SARS-CoV-2 variant emergence and spread
Trevor Bedford (@trvrb)
Fred Hutchinson Cancer Center / Howard Hughes Medical Institute
29 Aug 2022
SAVE Early Detection Meeting
NIH NIAID
1. SARS-CoV-2 evolution
2. SARS-CoV-2 variant frequency dynamics
3. BA.2.75 as case study
4. Emerging variants
5. Genomic surveillance
SARS-CoV-2 evolution
Genetic relationships of globally sampled SARS-CoV-2 to present
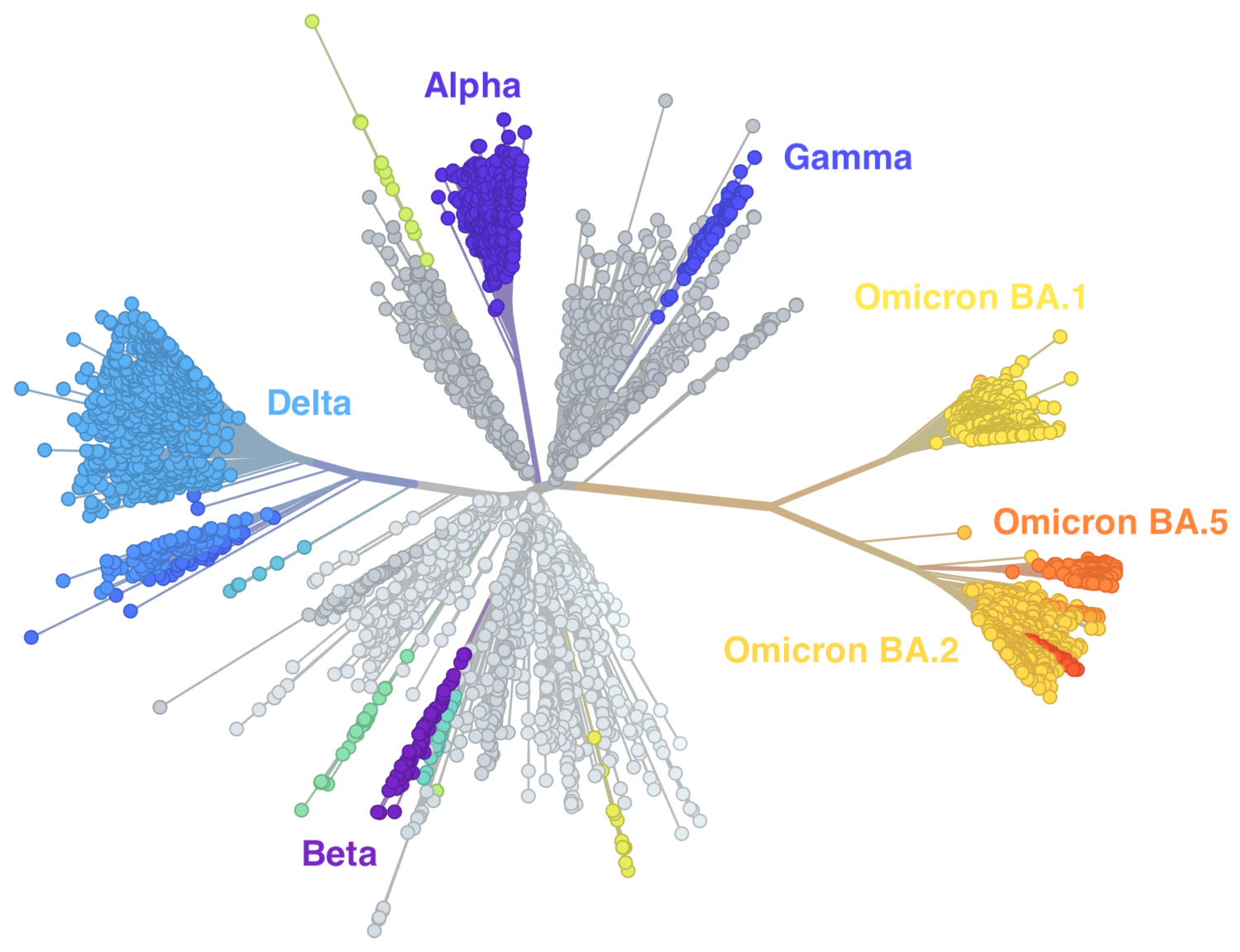
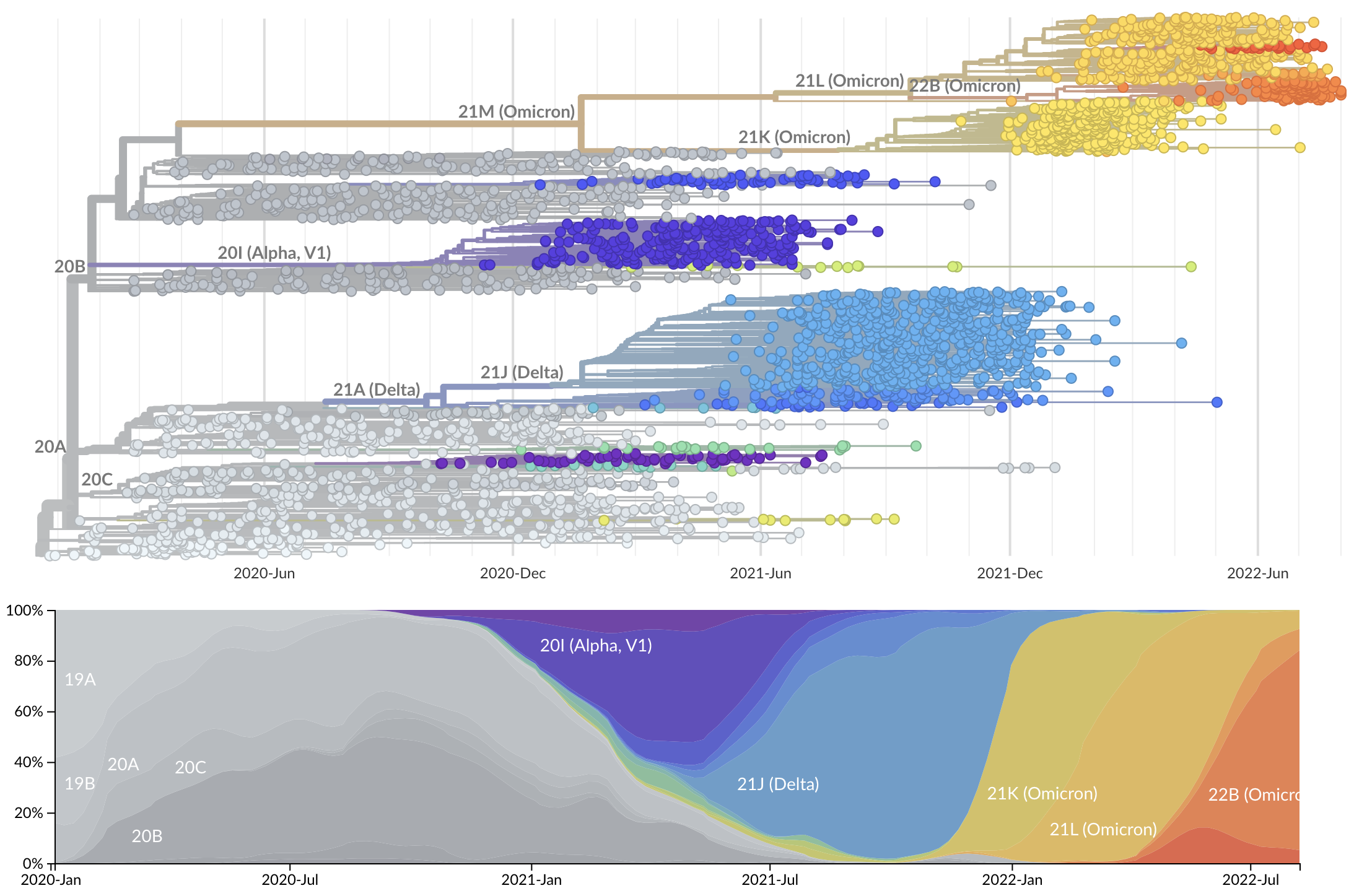
Rapid displacement of existing diversity by emerging variants
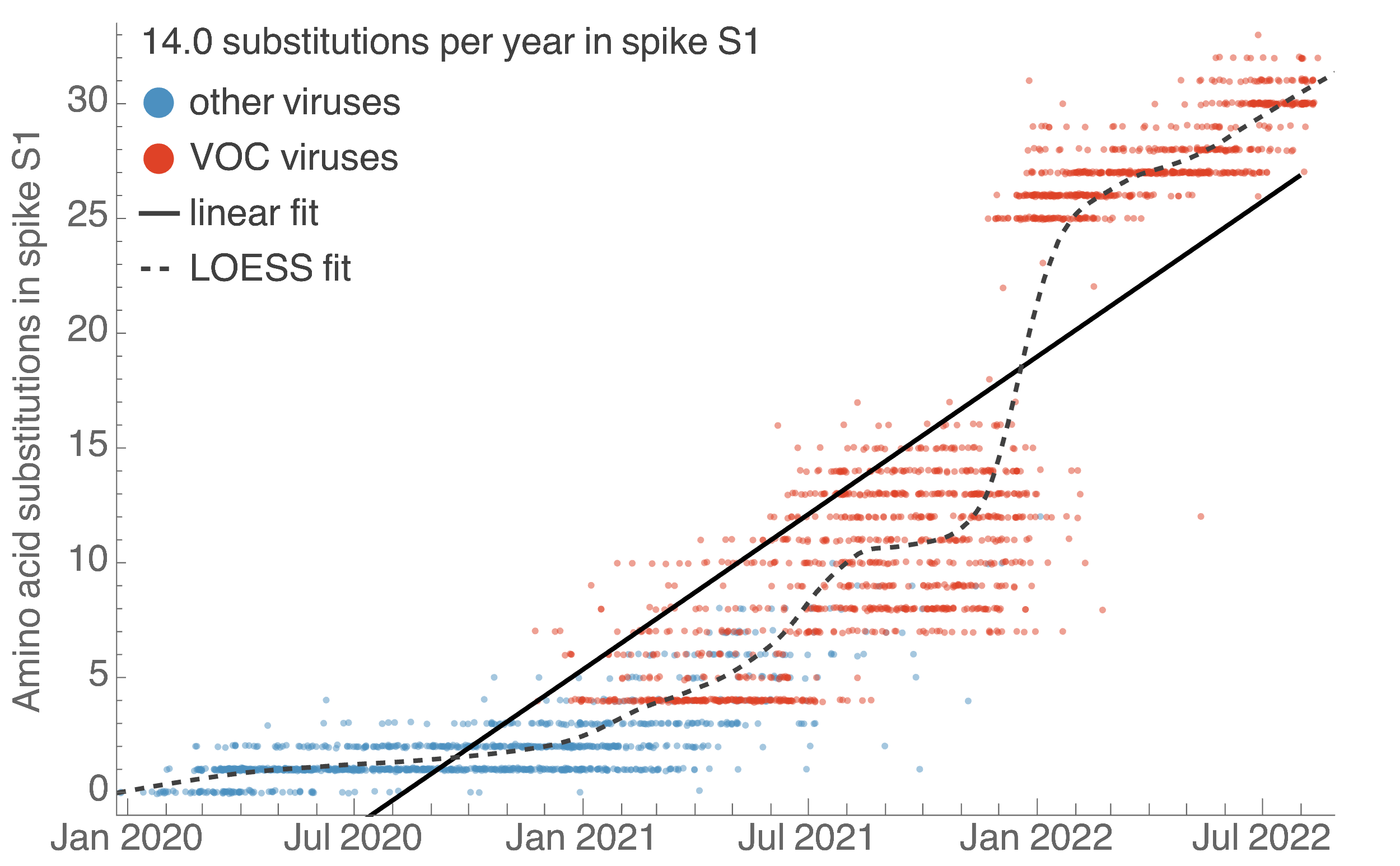
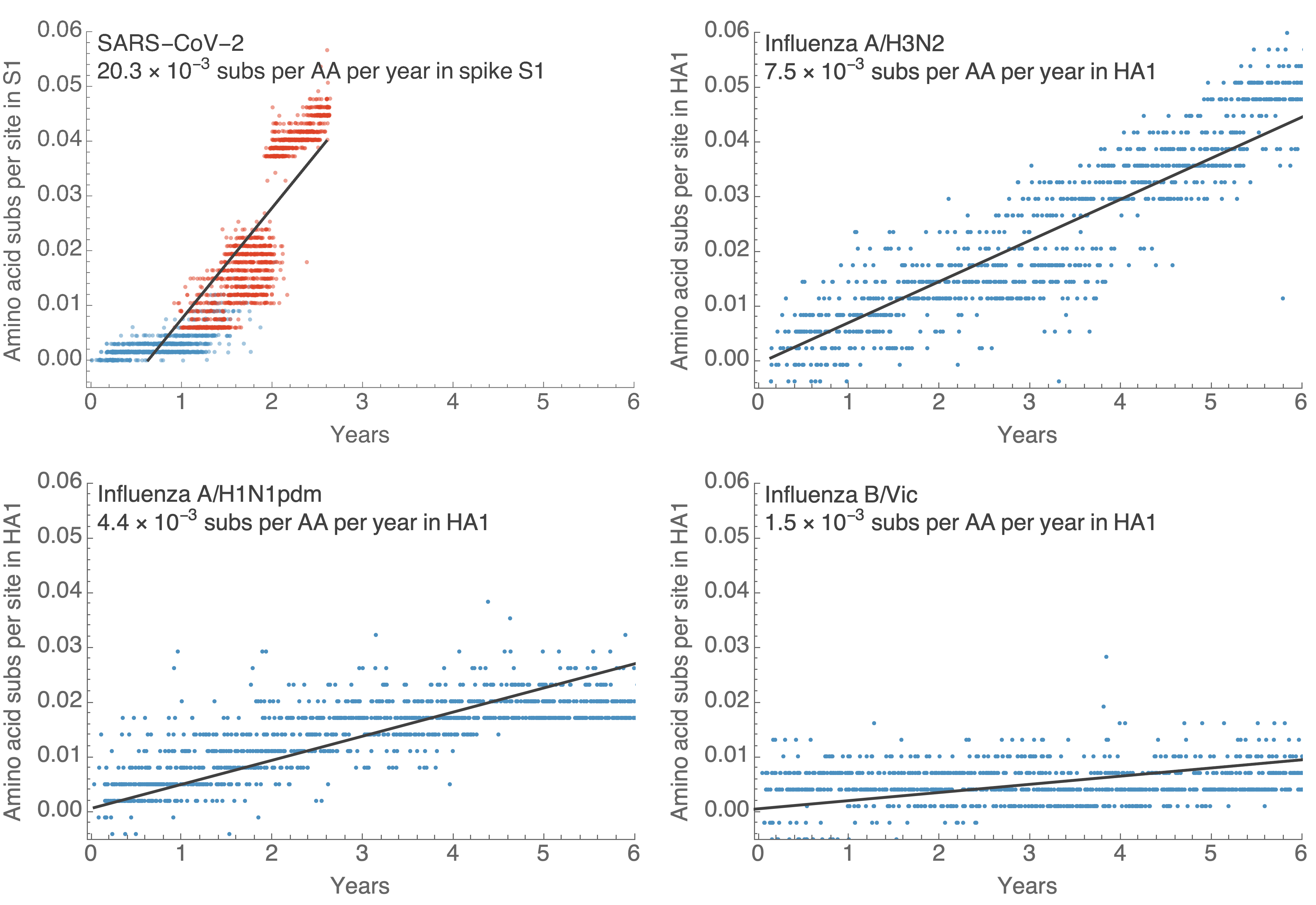
S1 evolved at a rate of 14 amino acid changes per year since pandemic start
