Continuing SARS-CoV-2 evolution and diversification post-Omicron
Trevor Bedford (@trvrb)
Fred Hutchinson Cancer Center / Howard Hughes Medical Institute
23 May 2022
Science for Resilience - Learnings from the Pandemic
Institute for Advanced Studies
Slides at: bedford.io/talks
1. SARS-CoV-2 evolution over the course of the pandemic
2. Omicron emergence and spread
3. Emerging variants
4. Expectations for endemicity
SARS-CoV-2 evolution over the course of the pandemic
Genetic relationships of SARS-CoV-2
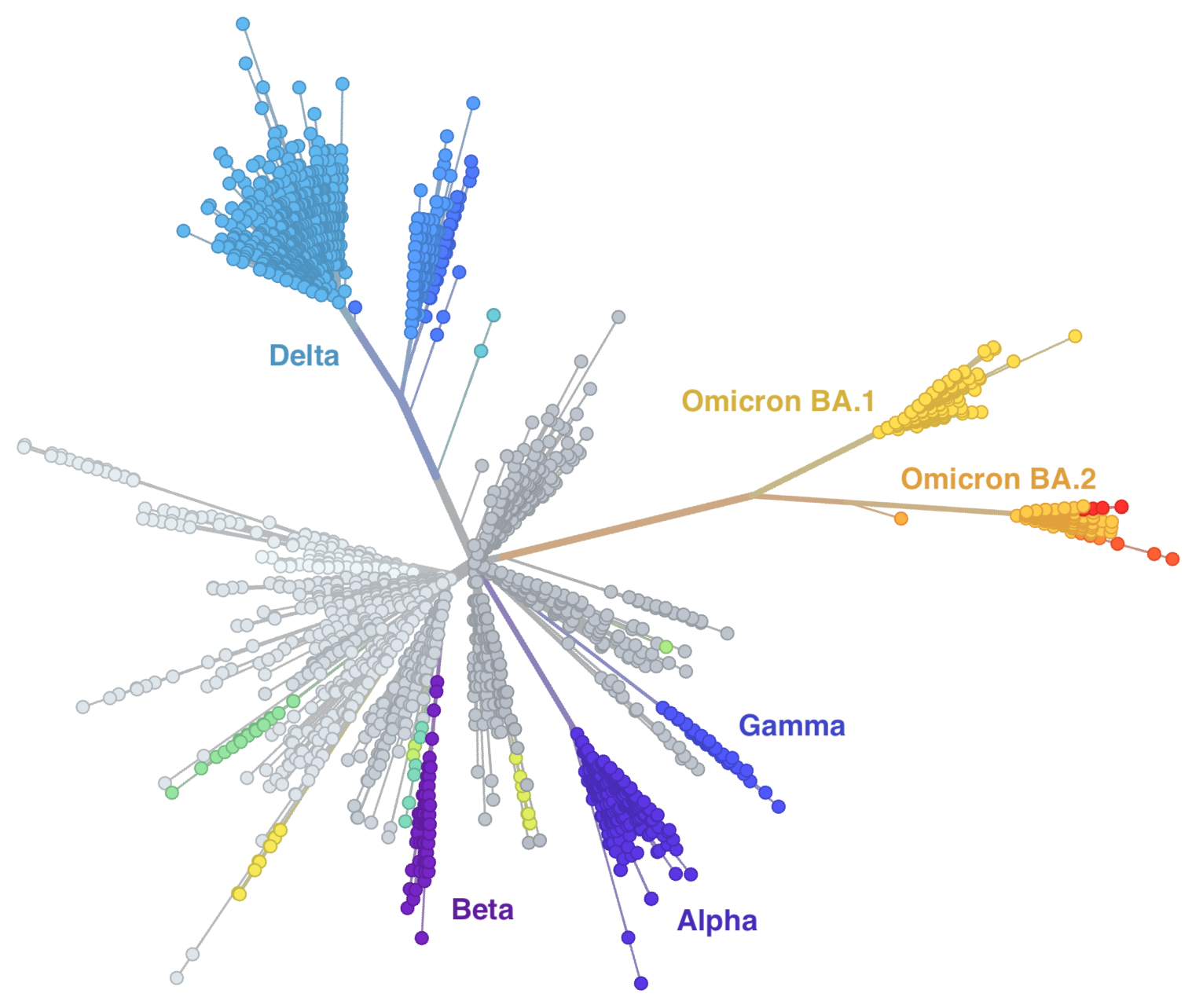
