Real-time tracking of virus evolution
Trevor Bedford (@trvrb)
29 Oct 2015
Evolution Seminar
JF Crow Institute, UW Madison
Slides at bedford.io/talks/
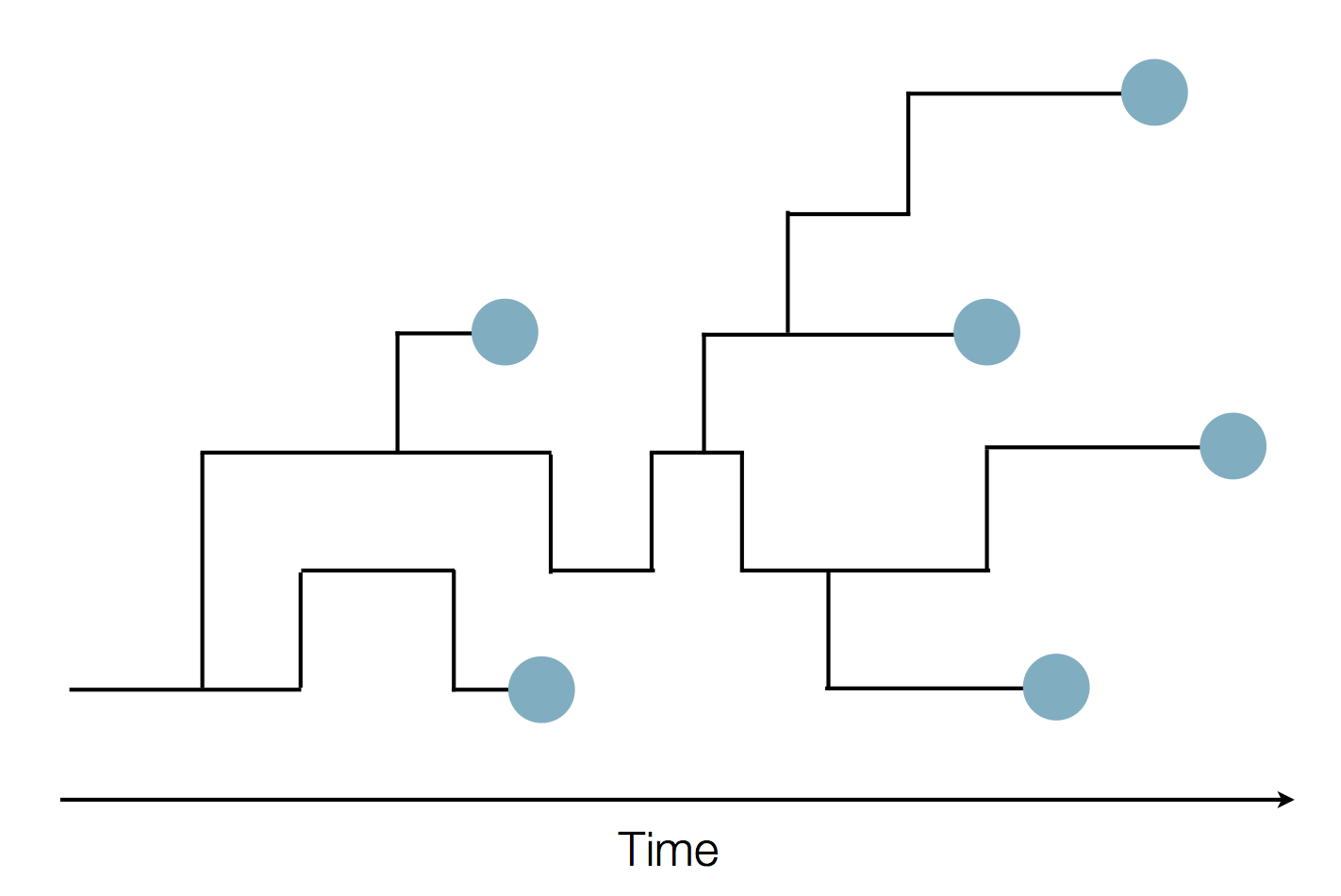
Phylogenies
Phylogenies describe history
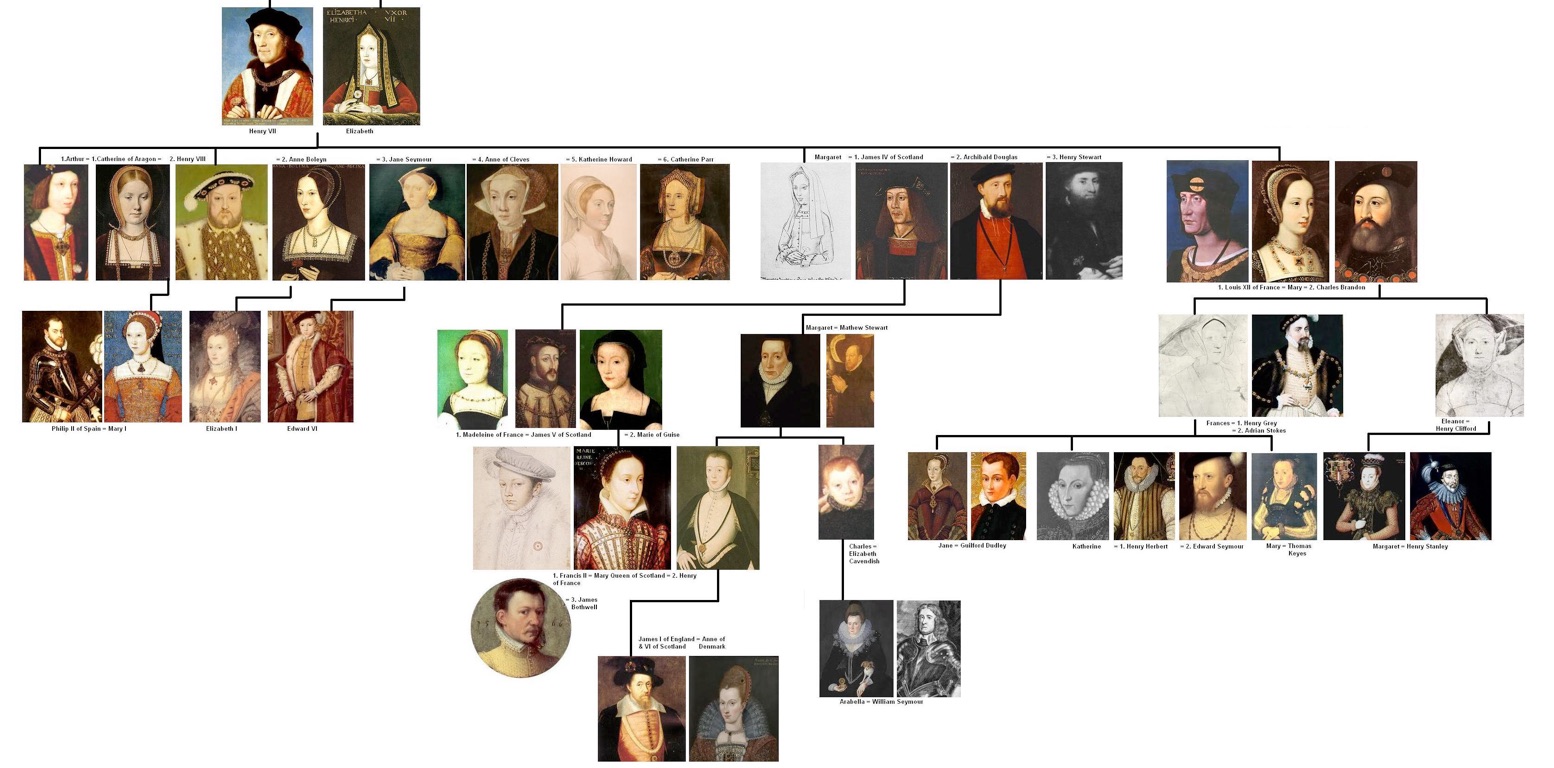
Phylogenies describe history
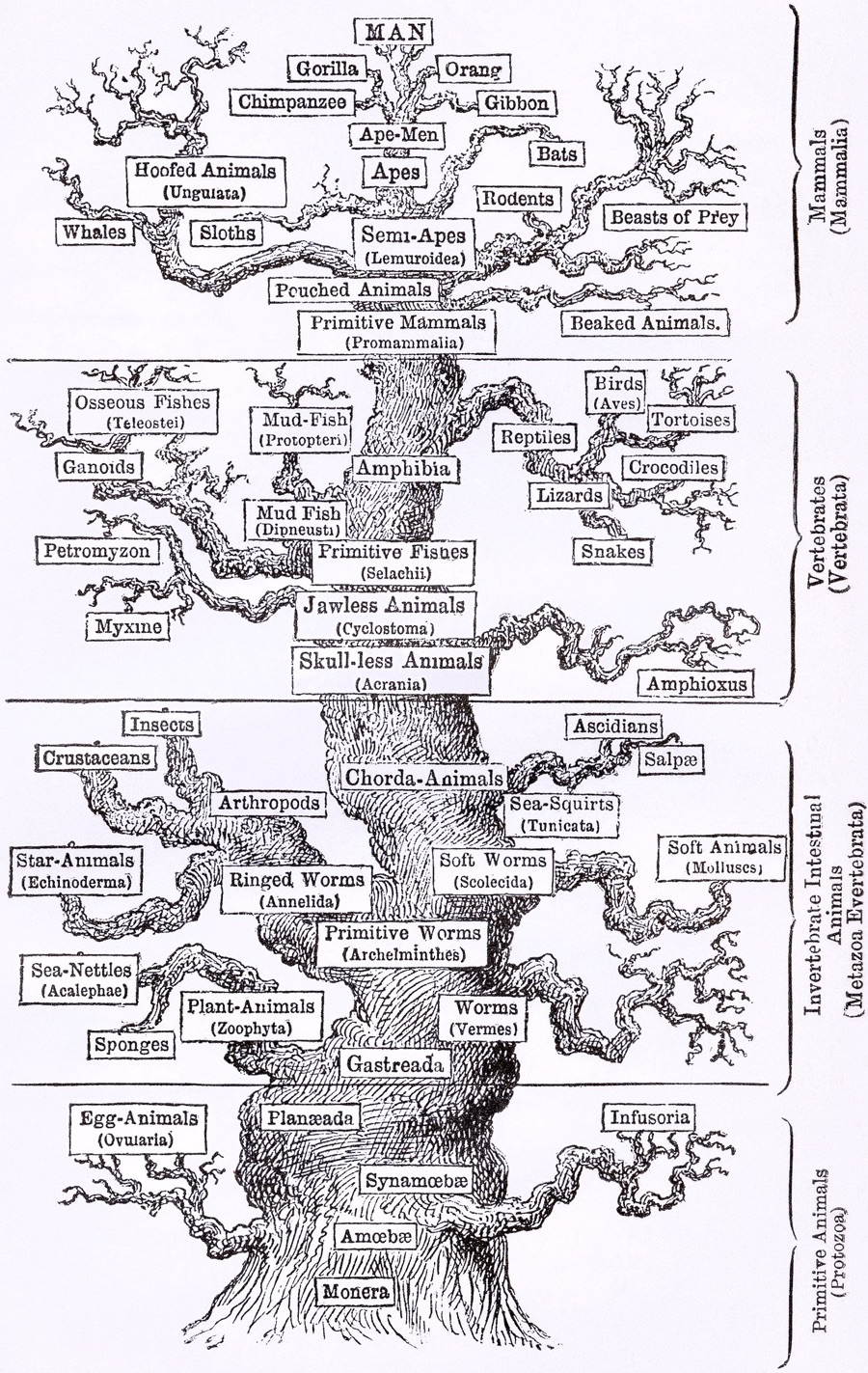
Phylogenies describe history
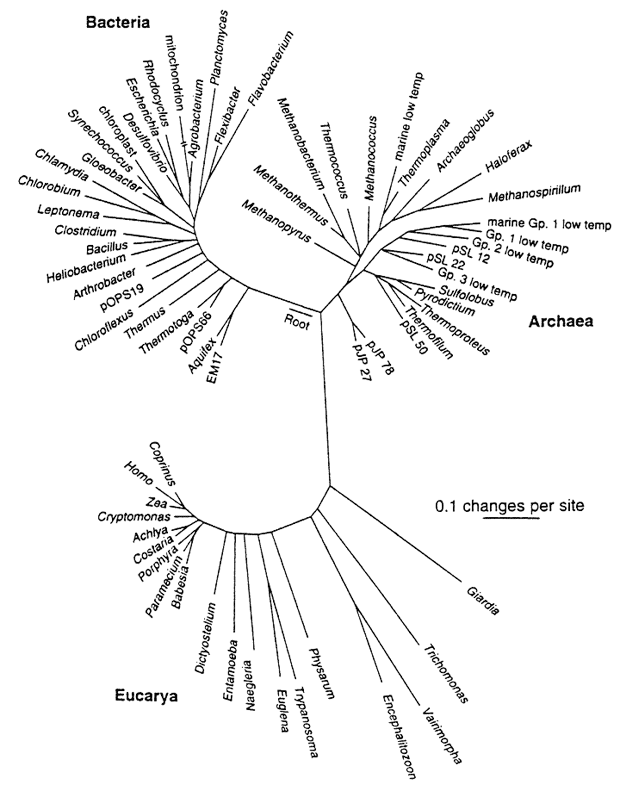
Phylogenies reveal process
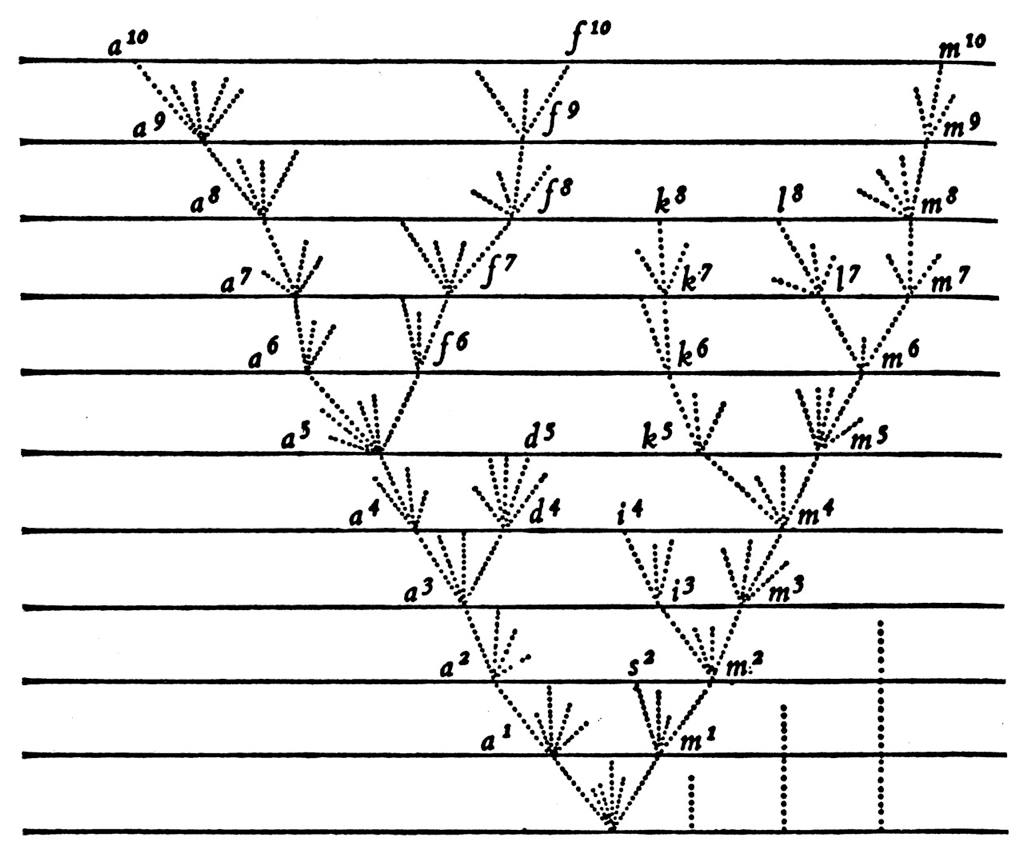
