High-resolution phylogenetic tracking of city-level spread of seasonal influenza
Trevor Bedford (@trvrb)
6 Dec 2019
Epidemics7
Charleston, SC
Okay to share on social media #SeattleFluStudy
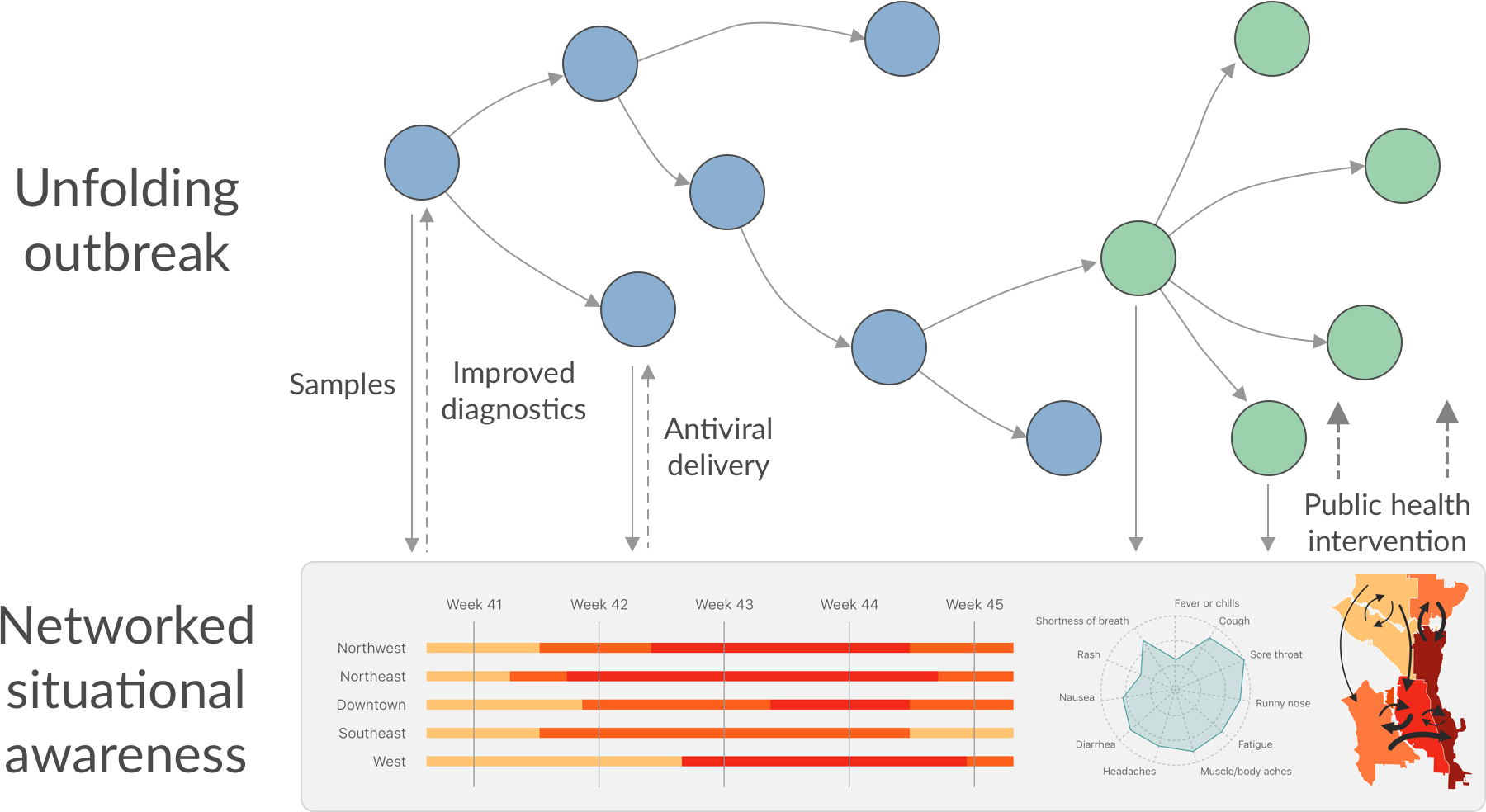
 Seattle Flu Study
Seattle Flu Study
Lead investigators: Helen Chu, Michael Boeckh, Janet Englund, Michael Famulare, Barry Lutz, Deborah Nickerson, Mark Rieder, Lea Starita, Matthew Thompson, Jay Shendure, Trevor Bedford