Global SARS-CoV-2 evolution over the course of the pandemic
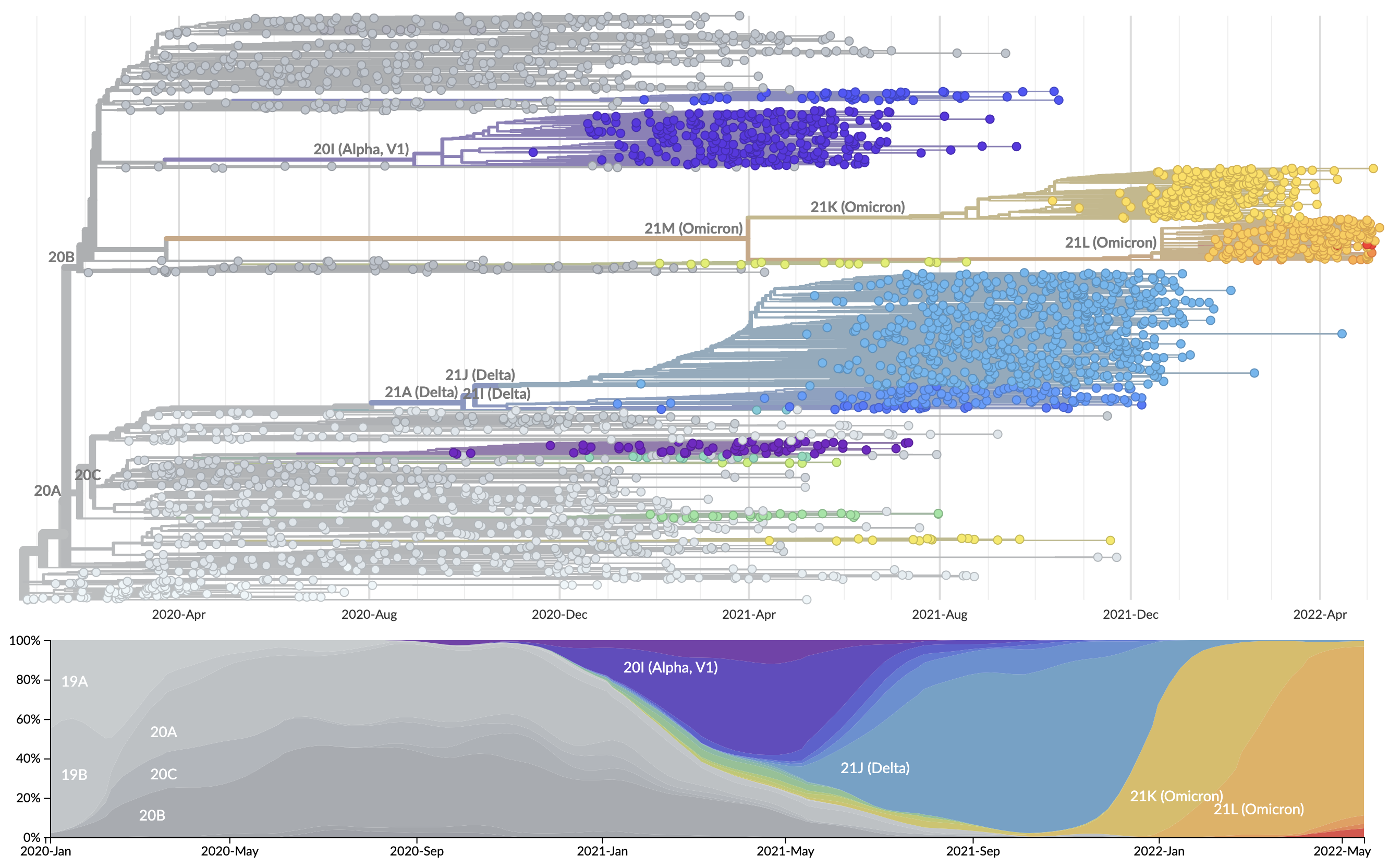
Rapid accumulation of spike mutations driving displacement
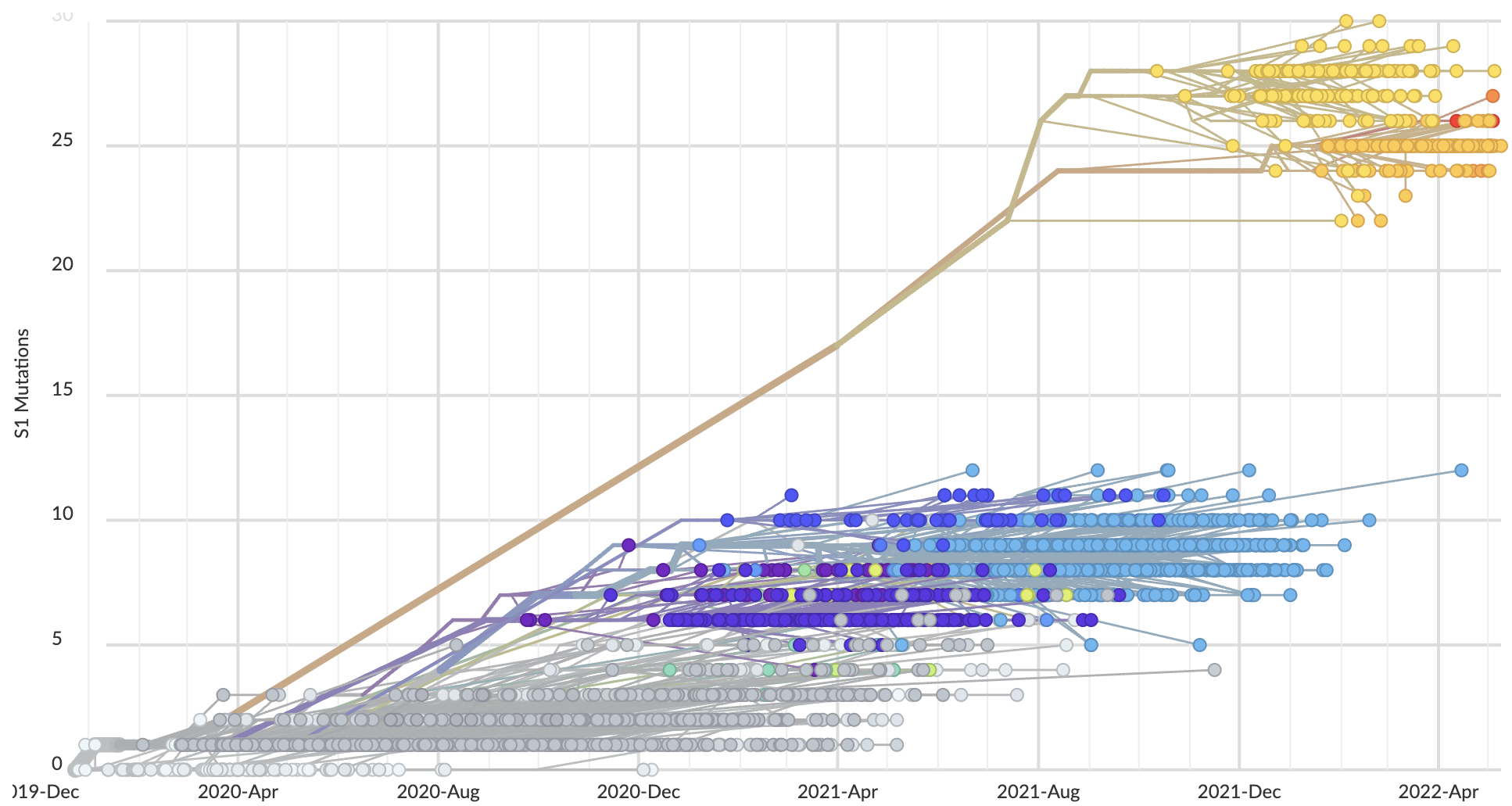
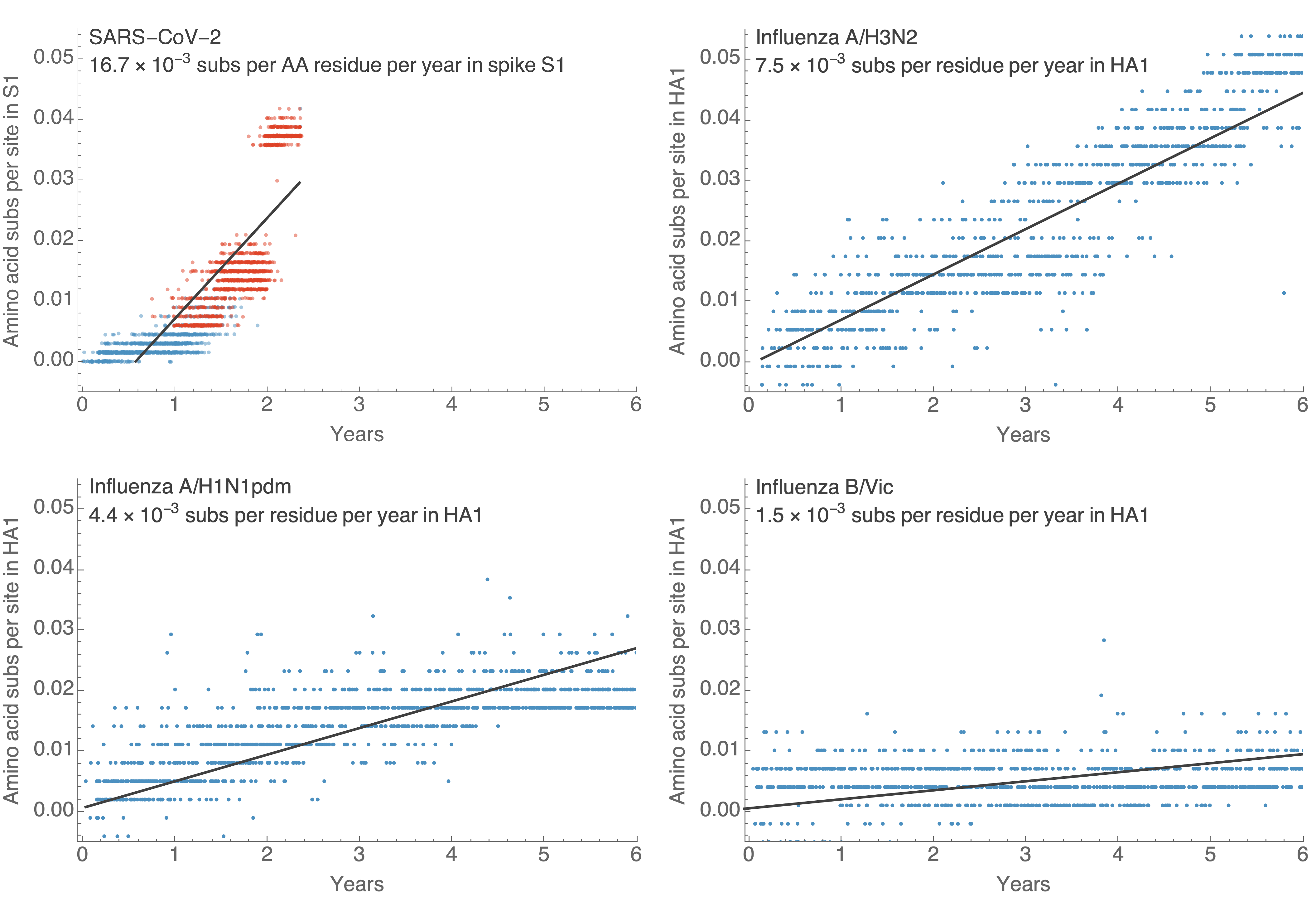
S1 evolved at a rate of 11 amino acid changes per year in 2021
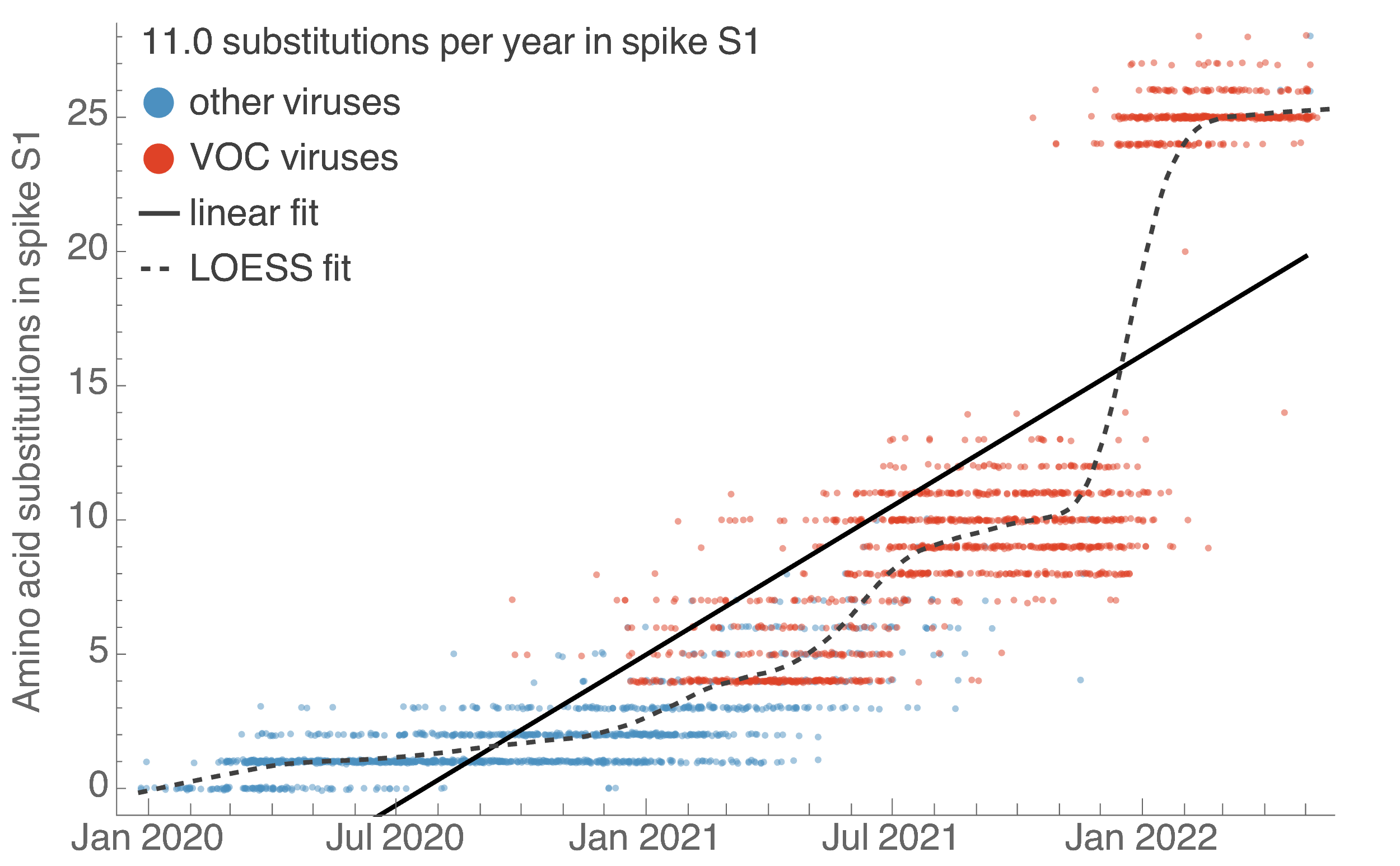
S1 evolution remarkably fast relative to seasonal influenza
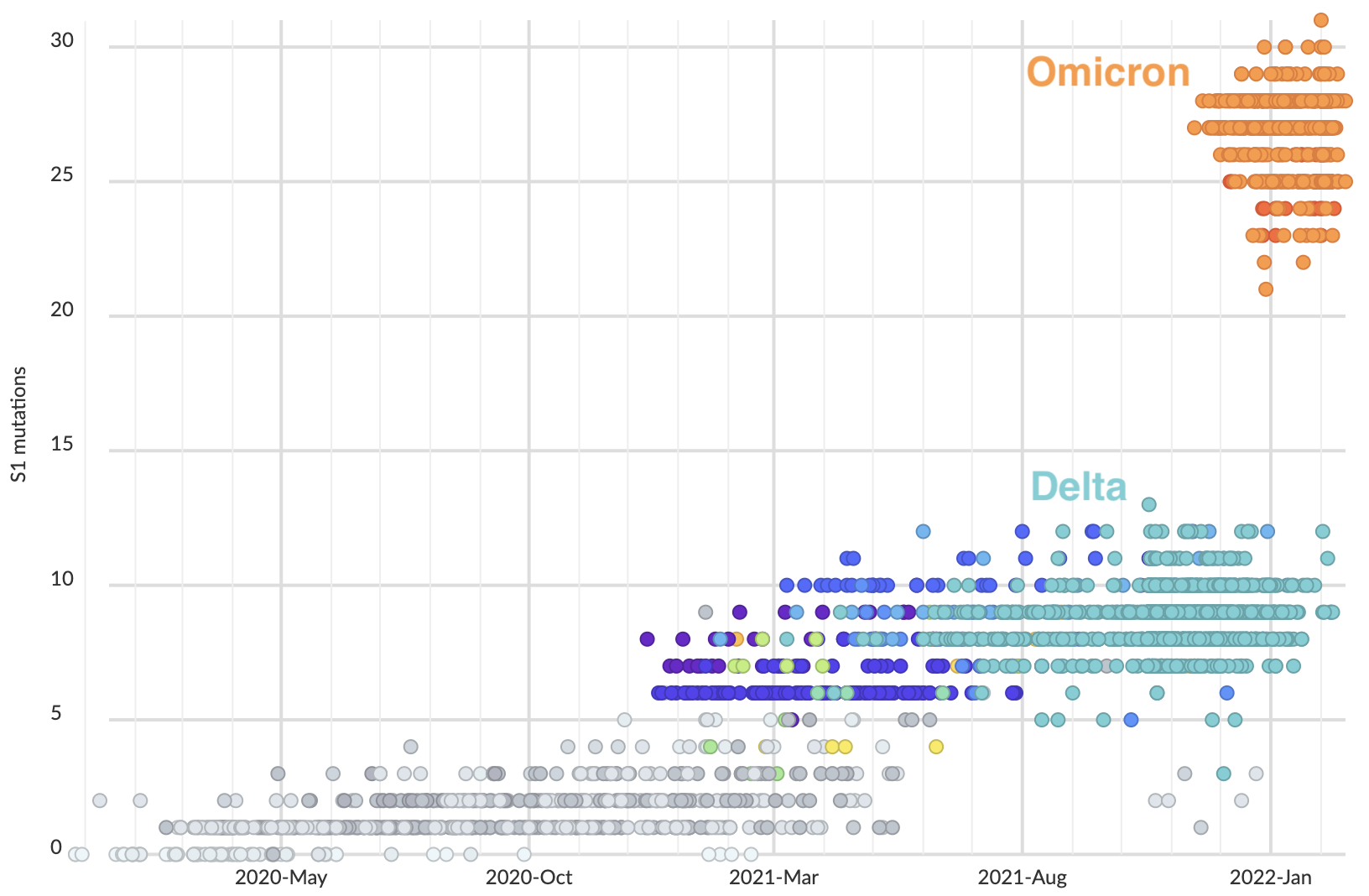
Omicron emergence and spread
Omicron viruses possess huge excess of mutations in S1
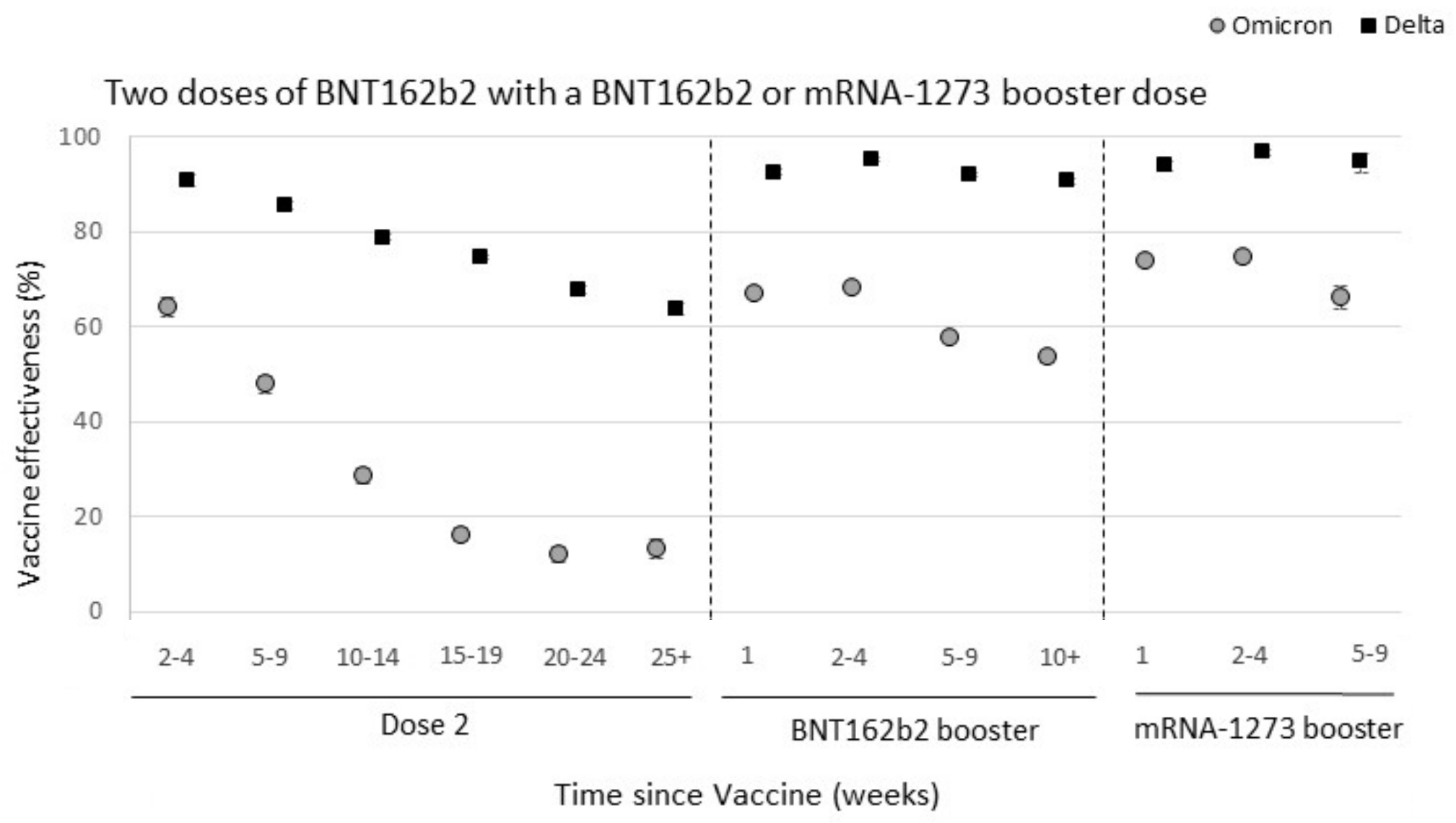