Demo
With source-sink dynamics, one deme is ancestral
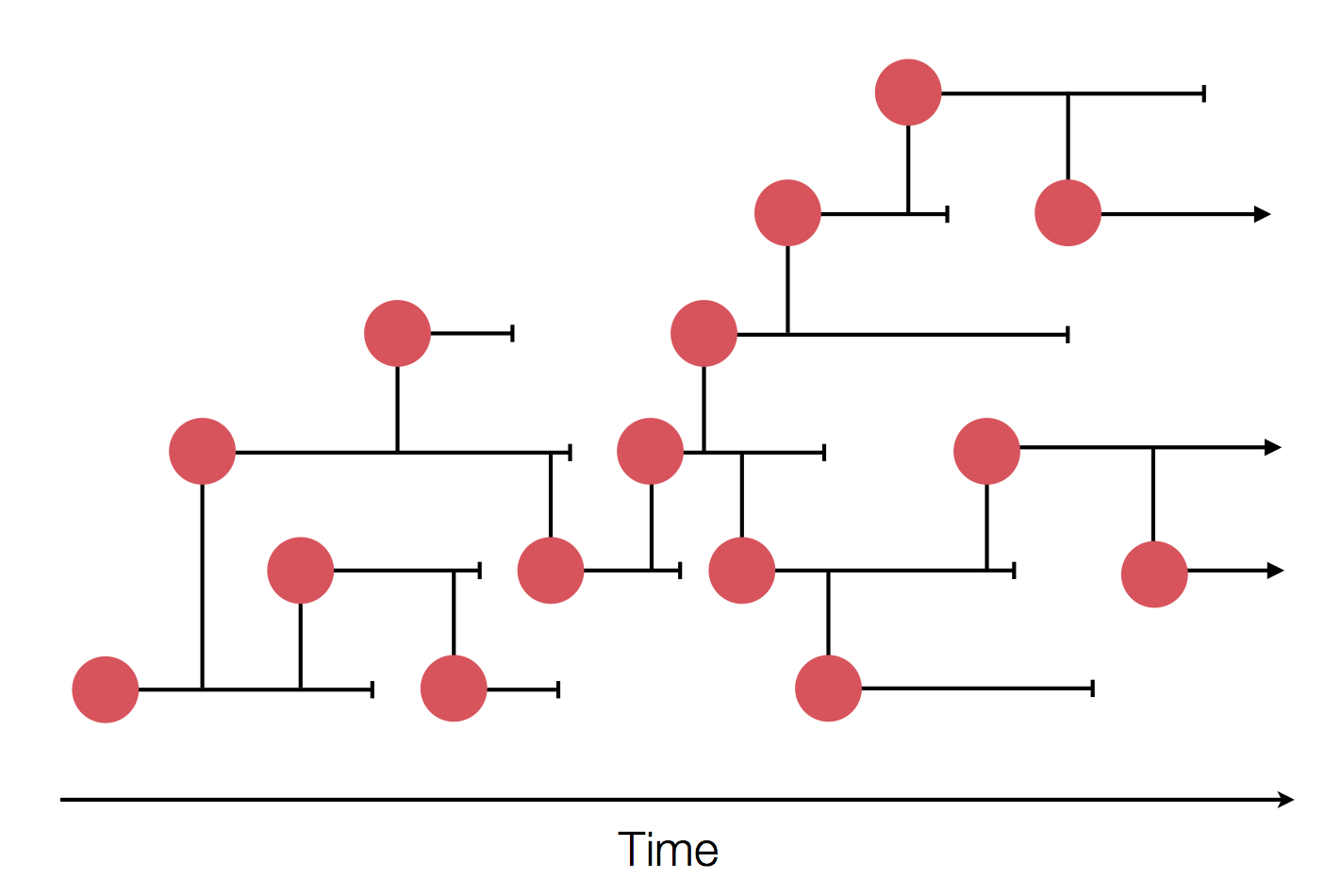
Epidemics
Epidemic process
Sample some individuals
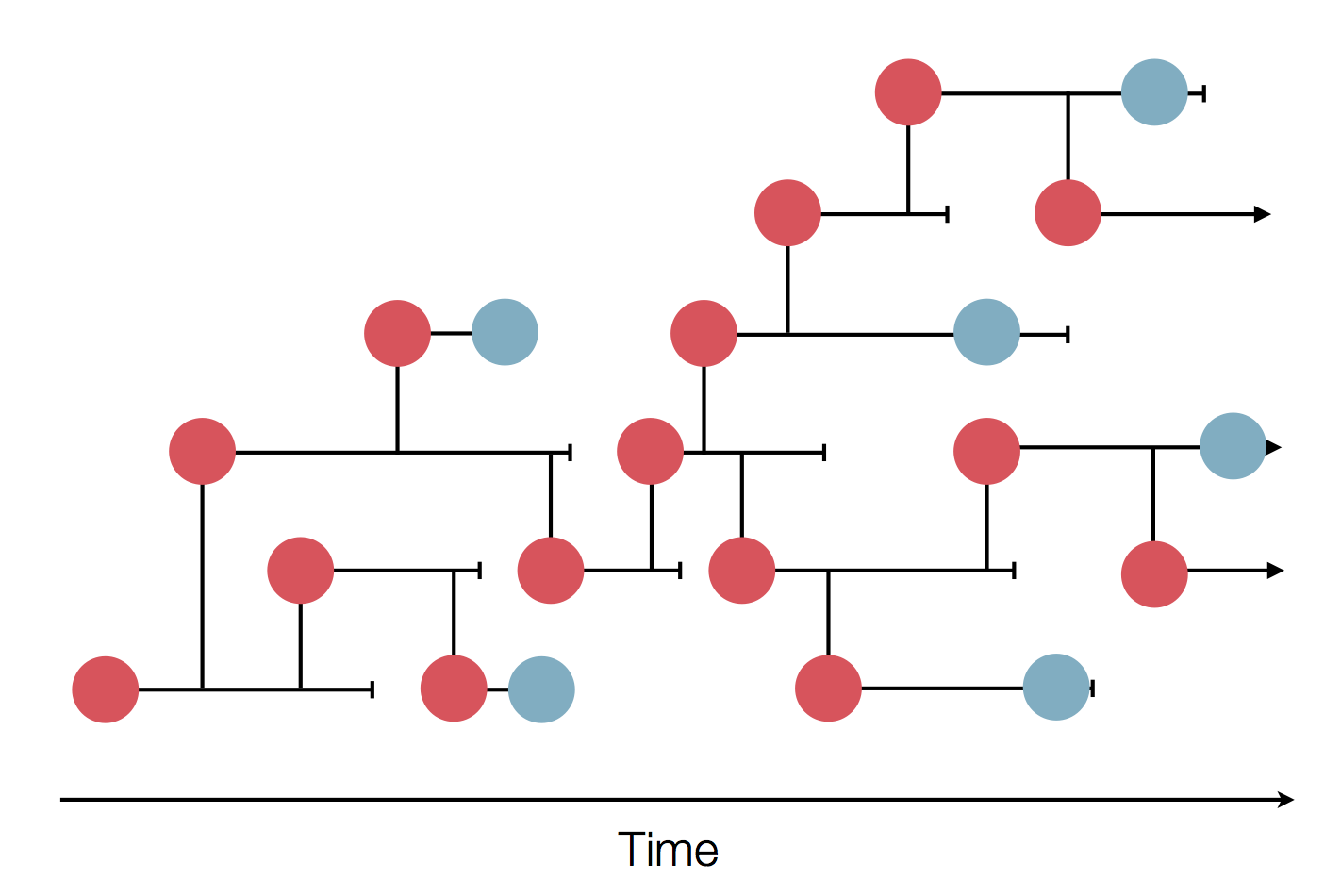
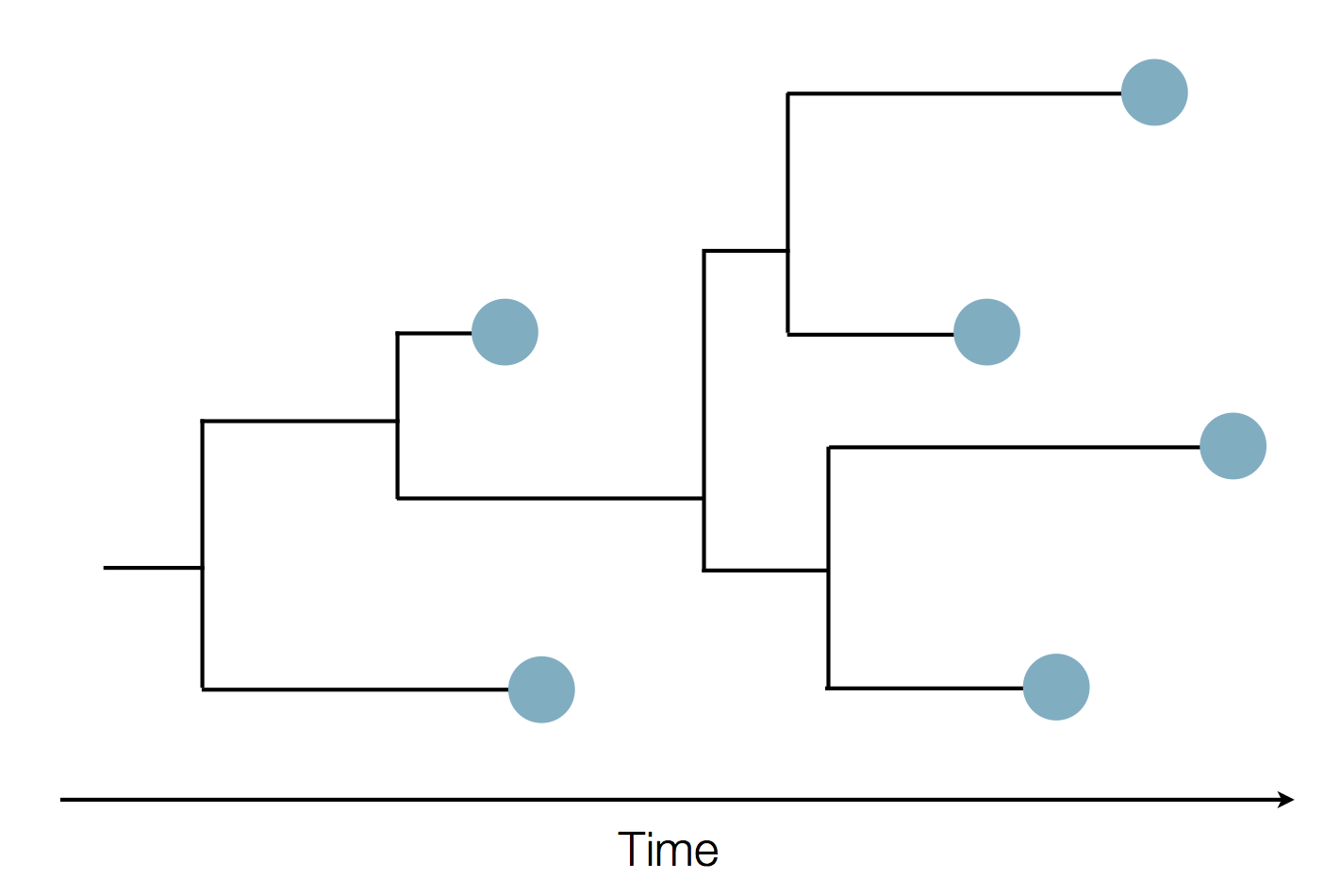
Sequence and determine phylogeny
Sequence and determine phylogeny
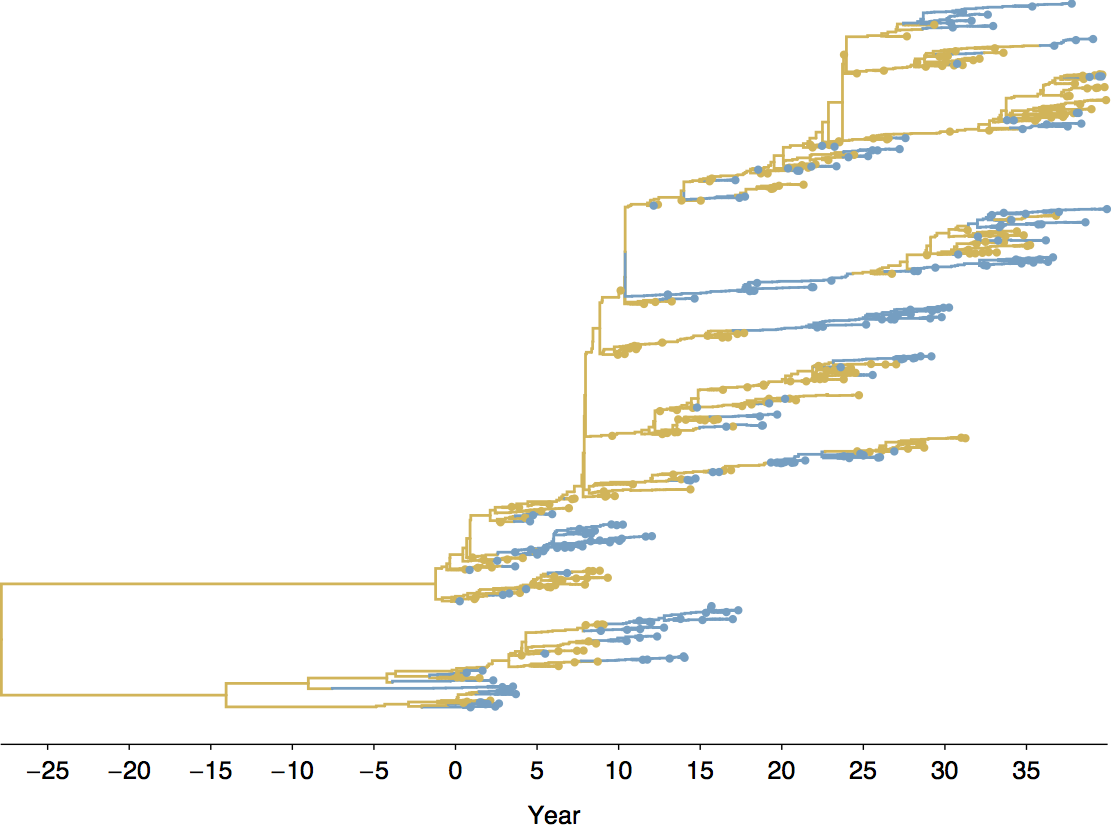
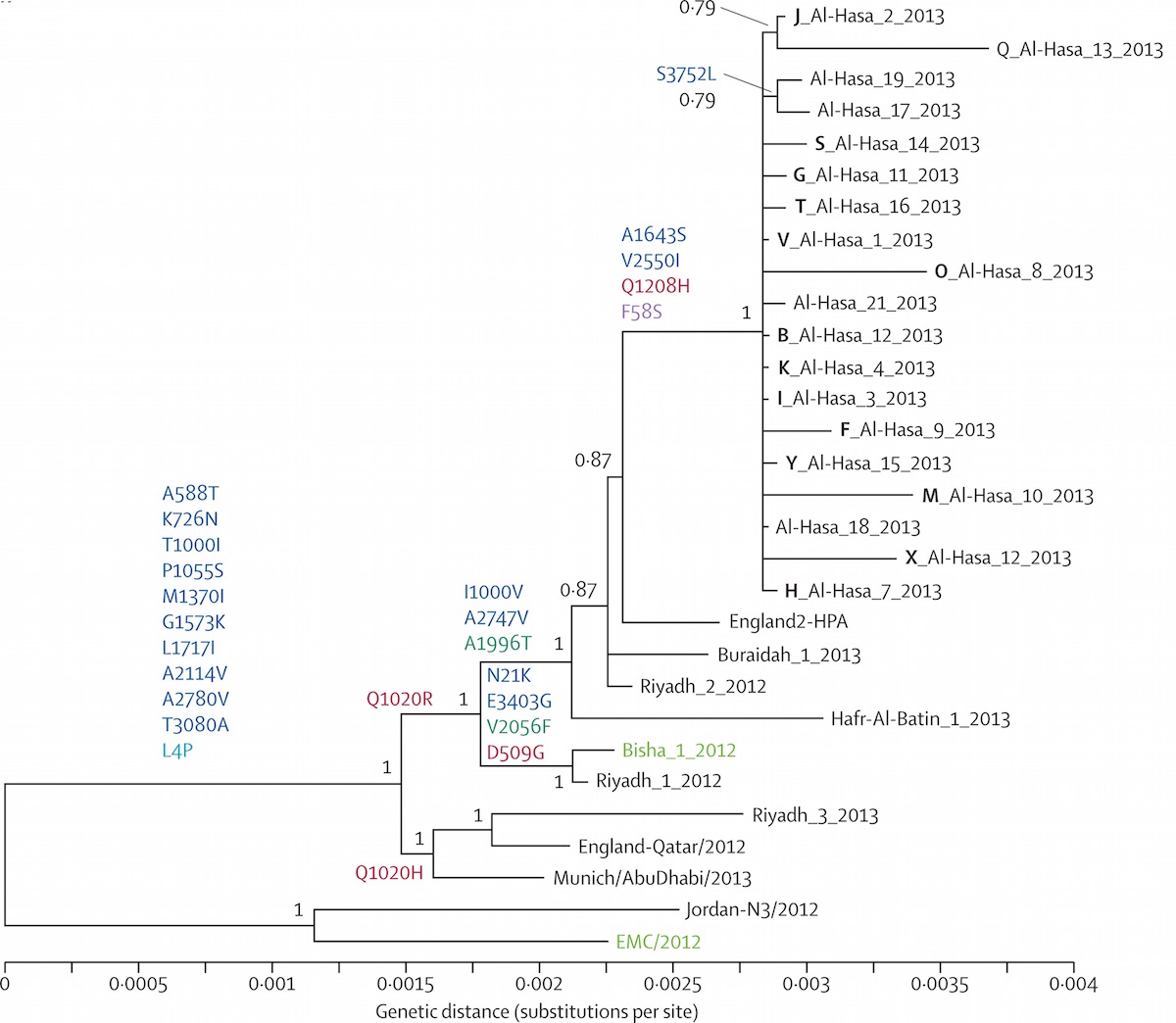
Middle Eastern MERS-CoV phylogeny
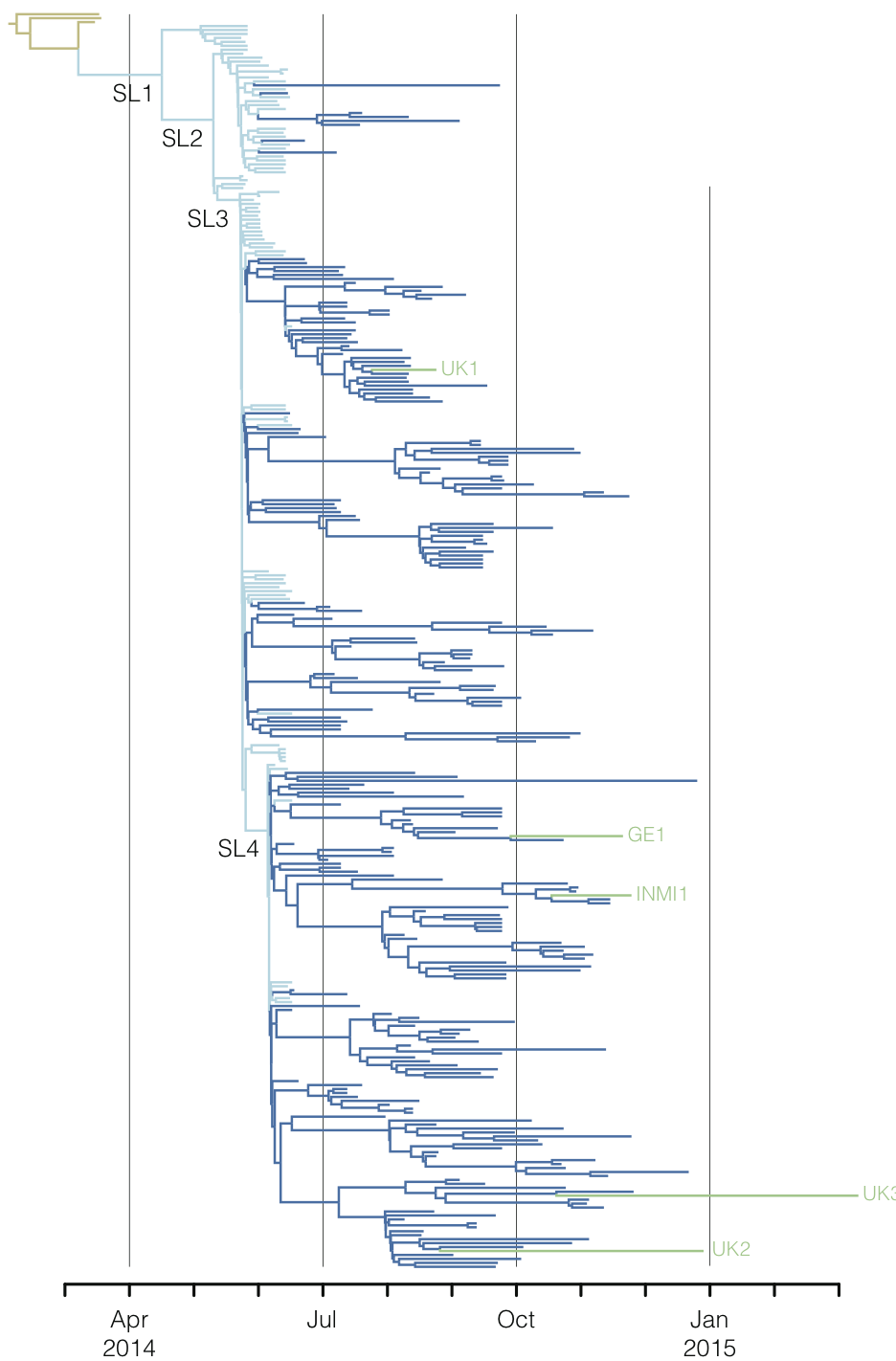