Co-investigators: Amanda Adler, Jeris Bosua, Elisabeth Brandstetter, Kairsten Fay, Chris Frazar, Peter Han, Reena Gulati, James Hadfield, ShiChu Huang, Misja Ilcisin, Michael Jackson, Anahita Kiavand, Louise Kimball, Enos Kline, Kirsten Lacombe, Jover Lee, Jennifer Logue, Victoria Lyon, Kira Newman, Miguel Paredes, Thomas Sibley, Monica Zigman Suchsland, Cassia Wagner, Caitlin Wolf




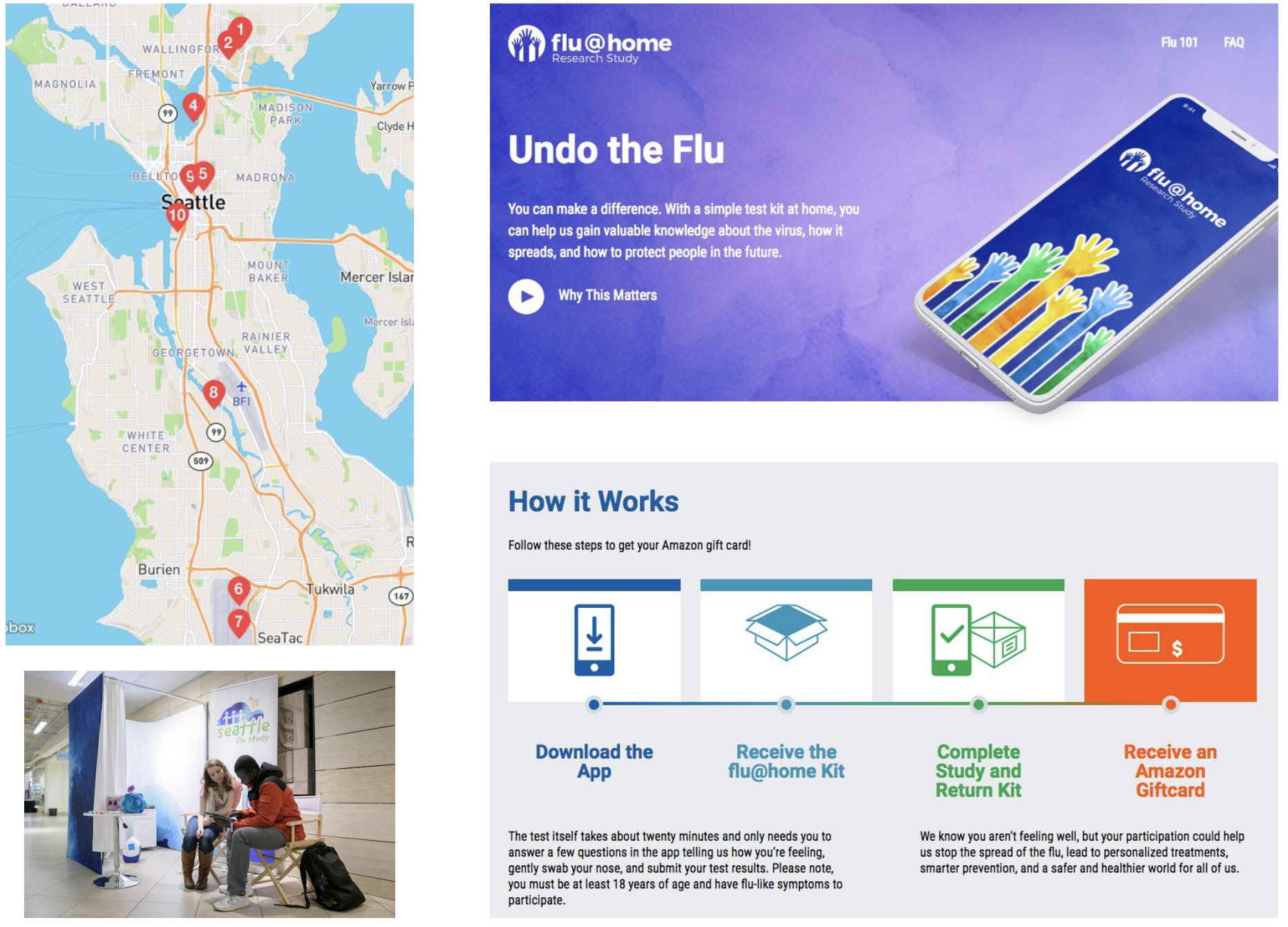
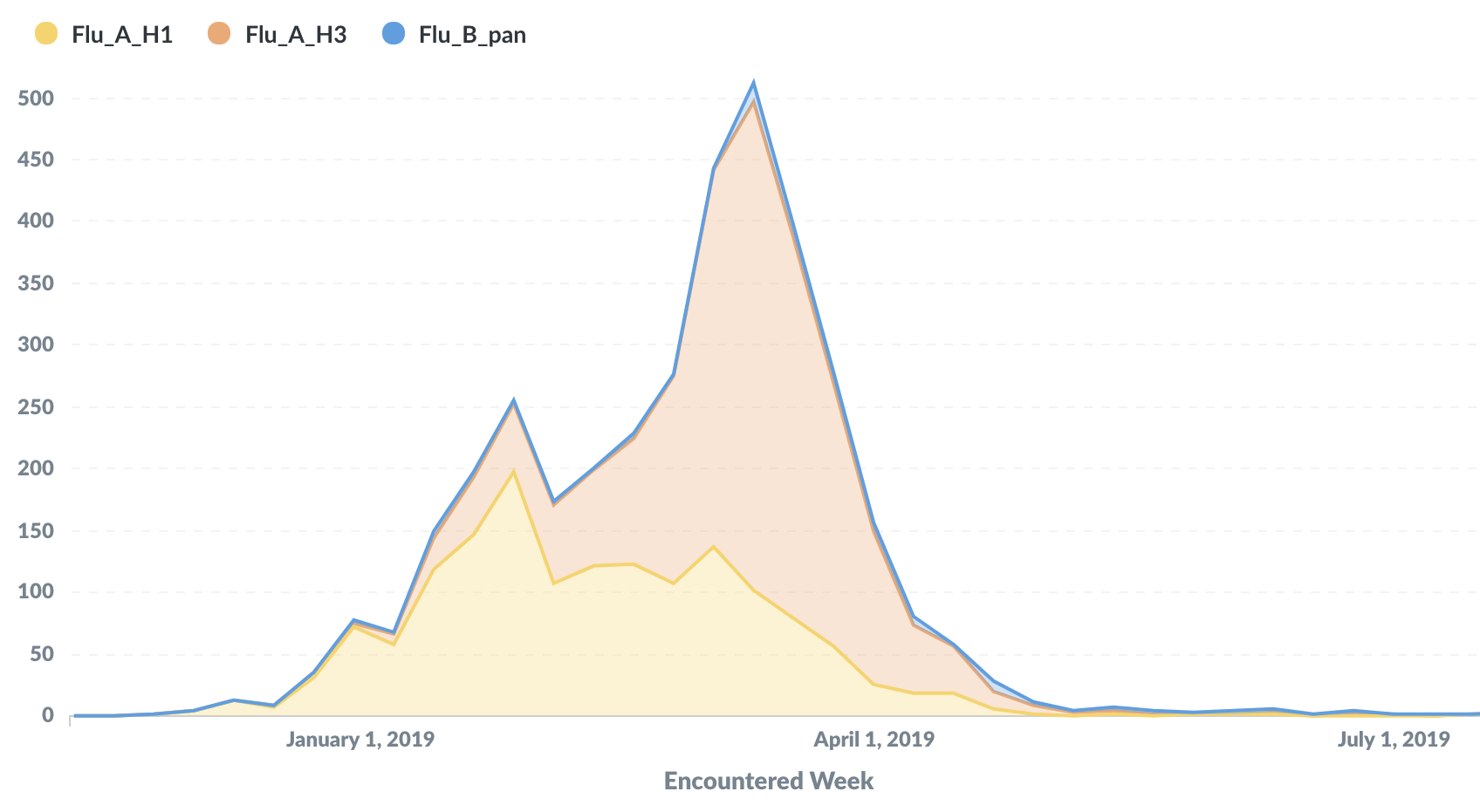
Sequenced ~2200 flu+ samples collected from the Seattle metro area