Omicron spike mutations substantially drop VE against infection
Significant immune escape drove large-scale epidemics
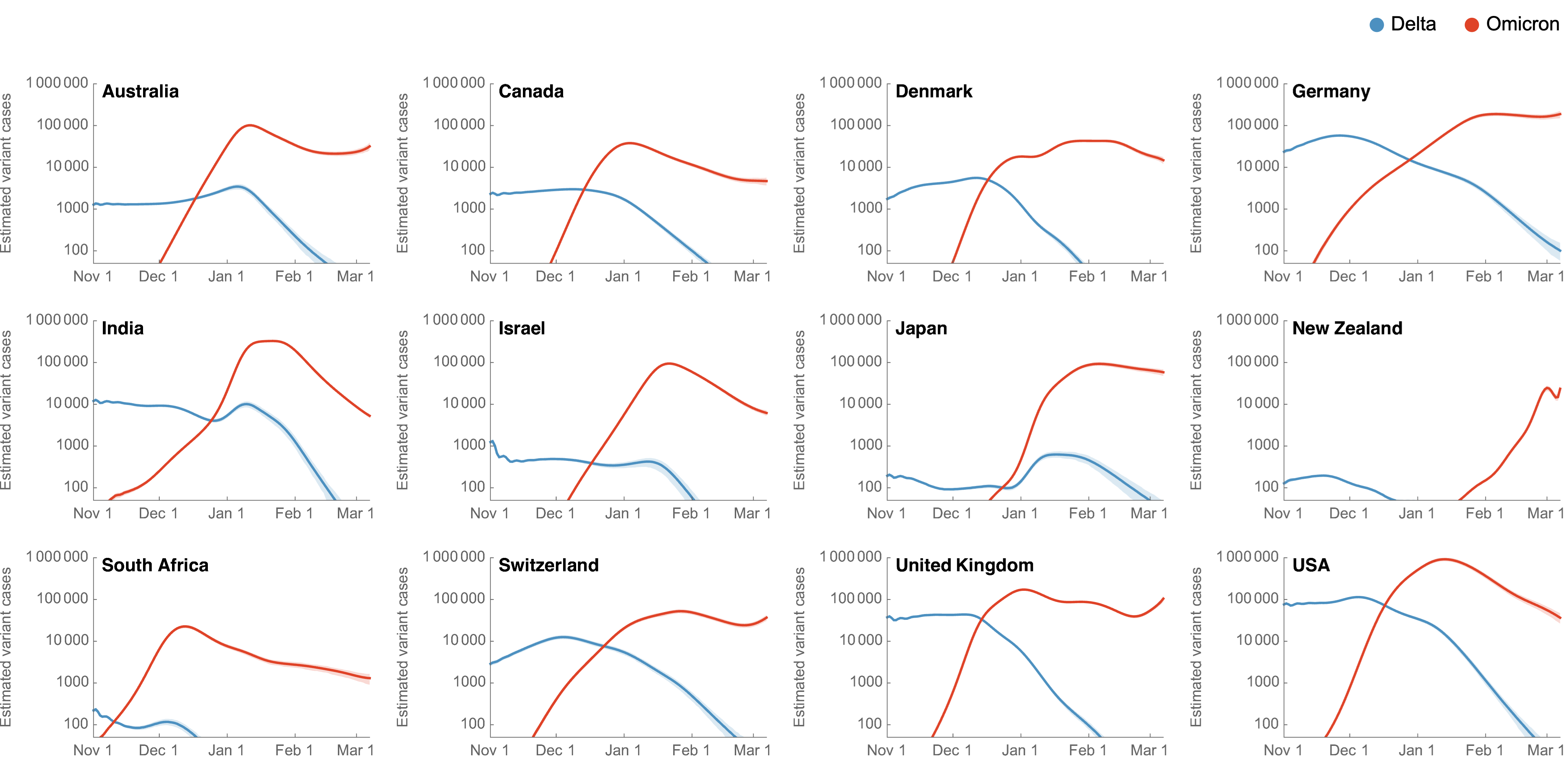
Starting Rt approached that of the initial wave in spring 2020
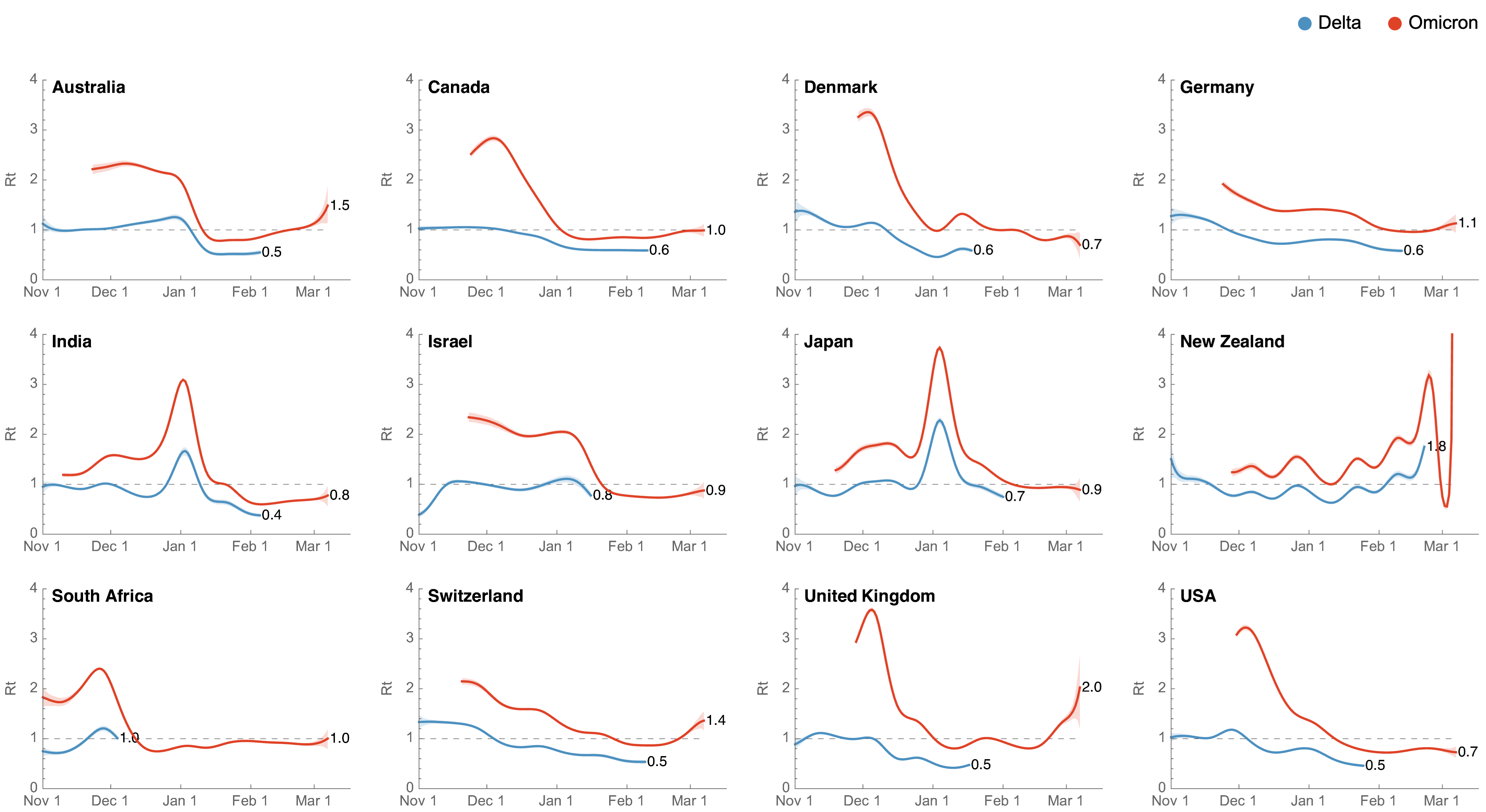
Omicron attack rate
- We estimate US has seen 9.8% of the population as confirmed cases of Omicron through Mar 3, with the large majority accumulating after Dec 15
- Assuming a case detection rate of 1 in 5 infections, we estimate almost 50% of the US infected with Omicron, most in the span of ~10 weeks
