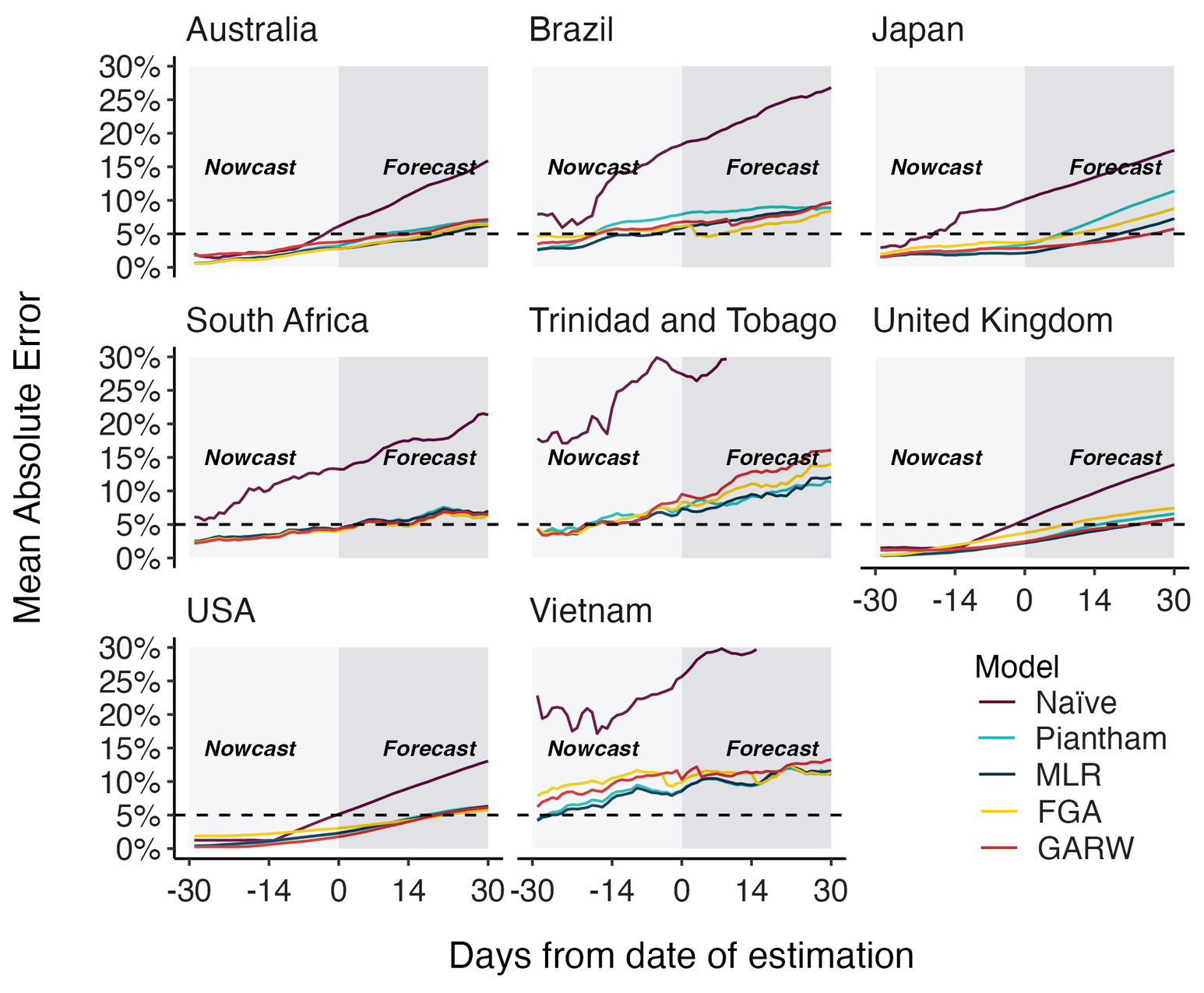
Fitness flux
Fitness flux is the rate of change of mean population fitness
With variant frequency $x_i(t)$ and constant variant fitness $f_i$
Mean population fitness $\bar{f}(t) = \sum_i x_i(t) \, f_i$ Fitness flux $\phi(t) = \Delta \bar{f}(t) / \Delta t$
 Mustonen and Lässig. 2010.
Mustonen and Lässig. 2010.
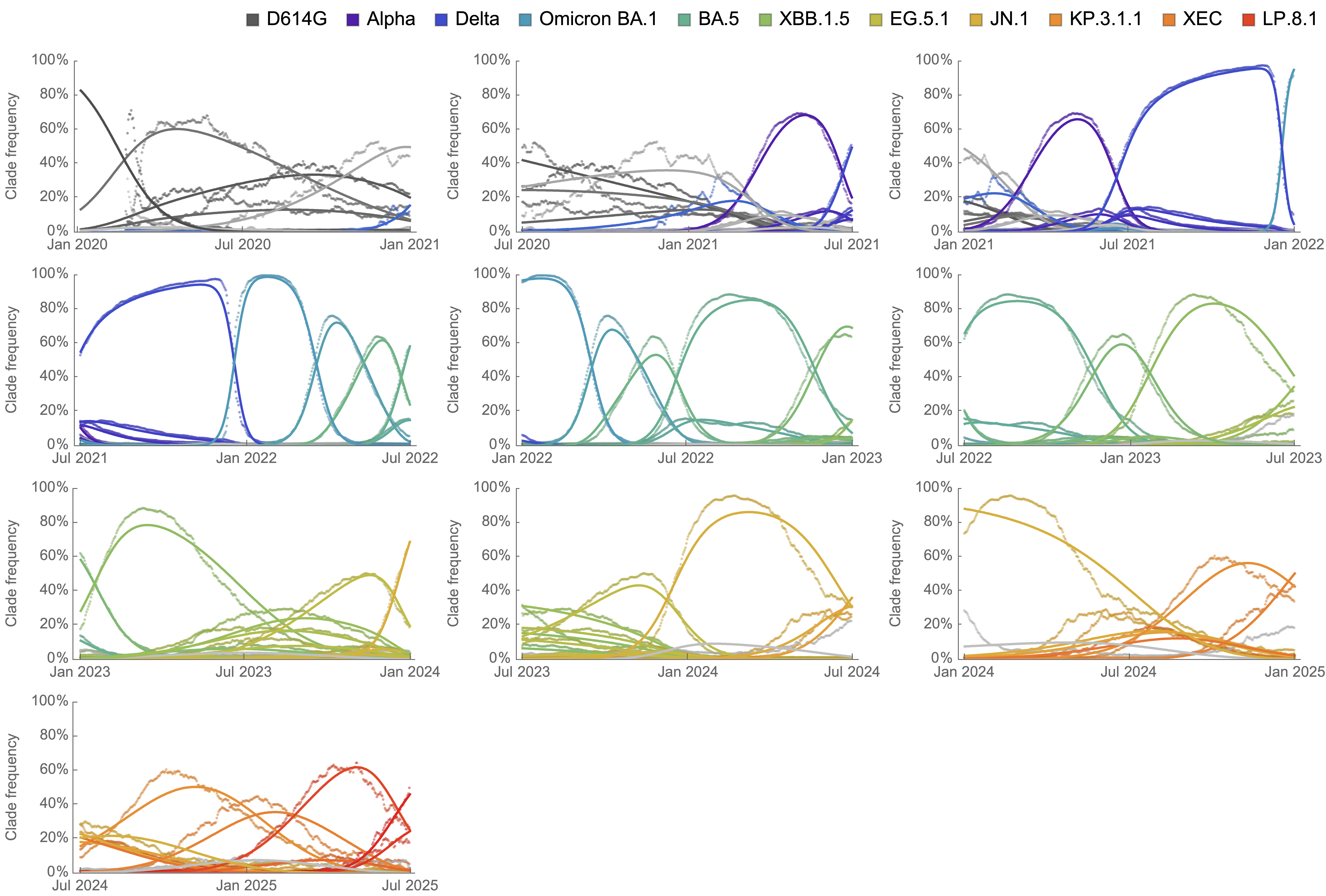
Clade-level frequency dynamics and MLR fits in sliding windows