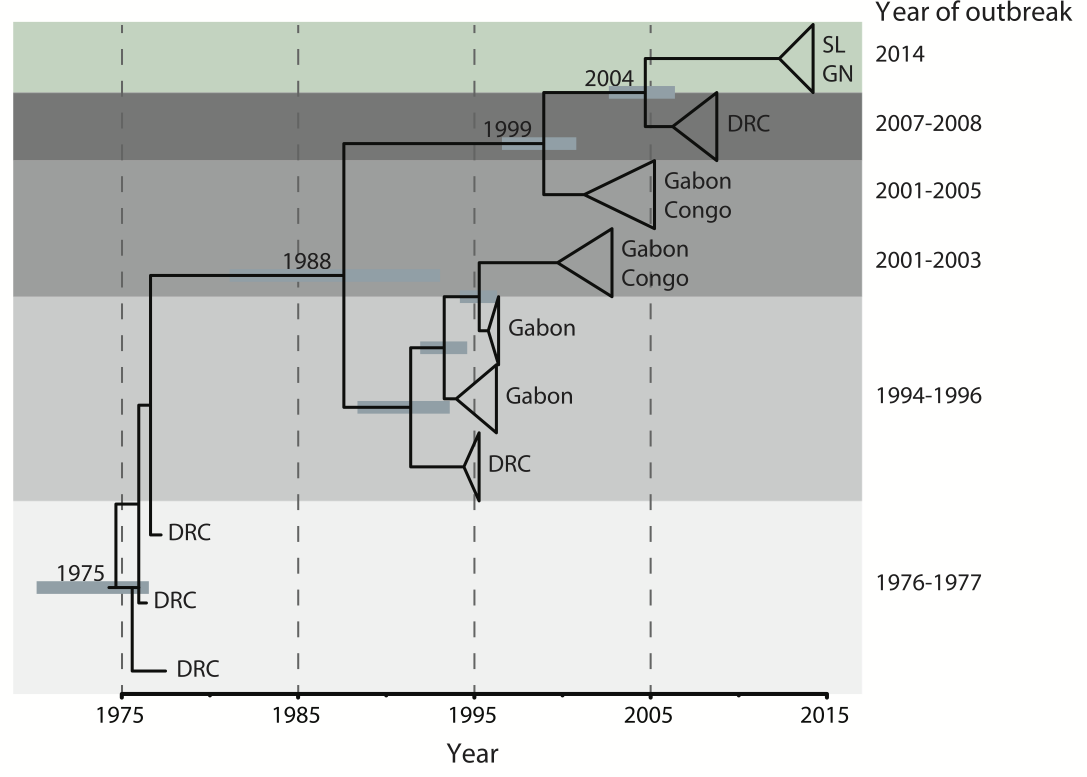
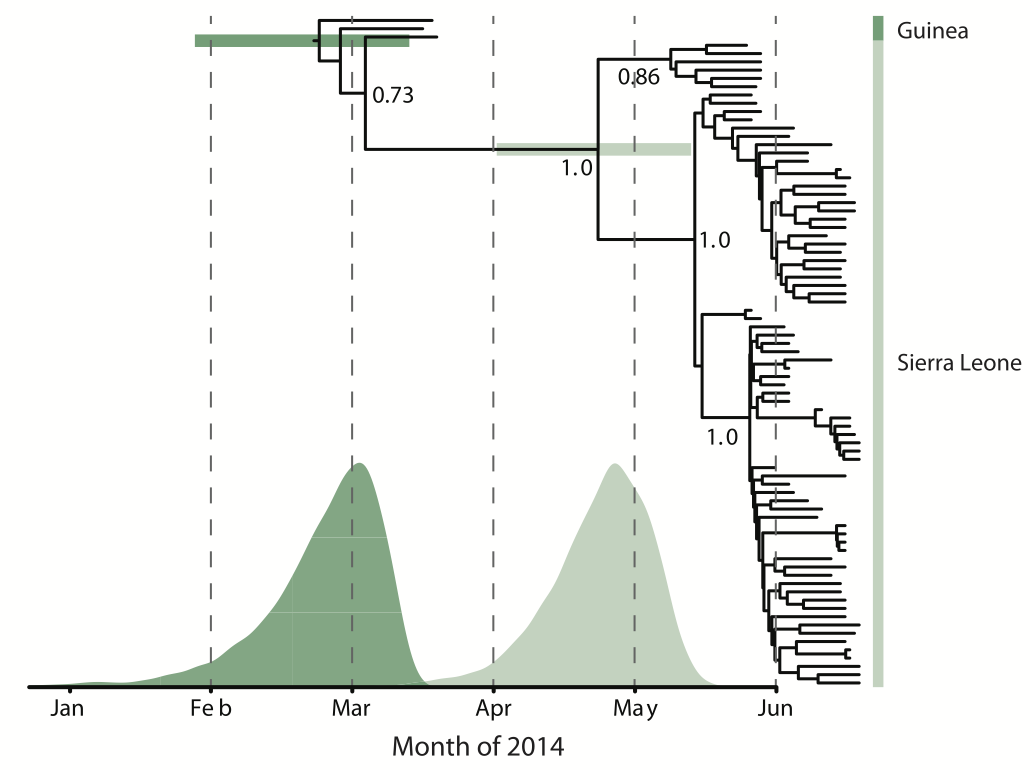
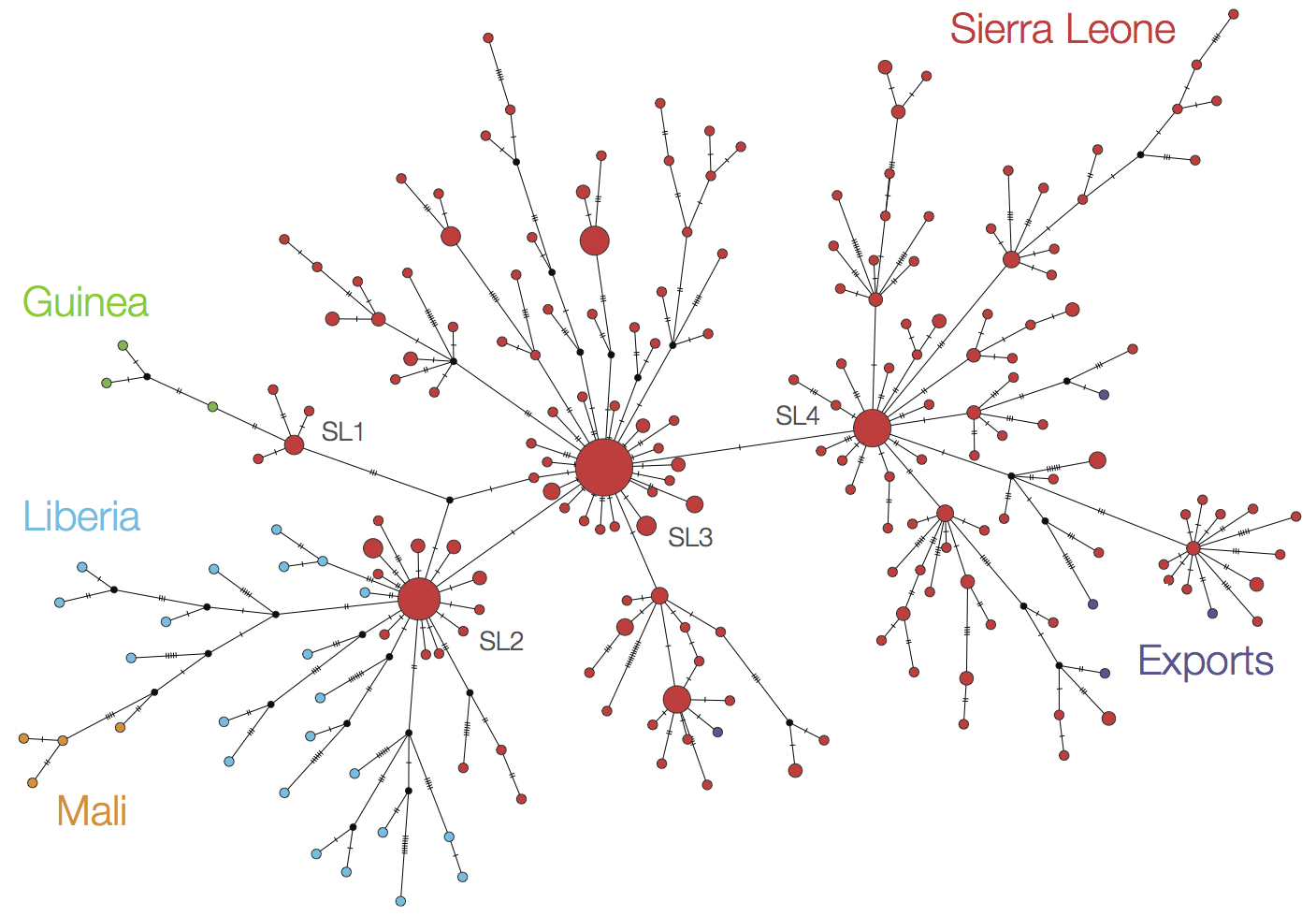
West African Ebola phylogeny
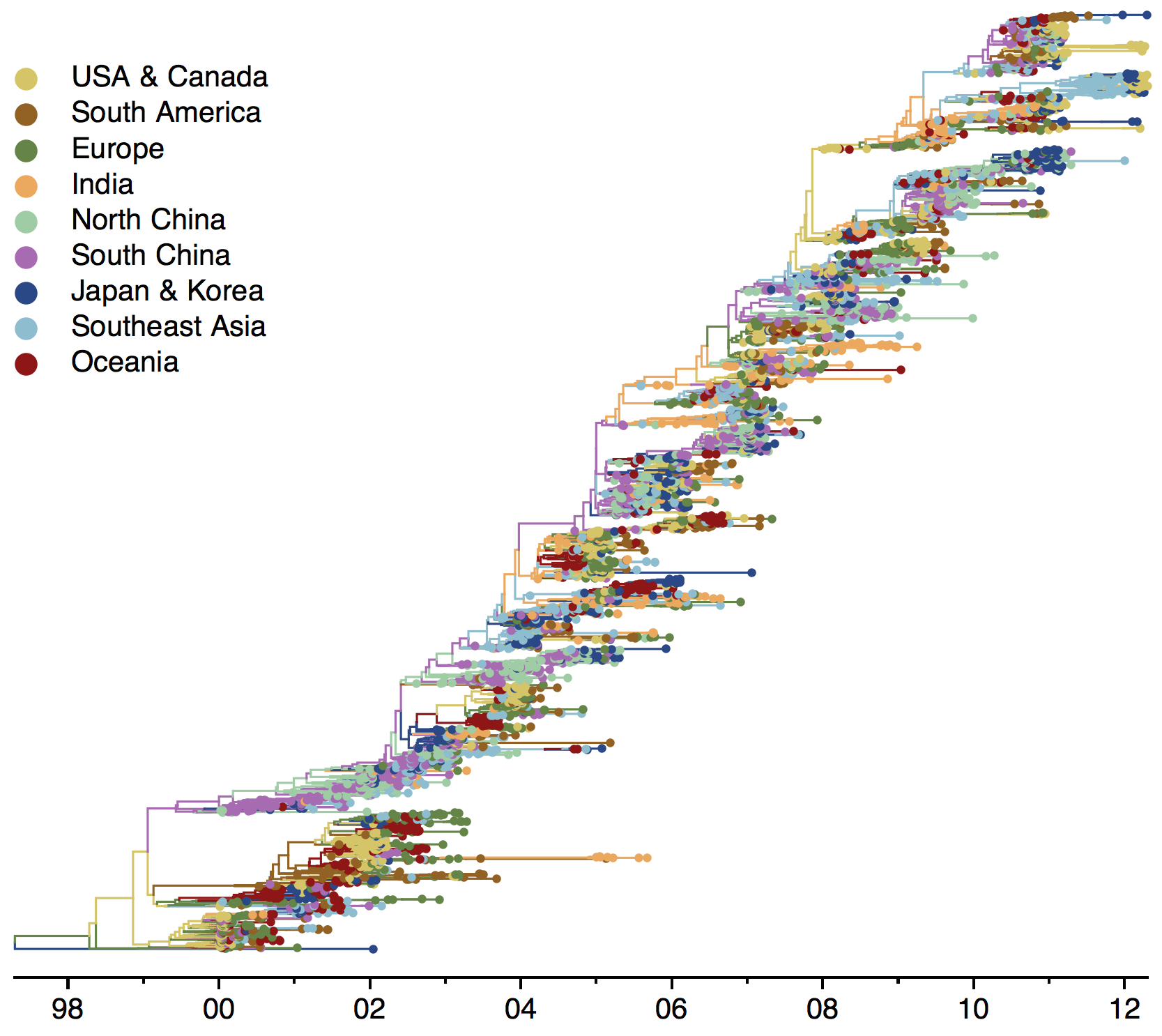
Global influenza phylogeny
Applications of evolutionary analysis for influenza vaccine strain selection and charting outbreak spread
Influenza
Influenza virion
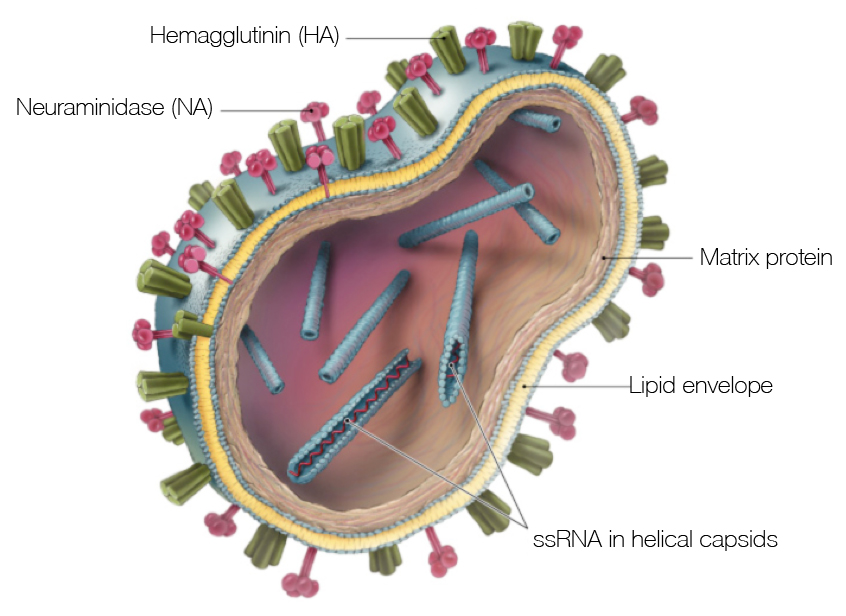
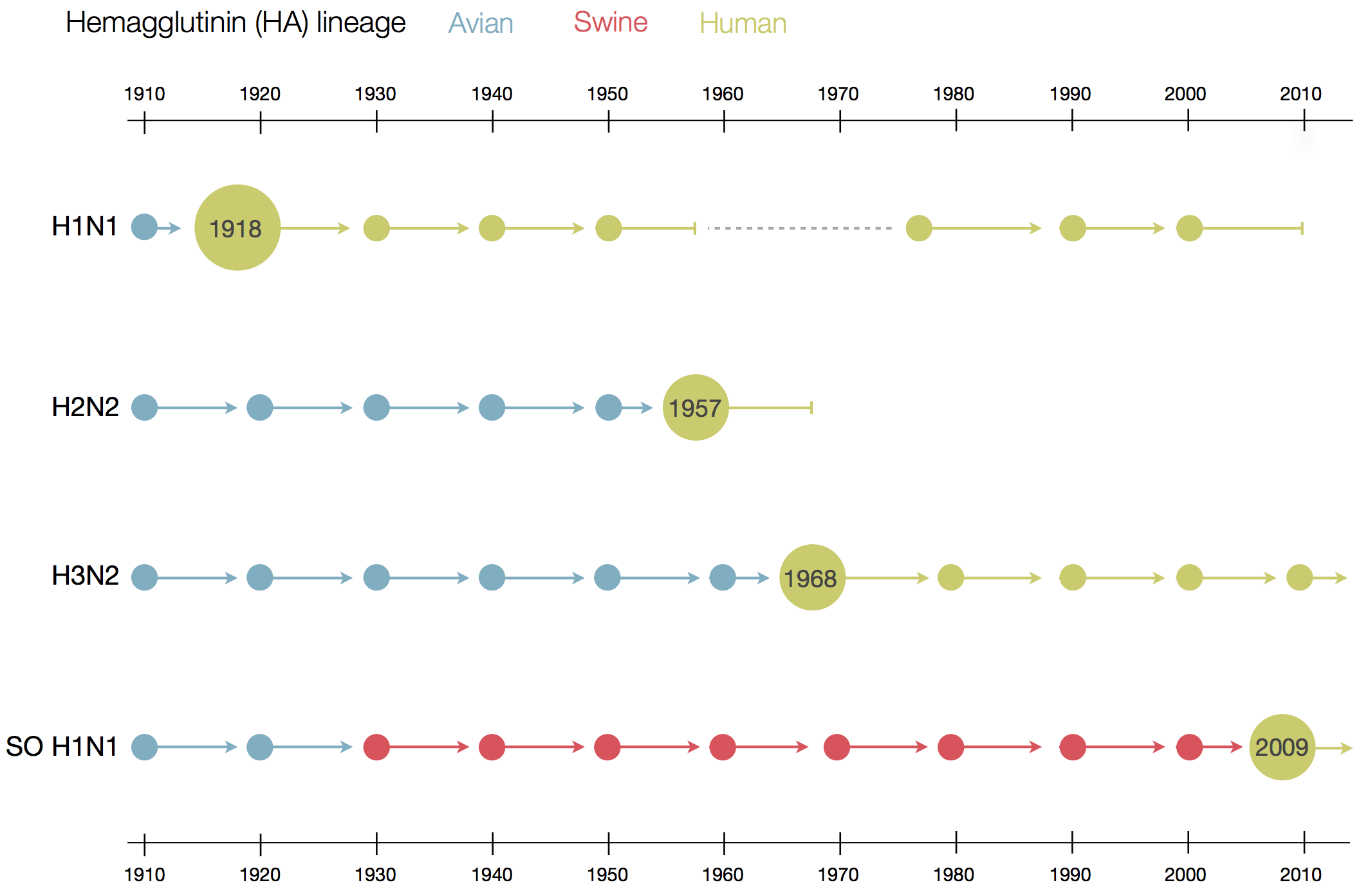
Flu pandemics caused by host switch events