Similar epidemics across US states with downturn driven by immunity
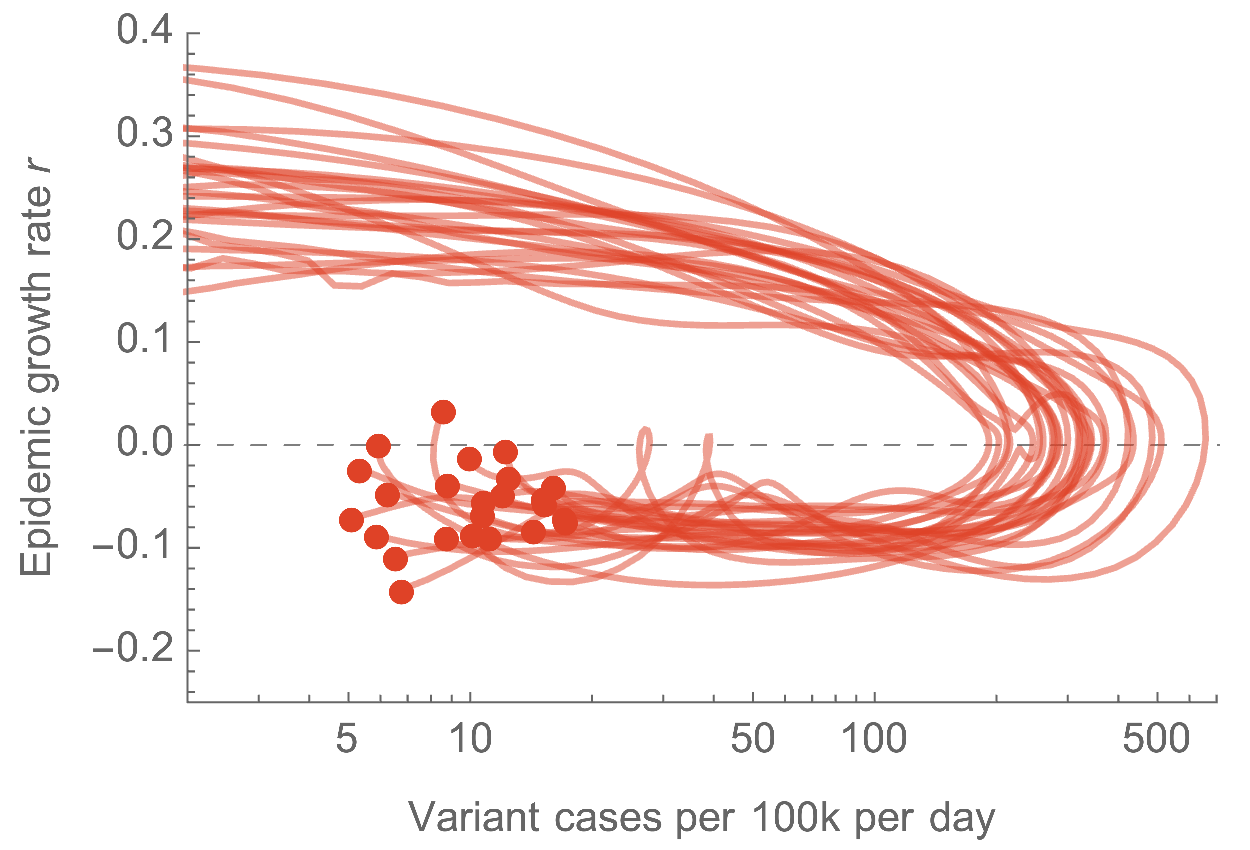
Emerging variants
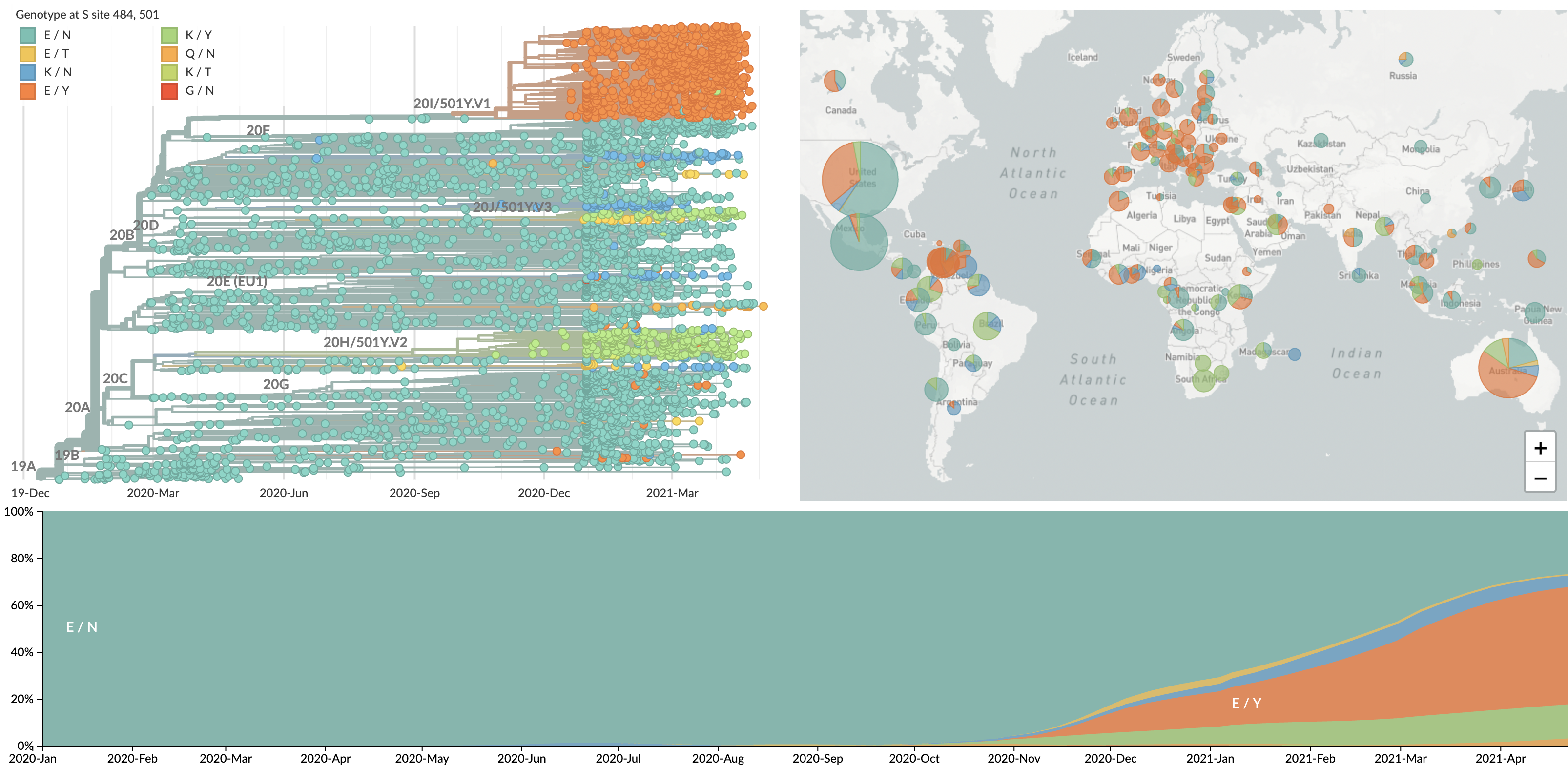
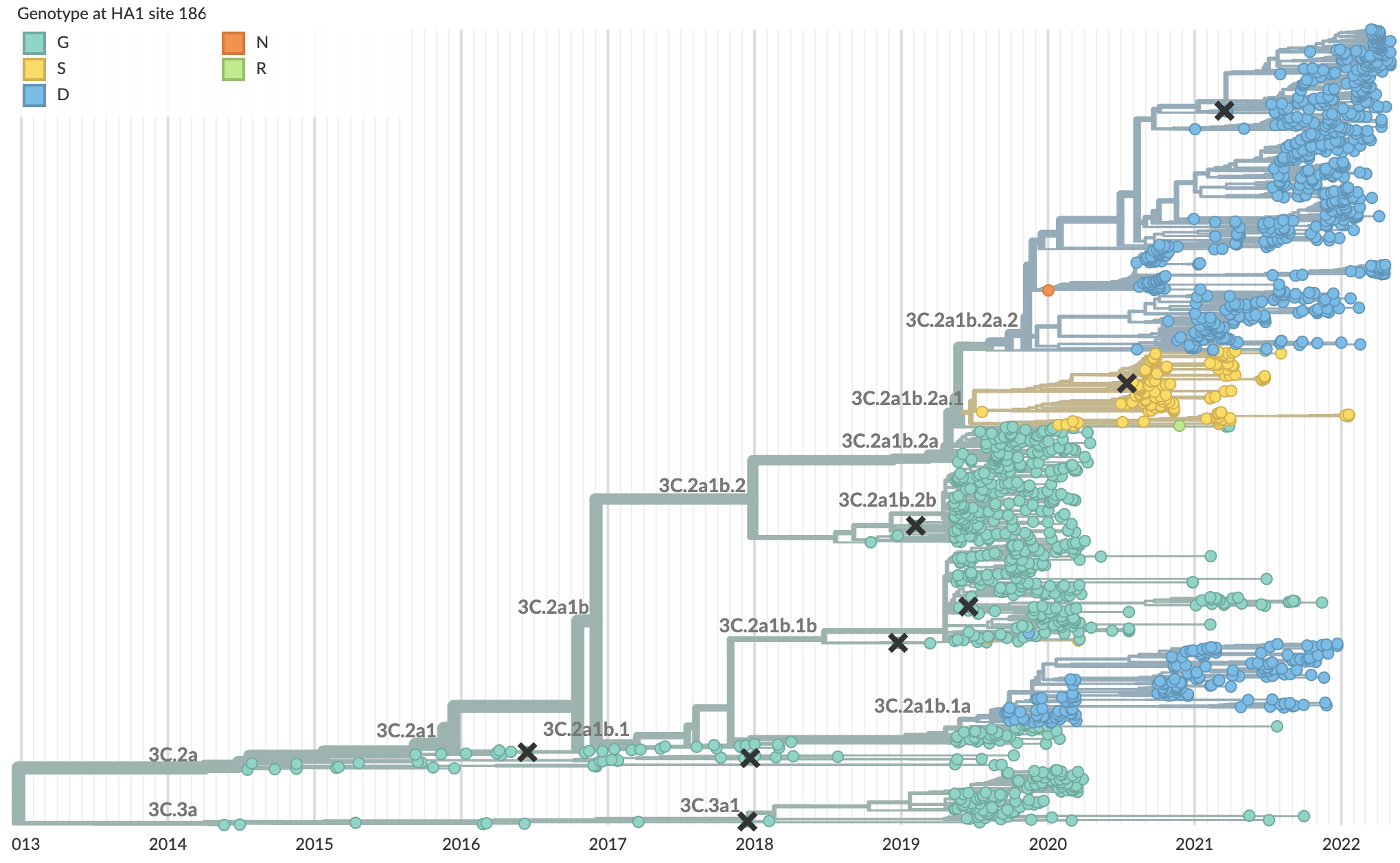
Initial variants characterized by repeated evolution at spike 484K and 501Y
We're now seeing repeated evolution at spike 452
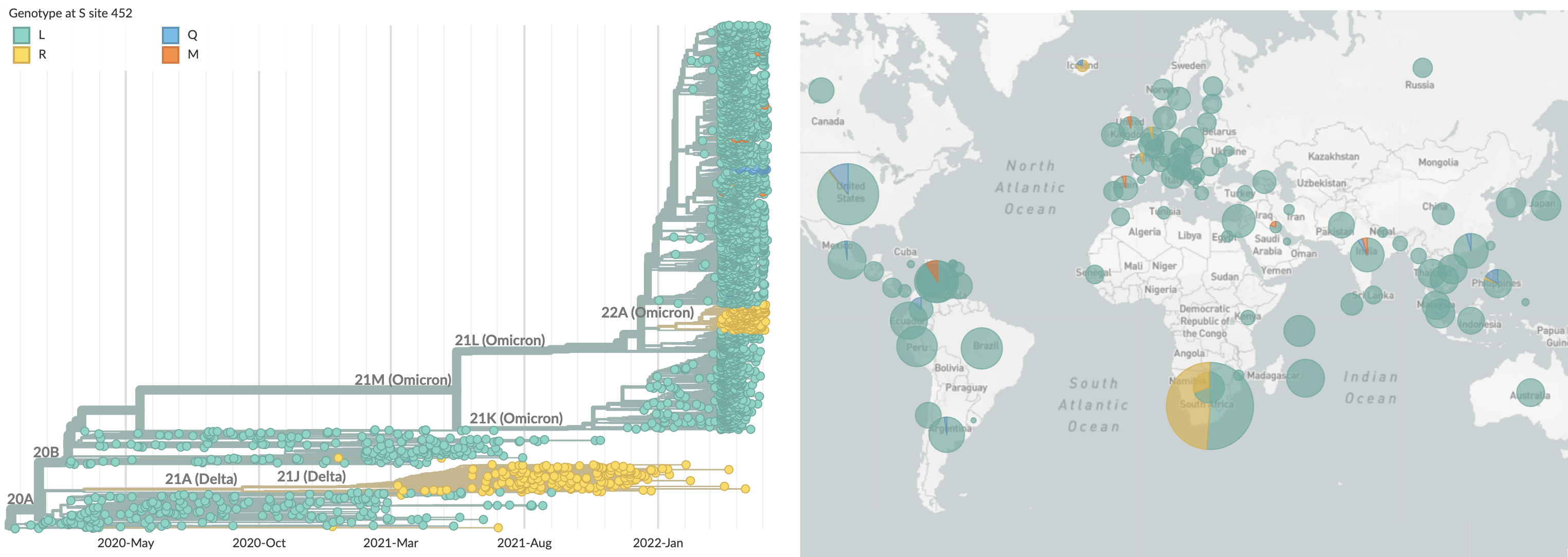
This pattern is familiar from seasonal influenza
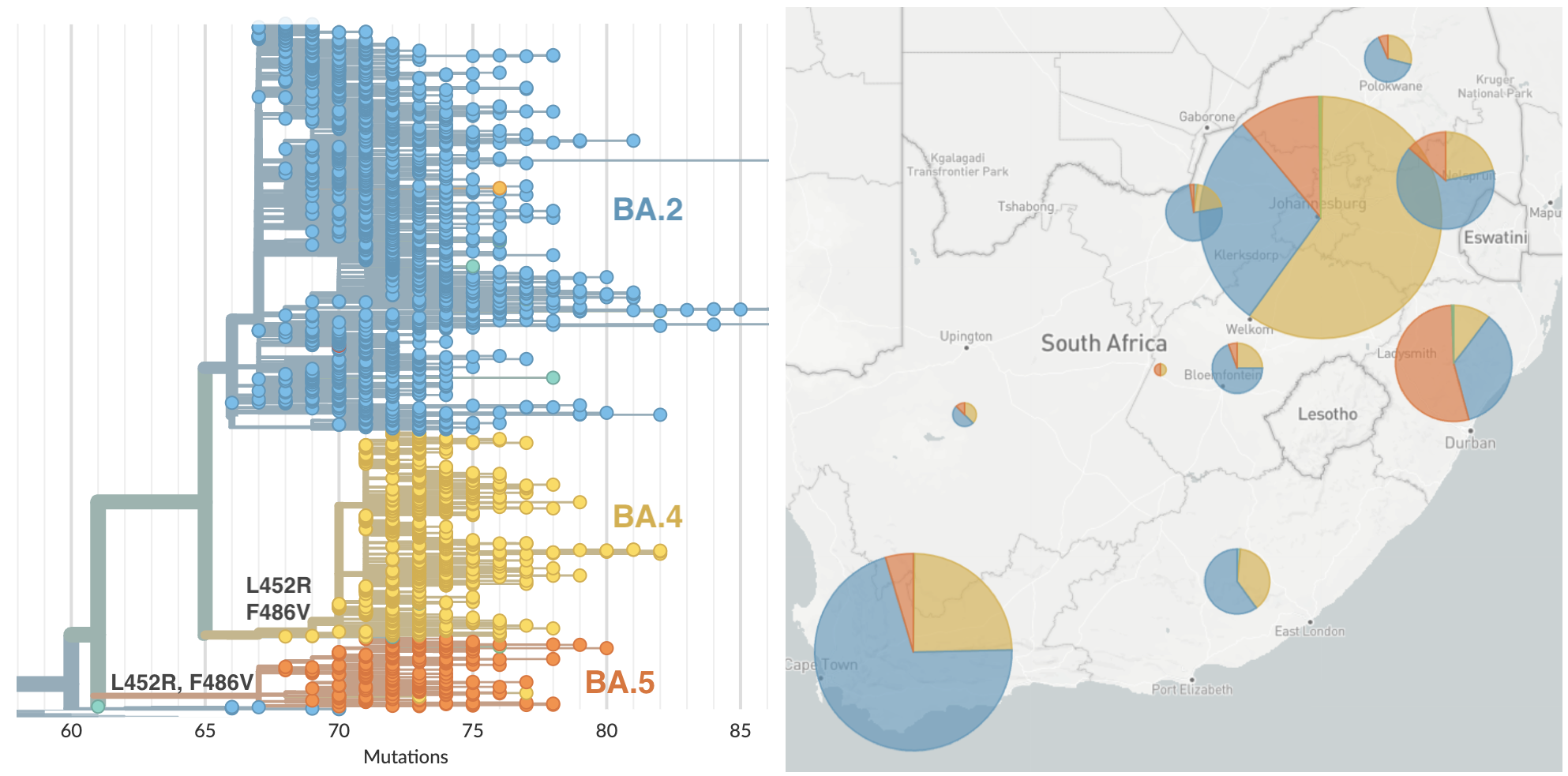
BA.4 and BA.5 emerging from South Africa with spike mutations 452R and 486V
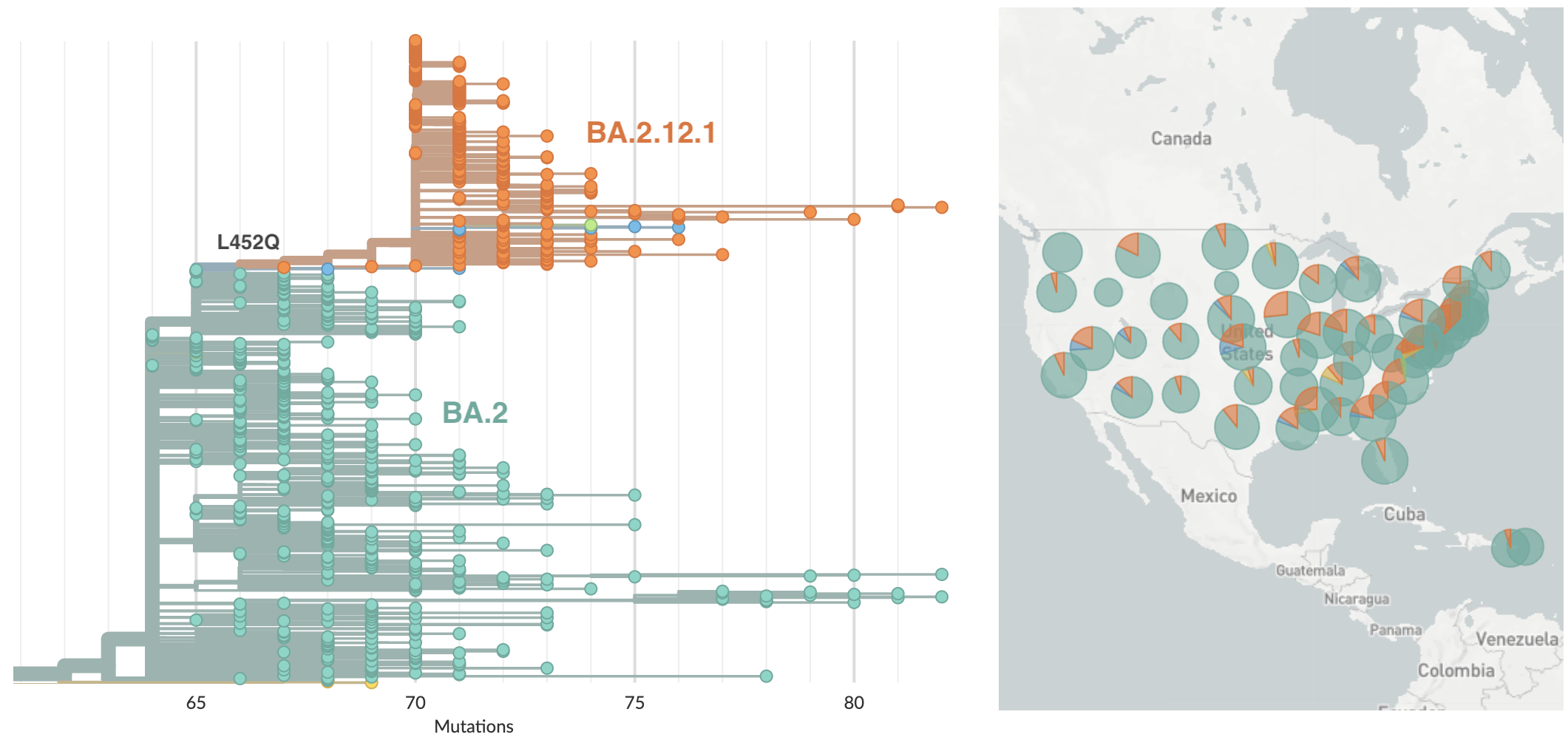