Full genomes substantially increase phylogenetic resolution
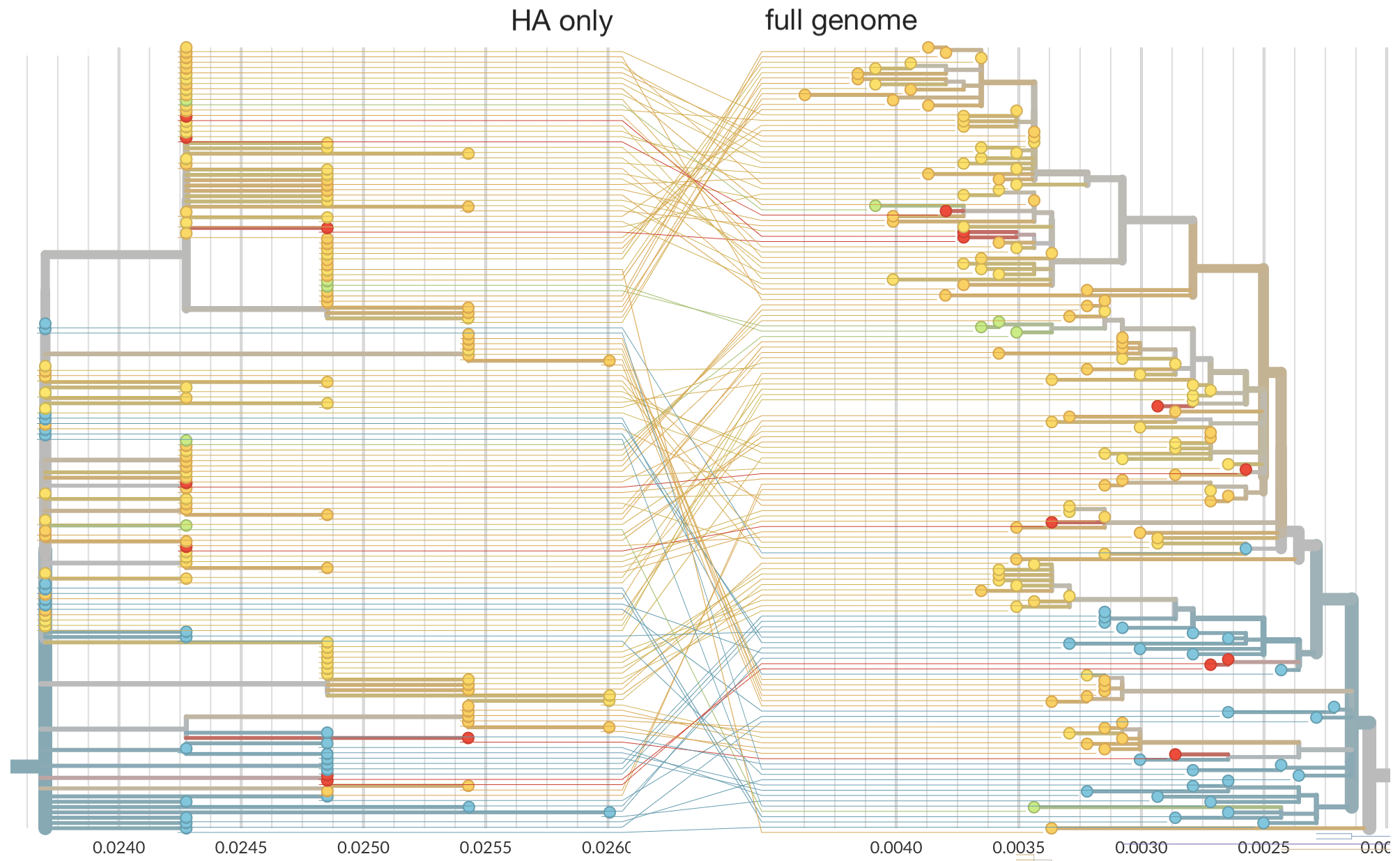
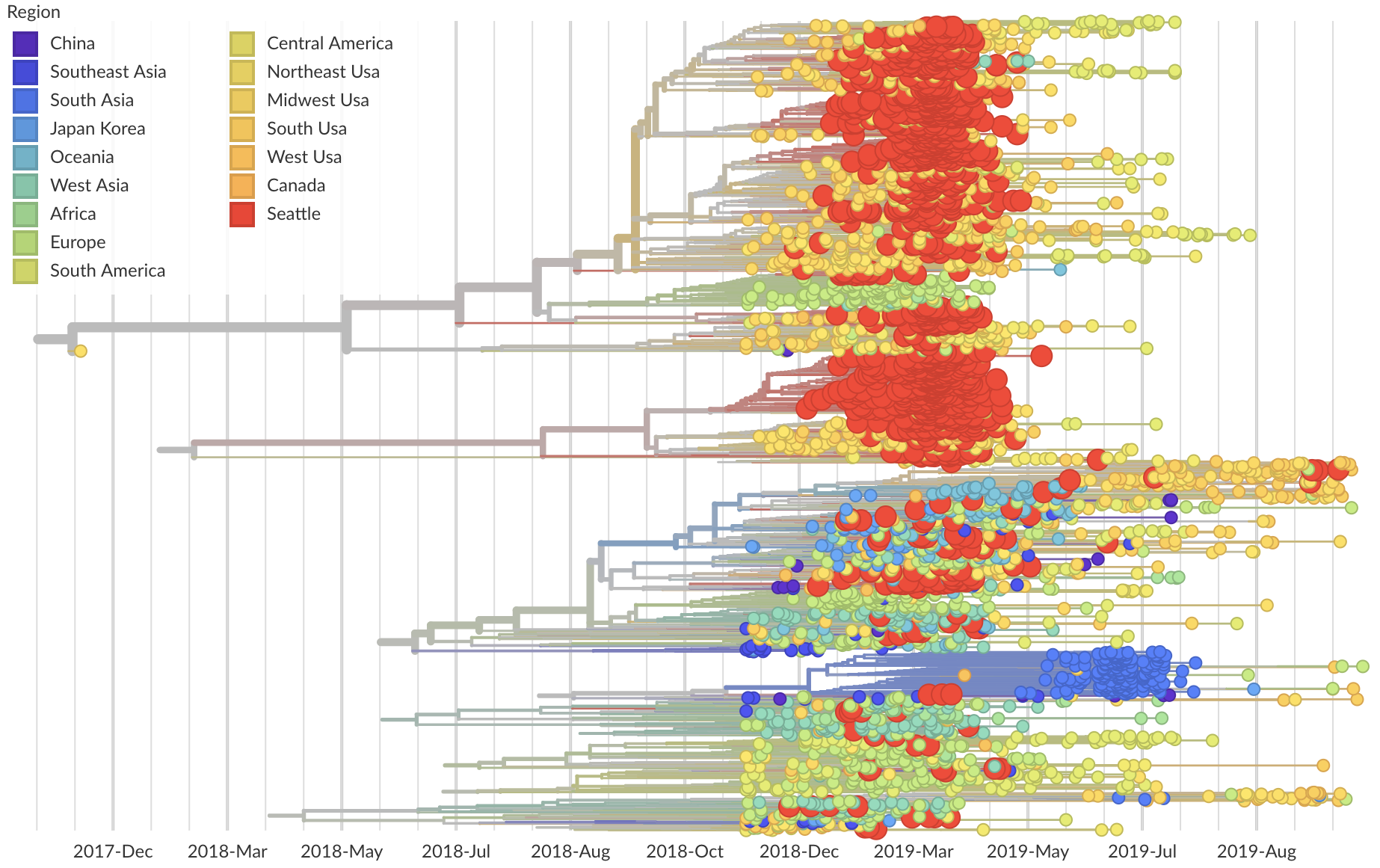
Phylogeny of 1150 Seattle H3N2 viruses
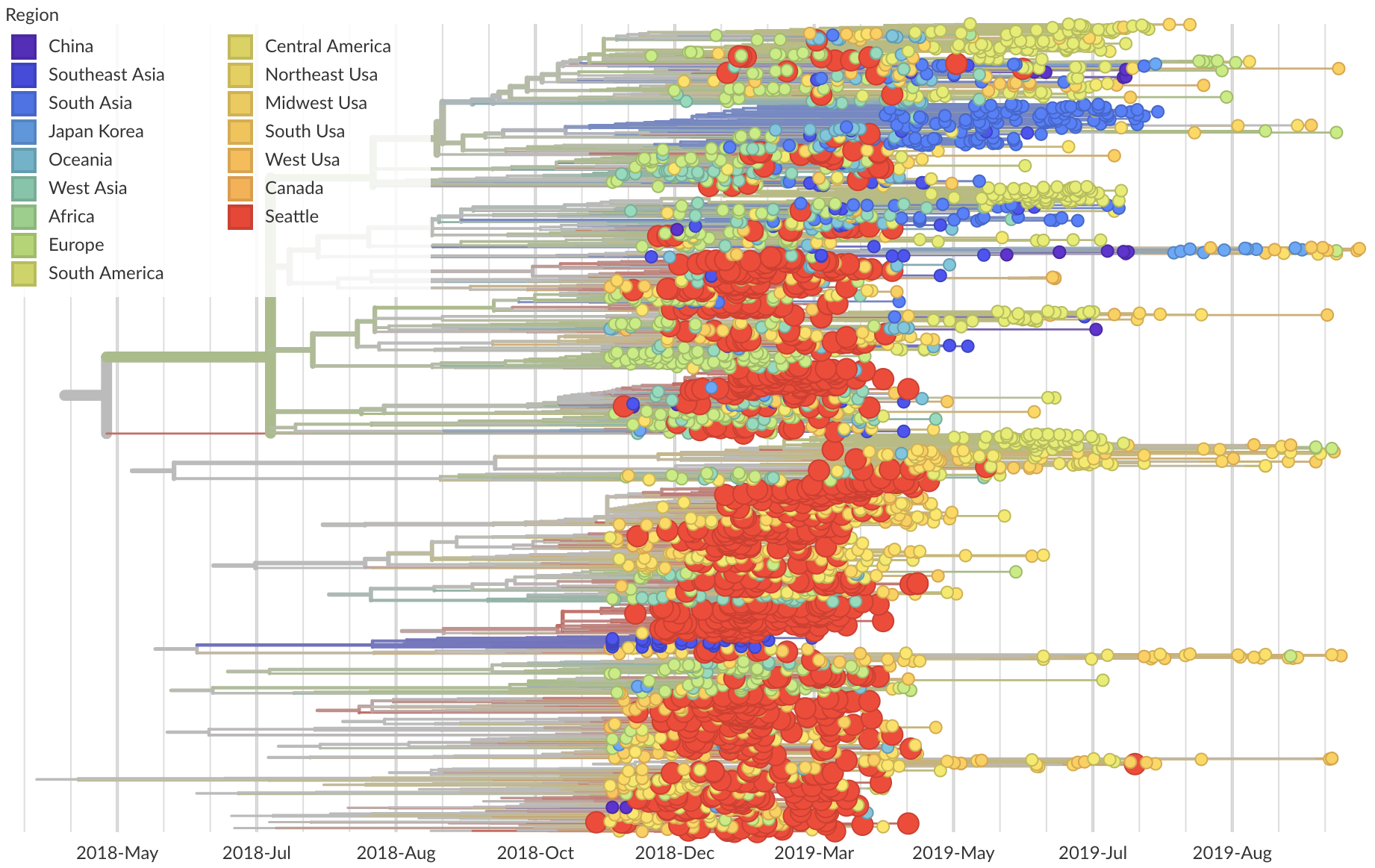
Phylogeny of 939 Seattle H1N1pdm viruses
Divide analyses:
- Partitioning separate introductions
- Tracing transmission chains after arrival
Introductions
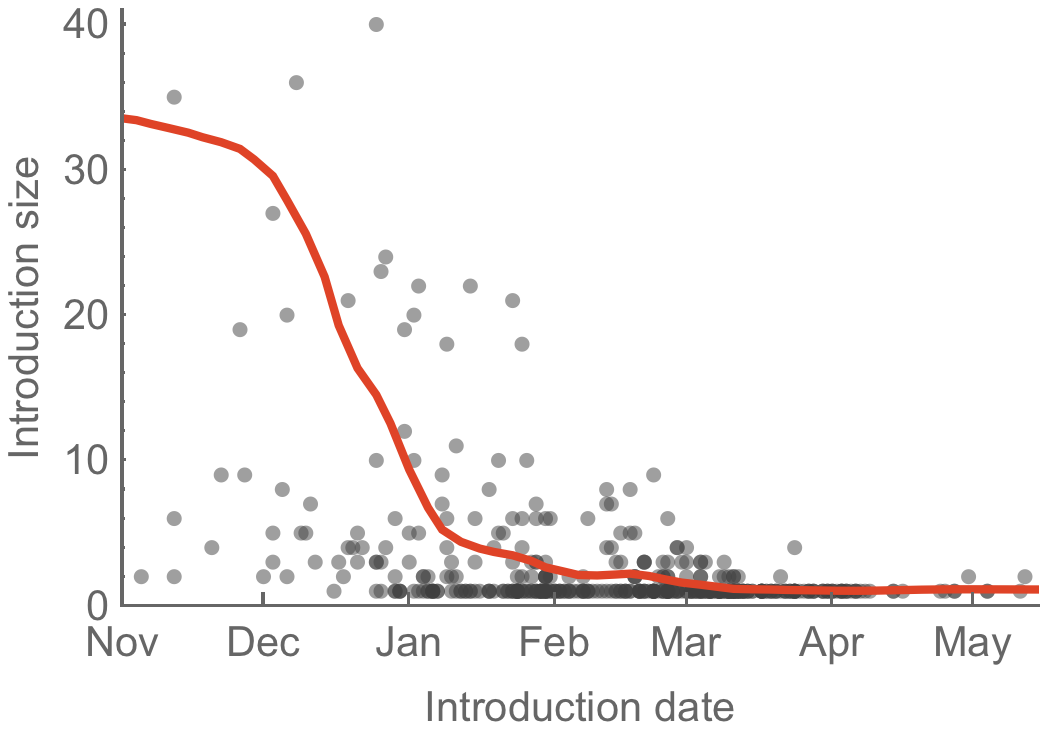
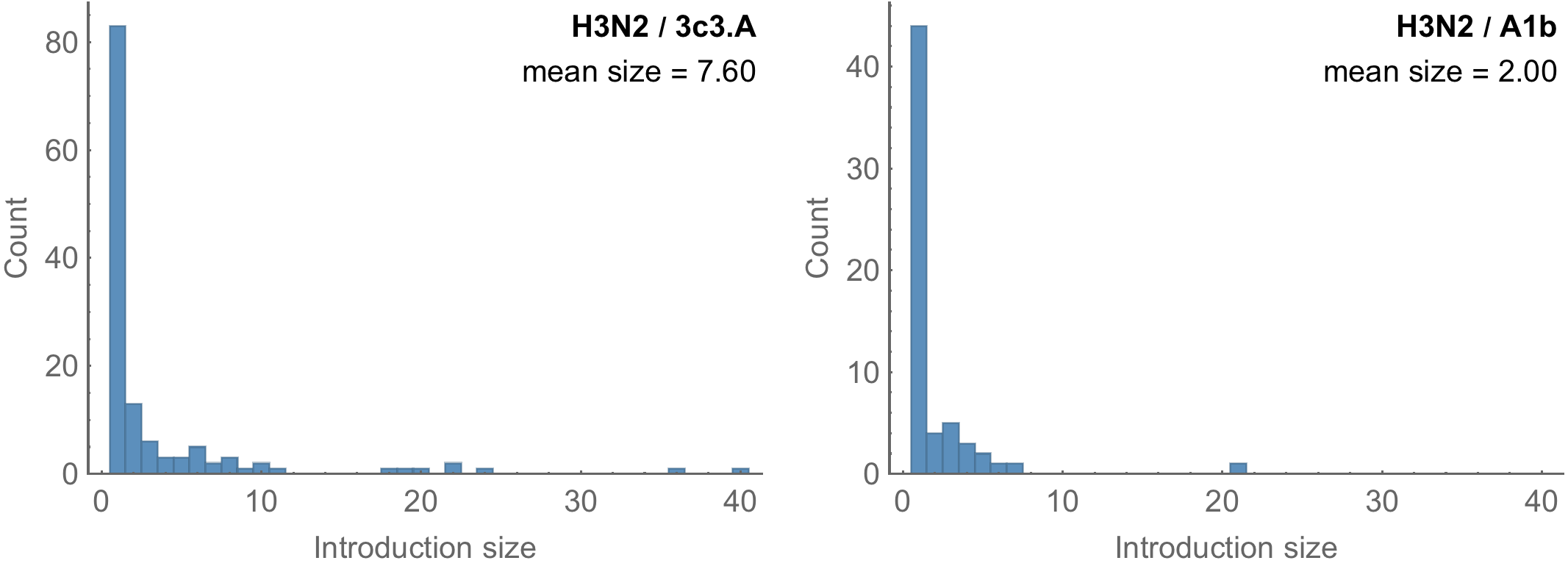
Introduction cluster size correlates with transmission potential R