Influenza B does not have pandemic potential
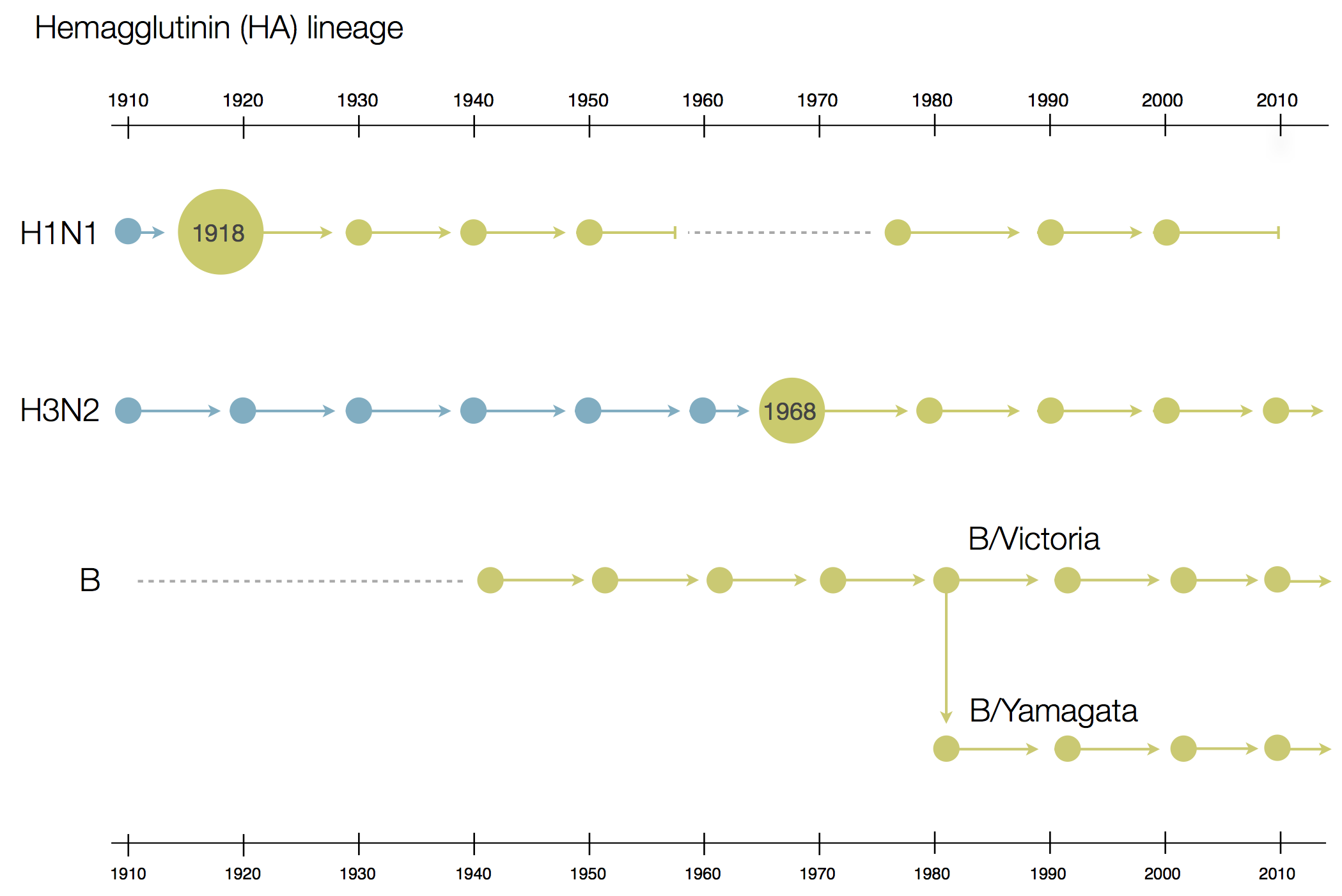
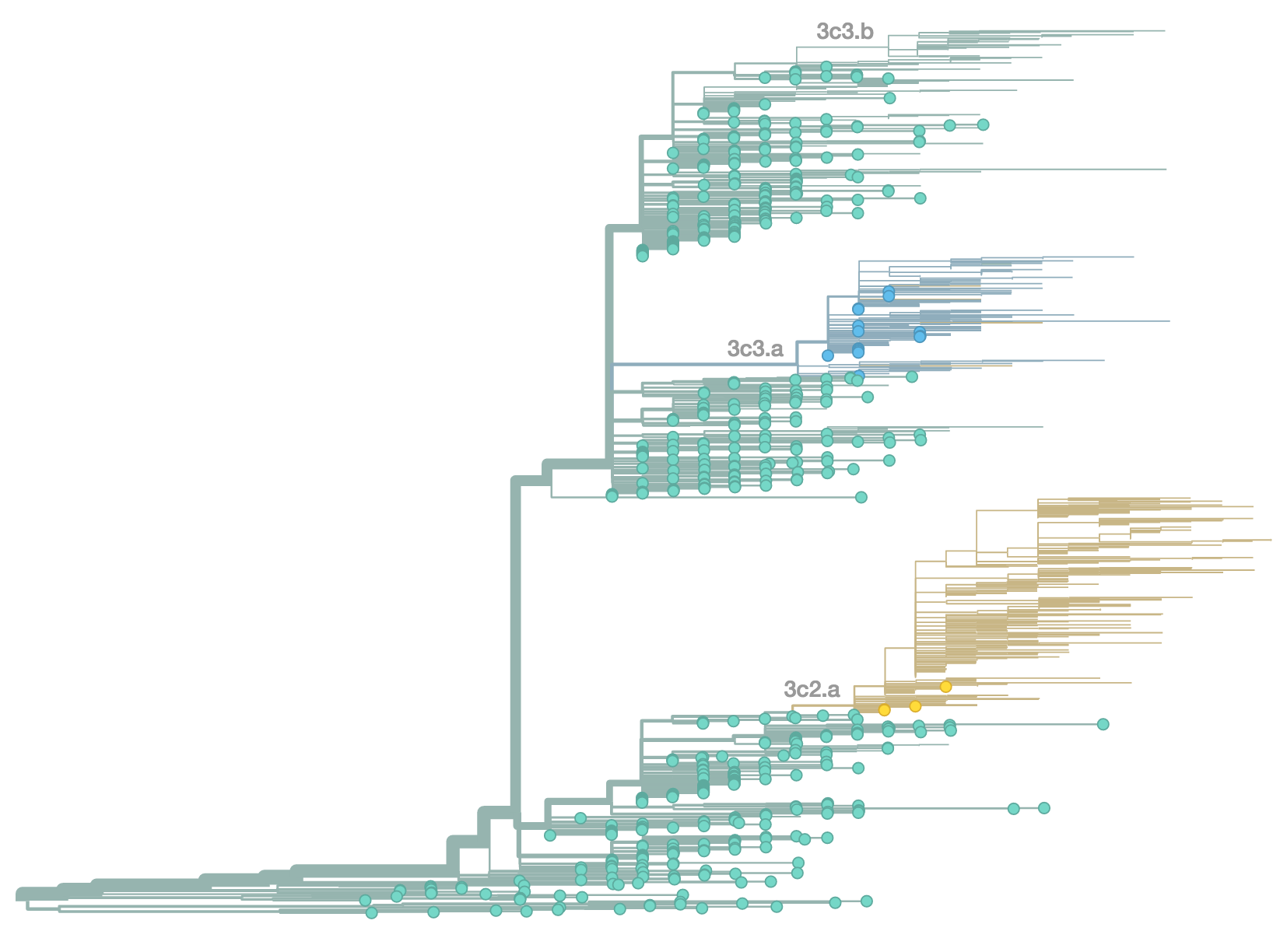
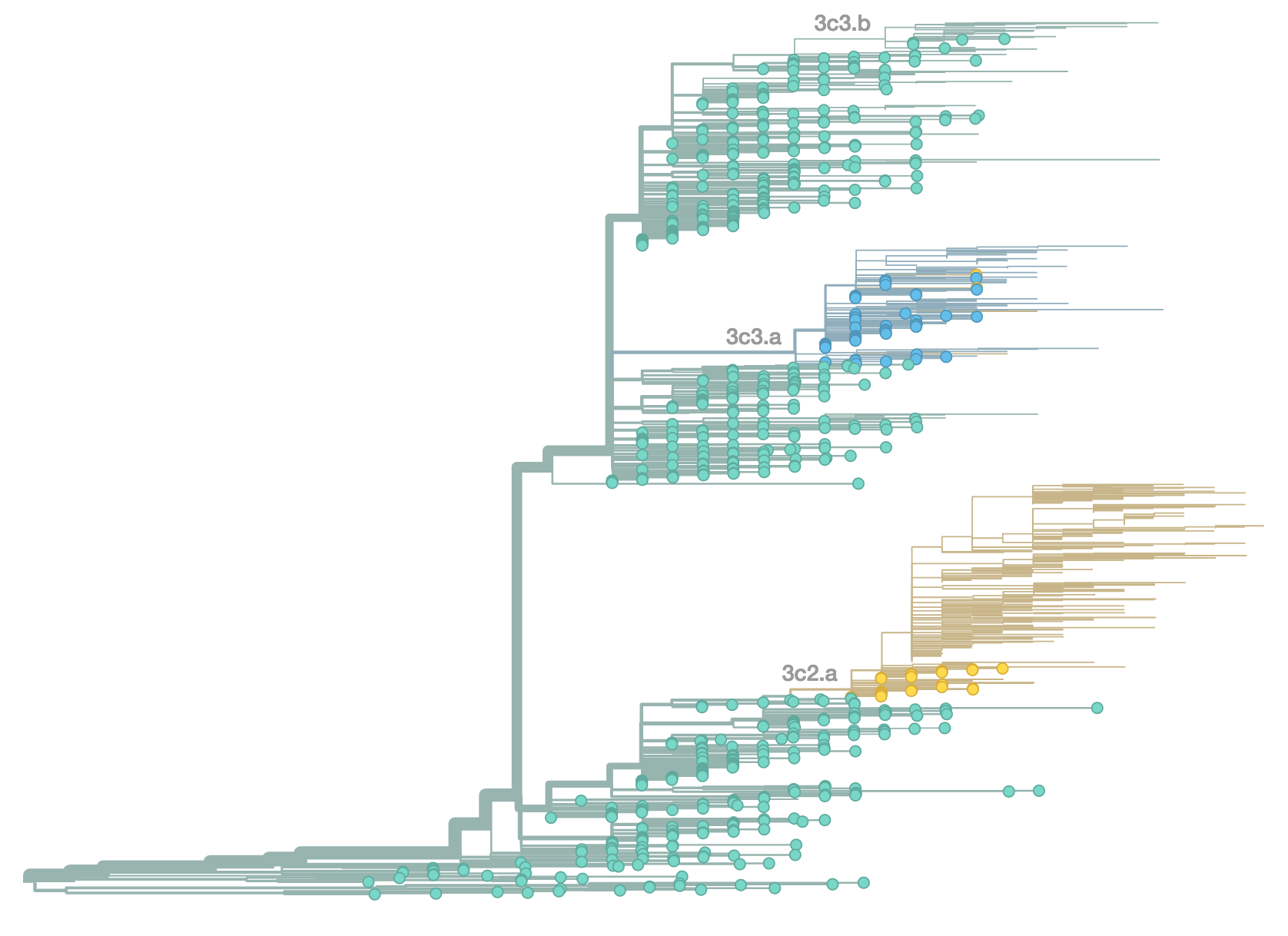
Phylogenetic trees of different influenza lineages
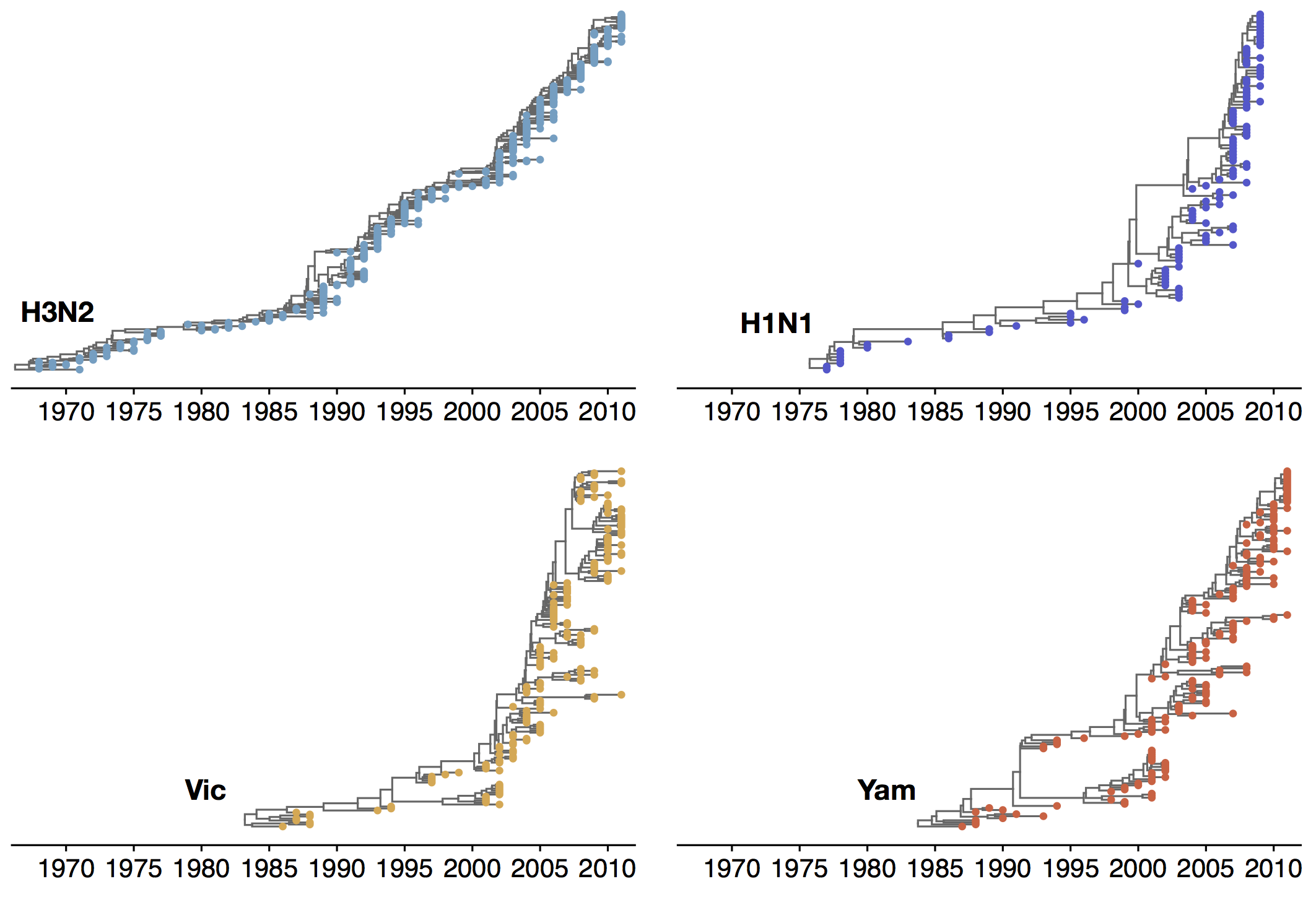
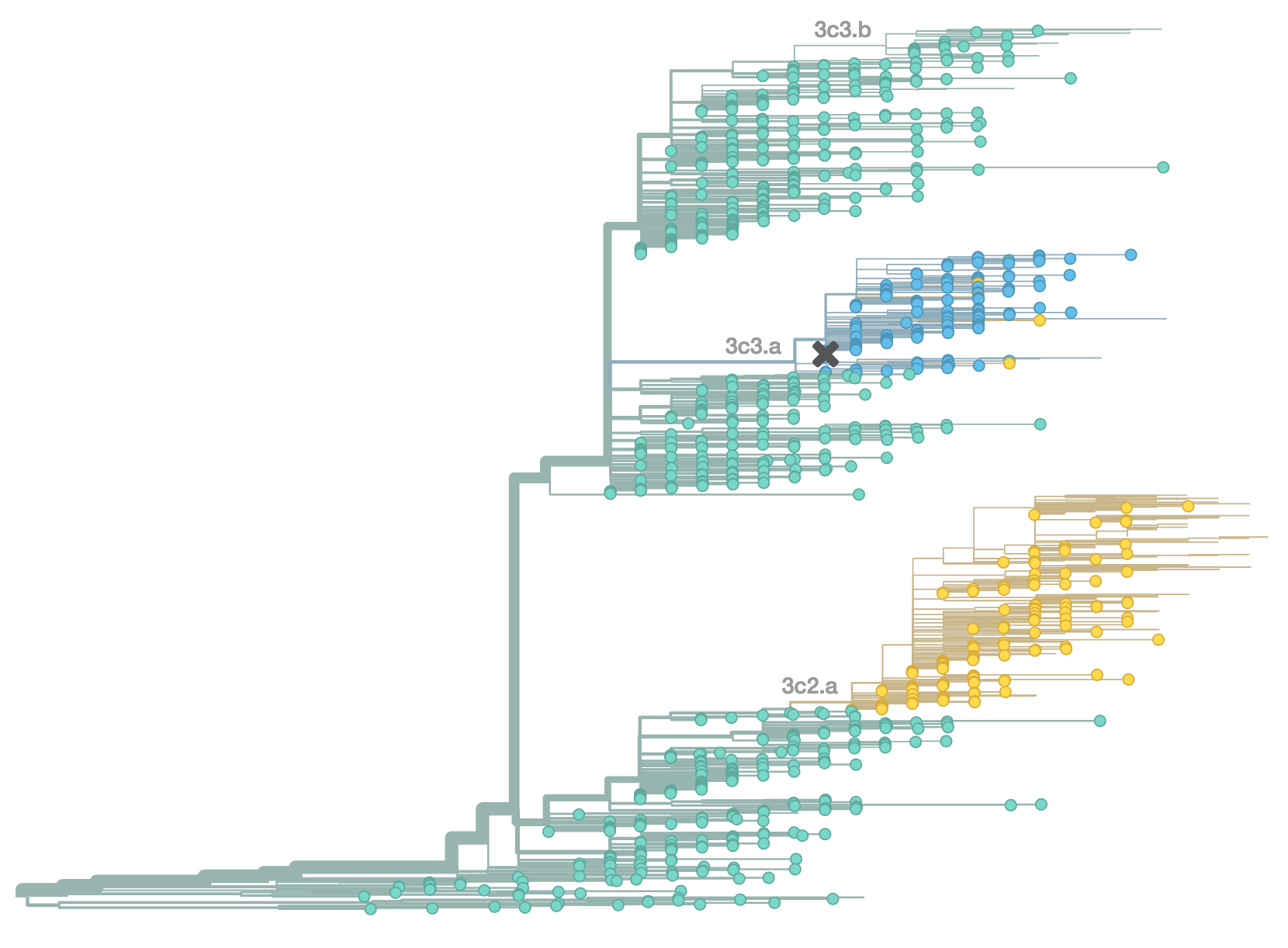
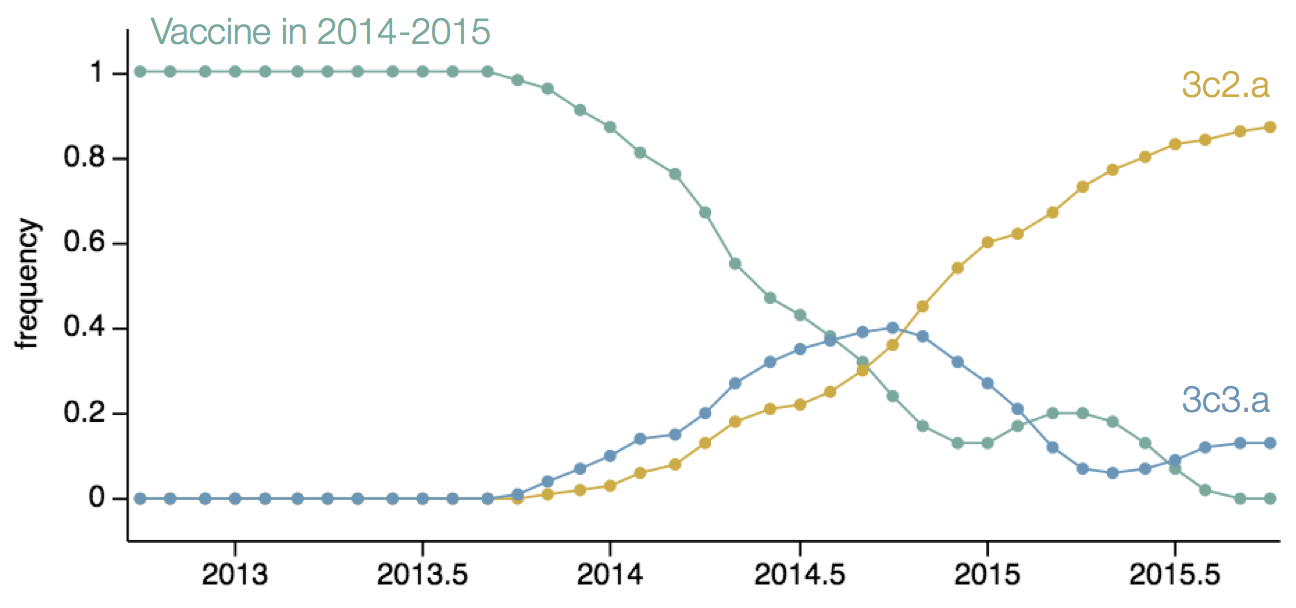
Antigenic evolution drives viral dynamics
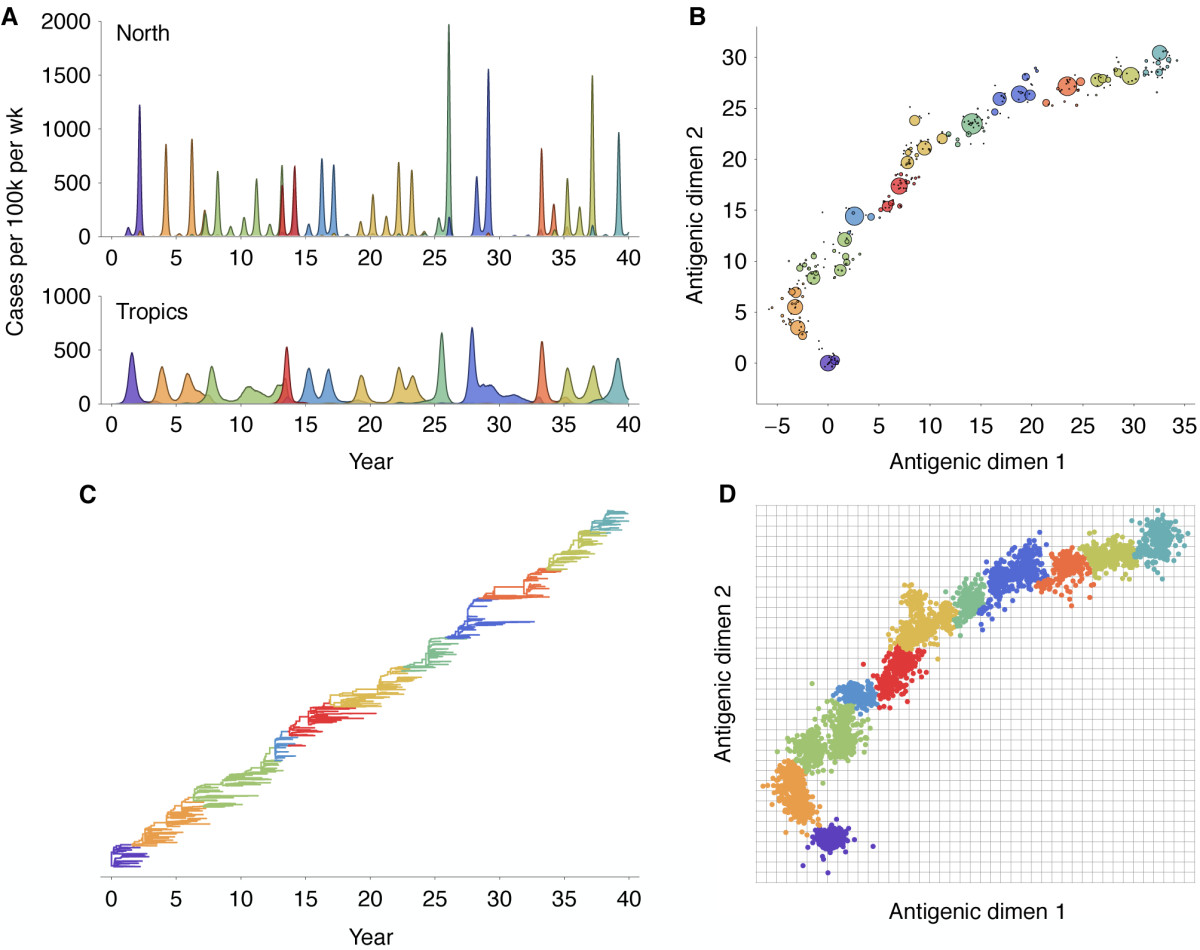
Antigenic evolution in H3N2
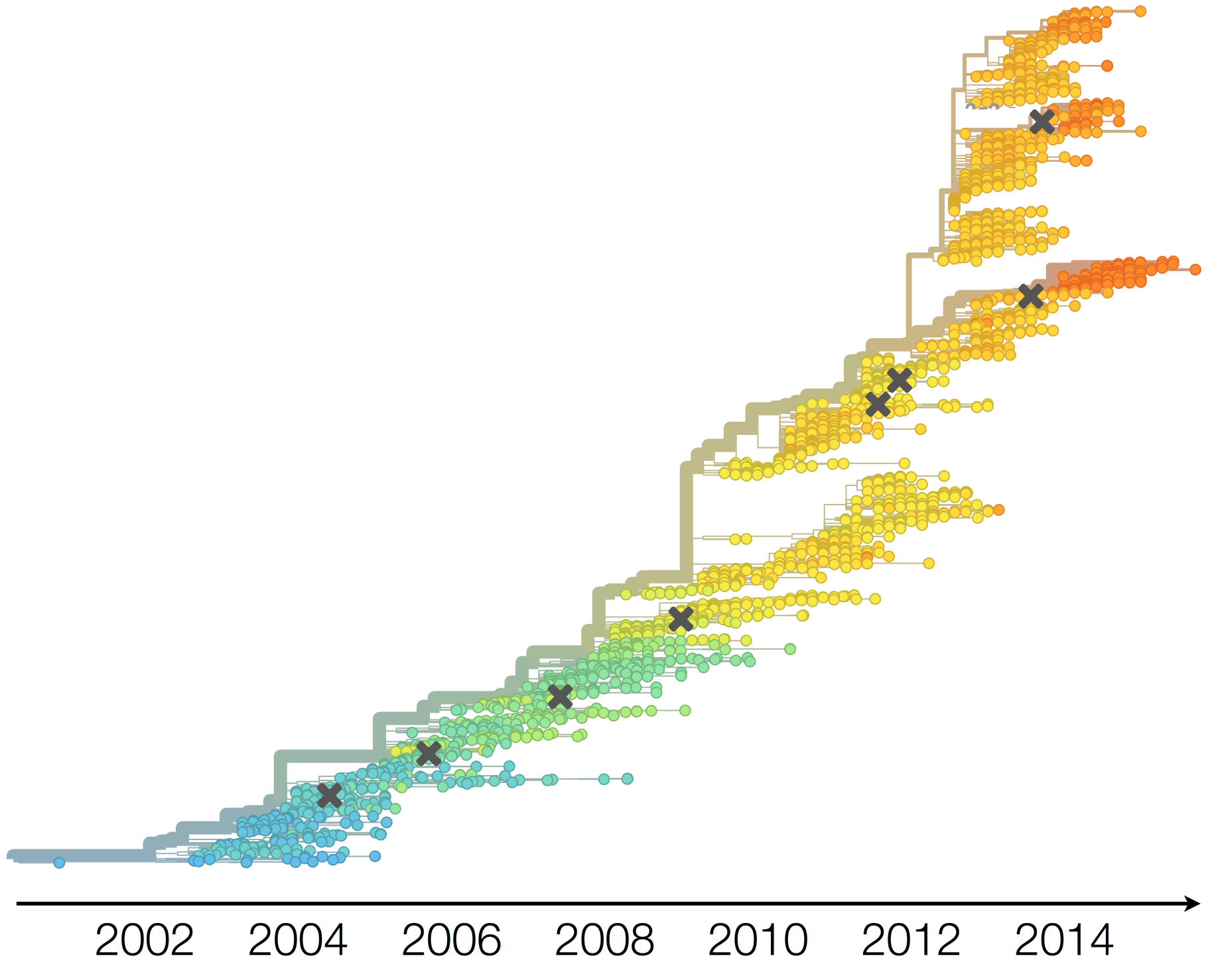