BA.2.12.1 emerging from Northeastern US bearing spike mutation 452Q
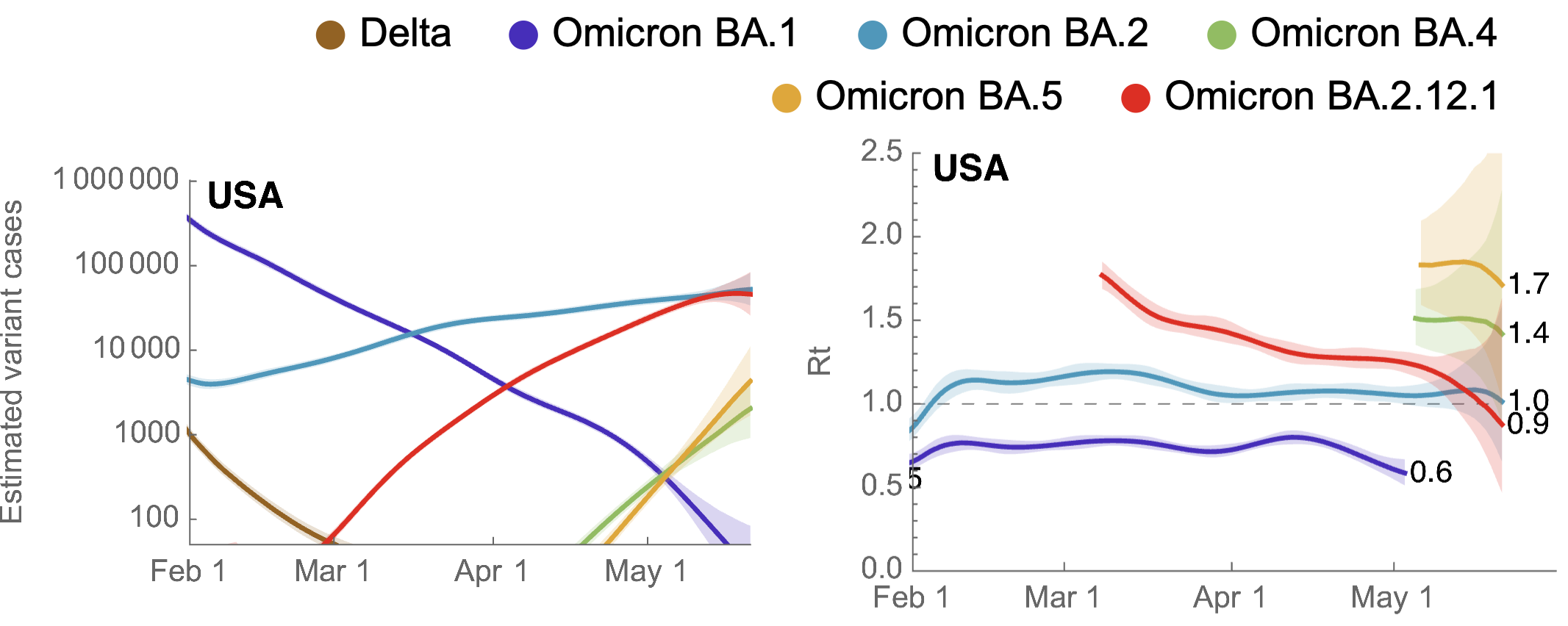
Variant-specific epidemics shown by partitioning case counts with variant frequency
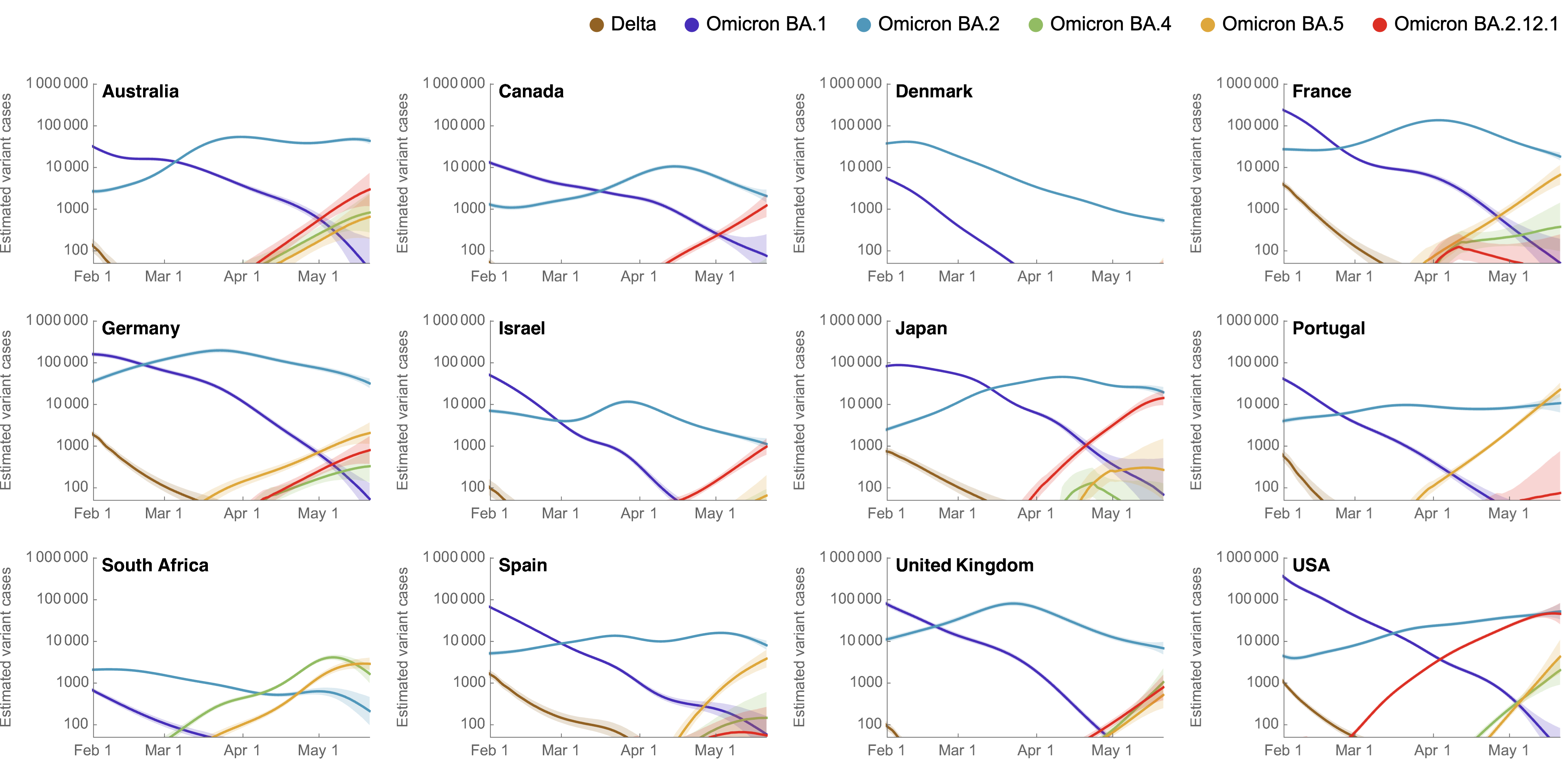
BA.4, BA.5, BA.2.12.1 driving cases across countries
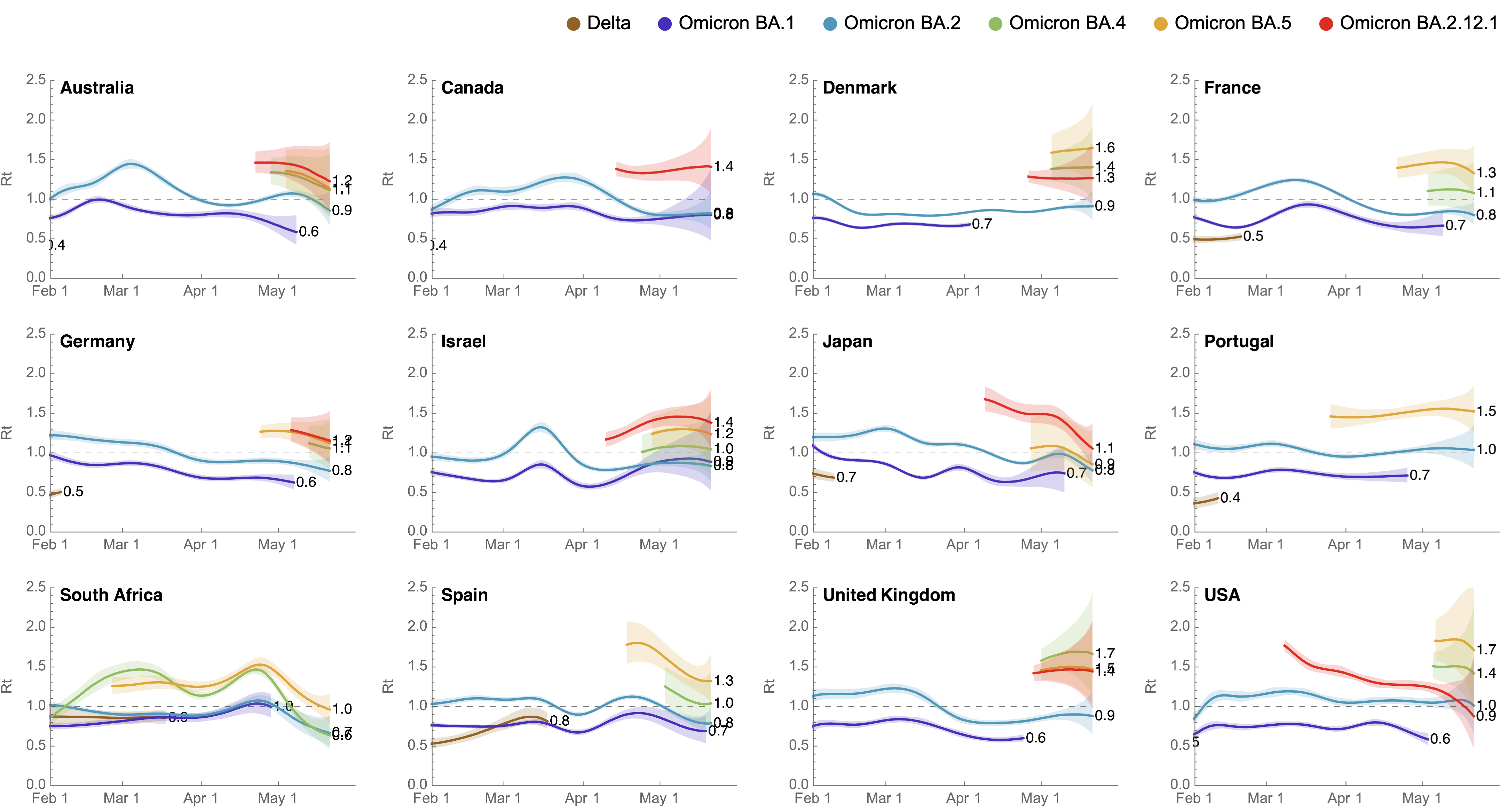
Rt for BA.2.12.1 greater than BA.2 and Rt for BA.4/5 greater still
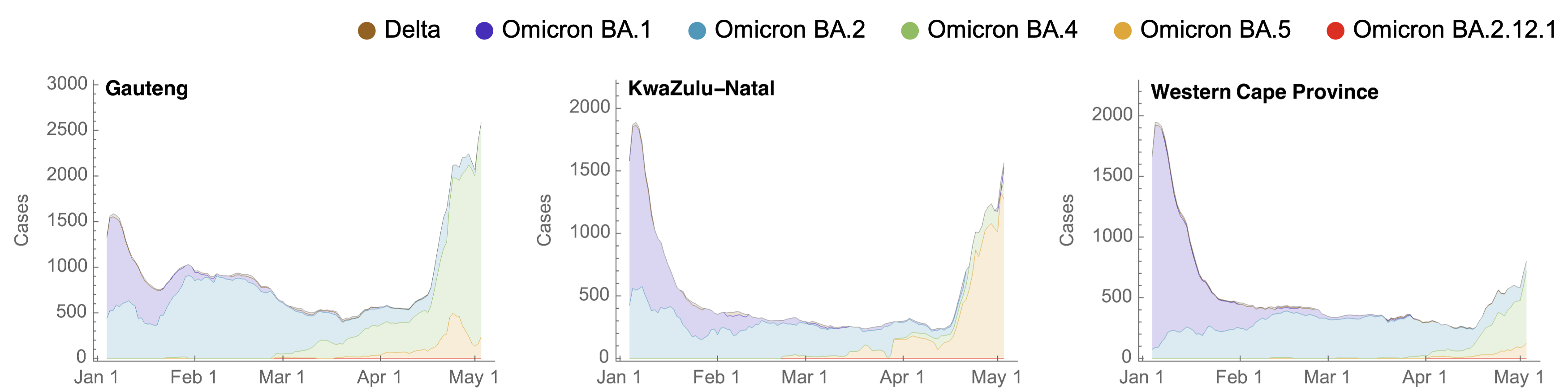
BA.4 and BA.5 driving substantial epidemics in South Africa
Expectations for endemicity
Immunity and reduced intrinsic severity has dropped CFR