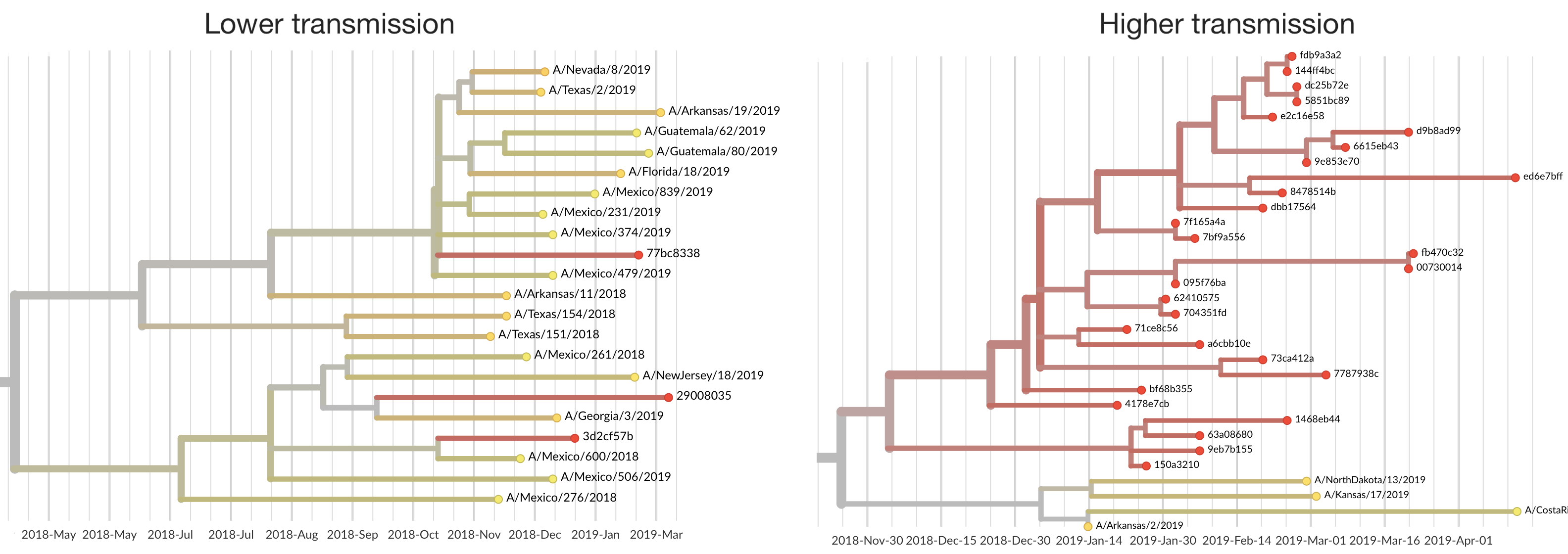
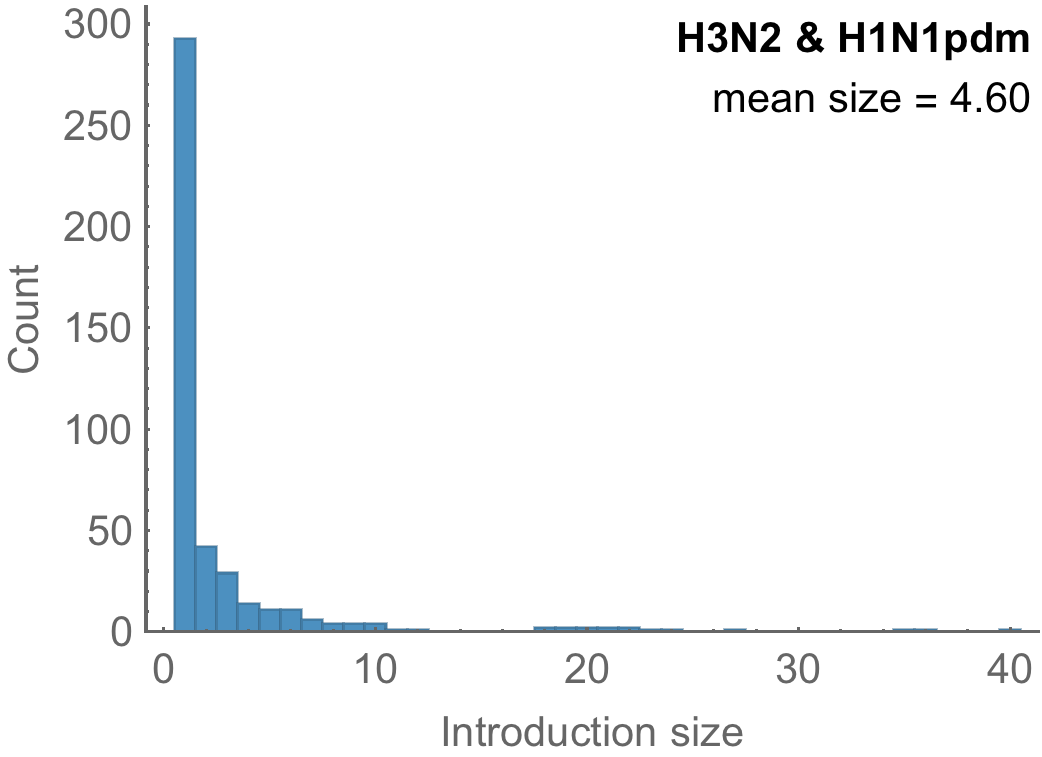
Highly skewed distribution of introduction sizes
Early season introductions more successful than late season introductions
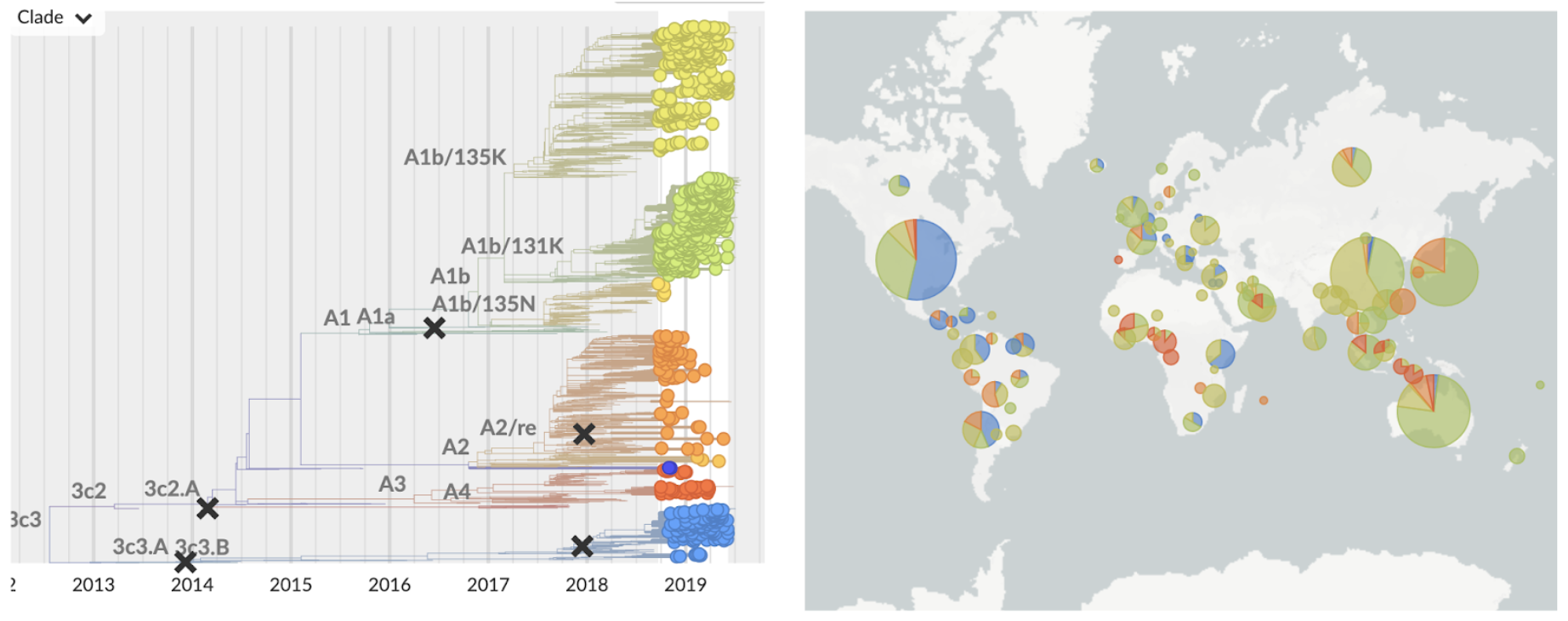
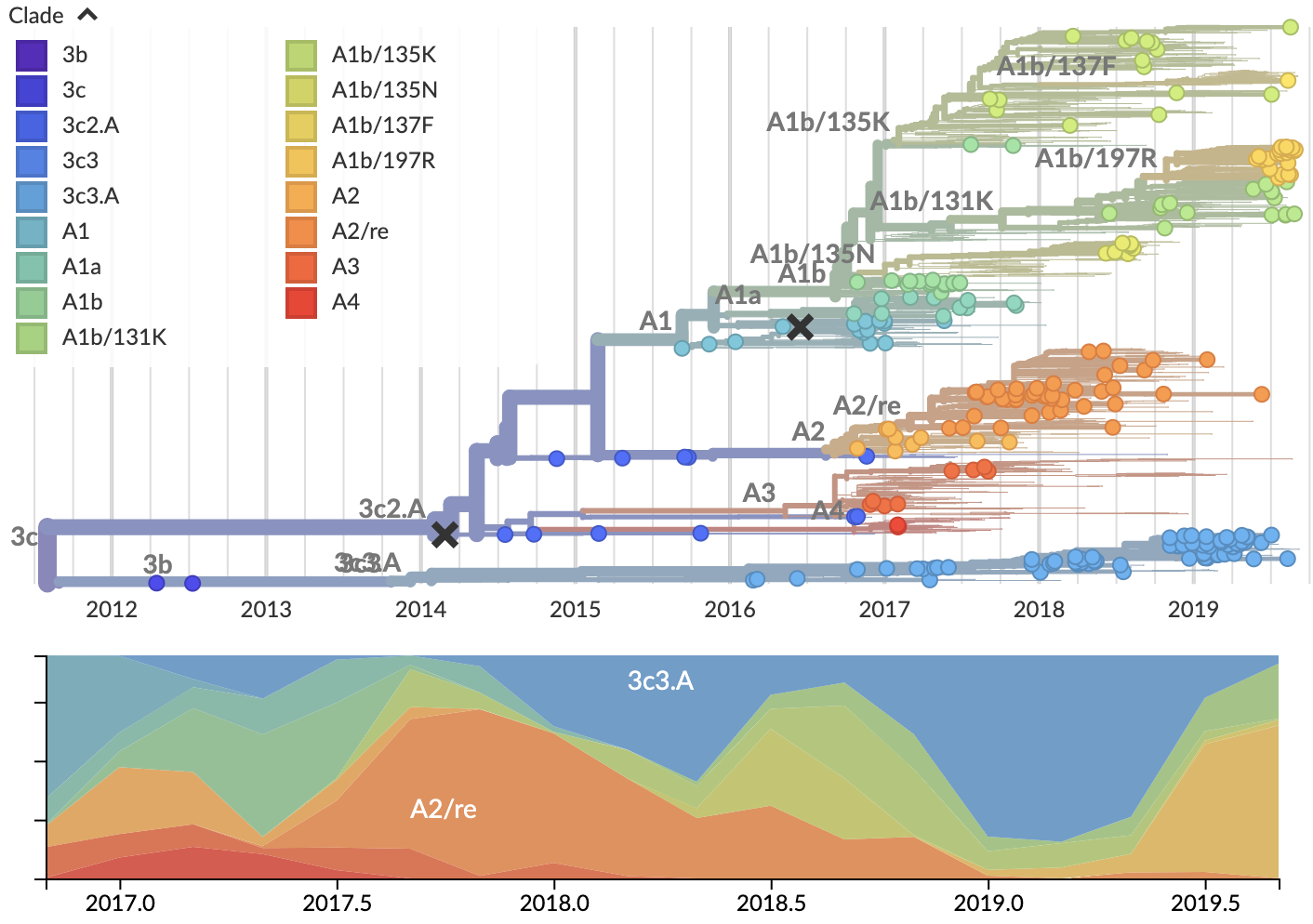
H3N2 viruses in the USA predominantly 3c3.A
Anomalous compared to the rest of the world
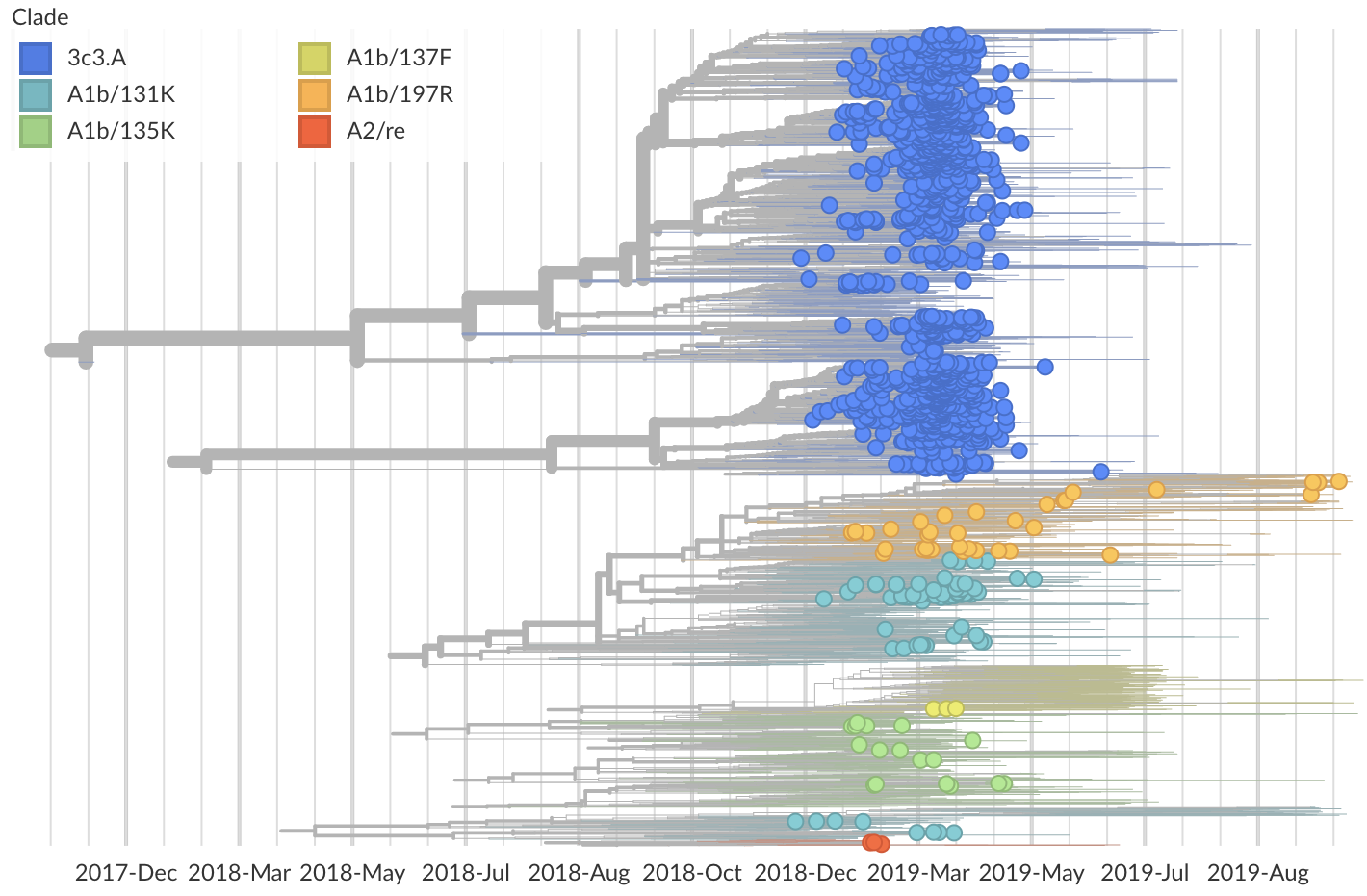
Seattle had a similarly focused 3c3.A season
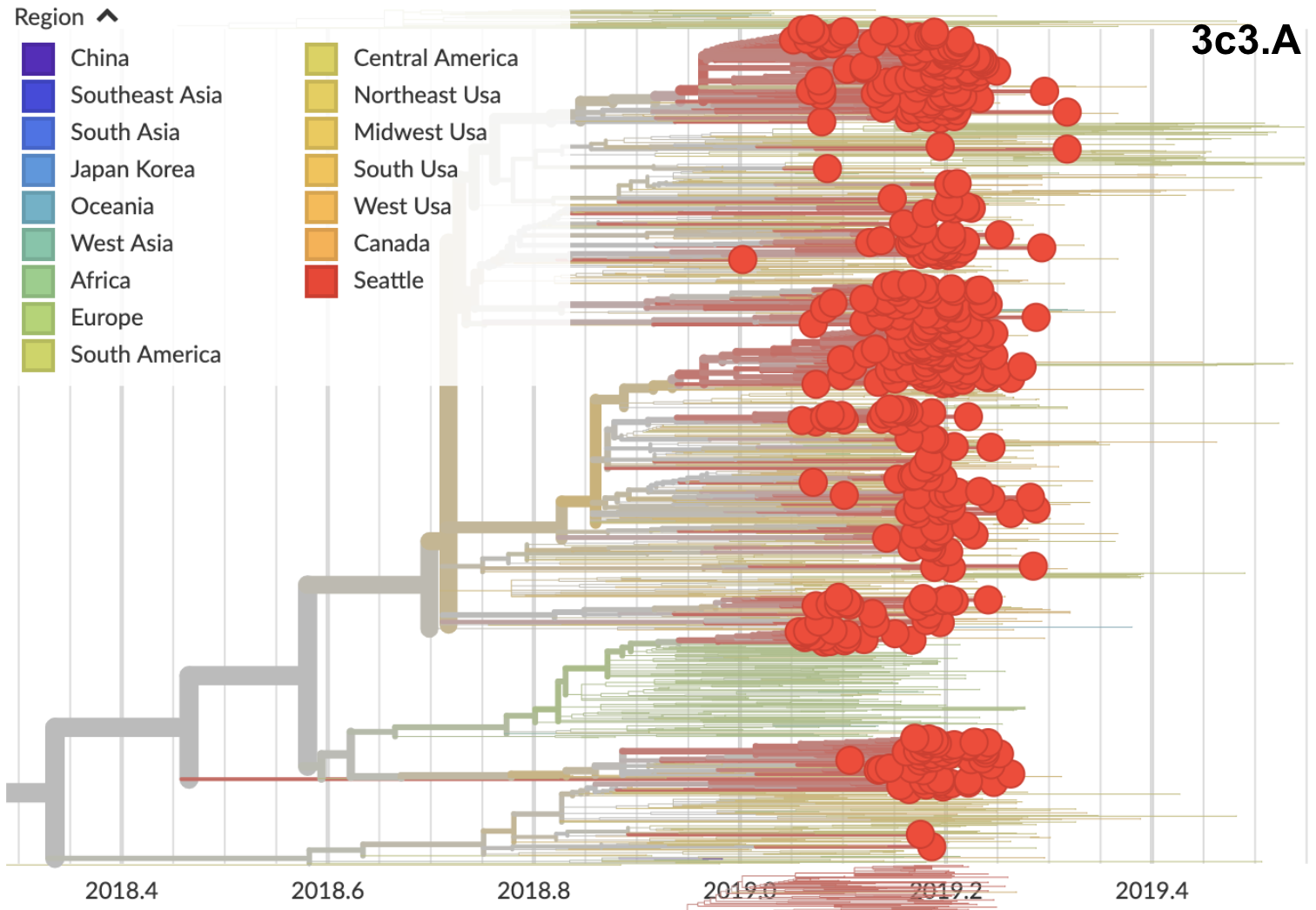