Constant clade fitness within each window, USA data only, ignores within-clade fitness variation
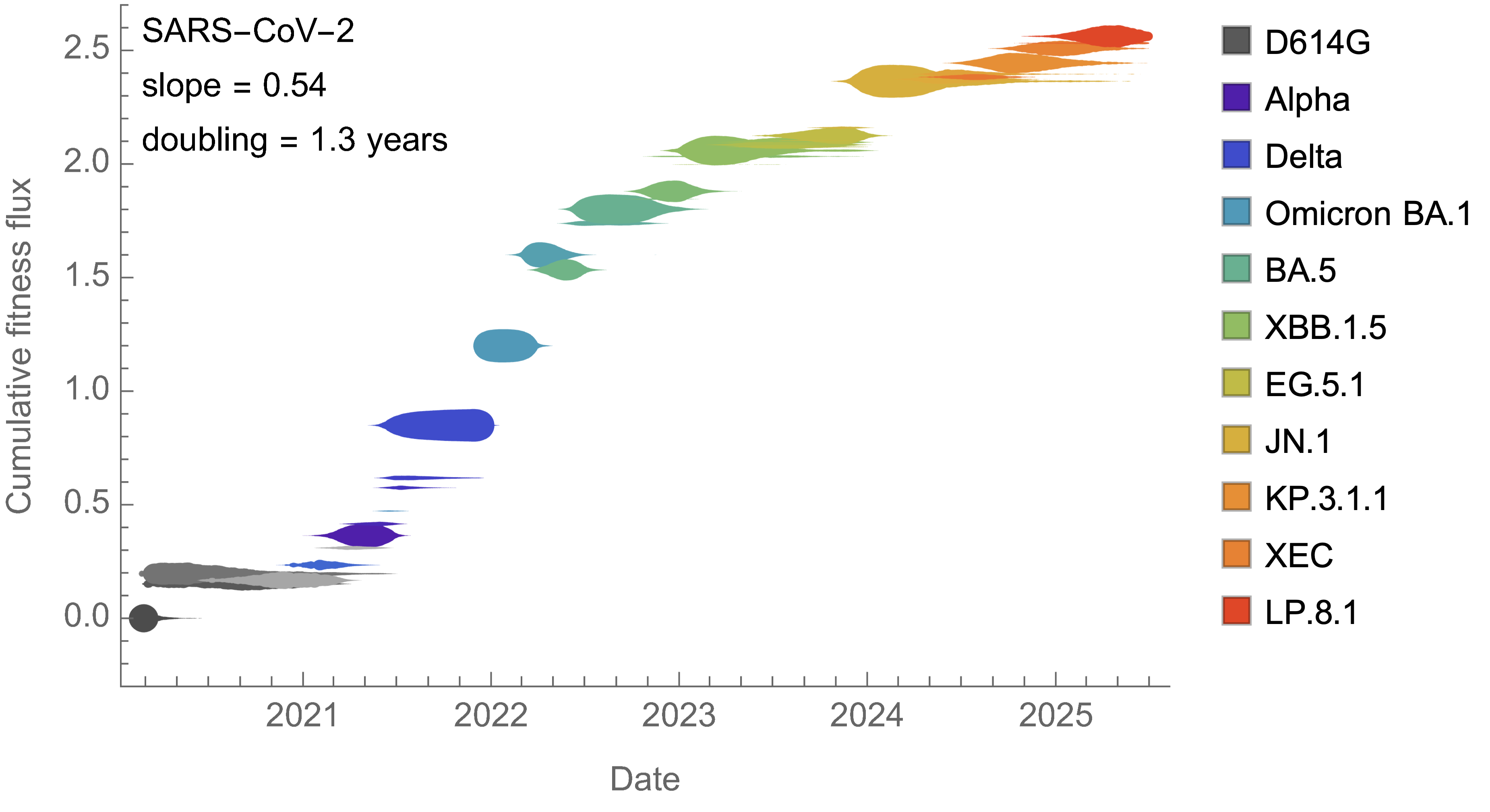
SARS-CoV-2 roughly doubled in fitness every year
Line thickness is proportional to variant frequency, 40 total variants
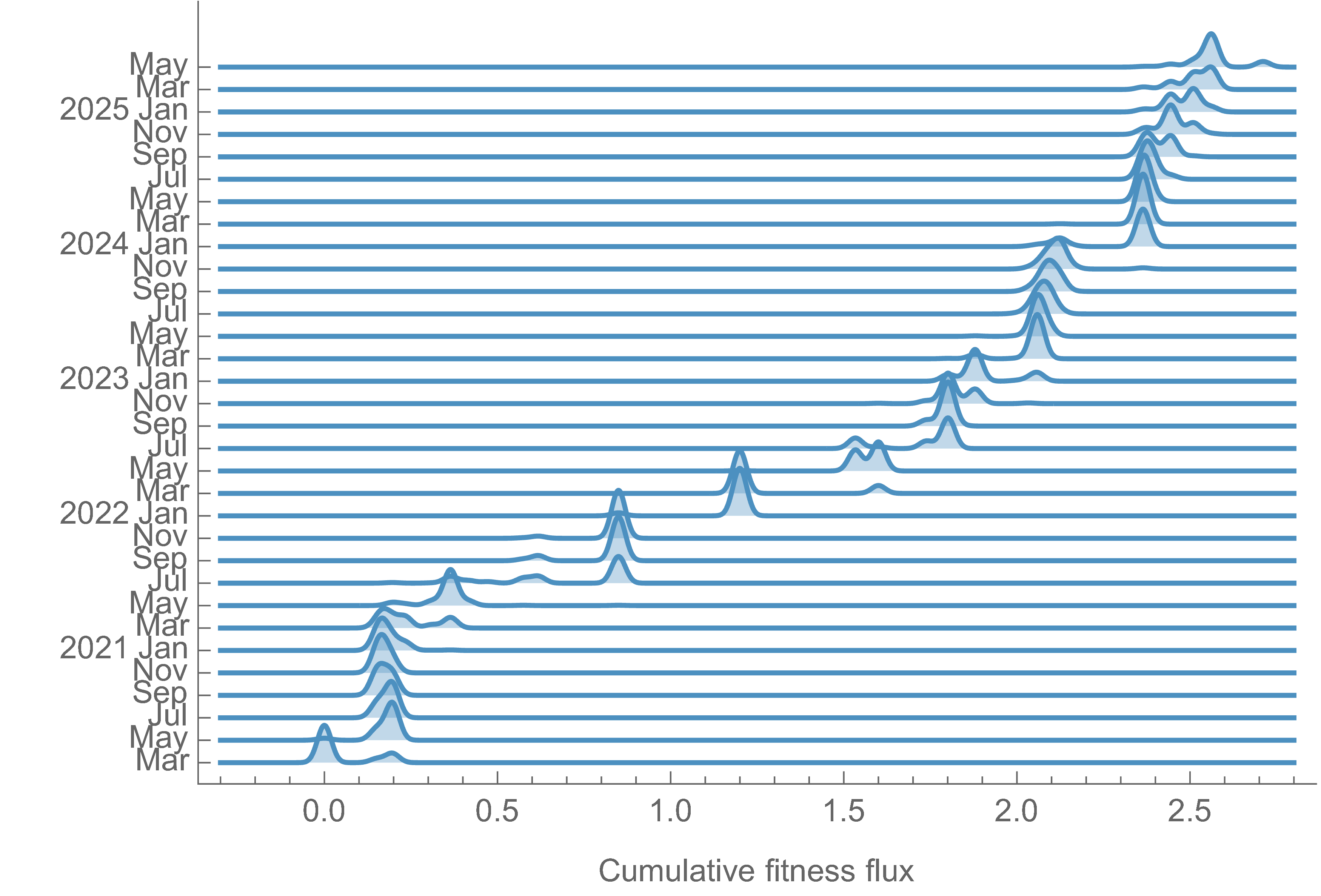
Traveling wave as SARS-CoV-2 population increases in fitness
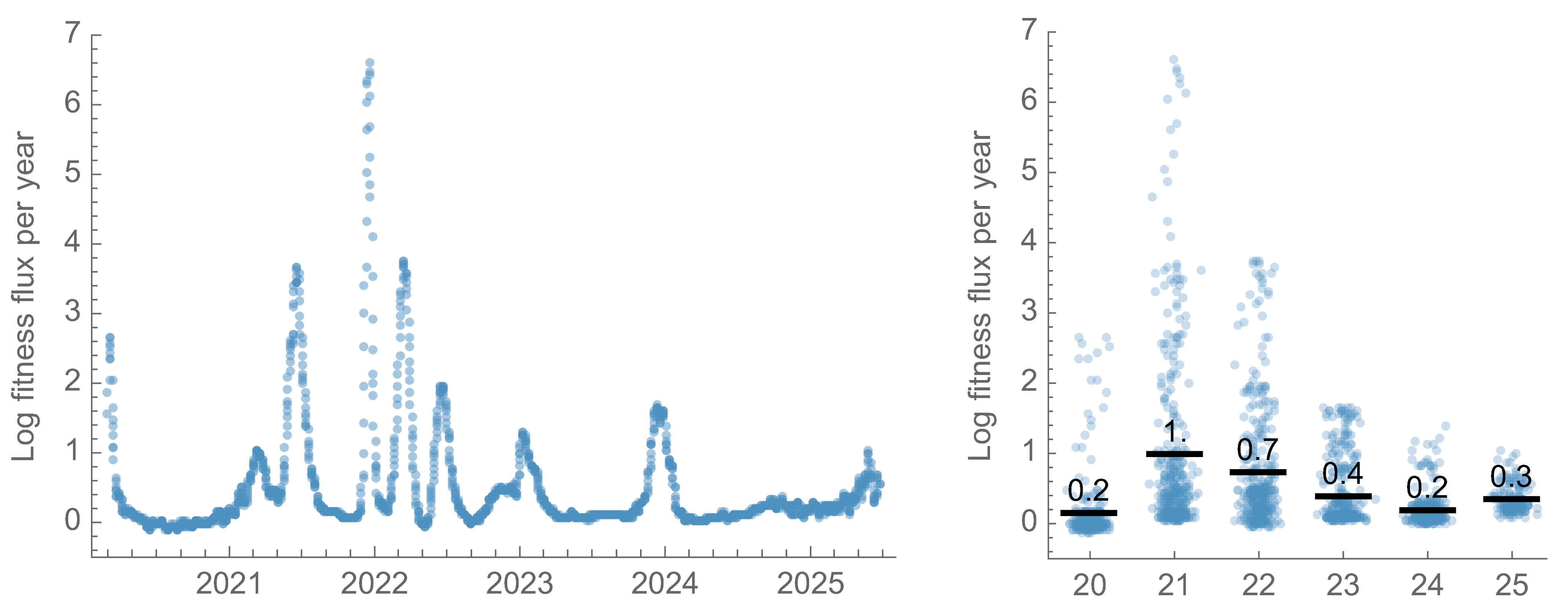