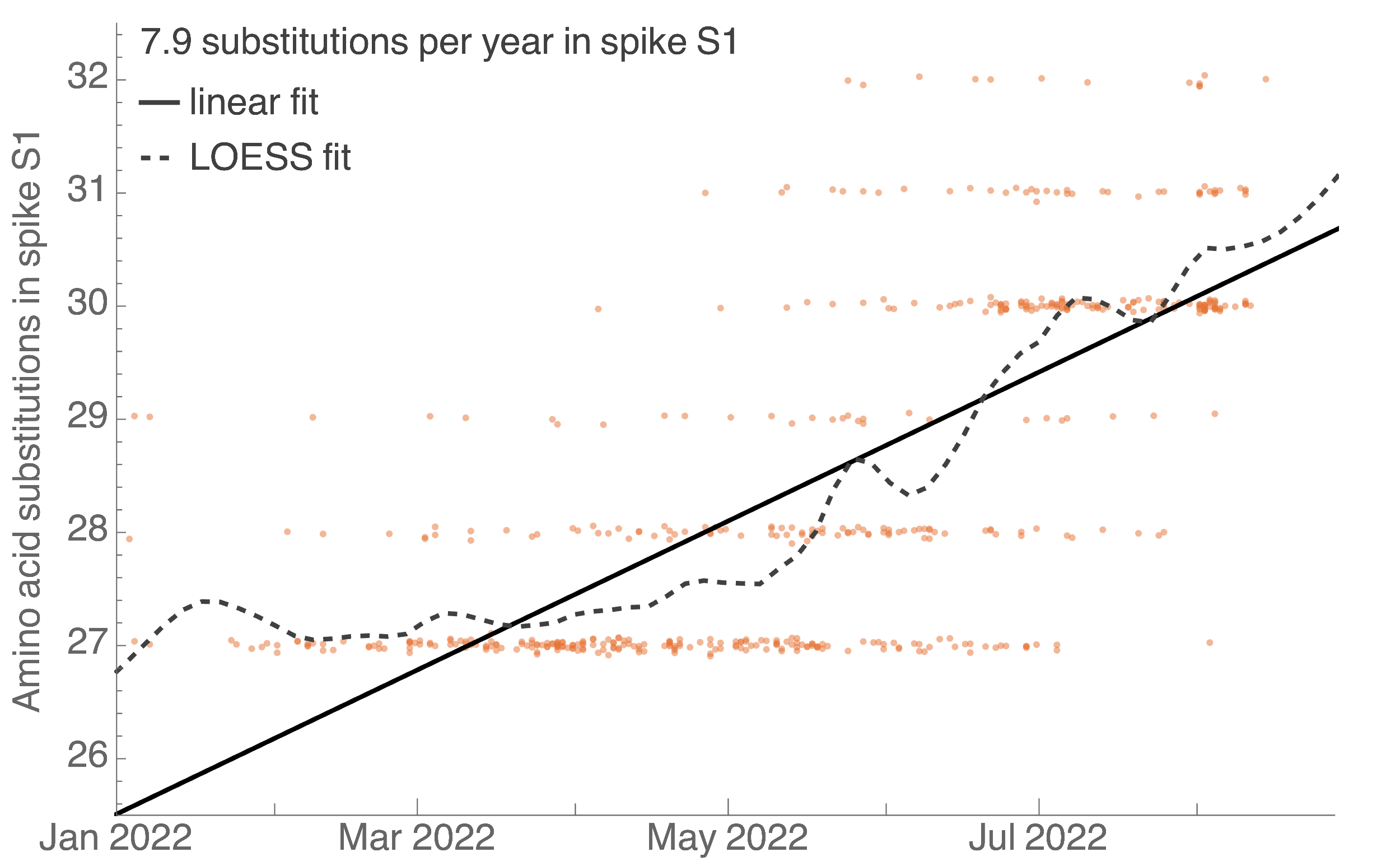
Continued rapid accumulation since BA.2 with 8 amino acid changes per year
S1 evolution remarkably fast relative to seasonal influenza
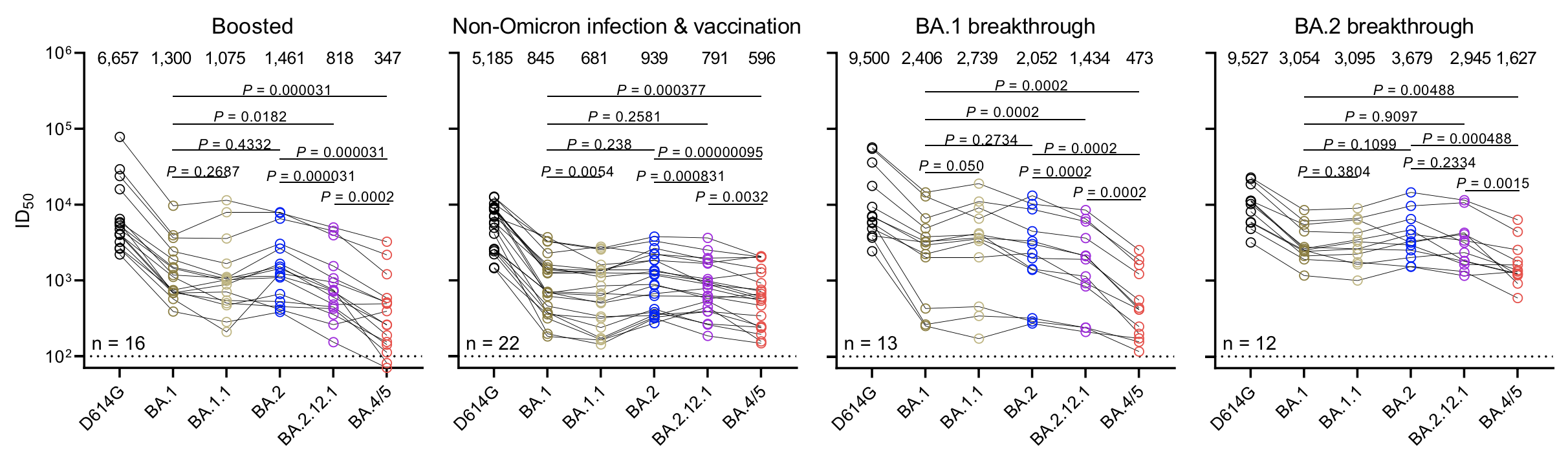
Continued escape from neutralization by existing population immunity
SARS-CoV-2 variant frequency dynamics
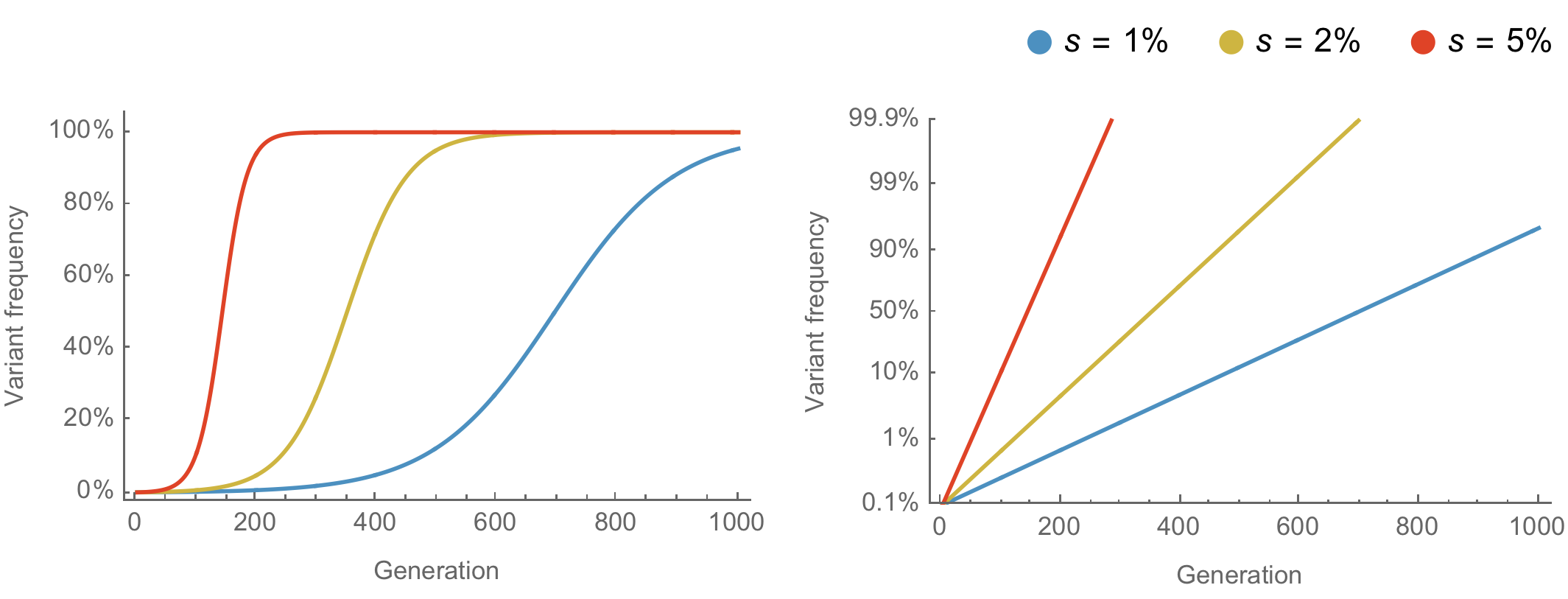
Population genetic expectation of variant frequency under selection
$x' = \frac{x \, (1+s)}{x \, (1+s) + (1-x)}$ for frequency $x$ over one generation with selective advantage $s$
$x(t) = \frac{x_0 \, (1+s)^t}{x_0 \, (1+s)^t + (1-x_0)}$ for initial frequency $x_0$ over $t$ generations
Trajectories are linear once logit transformed via $\mathrm{log}(\frac{x}{1 - x})$