3c3.A viruses spread after arrival
A1b viruses stuttered out after arrival
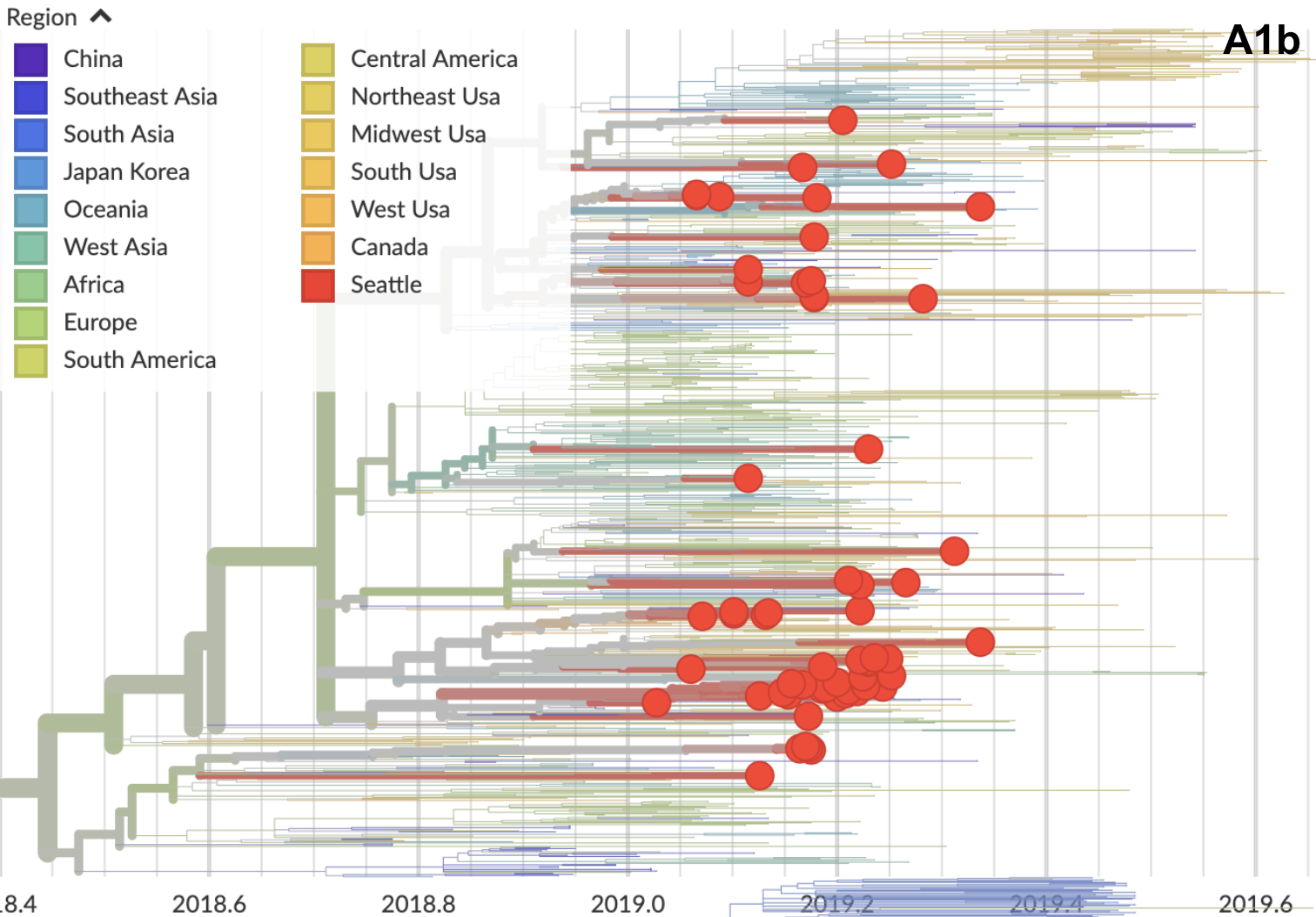
3c3.A introductions more successful
Suggests intrinsic advantage to 3c3.A, rather than stochastic early start
Fits a hypothesis of vaccine-induced immunity
USA saw repeated resurgance of vaccine mismatched 3c3.A viruses
Local transmission
Genomic mutation rate
Genomic mutation rate of 2.70 × 10-3 subs per site per year, which translates to 36.7 subs per genome per year, or 1 mutation per 10 days