Slow down of fitness flux after 2022, but still rapid evolution
Many mutations of small effect create traveling fitness waves
Richard Neher and others have analytically characterized these waves
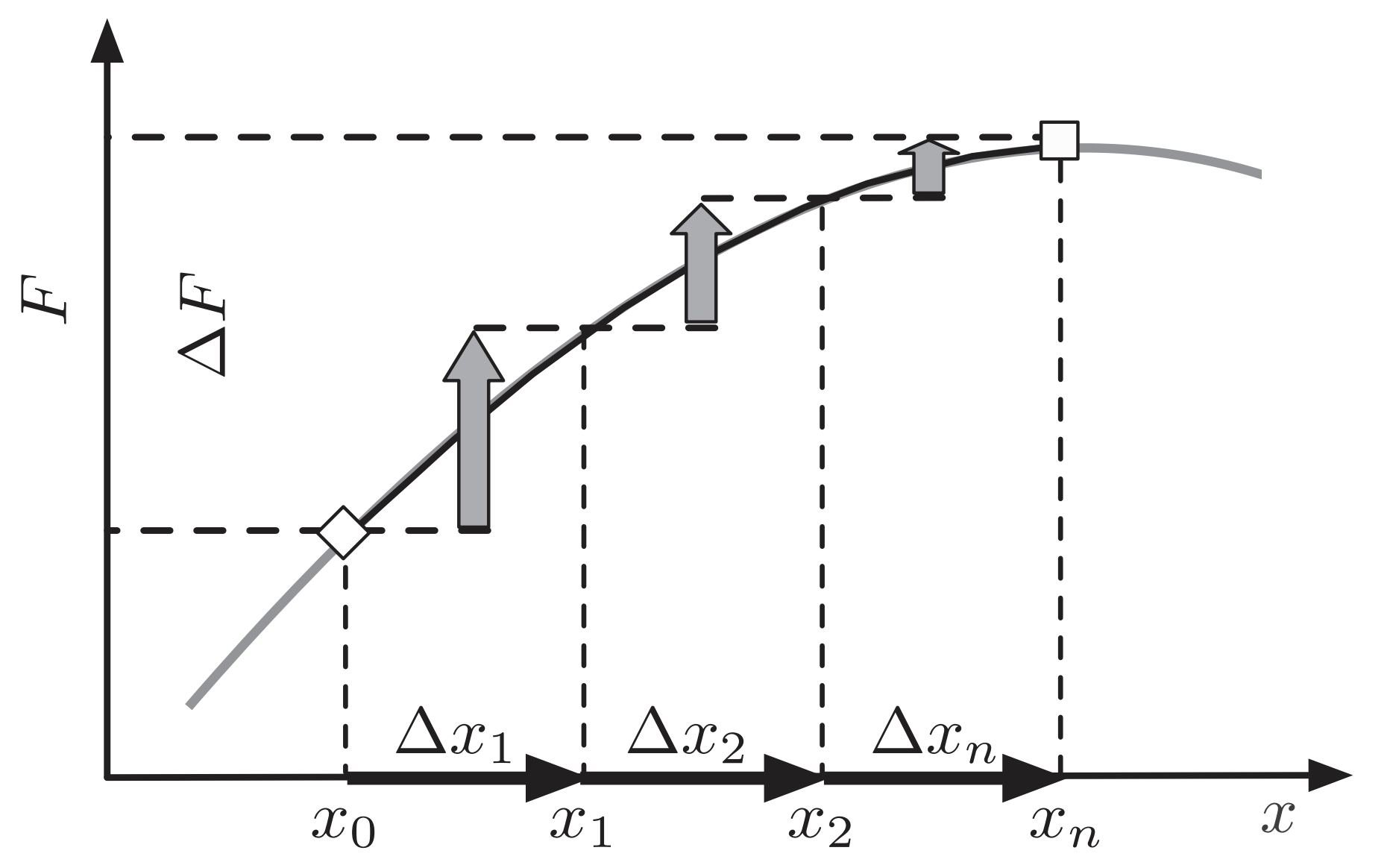
Diffusion constant $D = \mu \, \langle \delta^2 \rangle/2$, where the average $\langle \ldots \rangle$ is over the distribution of mutational effects $K(\delta)$
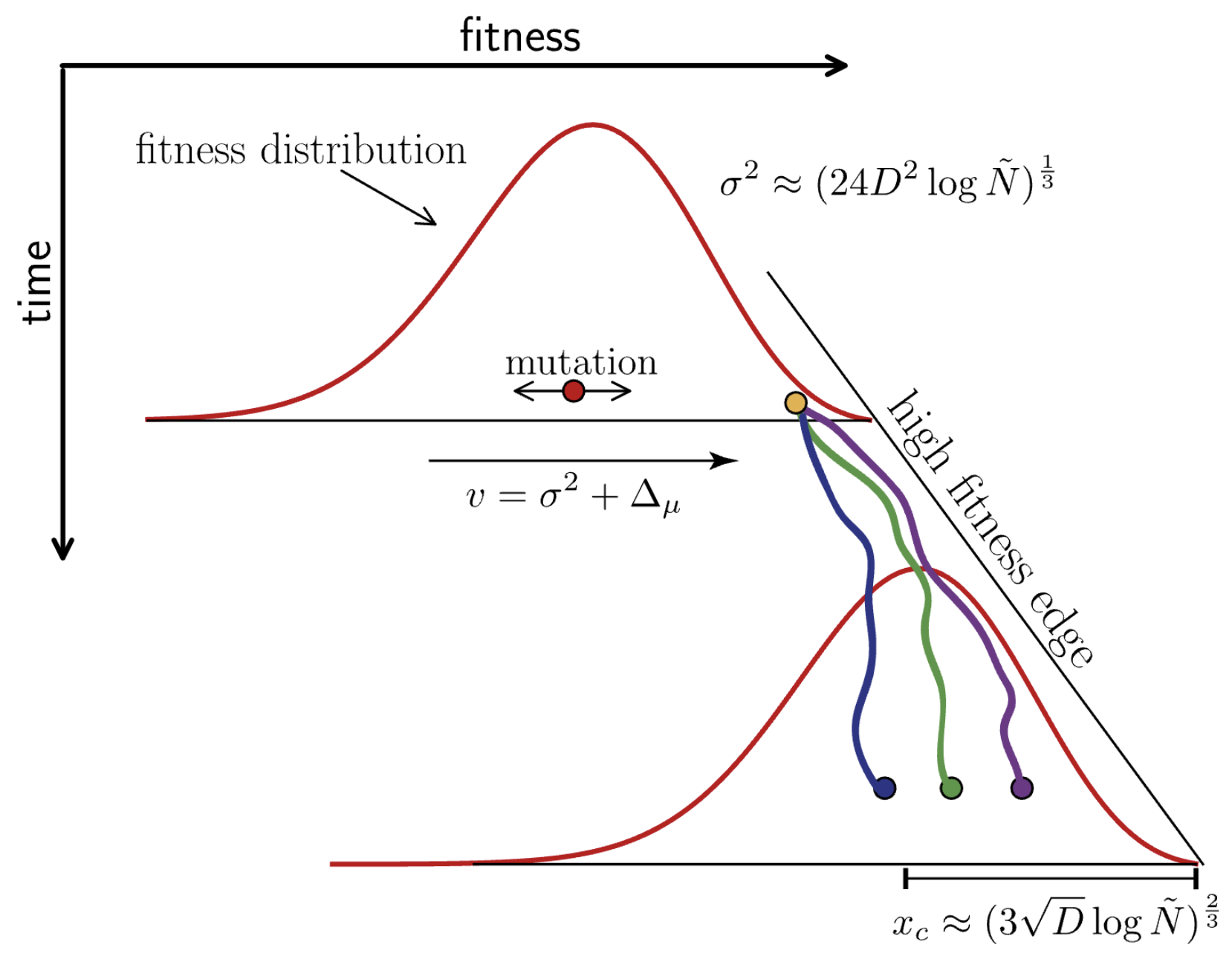 Neher and Hallatschek. 2013.