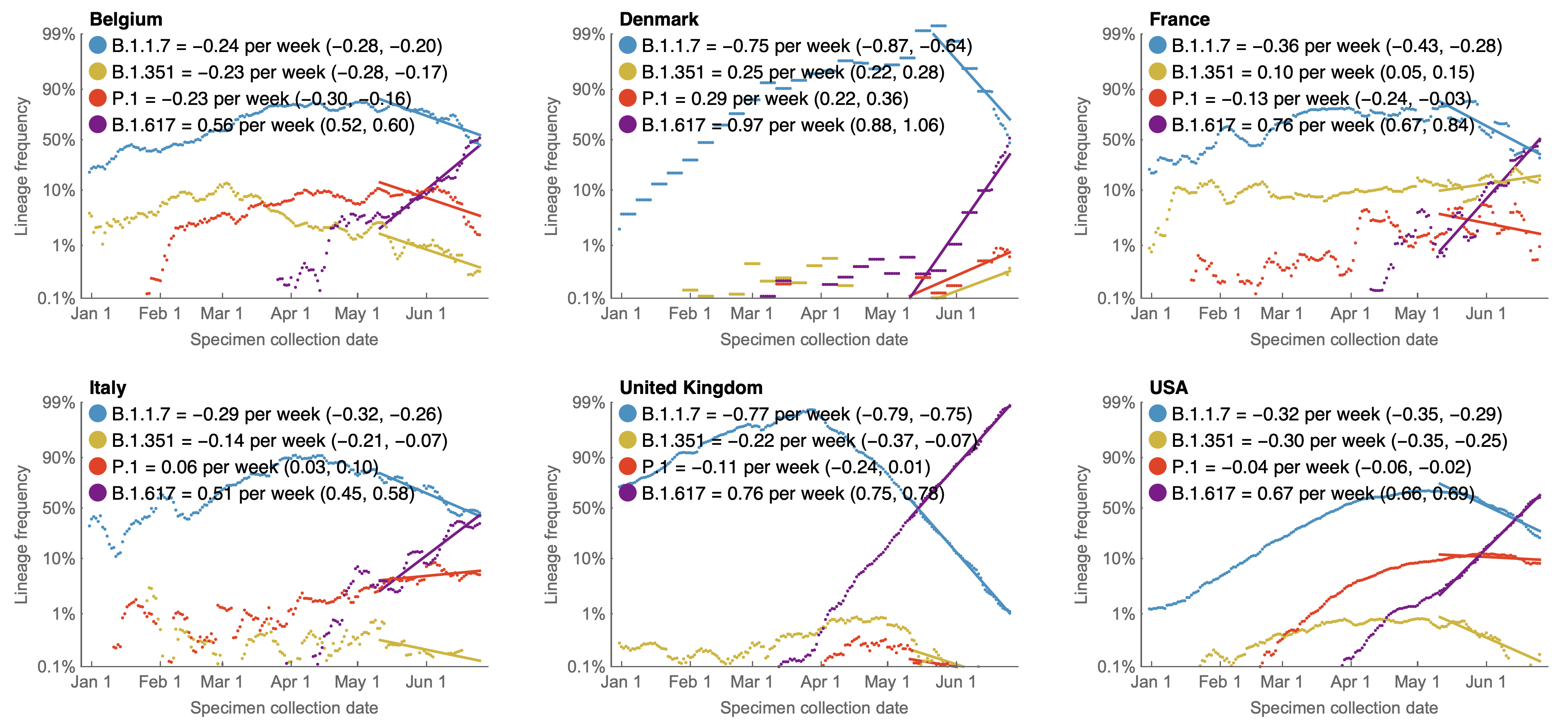
Variants show consistent frequency dynamics in logit space
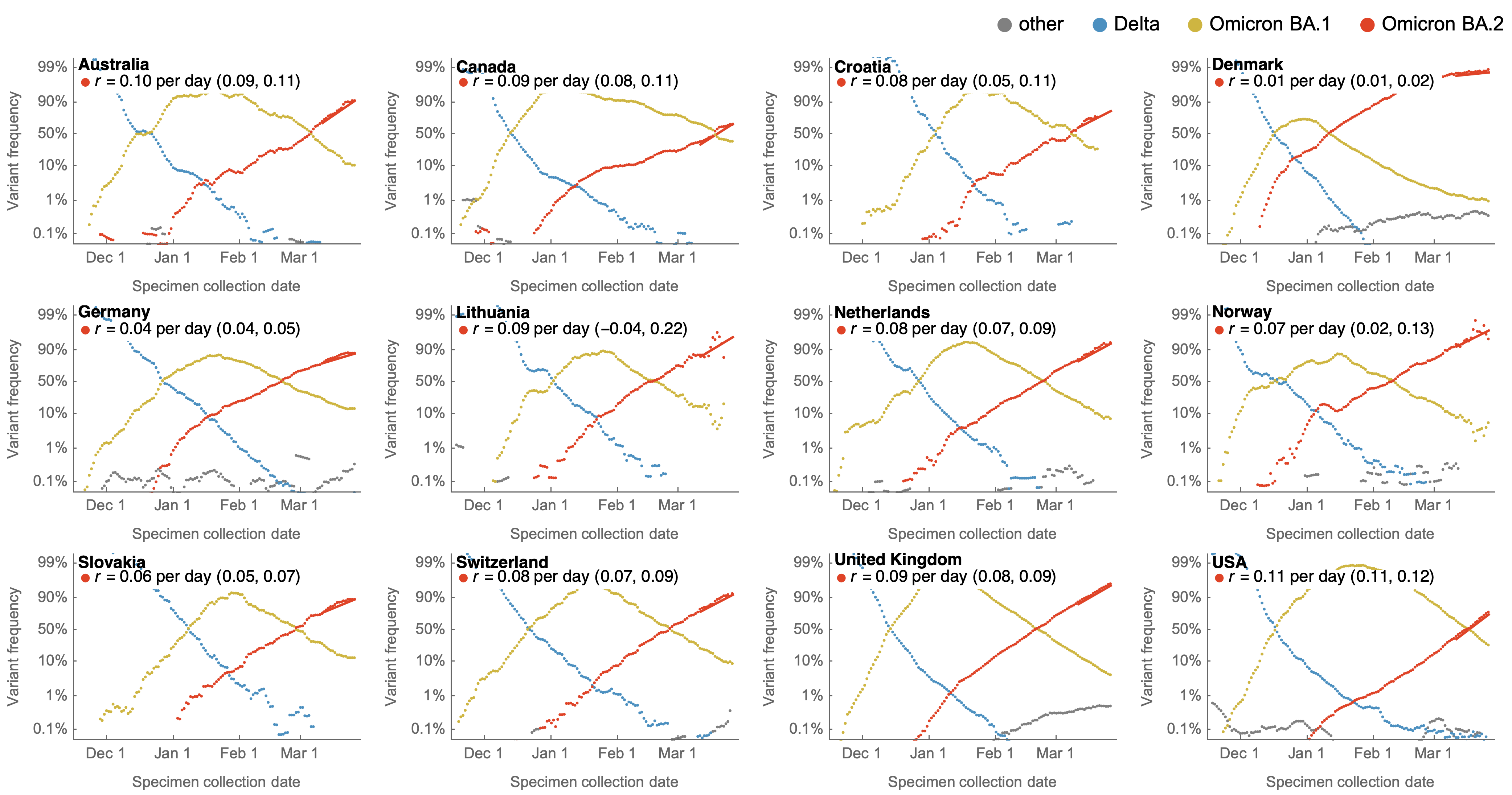
Variants show consistent frequency dynamics in logit space
Multinomial logistic regression
Multinomial logistic regression models the probability of a virus sampled at time $t$ belonging to variant $i$ as
$$\mathrm{Pr}(X = i) = x_i(t) = \frac{p_i \, \mathrm{exp}(f_i \, t)}{\sum_{1 \le j \le n} p_j \, \mathrm{exp}(f_j \, t) }$$
where the model has $2n$ parameters consisting of $p_i$ the frequency of variant $i$ at initial timepoint and $f_i$ the growth rate or fitness of variant $i$ for $n$ variants.