Influenza H3N2 vaccine updates
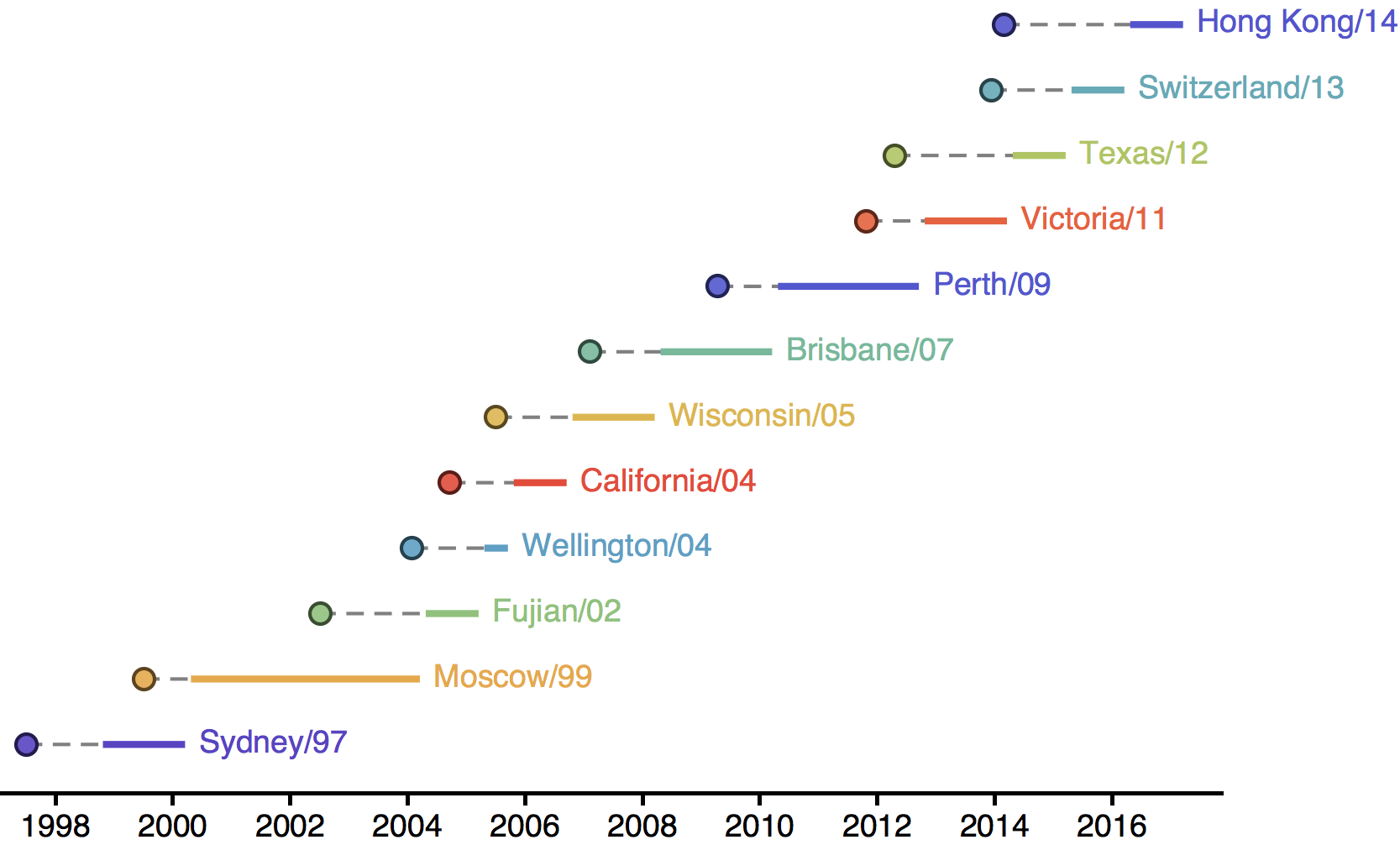
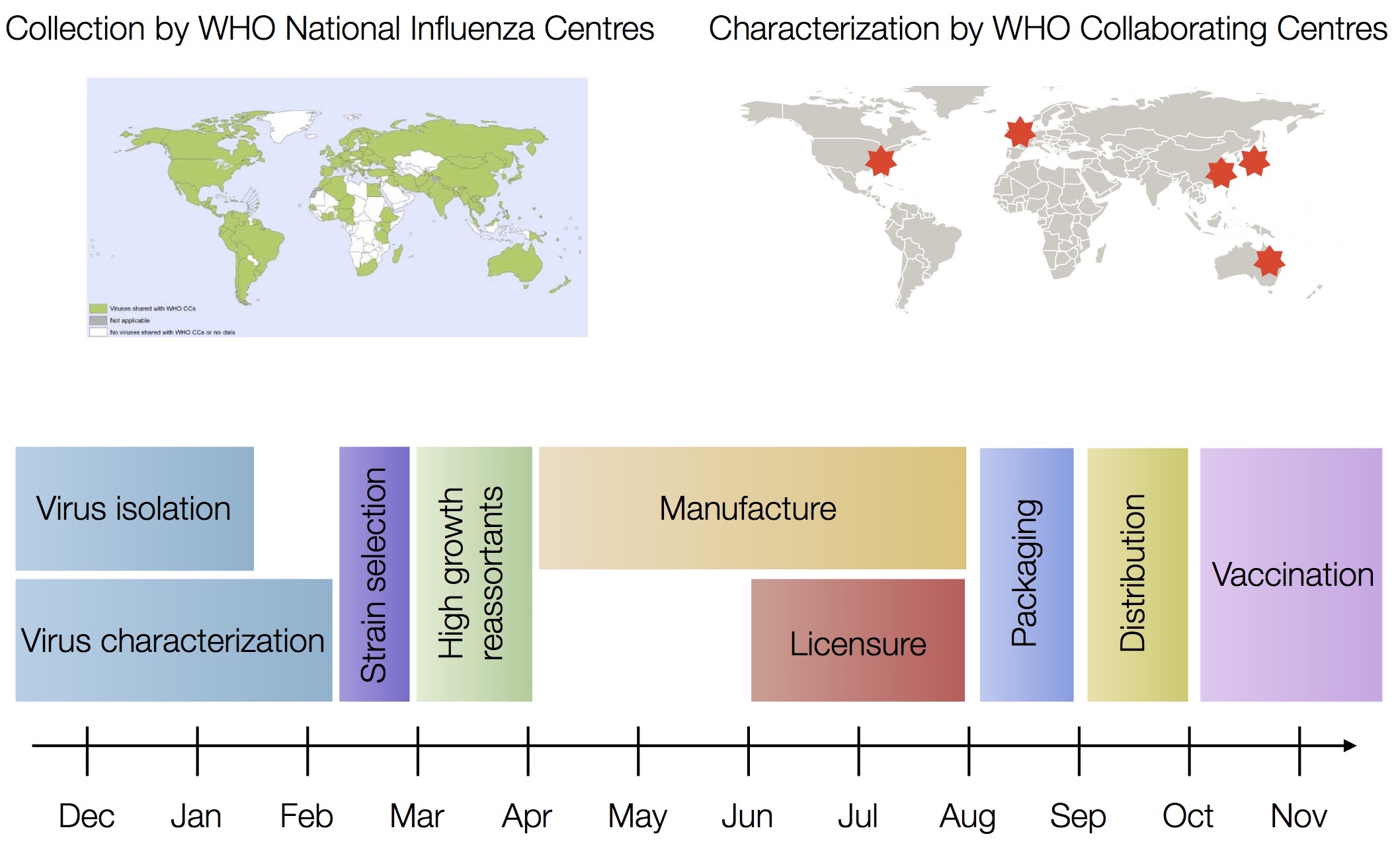
Vaccine strain selection timeline
Antigenic "match" of key importance
Antibodies by vaccination should effectively bind to circulating viruses. This requires:
- Identification of antigenically distinct clades of virus
- Prediction of clade growth/decline
Hemagglutination inhibition (HI) assays measure binding
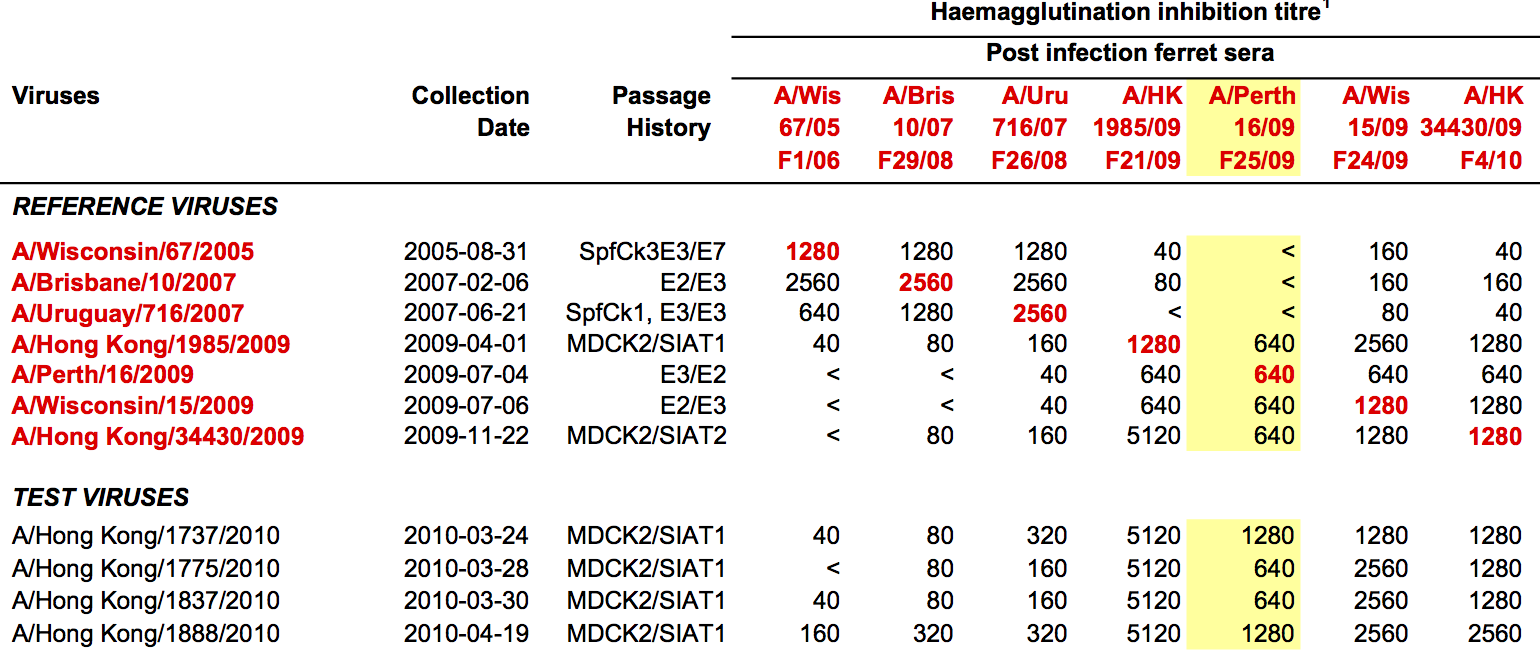