Neher and Hallatschek. 2013.
Fitness variance correlates well with fitness flux
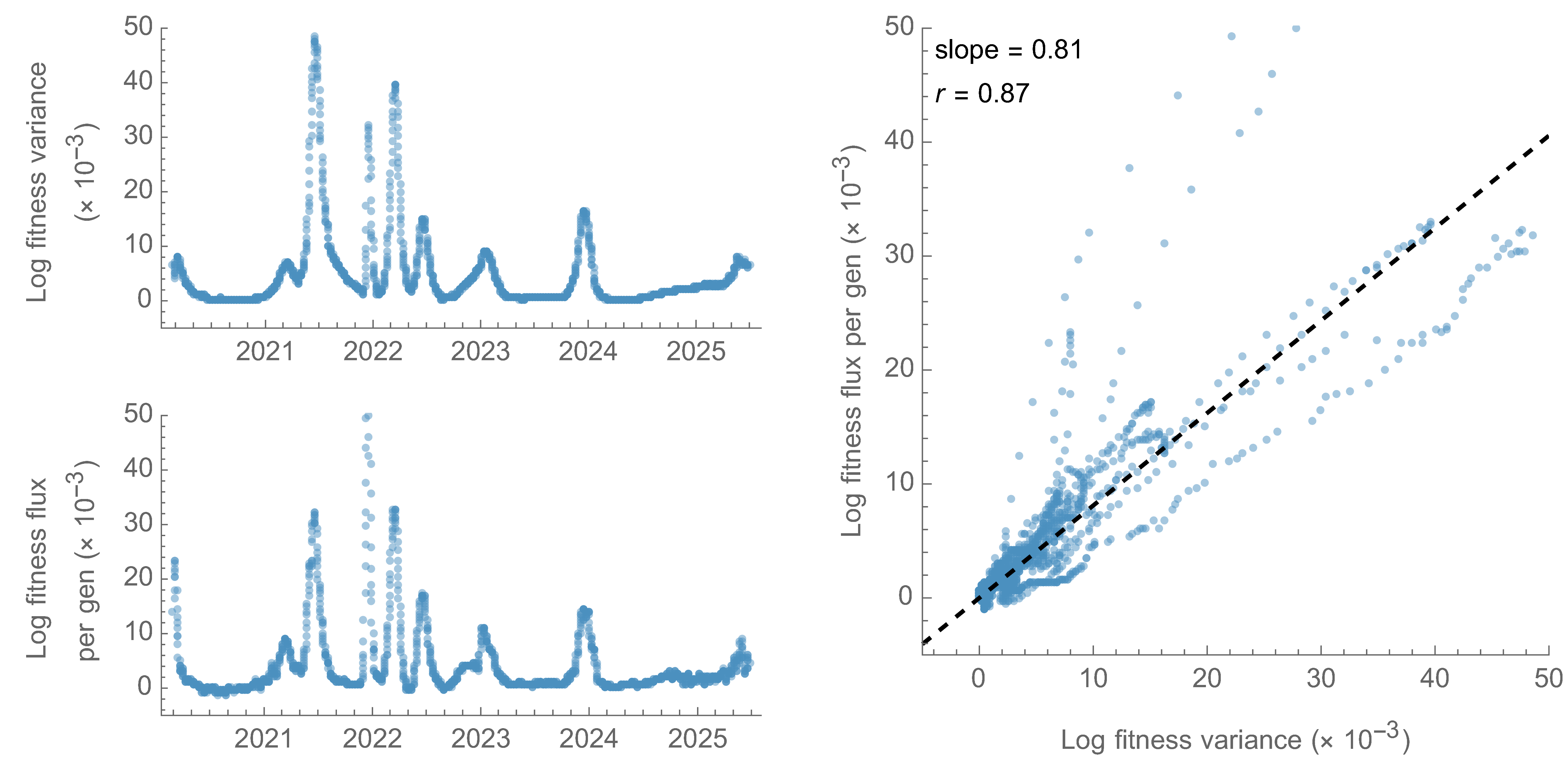
This is a specific example of Fisher's fundamental theorem
"The rate of increase in fitness of any organism at any time is equal to
its genetic variance in fitness at that time," ie
$$\frac{d\bar{f}}{dt} = Var(f)$$
Mutational fitness effects