The model is fit to minimize "log loss" of predicted variant vs observed variant across observations in dataset.
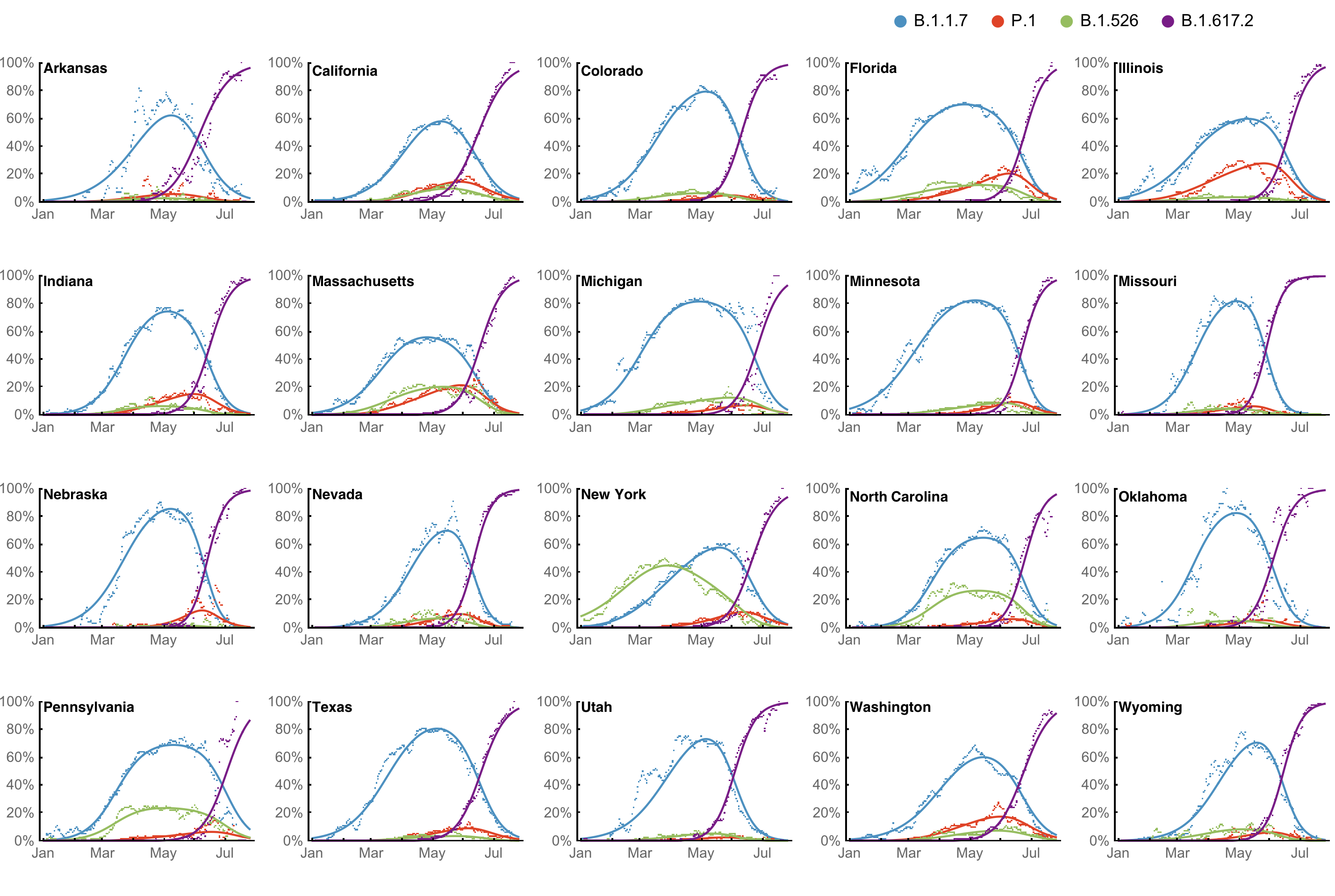
Multinomial logistic regression fits variant frequencies well
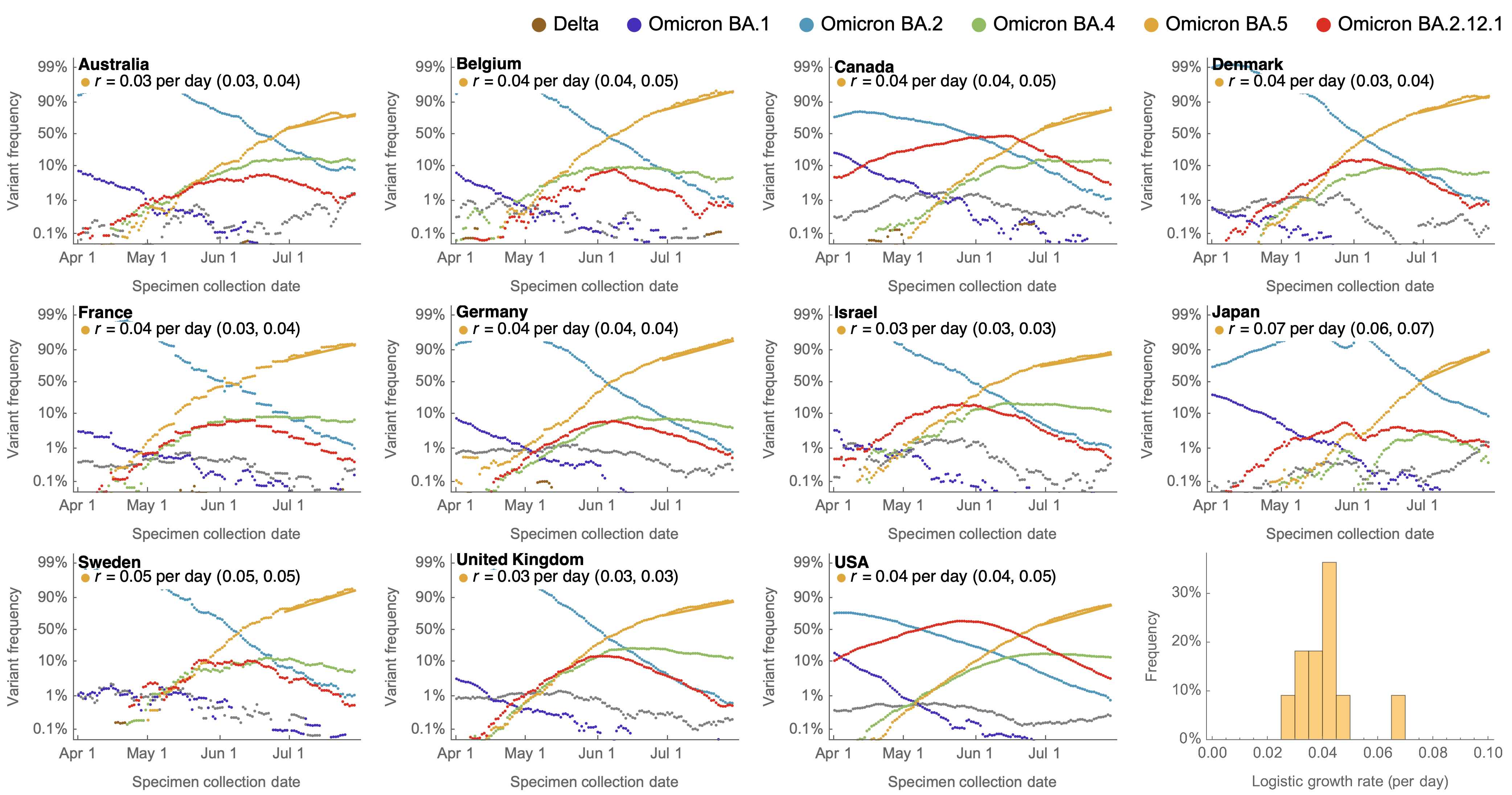
Consistent fitness advantage of BA.5 across countries
Despite similar rates of displacement, BA.5 epidemics vary
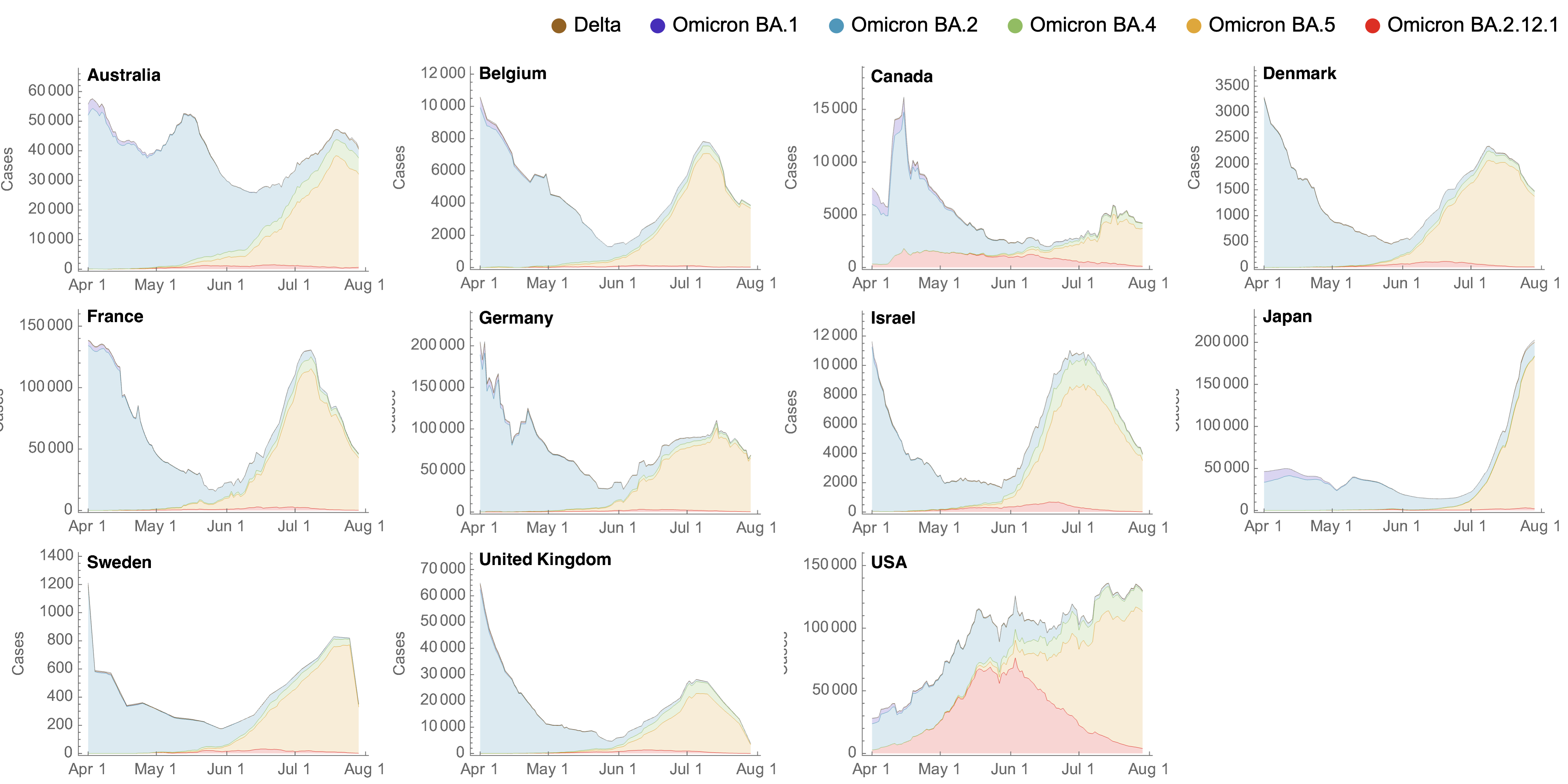
Despite similar rates of displacement, BA.5 epidemics vary
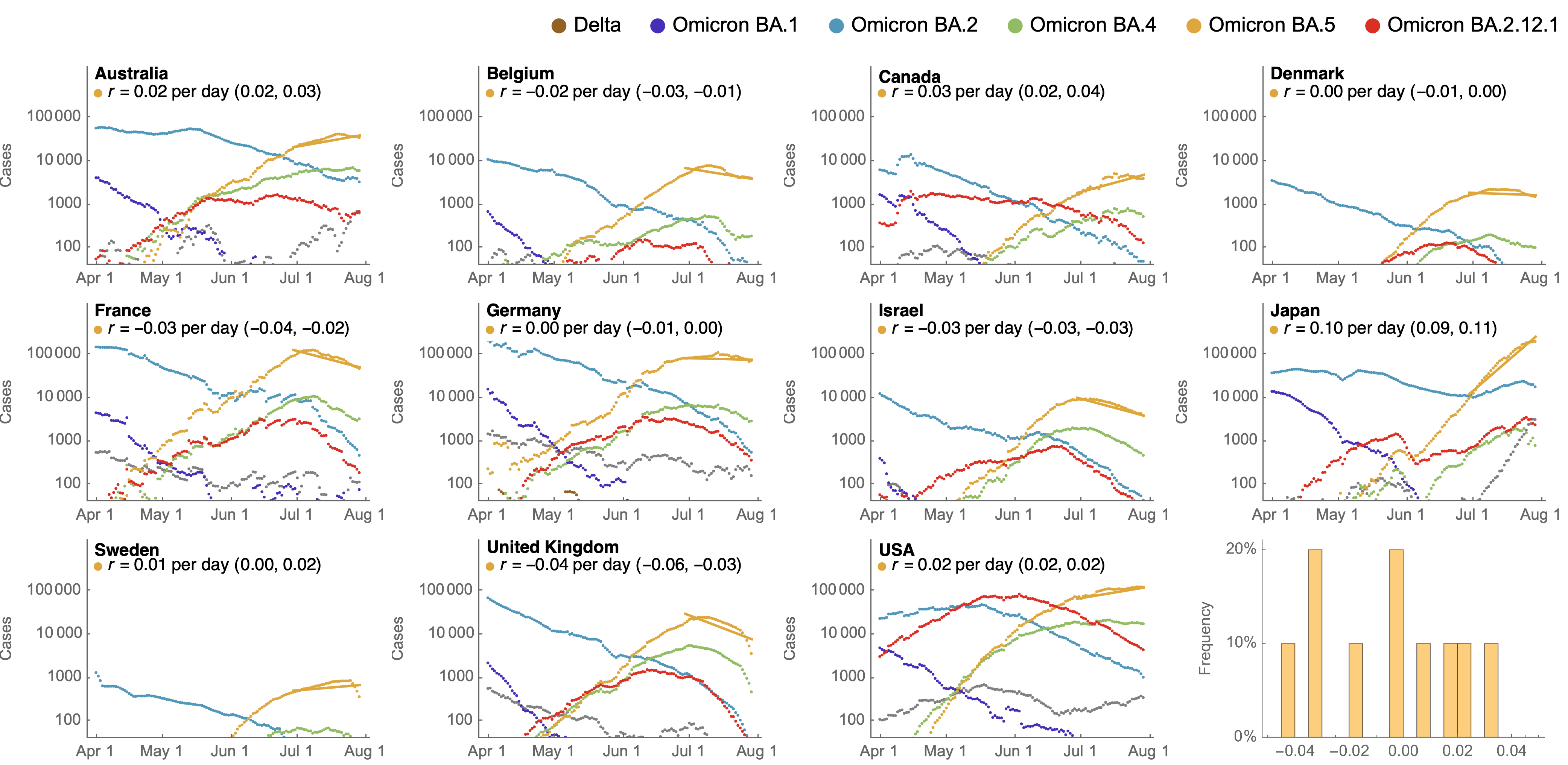
These differences can be explained by consistent growth advantage of BA.5, but different baseline Rt across countries
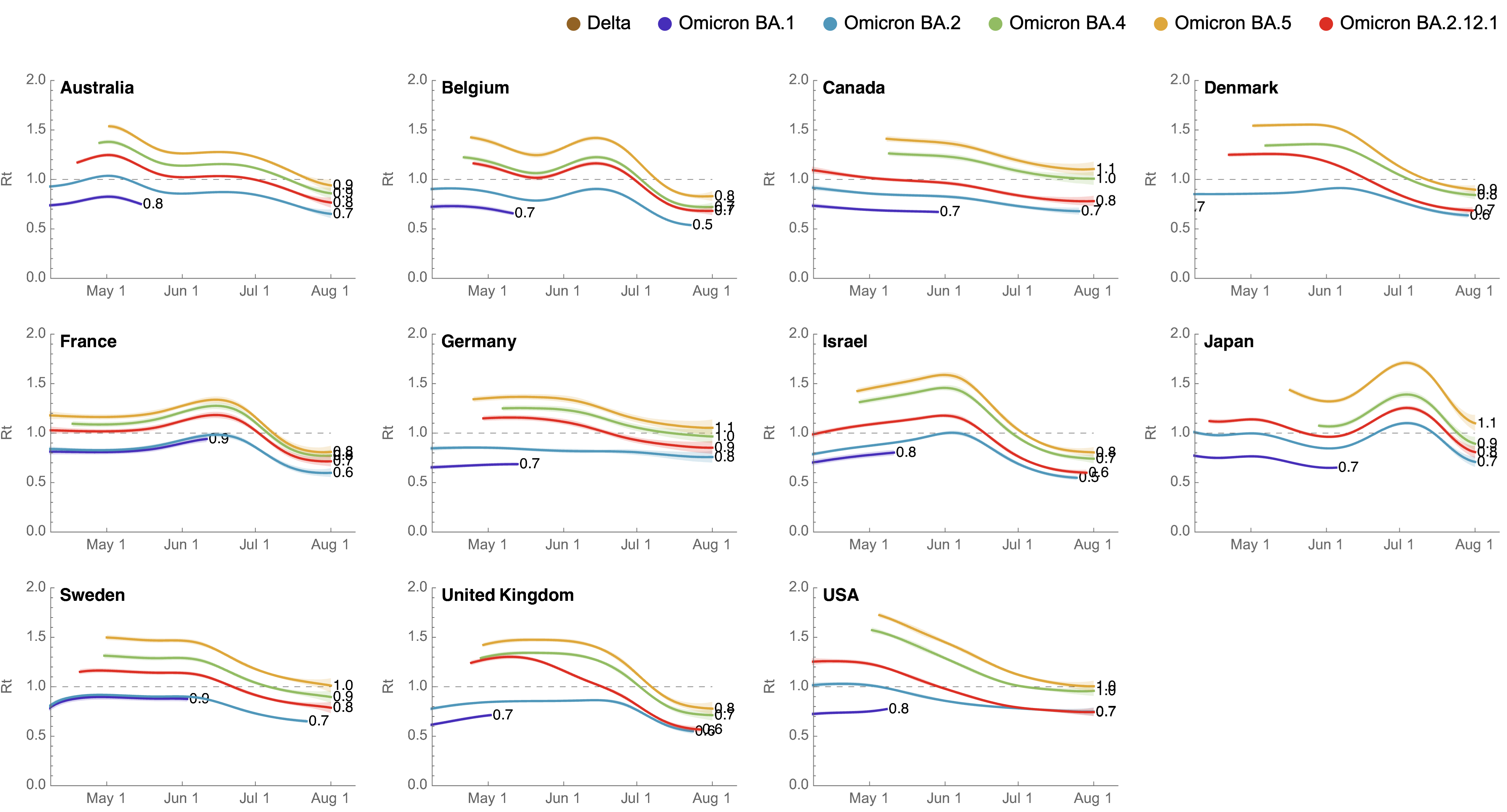