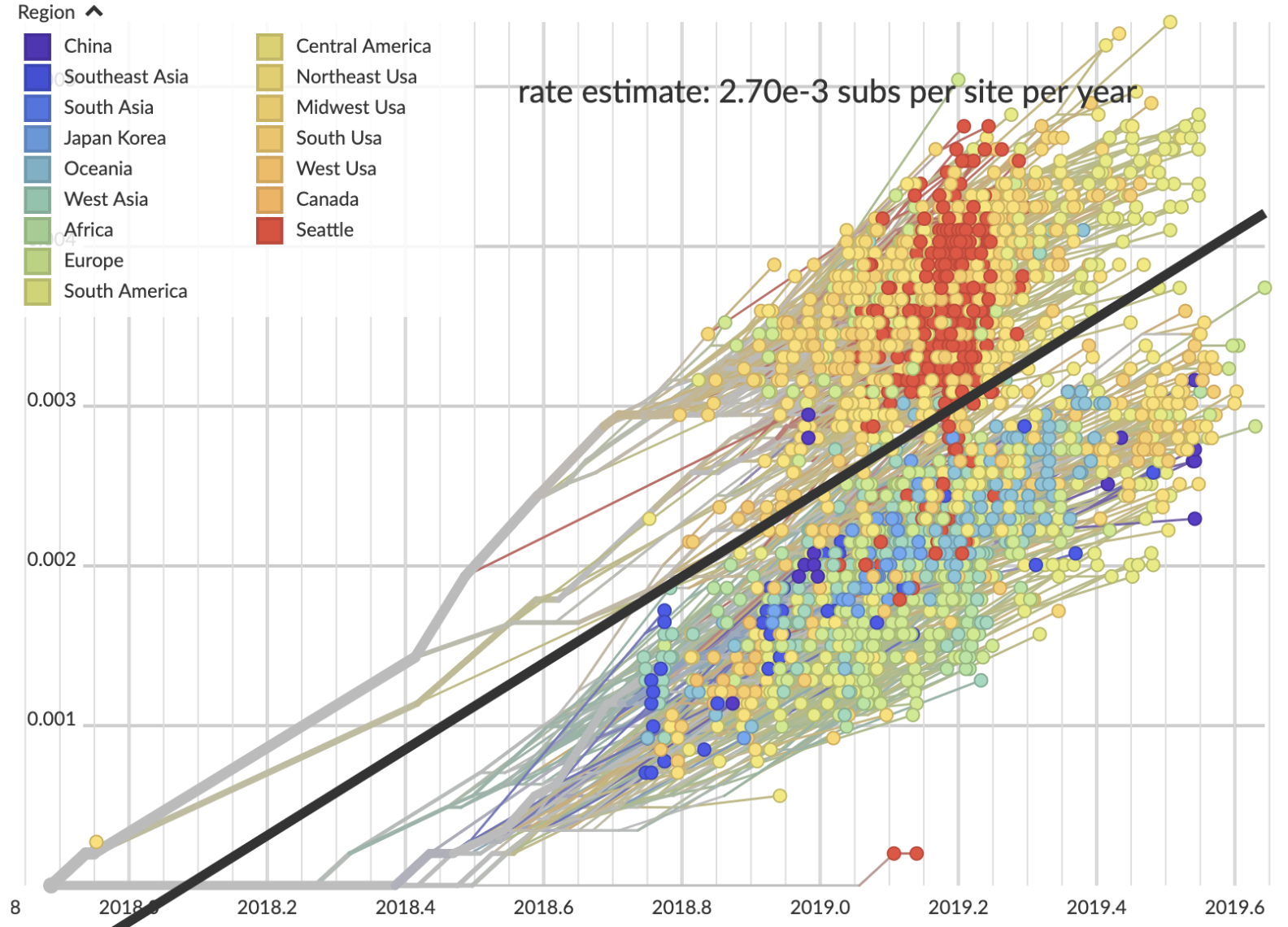
Closest genomic neighbors often <3 mutations distant
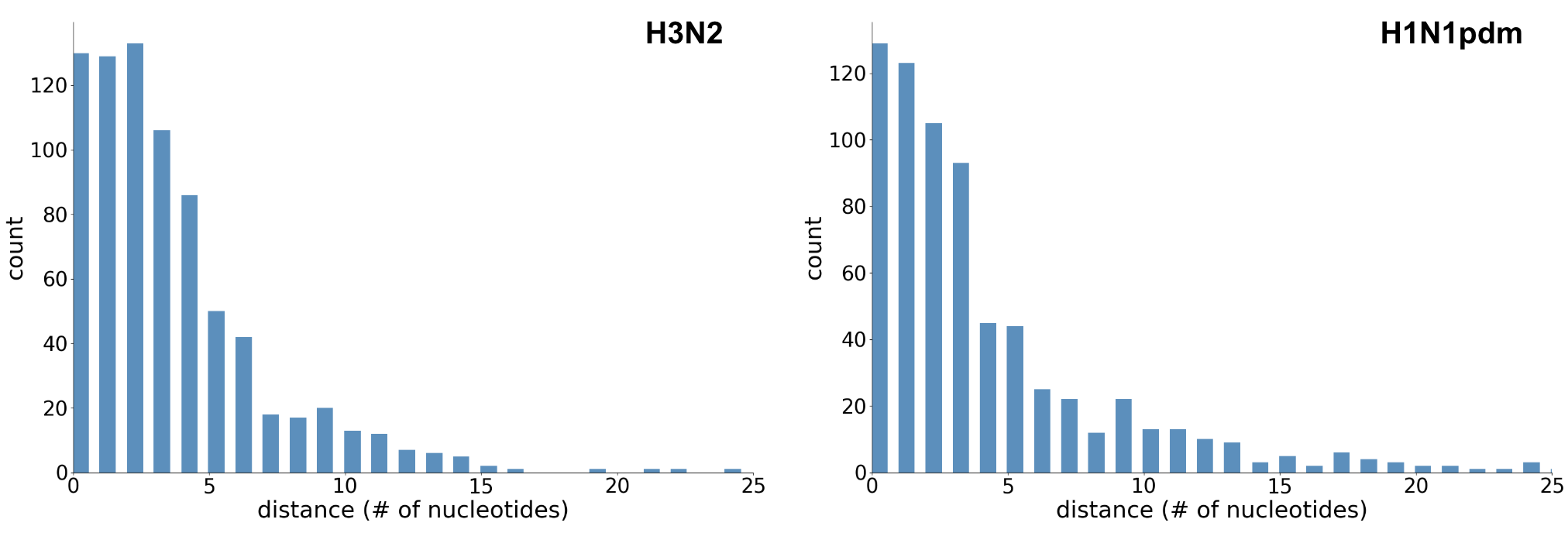
Allows tracking of individual transmission chains
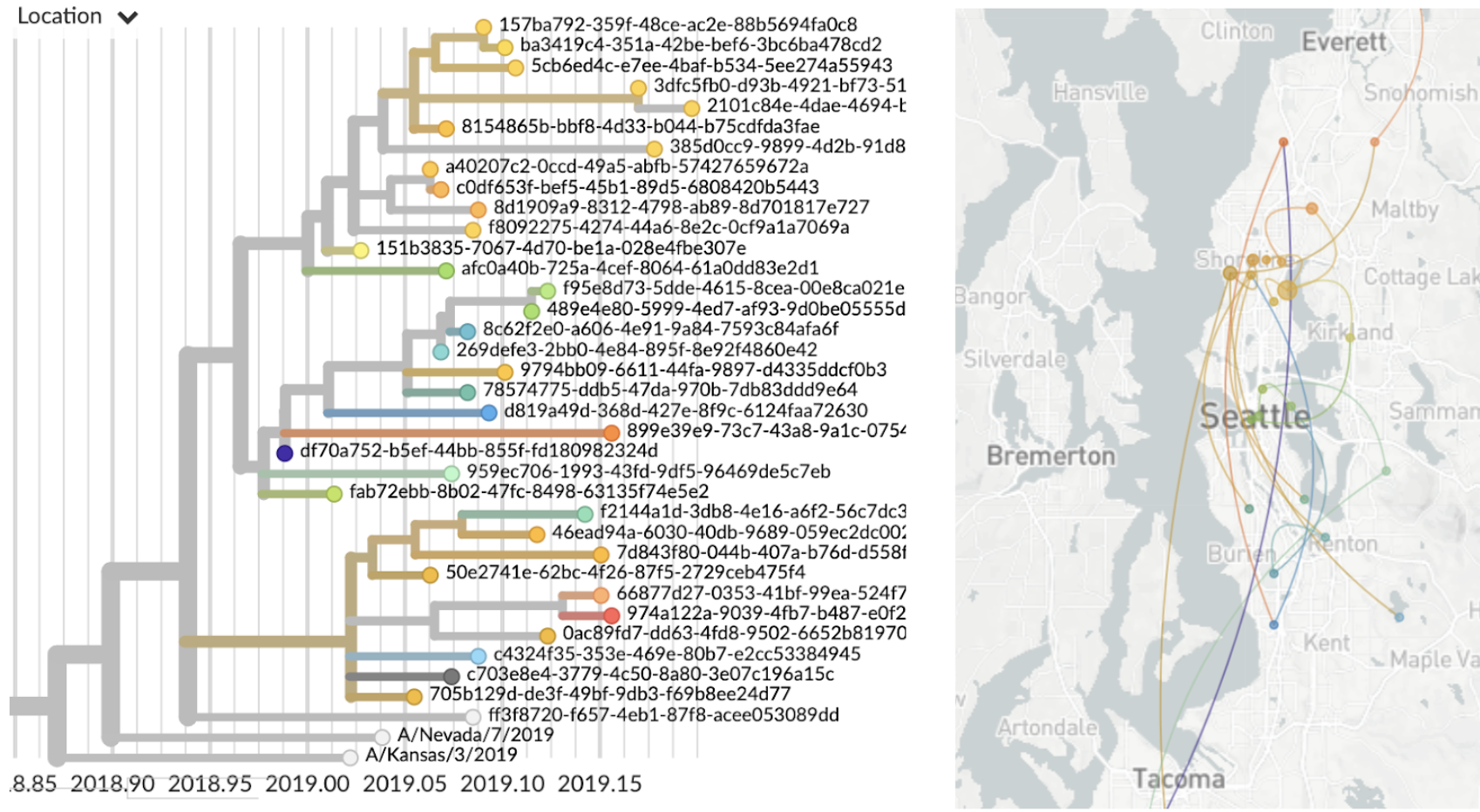
Allows tracking of individual transmission chains

Genetically similar viruses cluster geographically
Signal decays at ~1 mutation or ~10 days
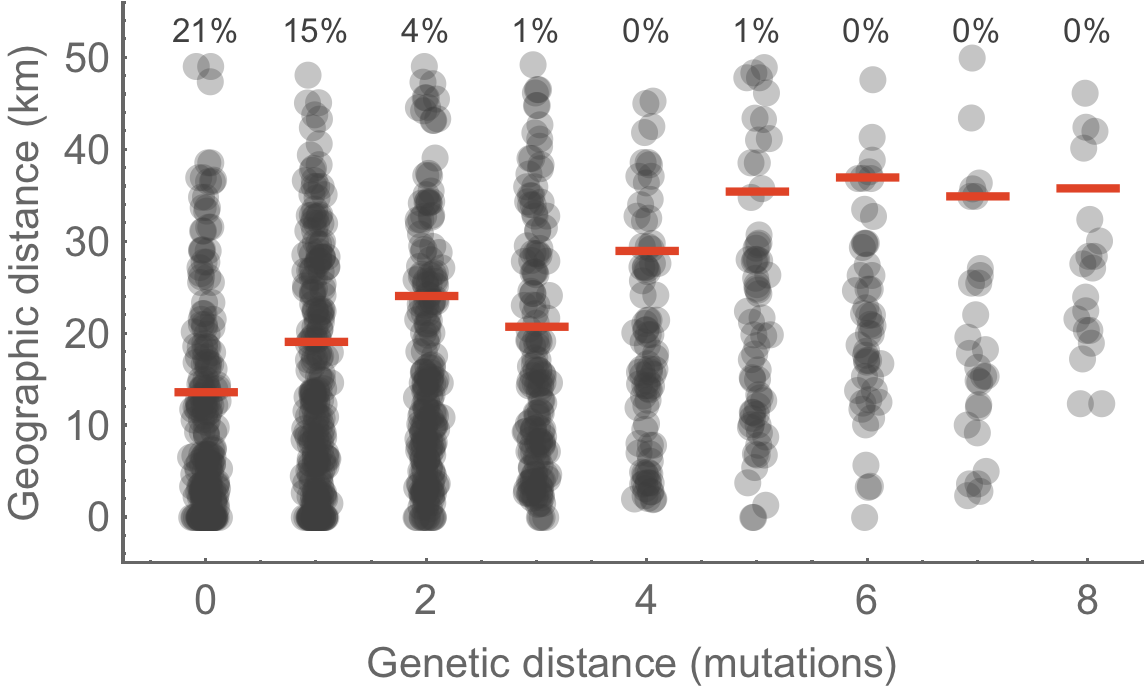
Some future directions
- Strain-specific local transmission potential to estimate viral fitness
- Demographic and viral characteristics associated with transmission
- Geographic and age mixing matrices
Acknowledgements
Bedford Lab:
![]() Alli Black,
Alli Black,
![]() John Huddleston,
John Huddleston,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Louise Moncla,
Louise Moncla,
![]() Maya Lewinsohn,
Maya Lewinsohn,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Kairsten Fay,
Kairsten Fay,
![]() Misja Ilcisin,
Misja Ilcisin,
![]() Nicola Müller
Nicola Müller
Seattle Flu Study: Helen Chu, Michael Boeckh, Janet Englund, Michael Famulare, Barry Lutz, Deborah Nickerson, Mark Rieder, Lea Starita, Matthew Thompson, Jay Shendure, Amanda Adler, Jeris Bosua, Elisabeth Brandstetter, Kairsten Fay, Chris Frazar, Peter Han, Reena Gulati, James Hadfield, ShiChu Huang, Misja Ilcisin, Michael Jackson, Anahita Kiavand, Louise Kimball, Enos Kline, Kirsten Lacombe, Jover Lee, Jennifer Logue, Victoria Lyon, Kira Newman, Miguel Paredes, Thomas Sibley, Monica Zigman Suchsland, Cassia Wagner, Caitlin Wolf