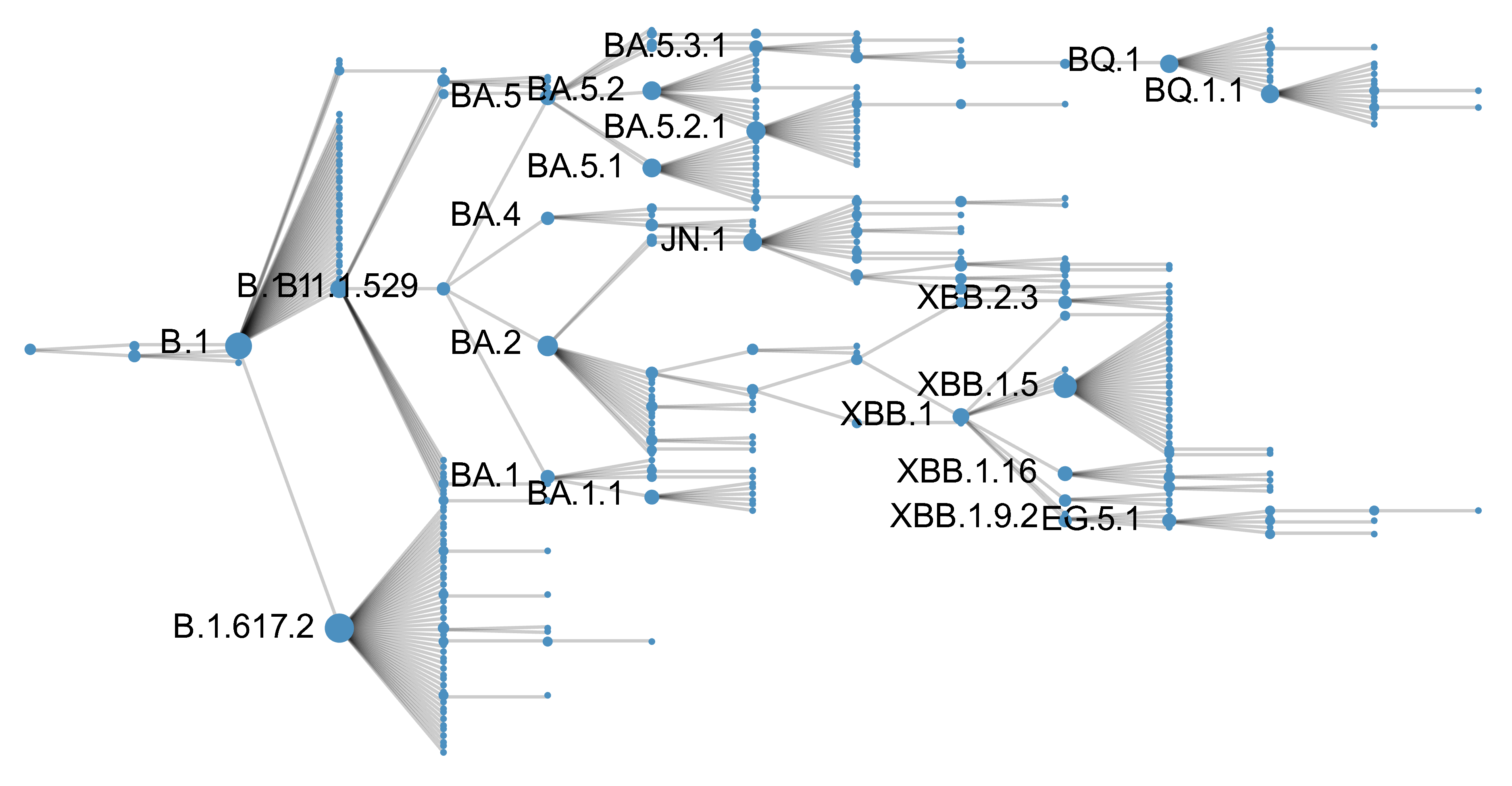
Analyze MLR fitness between parent/child lineages
Expand to 367 Pango lineages with at least 1000 sequence counts in the US from 2020 to 2025
Similar concept to Obermeyer et al
Most Pango branches have 0-1 spike mutations and change log fitness by ±0.1
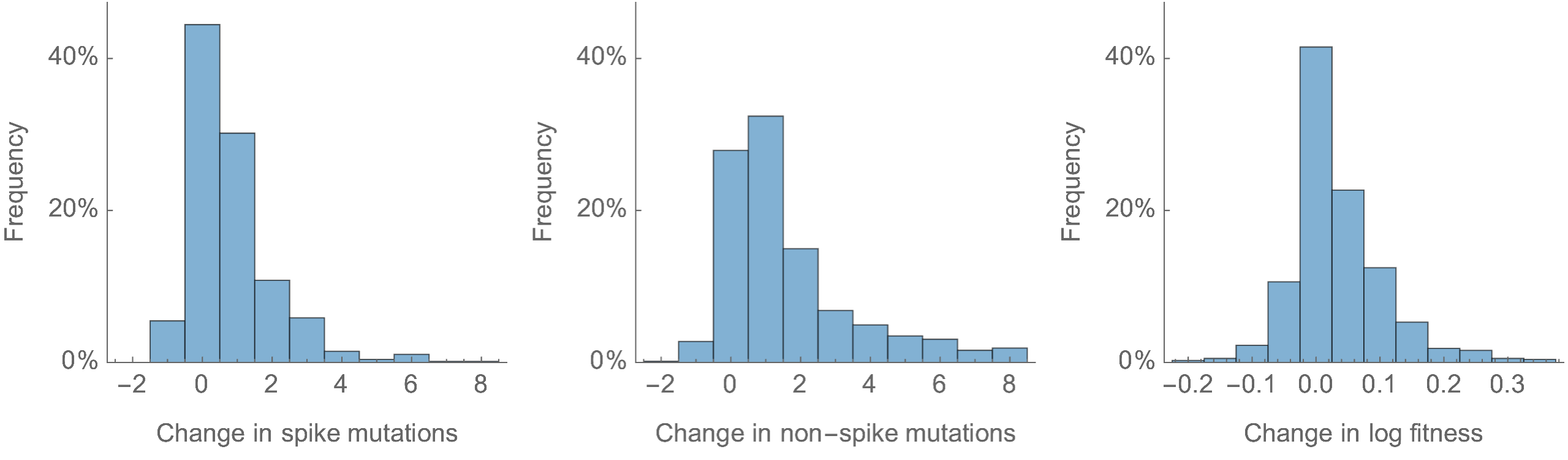
Spike mutations tend to increase fitness
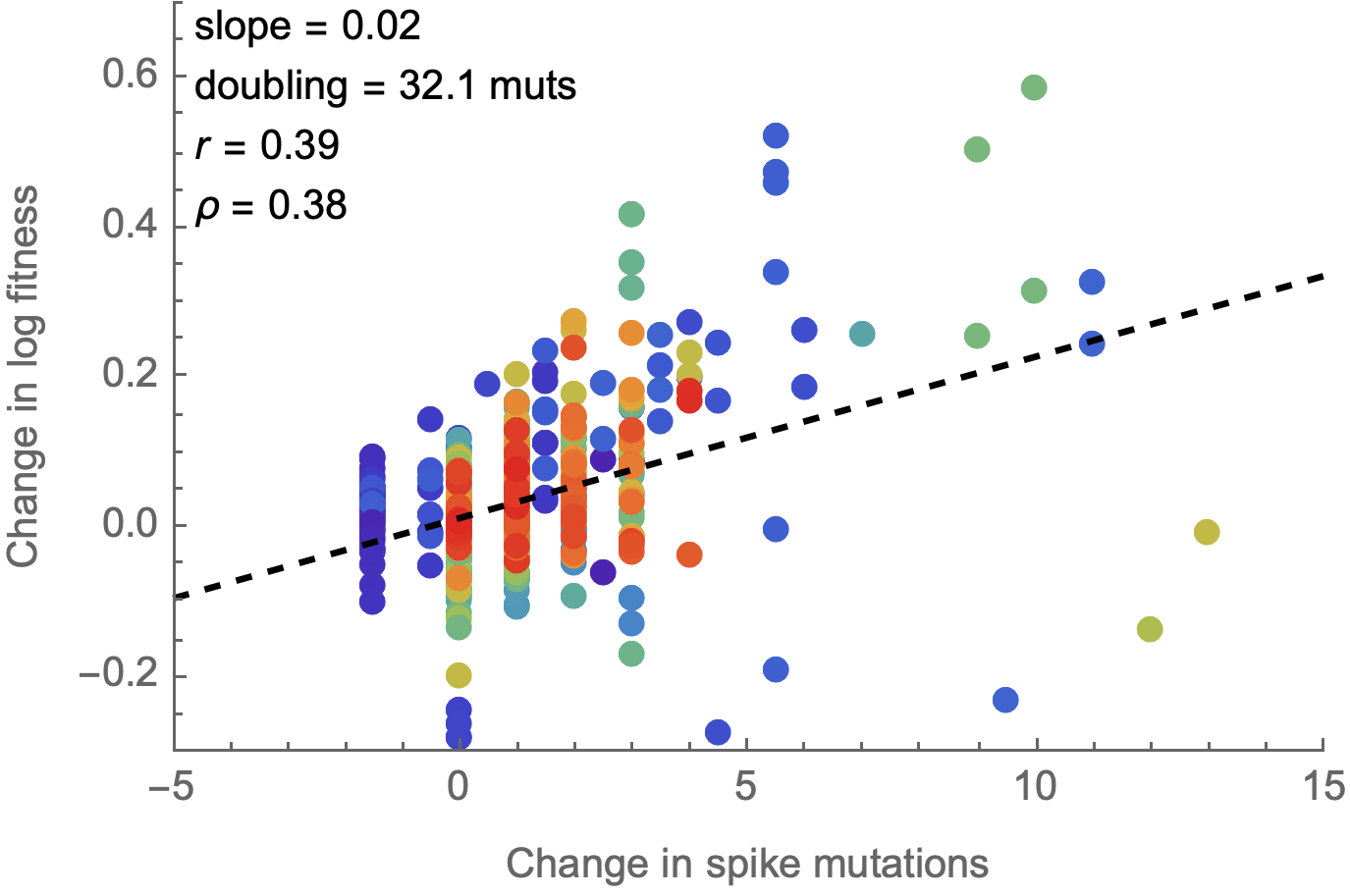