Multinomial logistic regression should work well for SARS-CoV-2 prediction, except new variants have been emerging fast enough that the prediction horizon is really quite short
BA.2.75 as case study
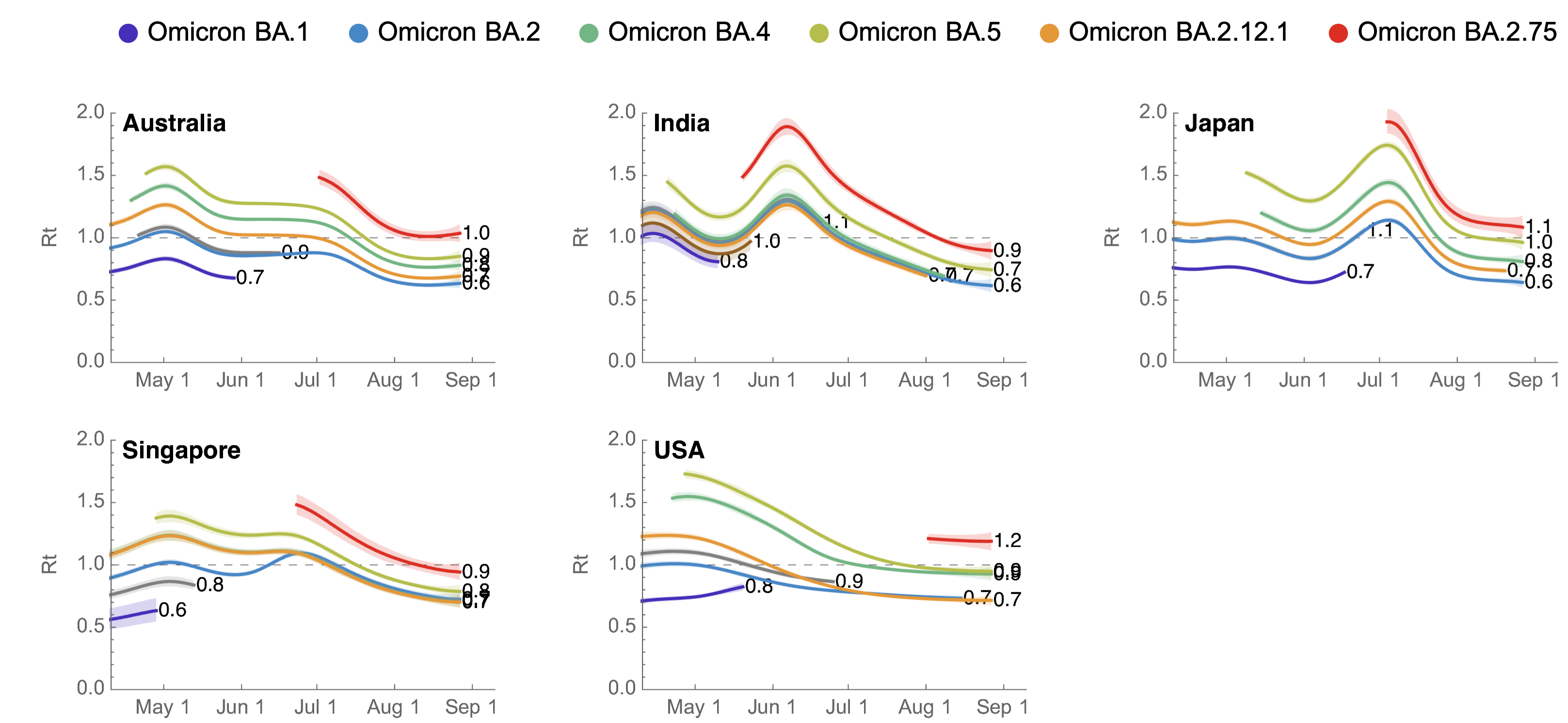
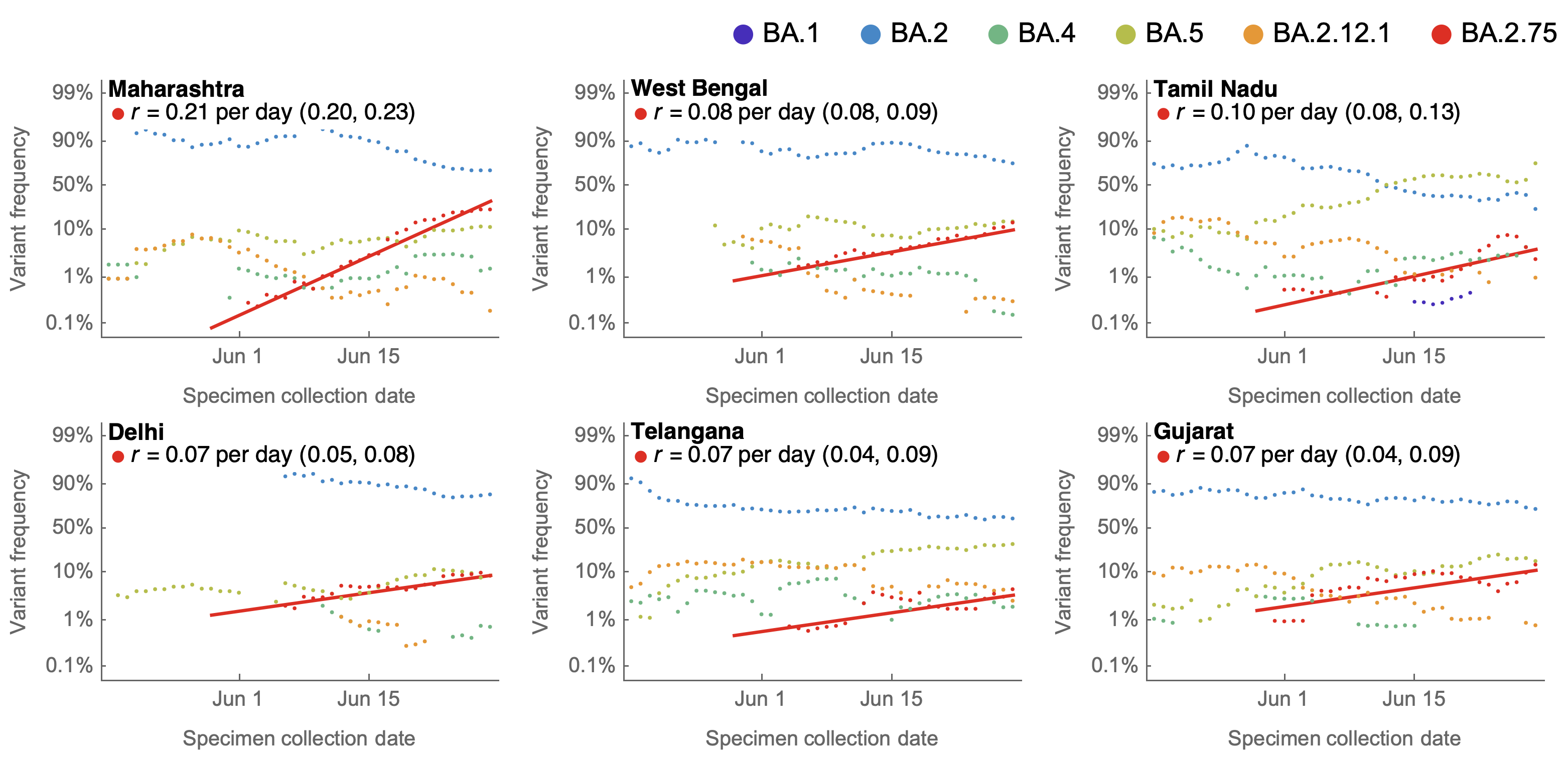
BA.2.75 shows consistent selective advantage across countries
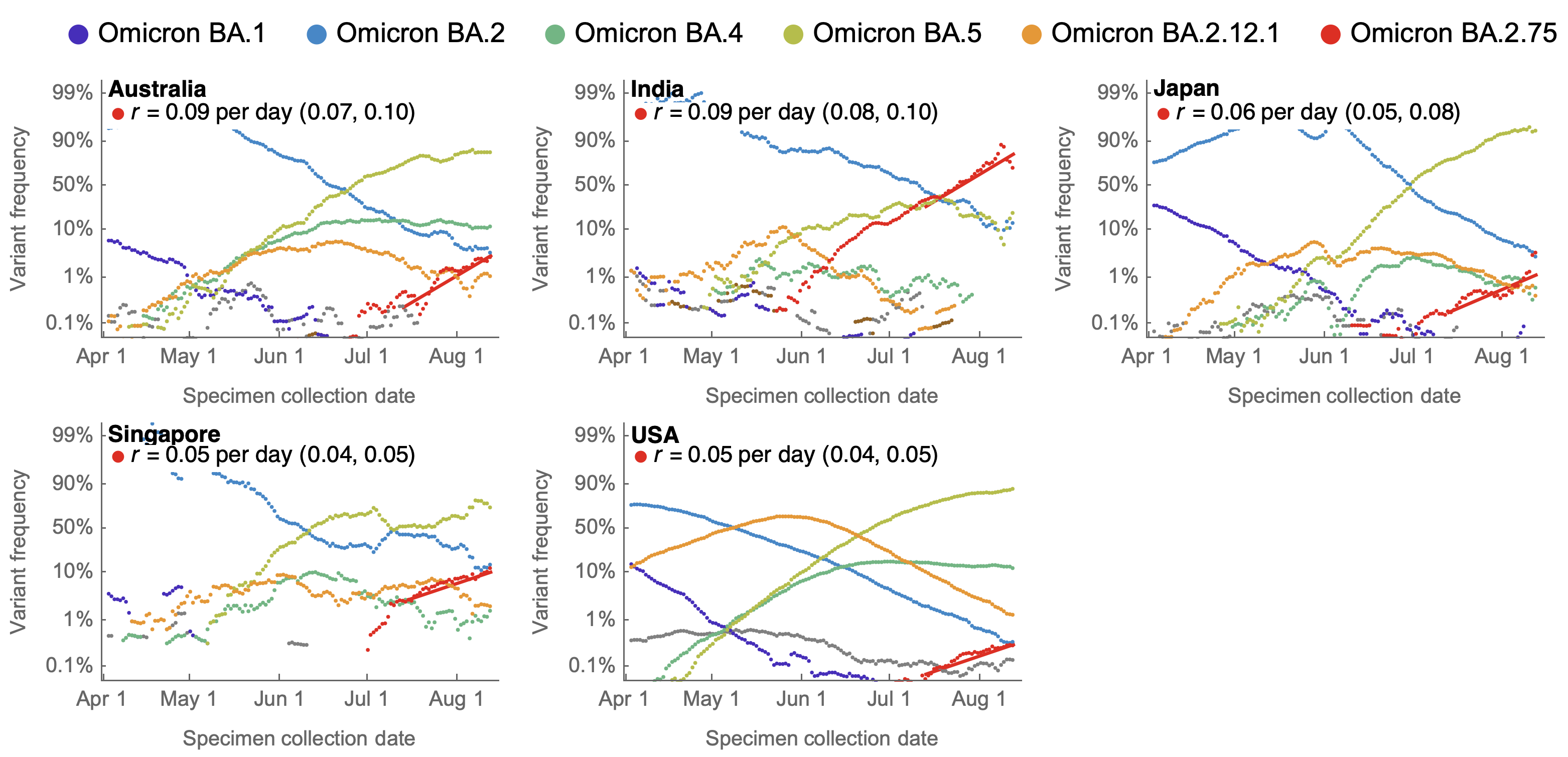
BA.2.75 did not drive large increase in total case loads in India
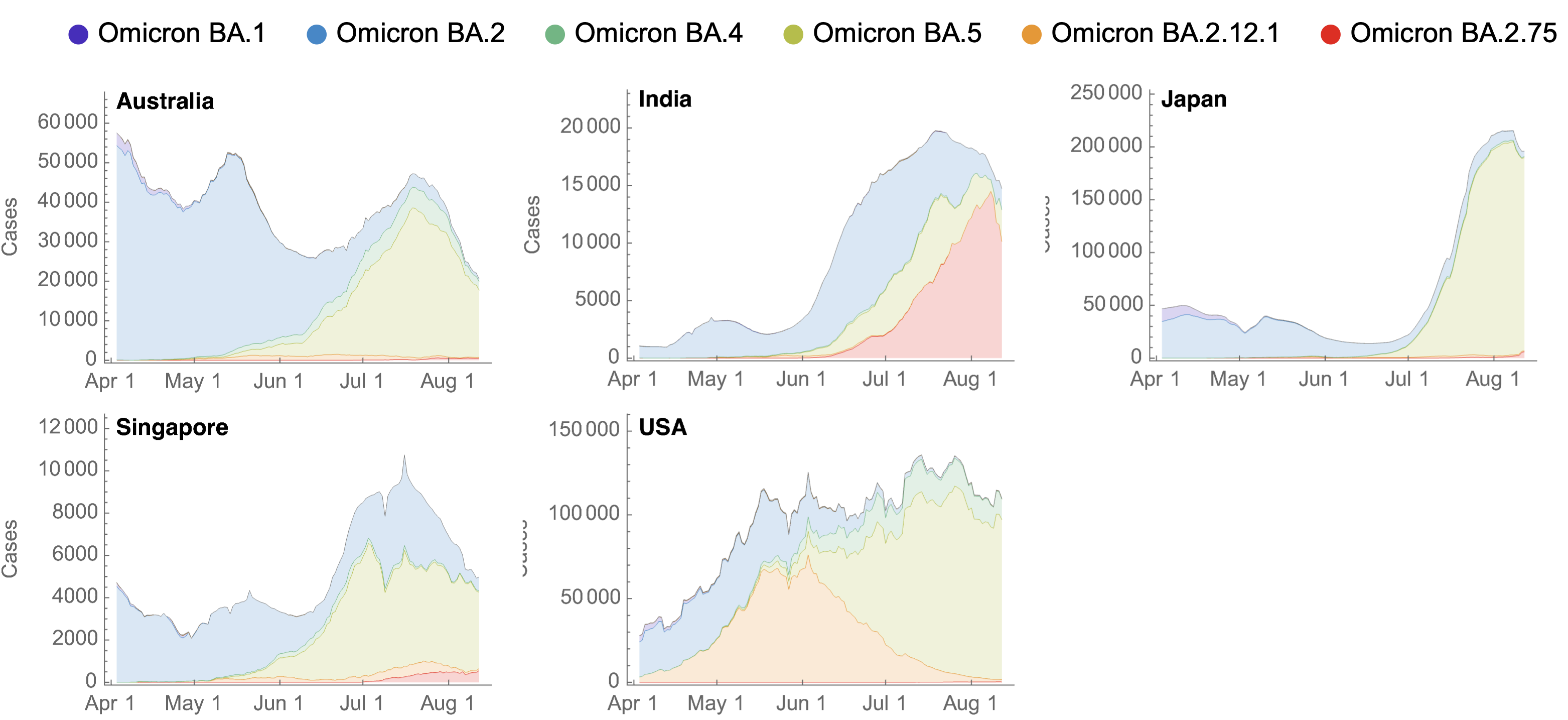
BA.2.75 current Rt greater than 1 in the US, but lower than initial Rt of BA.5
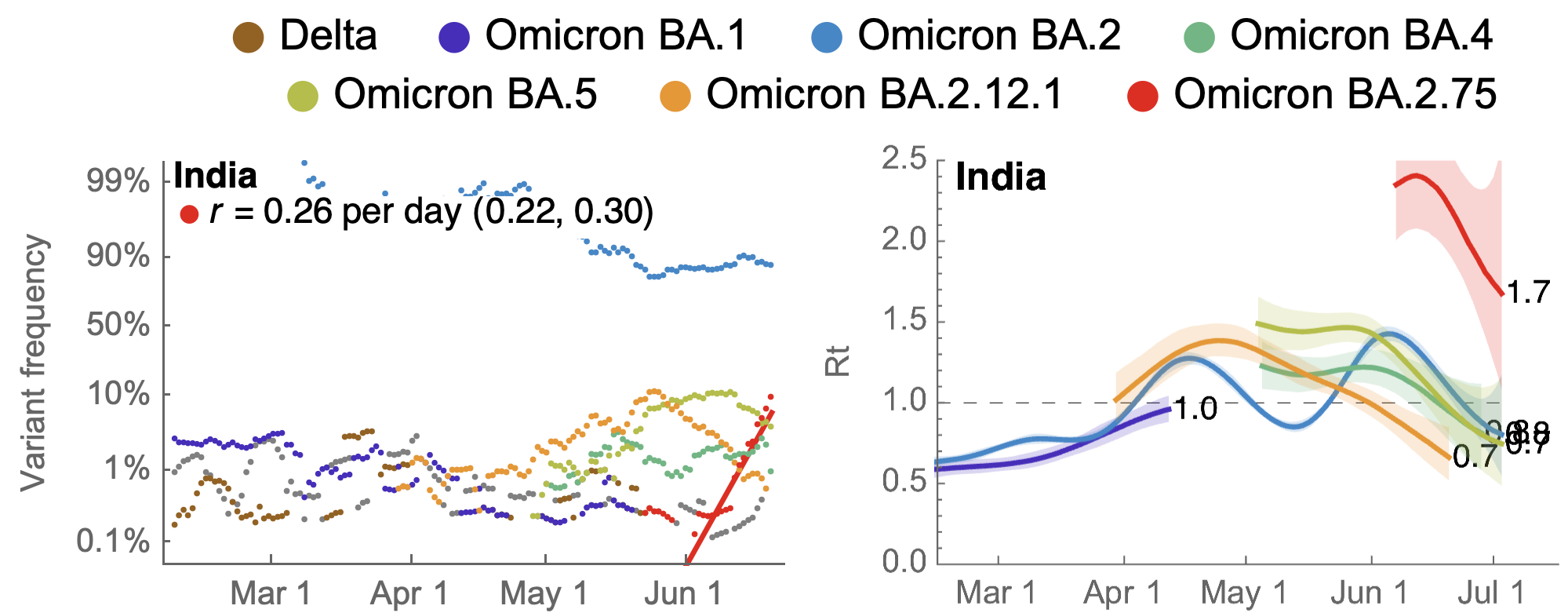
July 4: Initial frequency data from India were quite alarming
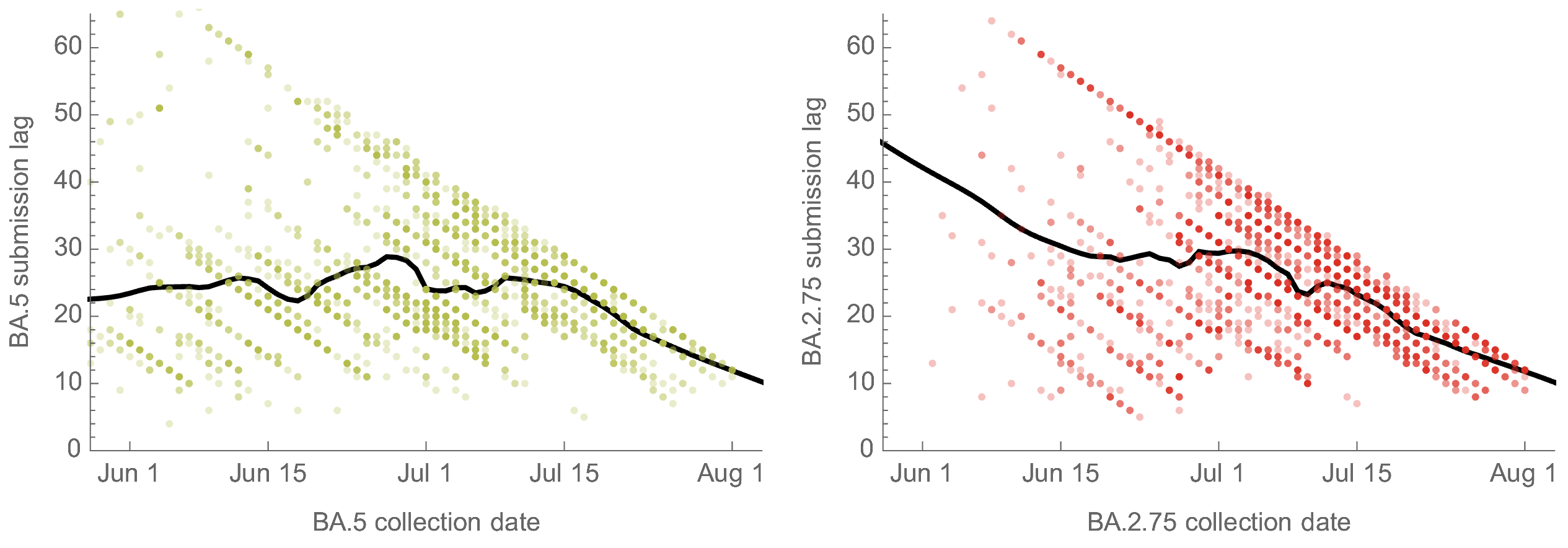
Differential submission lags don't explain discordance