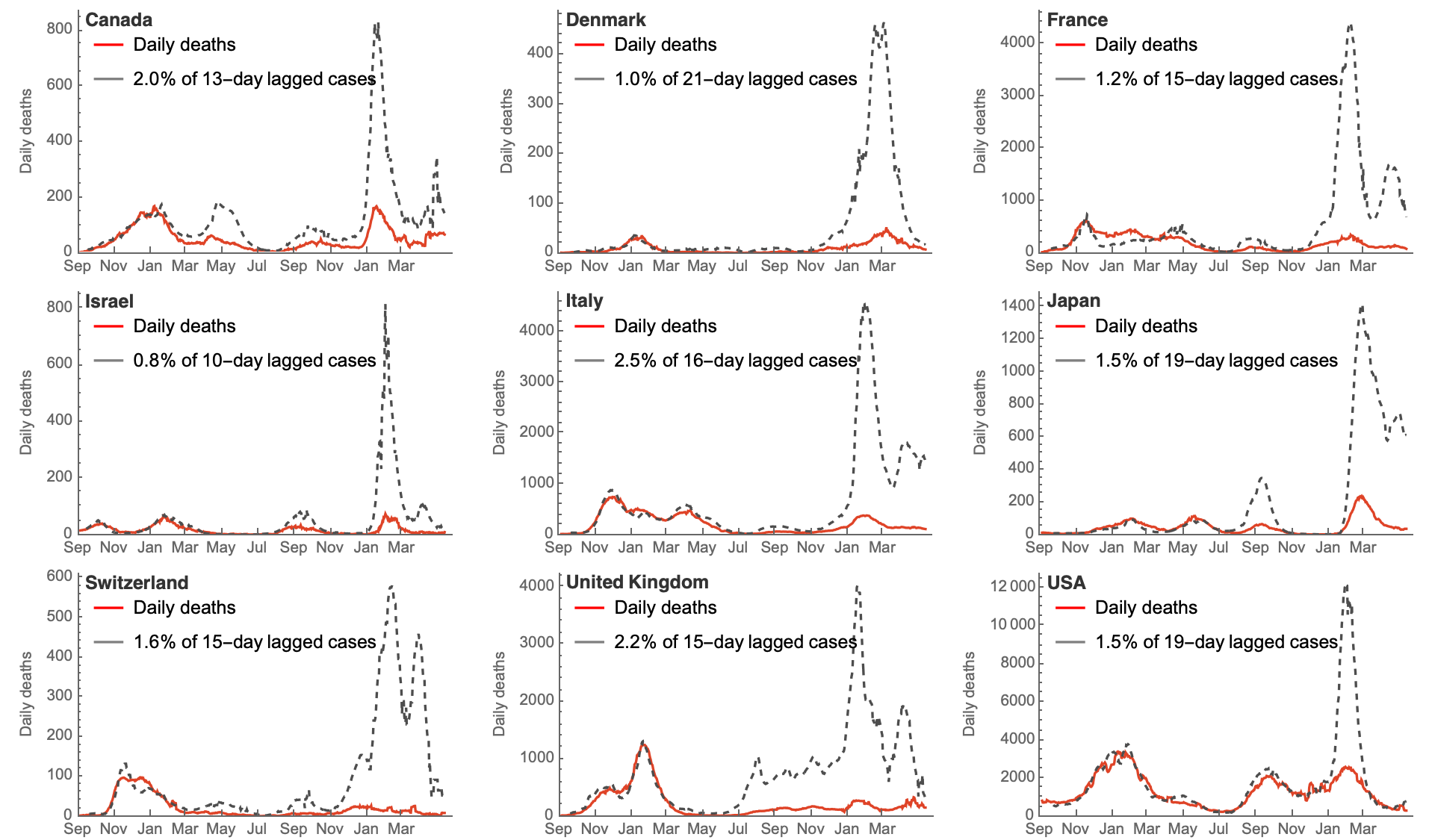
Broad expectations of endemicity based on comparison with flu
- R0 of Delta/Omicron is perhaps ~6 compared to R0 of flu of ~2. At same rates of evolution and waning expect more COVID circulation.
- Before Omicron rates of evolution in SARS-CoV-2 S1 had been roughly equal to H3N2 flu. Omicron equivalent to 5 years of H3N2 evolution.
- Infection fatality rate (IFR) of COVID is roughly comparable to flu once you have prior immunity.
1, 2 and 3 suggest a virus that circulates at higher levels than flu, but individual infections aren't much more severe in terms of mortality than H3N2 influenza.
Seasonality suggests winter "COVID season", but Omicron-like events could overcome seasonality.
Key parameters are seasonal attack rate and IFR
- Assume seasonal attack rate of 40%, with Omicron-like emergence driving 80% some years.
- Assume IFR of 0.05% (compared to early pandemic IFR of 0.5%), but new variant could push this higher.
- This would give somewhere around 20 per 100k population yearly deaths at endemicity, but potential for variant emergence to push this over 40 per 100k population.
Acknowledgements
SARS-CoV-2 genomic epi: Data producers from all over the world, GISAID and the Nextstrain team
Bedford Lab:
![]() John Huddleston,
John Huddleston,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Louise Moncla,
Louise Moncla,
![]() Maya Lewinsohn,
Maya Lewinsohn,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Cassia Wagner,
Cassia Wagner,
![]() Miguel Paredes,
Miguel Paredes,
![]() Nicola Müller,
Nicola Müller,
![]() Marlin Figgins,
Marlin Figgins,
![]() Denisse Sequeira,
Denisse Sequeira,
![]() Victor Lin,
Victor Lin,
![]() Jennifer Chang
Jennifer Chang