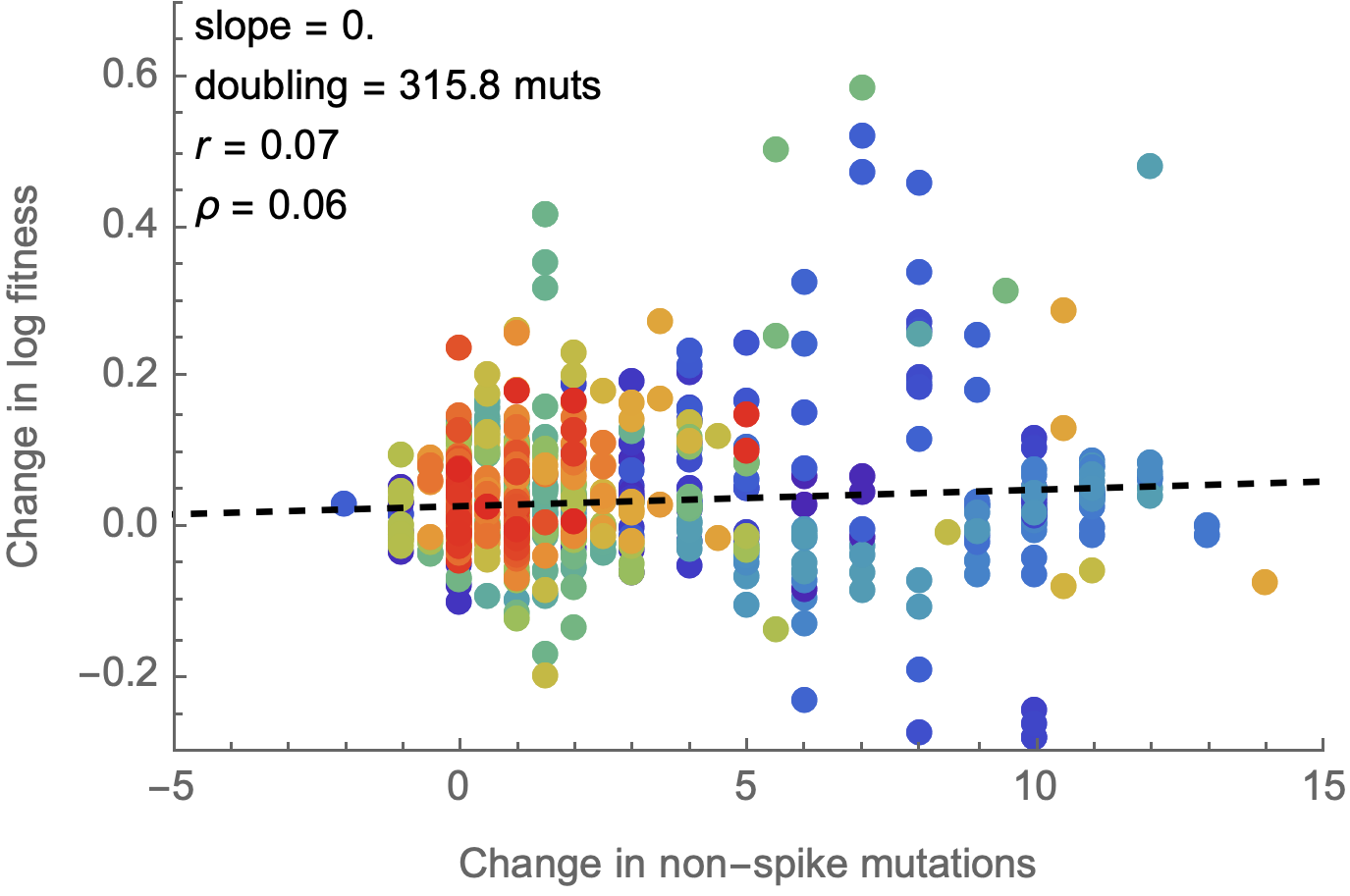
Non-spike mutations do not impact fitness on average
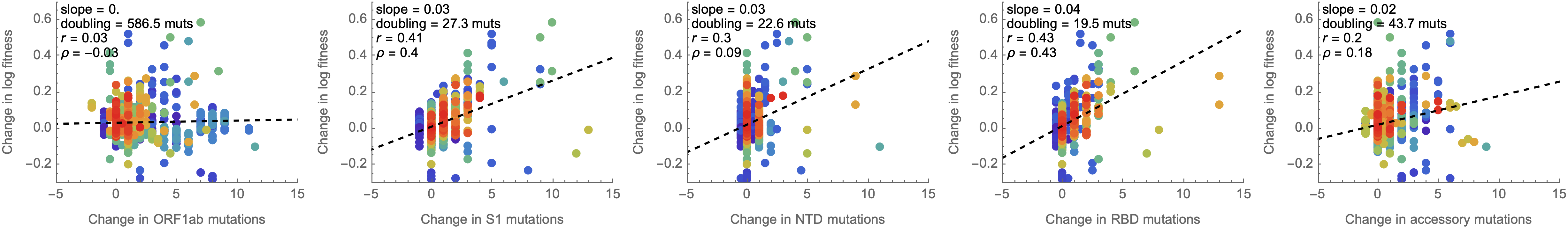
Looking across the genome shows that spike is the focus for positive selection, but accessory genes have some signal

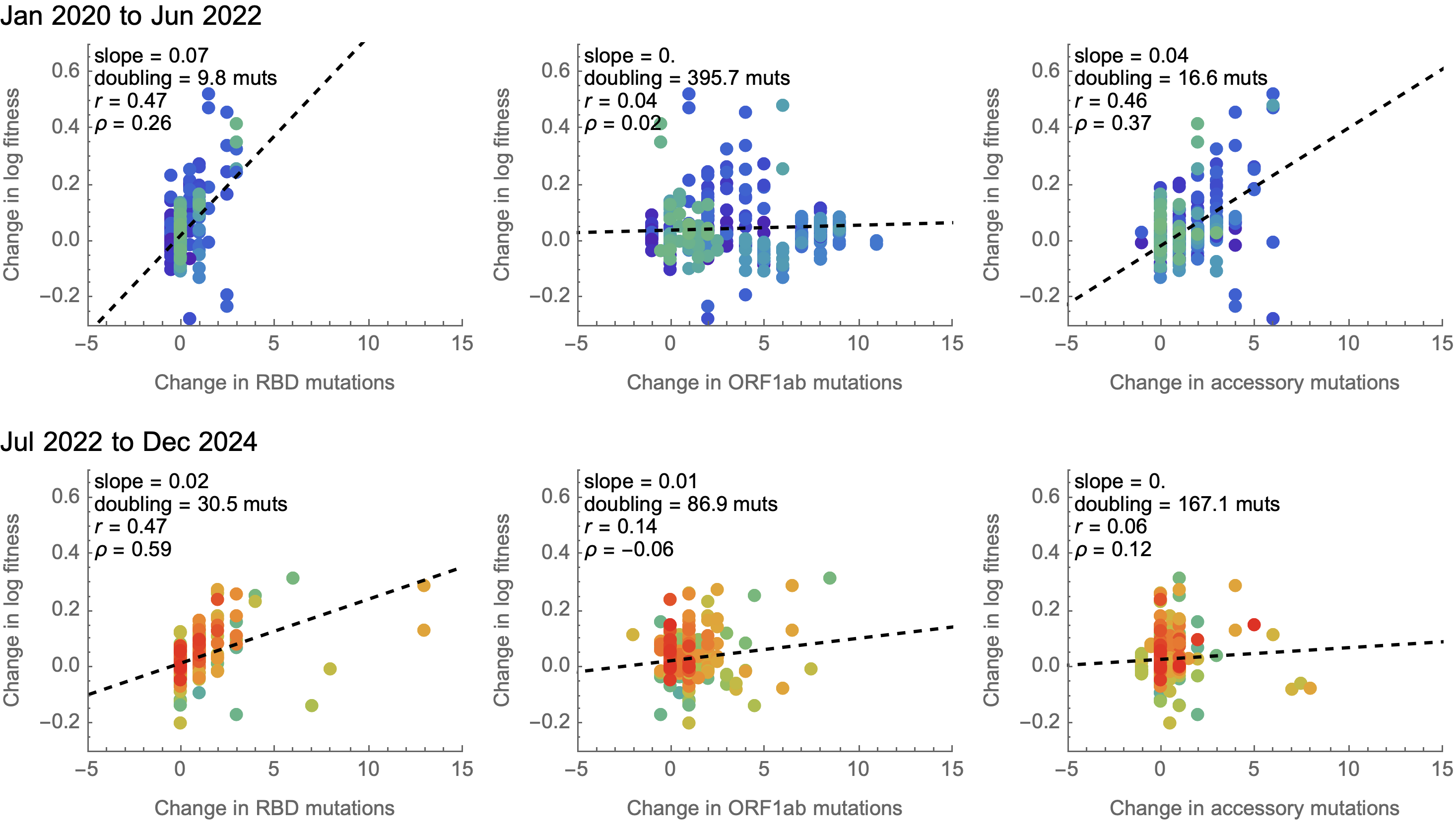
Some attenuation of fitness effects over time