Antigenic cartography
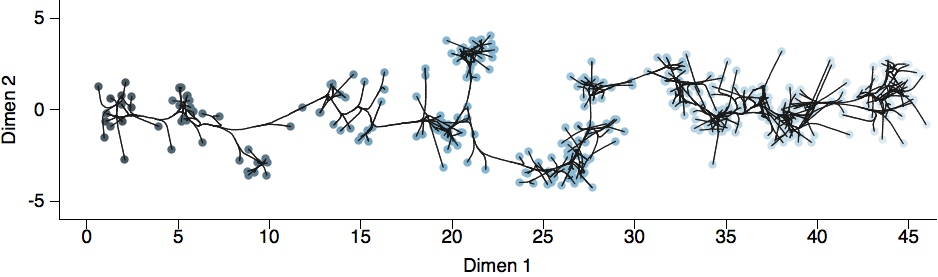
H3N2 population in Feb 2014
H3N2 population in Jun 2014
H3N2 population in Oct 2014
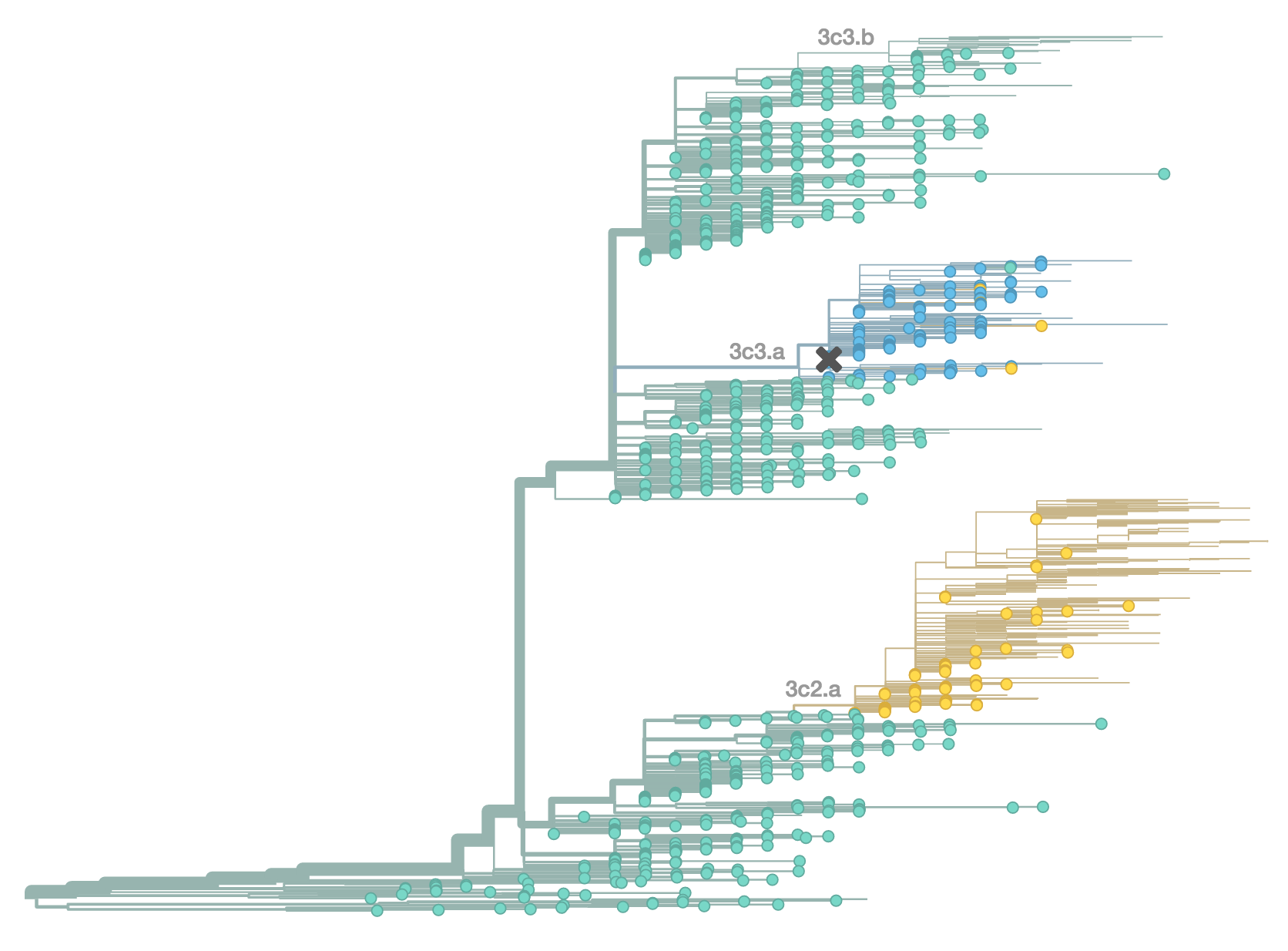
H3N2 population in Feb 2015
Resulted in a mismatched 2014-2015 vaccine
nextflu
Project to provide a real-time view of the evolving influenza population
All in collaboration with Richard Neher

nextflu pipeline
- Download all recent HA sequences from GISAID
- Filter to remove outliers
- Align sequences
- More filtering
- Build tree
- Estimate frequencies
- Export JSON for visualization
nextflu.org
Predictive models
A simple predictive model estimates the fitness $f$ of virus $i$ as
$$\hat{f}_i = \beta^\mathrm{ep} \, f_i^\mathrm{ep} + \beta^\mathrm{ne} \, f_i^\mathrm{ne}$$
where $f_i^\mathrm{ep}$ measures cross-immunity via substitutions at epitope sites and $f_i^\mathrm{ep}$ measures mutational load via substitutions at non-epitope sites.