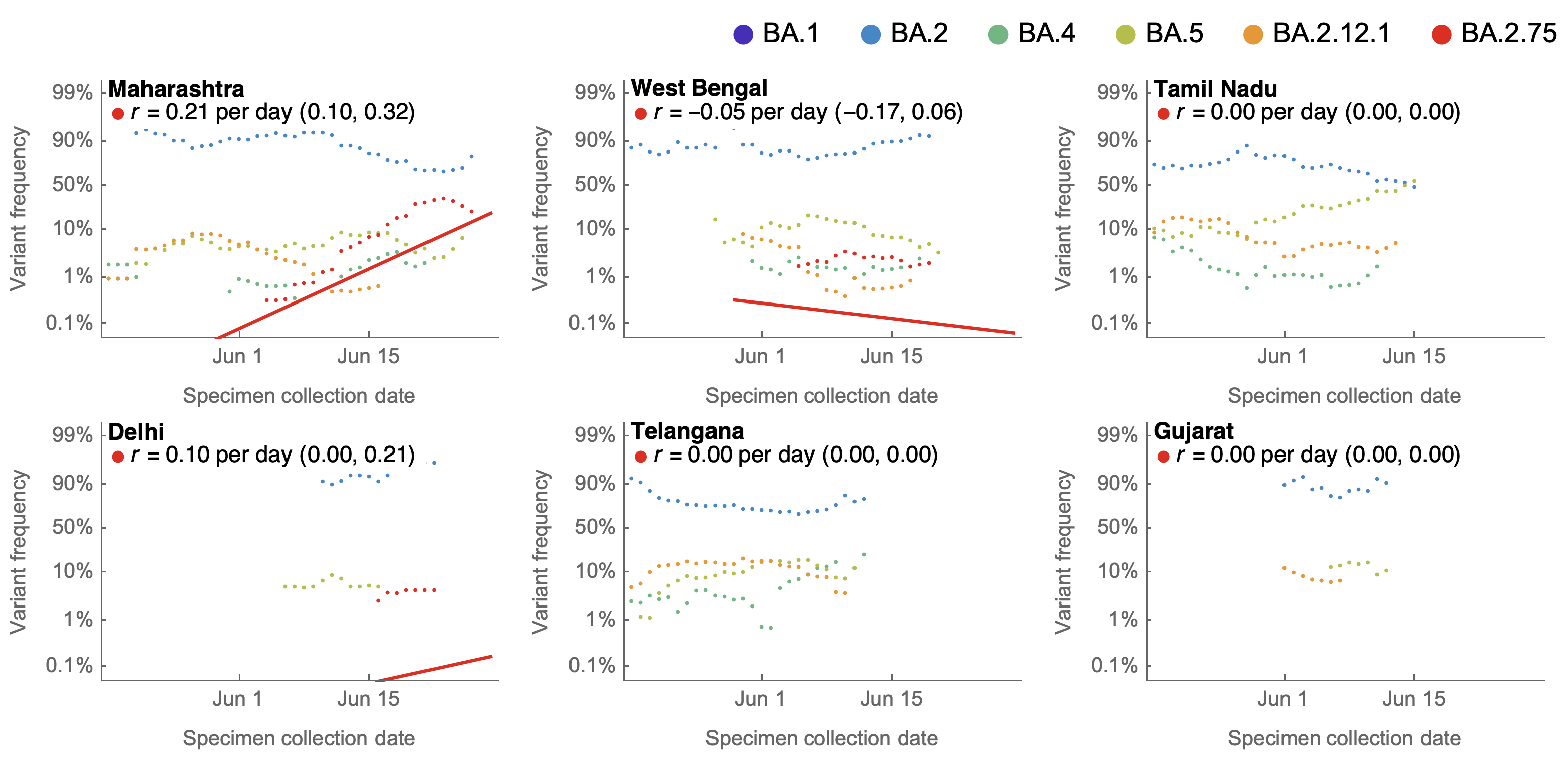
July 4: Early data is geographically focused on Maharashtra
Aug 27: Further sequencing fills out geography
Emerging variants
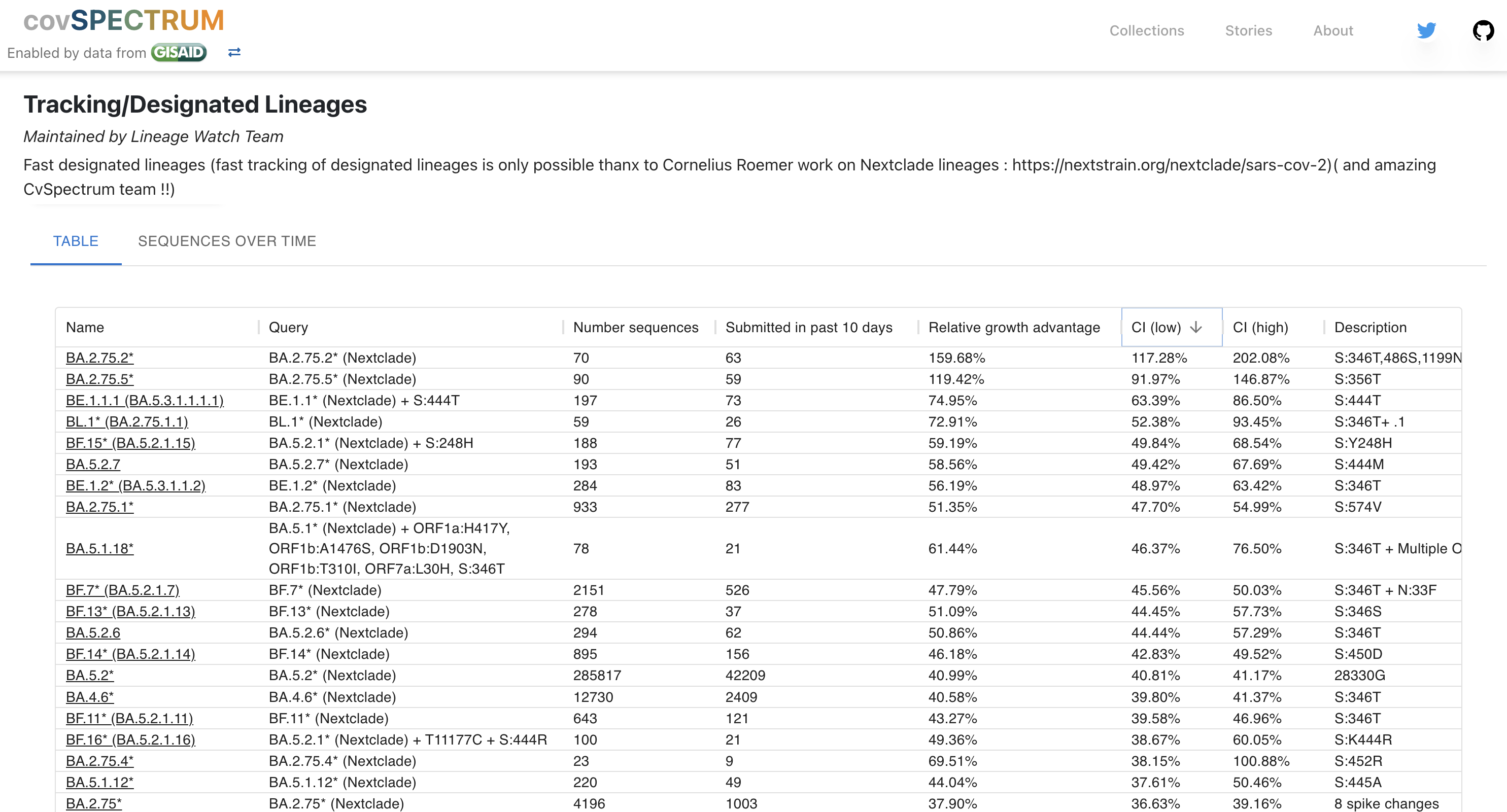
Watch lists for new Pango lineages rising quickly in frequency
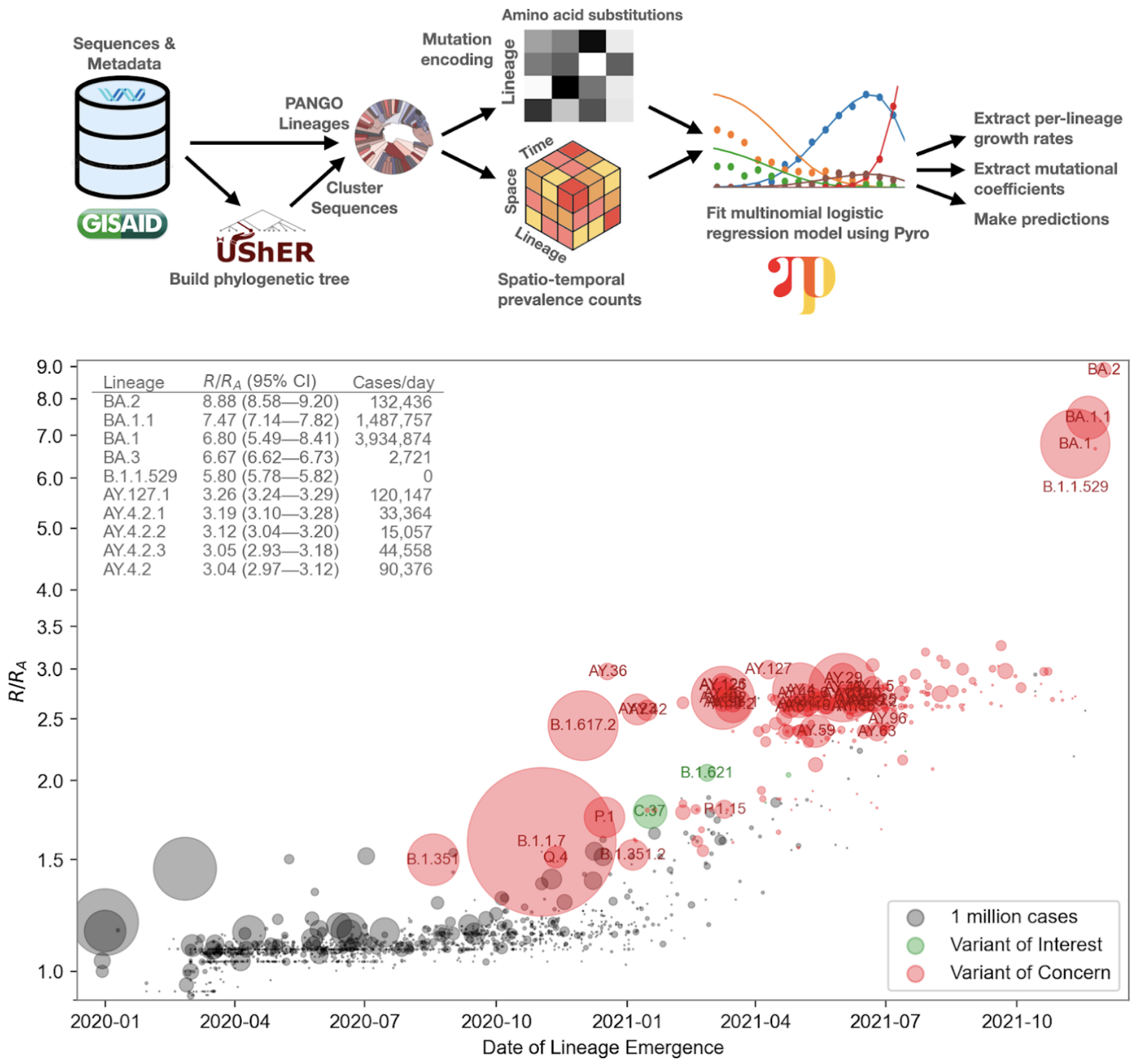
Hierarchical MLR models should work well to regularize and pool data
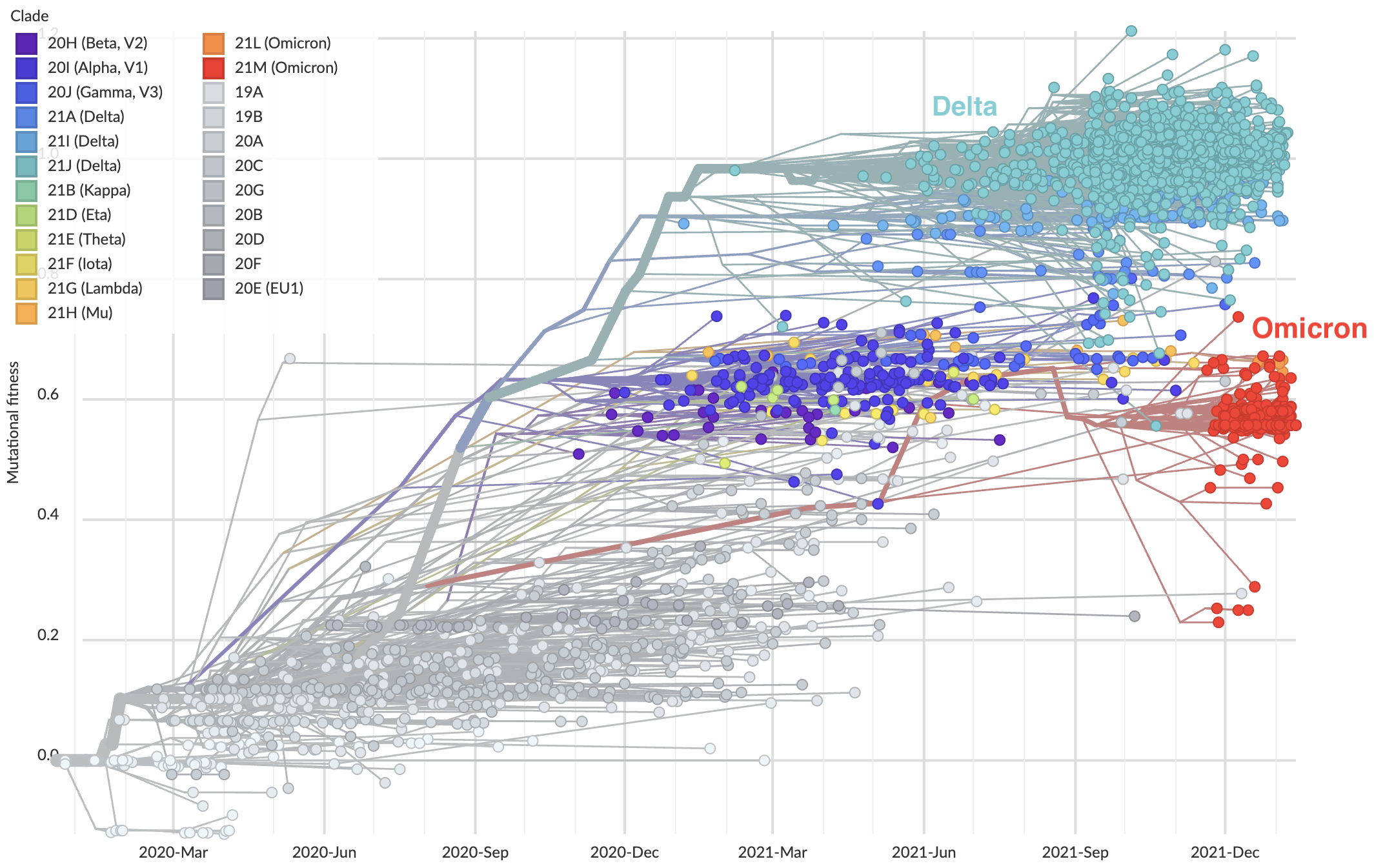
However, mutational makeup did not predict Omicron fitness
Genomic surveillance
Sequences generated and shared at an unprecedented pace with >5M in 2022 (compare to 8k flu genomes in 2022)
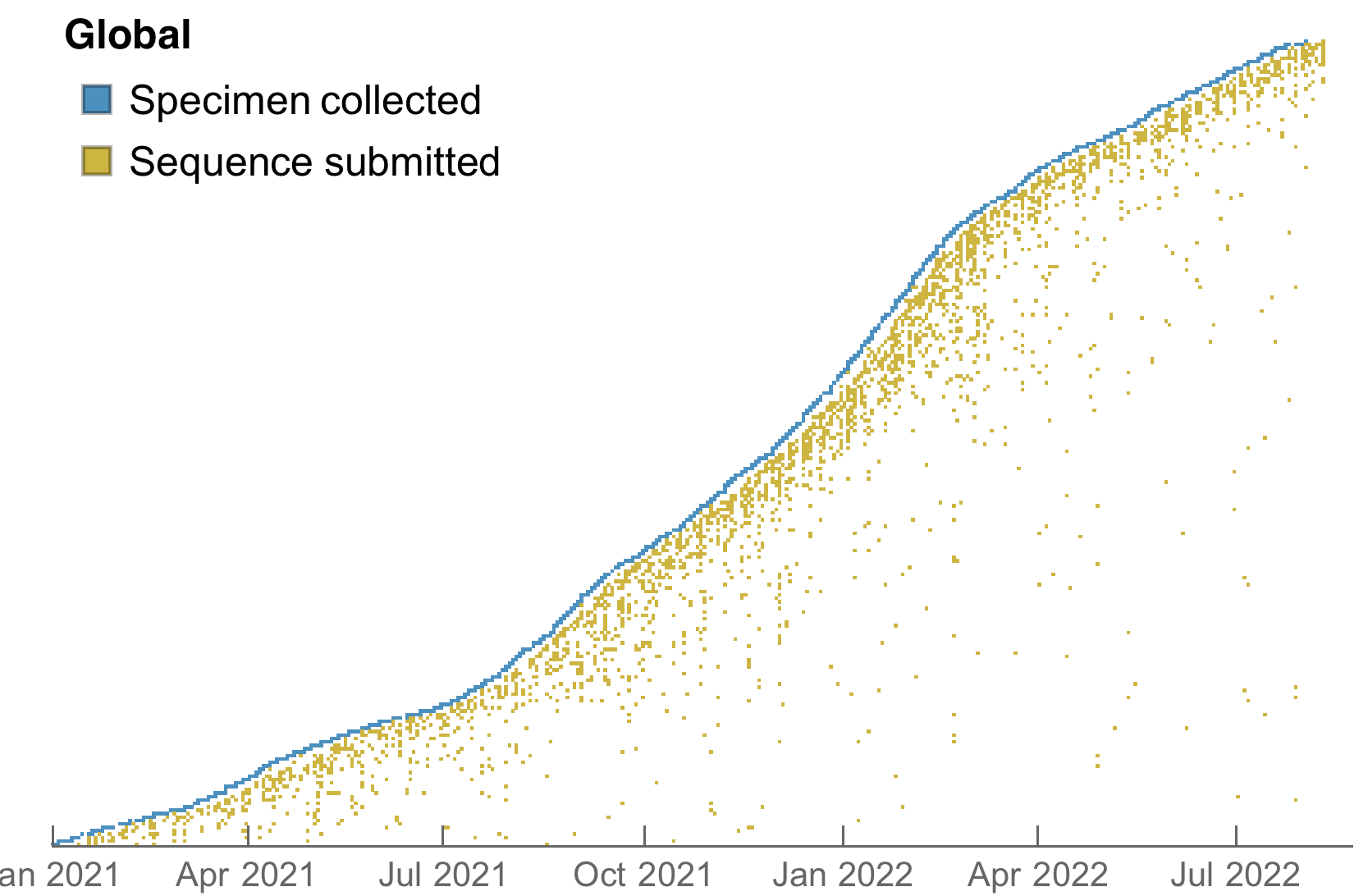
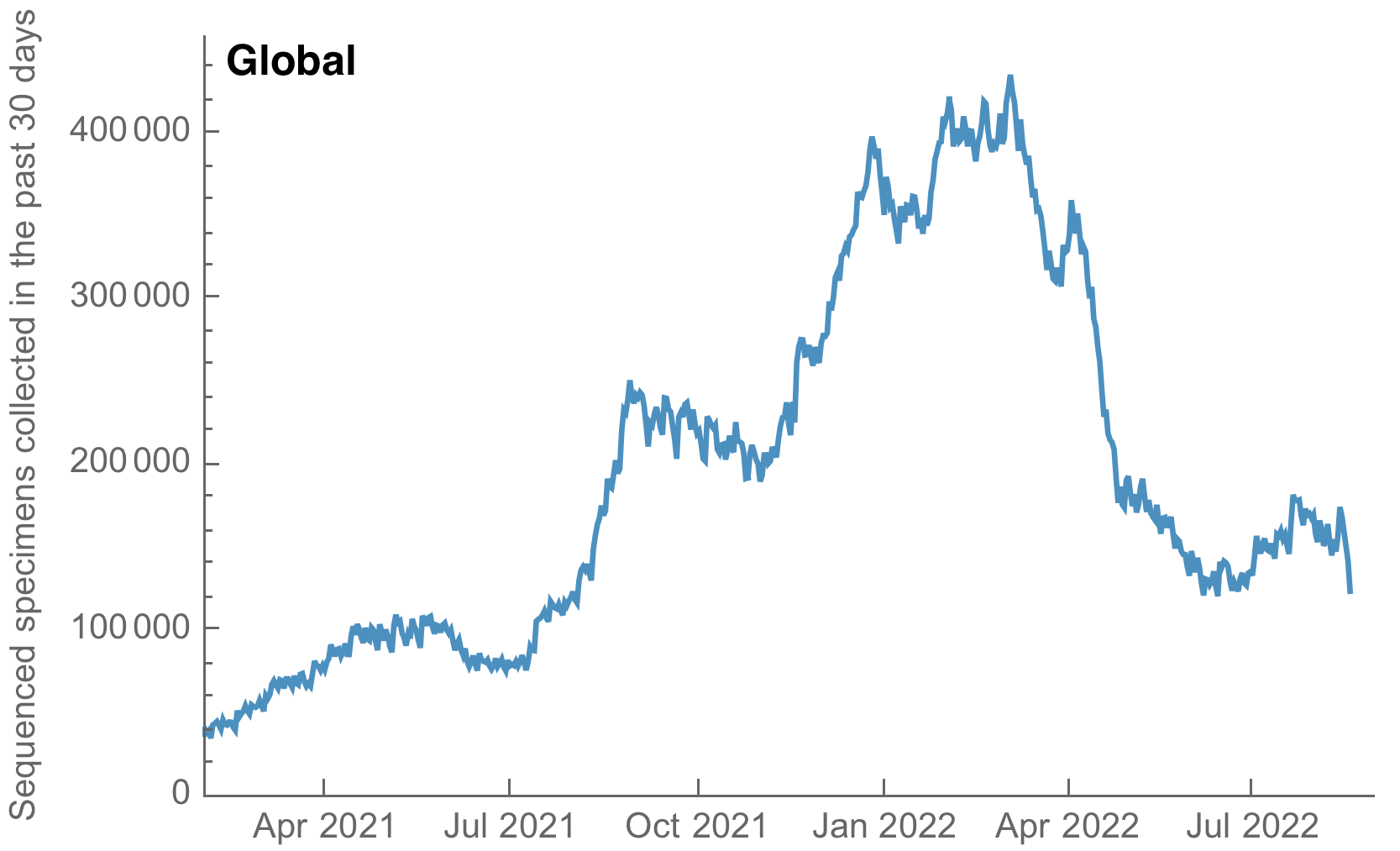