Framework to compare predictors of fitness
EvEscape combines a variational autoencoder for mutation effect + antibody accessibility + biochemical dissimilarity
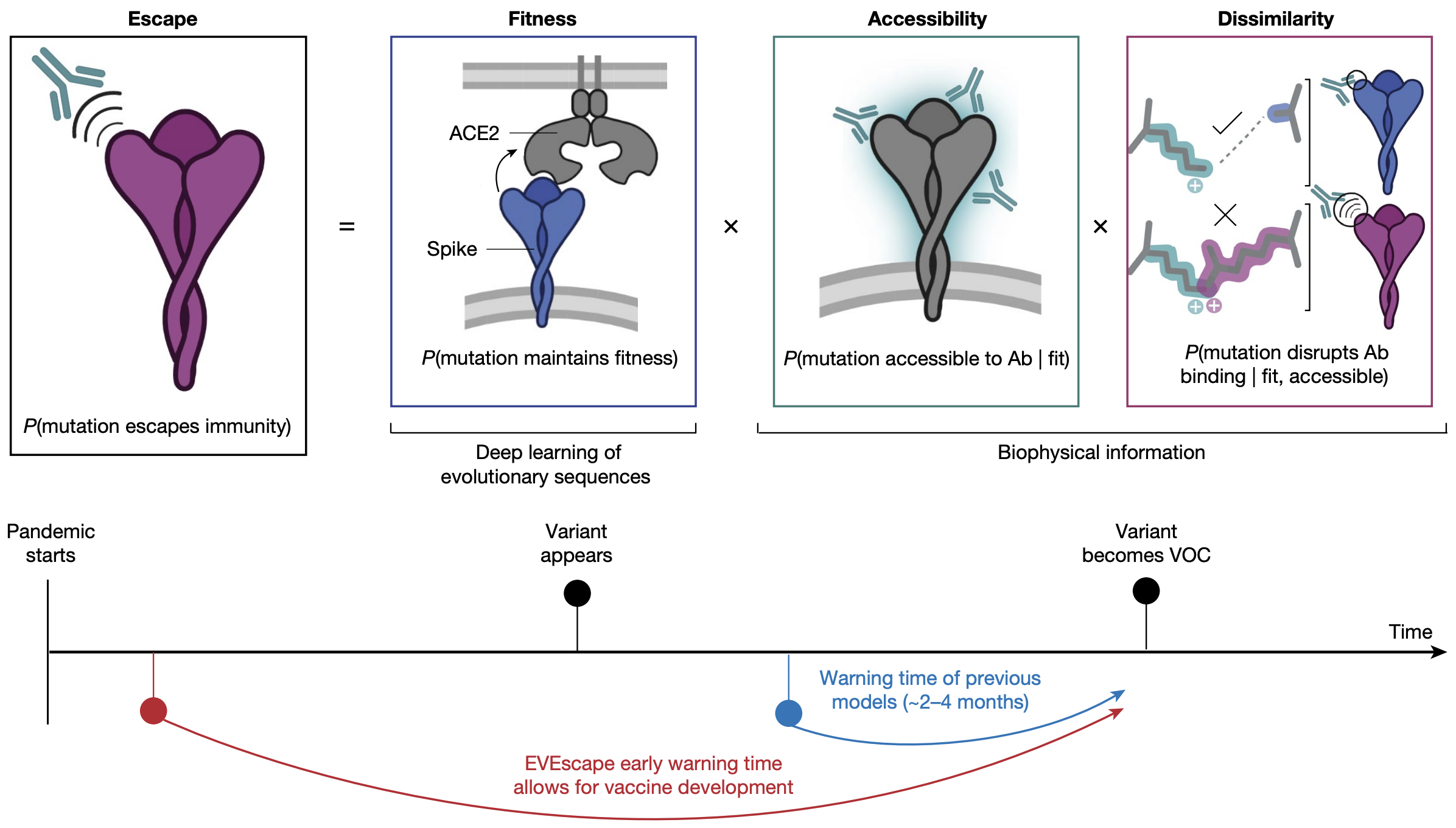 EvEscape from Thadani et al. 2023.
EvEscape from Thadani et al. 2023.
EvEscape does no better than counting spike mutations
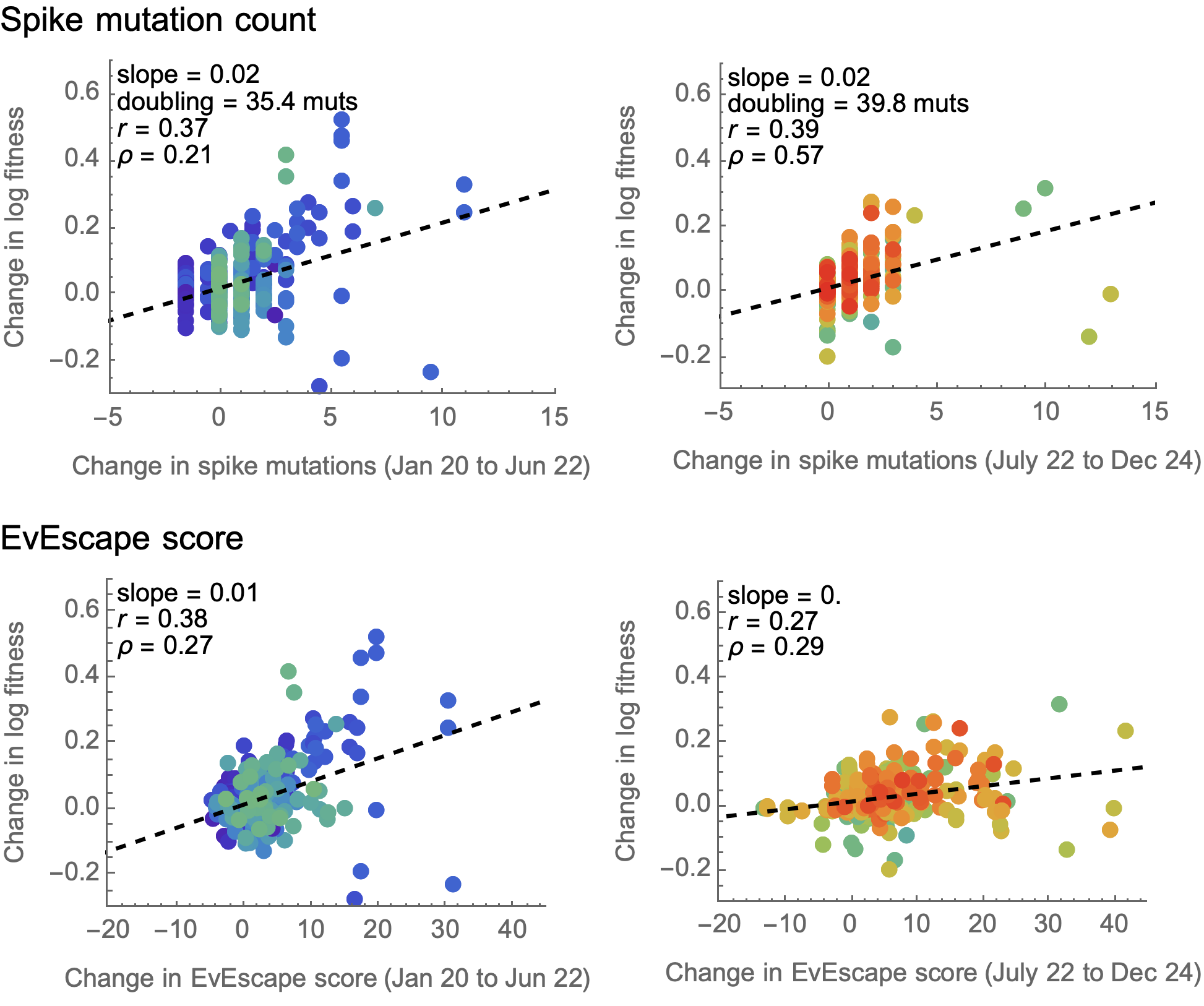 EvEscape from Thadani et al. 2023.
EvEscape from Thadani et al. 2023.
CoVFit uses spike protein embeddings from ESM-2 to predict MLR fitnesses
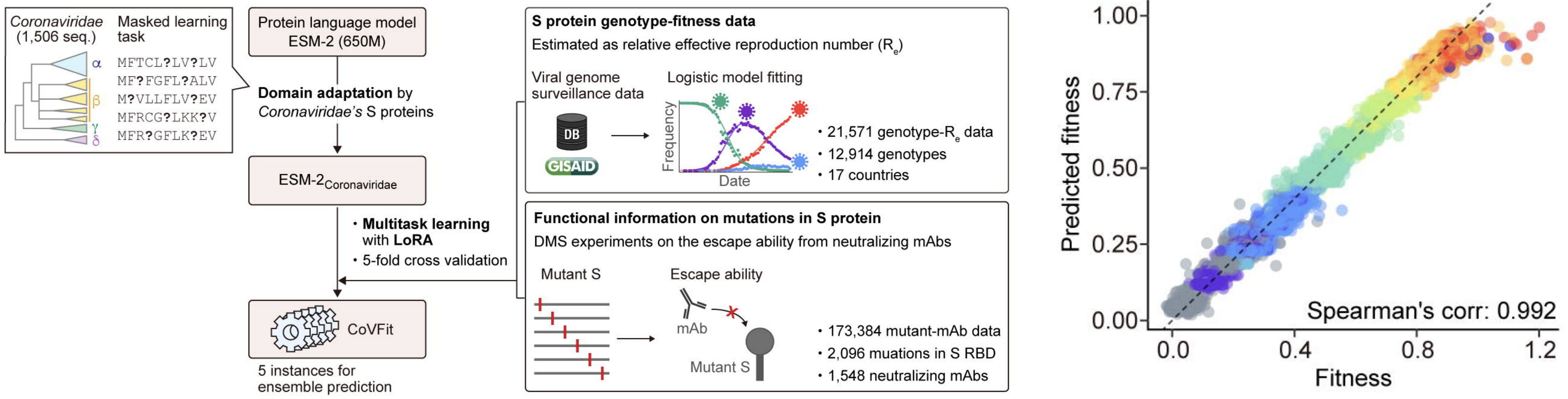 CoVFit from Ito et al. 2025.
CoVFit from Ito et al. 2025.
CoVFit performs poorly outside of training window
Note also the importance of properly assessing independent parent/child lineage deltas
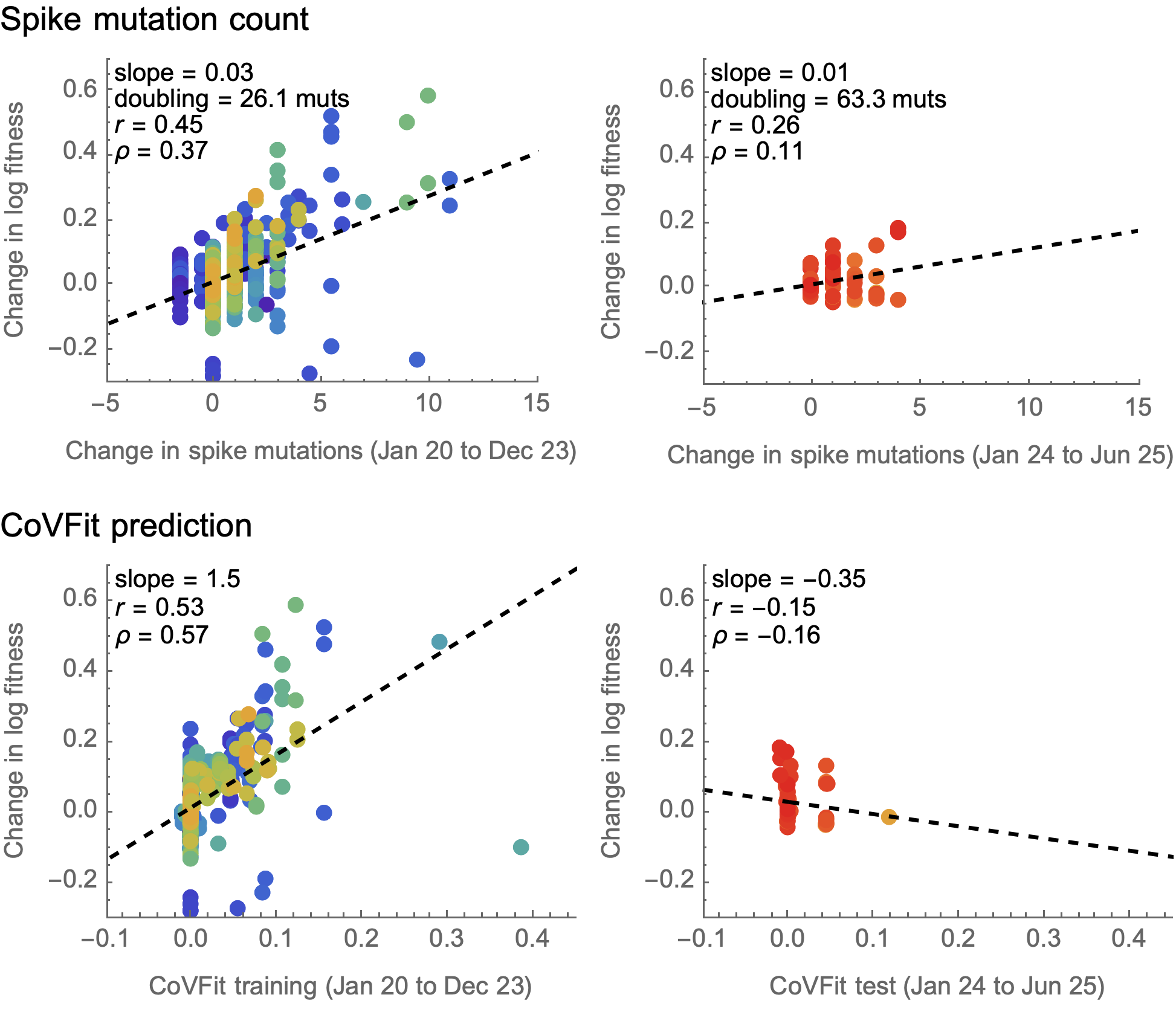 CoVFit from Ito et al. 2025.
CoVFit from Ito et al. 2025.
- Estimate fitness of circulating lineages and predict their short-term frequencies
- Predict fitness of proximal out-of-sample lineages
- Predict emergence and spread of out-of-sample lineages
Acknowledgements
Seasonal influenza and SARS-CoV-2 genomics: Data producers from all over the world, GISAID
Nextstrain: Richard Neher, Ivan Aksamentov, John SJ Anderson, Kim Andrews, Jennifer Chang, James Hadfield, Emma Hodcroft, John Huddleston, Jover Lee, Victor Lin, Cornelius Roemer, Thomas Sibley
MLR and fitness modeling: Marlin Figgins, Eslam Abousamra, Jover Lee, James Hadfield, John Huddleston, Philippa Steinberg, Jesse Bloom, Cornelius Roemer, Richard Neher
Bedford Lab:
![]() John Huddleston,
John Huddleston,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Jover Lee,
Jover Lee,
![]() Marlin Figgins,
Marlin Figgins,
![]() Victor Lin,
Victor Lin,
![]() Jennifer Chang,
Jennifer Chang,
![]() Nashwa Ahmed,
Nashwa Ahmed,
![]() Cécile Tran Kiem,
Cécile Tran Kiem,
![]() Kim Andrews,
Kim Andrews,
![]() Philippa Steinberg,
Philippa Steinberg,
![]() Jacob Dodds,
Jacob Dodds,
![]() Amin Bemanian,
Amin Bemanian,
![]() Carlos Avendano,
Carlos Avendano,
![]() Aayush Verma
Aayush Verma