My favorite metric is number of sequences available from samples collected in the past 30 days
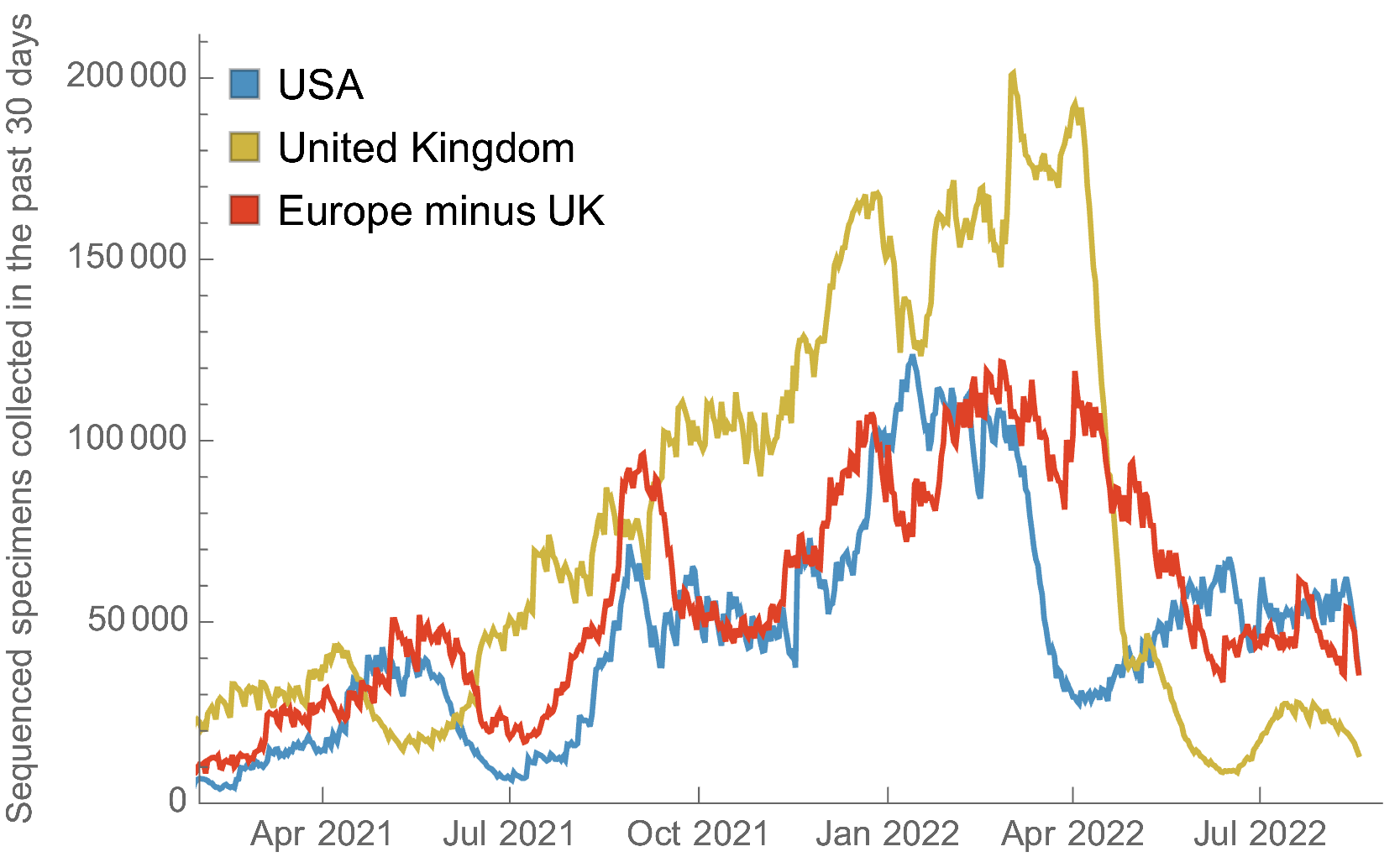
Largely consistent sequence counts for US and Europe, though recent decline in UK
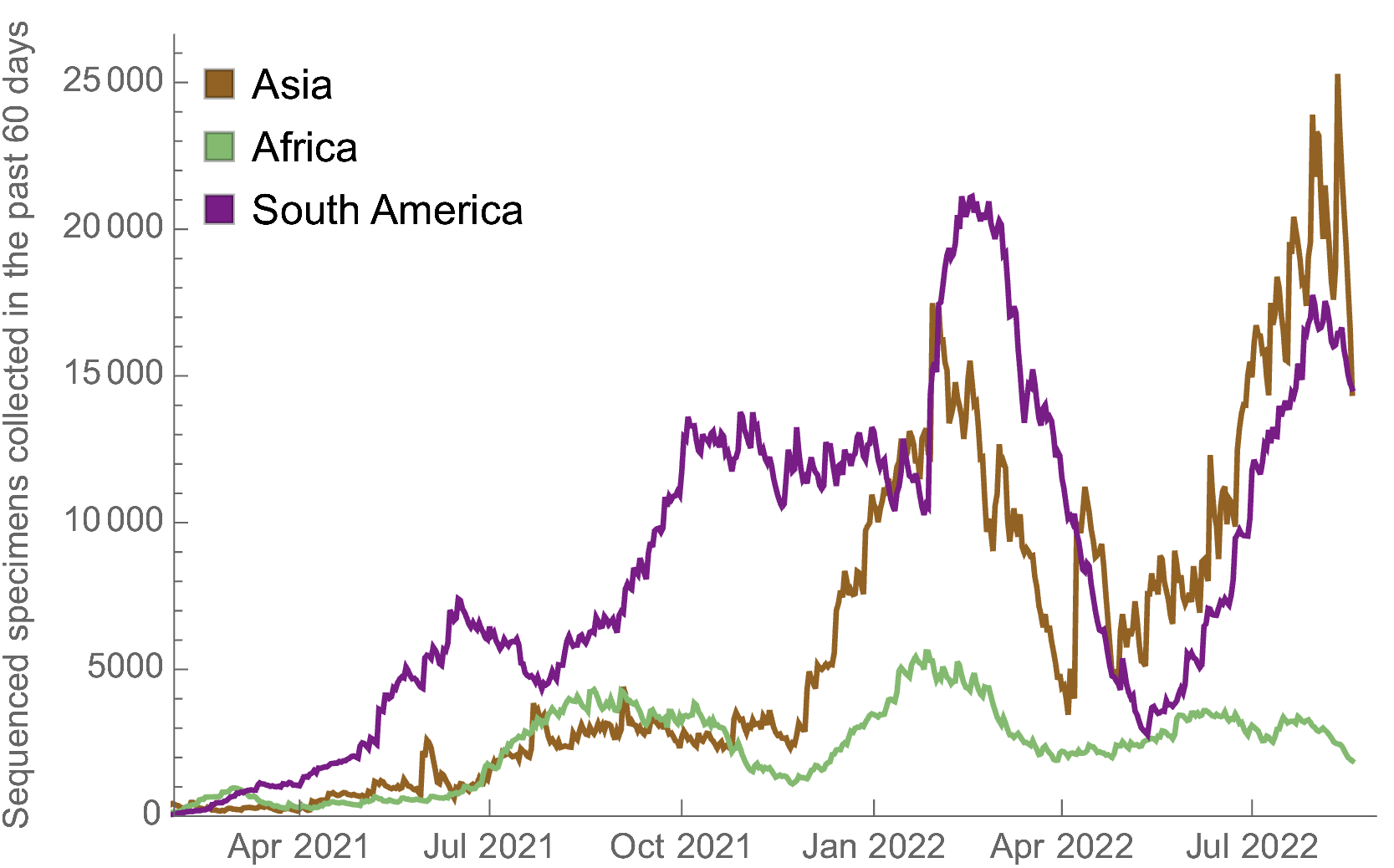
Investments in genomic surveillance should focus on improving latency and throughput in LMICs
Genomic surveillance and modeling to track SARS-CoV-2 variants
- Global genomic surveillance with rapid turnaround times essential, as this allows spotting variant early and provides geographic replication
- With detailed frequency data, MLR models work well to estimate local growth rates
- Fitness estimates become more reliable with multiple geographies
- Automated systems for near-term fitness estimation and forecasting should be continued research priority
Acknowledgements
SARS-CoV-2 genomic epi: Data producers from all over the world, GISAID and the Nextstrain team
Bedford Lab:
![]() John Huddleston,
John Huddleston,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Louise Moncla,
Louise Moncla,
![]() Maya Lewinsohn,
Maya Lewinsohn,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Cassia Wagner,
Cassia Wagner,
![]() Miguel Paredes,
Miguel Paredes,
![]() Nicola Müller,
Nicola Müller,
![]() Marlin Figgins,
Marlin Figgins,
![]() Denisse Sequeira,
Denisse Sequeira,
![]() Victor Lin,
Victor Lin,
![]() Jennifer Chang
Jennifer Chang