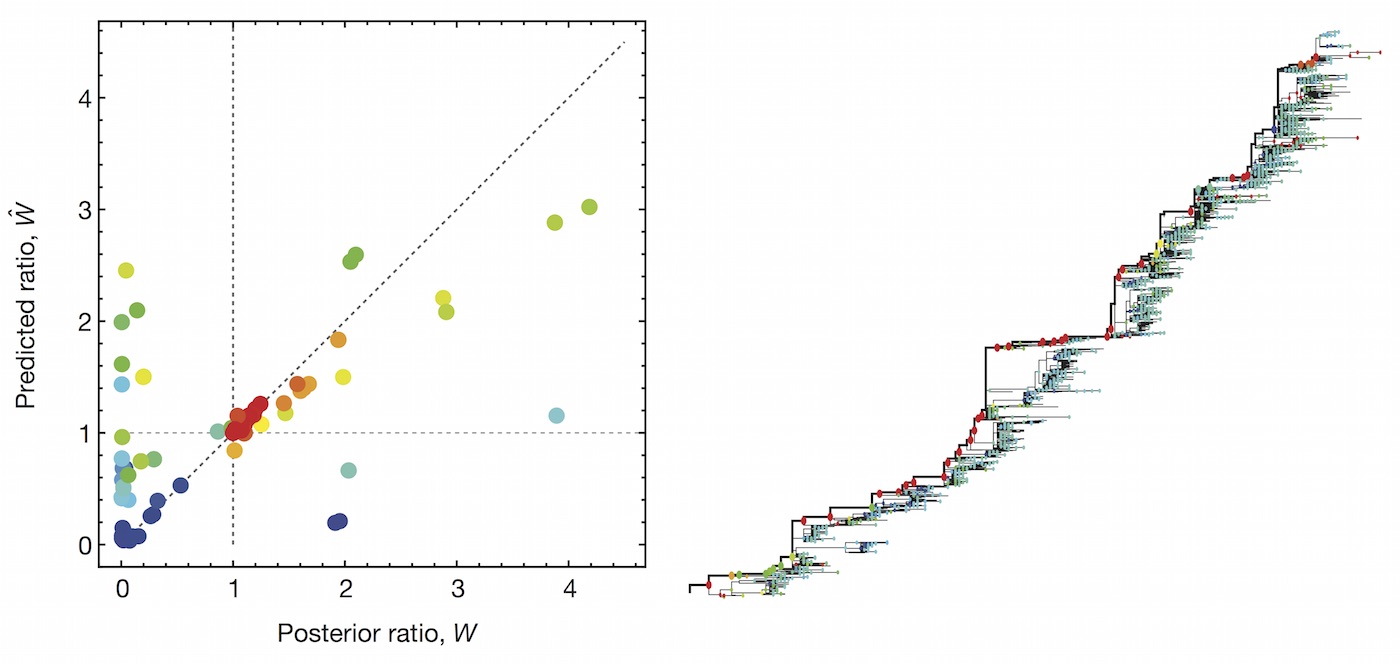
Predictive models
Another approach quantifies phylogenetic branching patterns
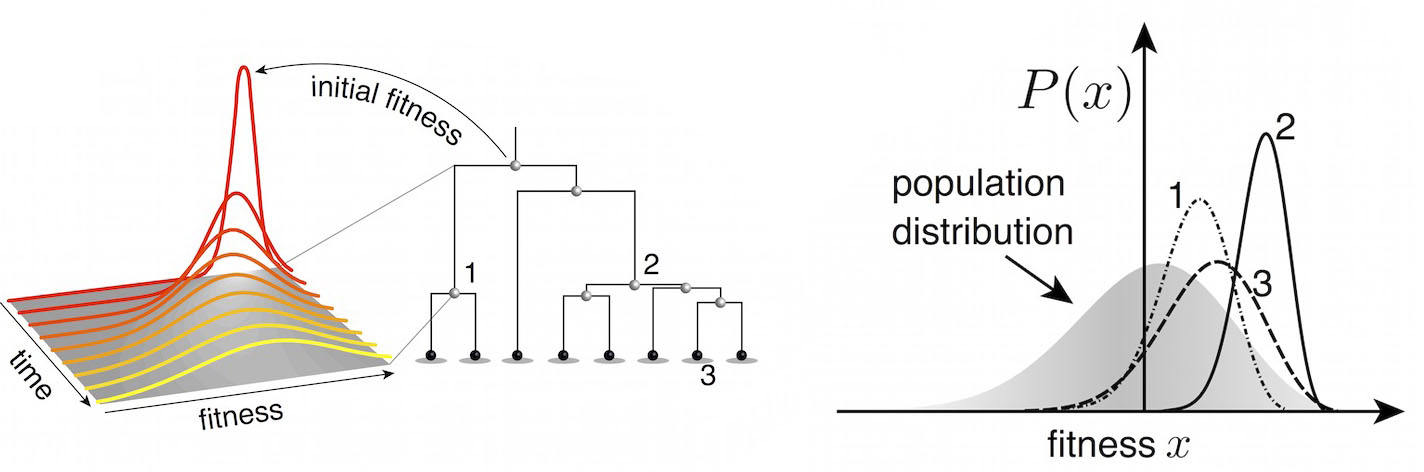
We're now working to include quantitative predictions of future clade behavior in nextflu
And also including other predictors, like geography:
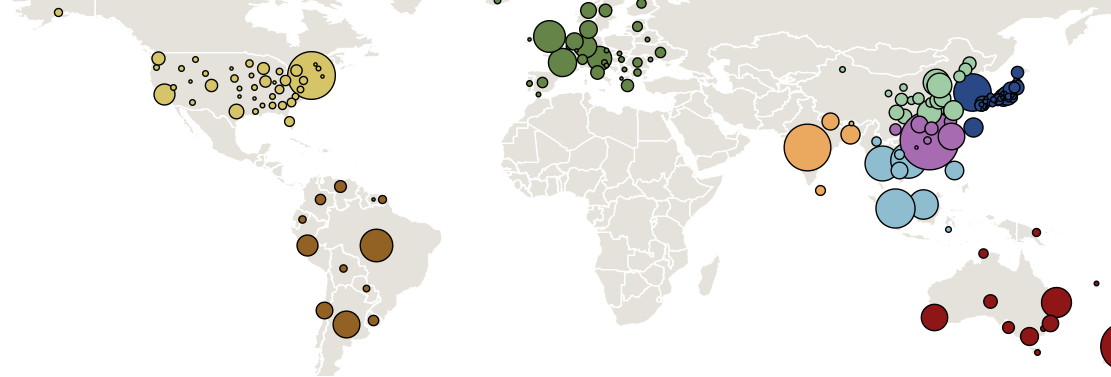
Global influenza phylogeny

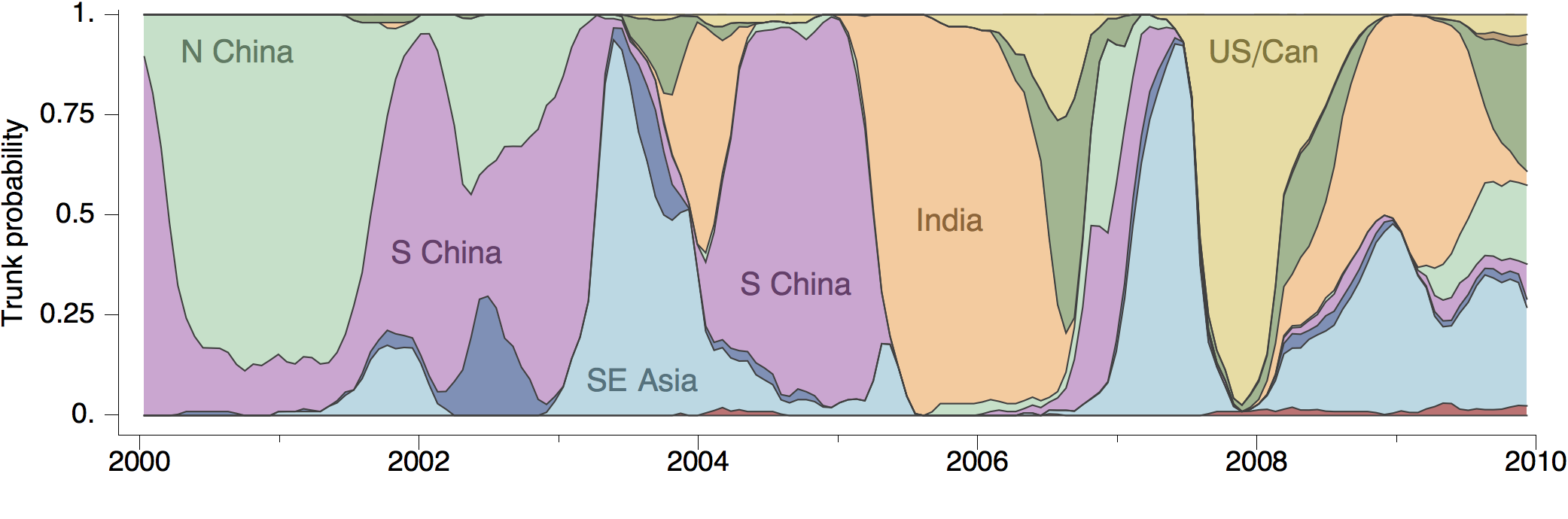
Track geographic transition events
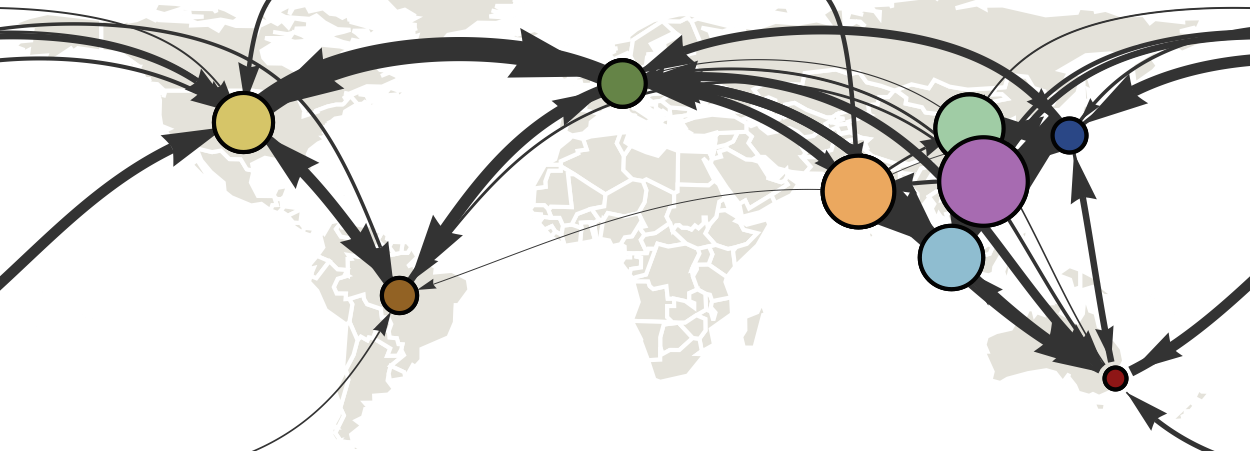
Global influenza circulation shows source-sink dynamics
Evolutionary analyses can inform influenza vaccine strain selection
Ebola
Epidemic nearly contained, but resulted in >28,000 confirmed cases and >11,000 deaths
Outbreaks are independent spillovers from the animal reservoir
Person-to-person spread in the early West African outbreak
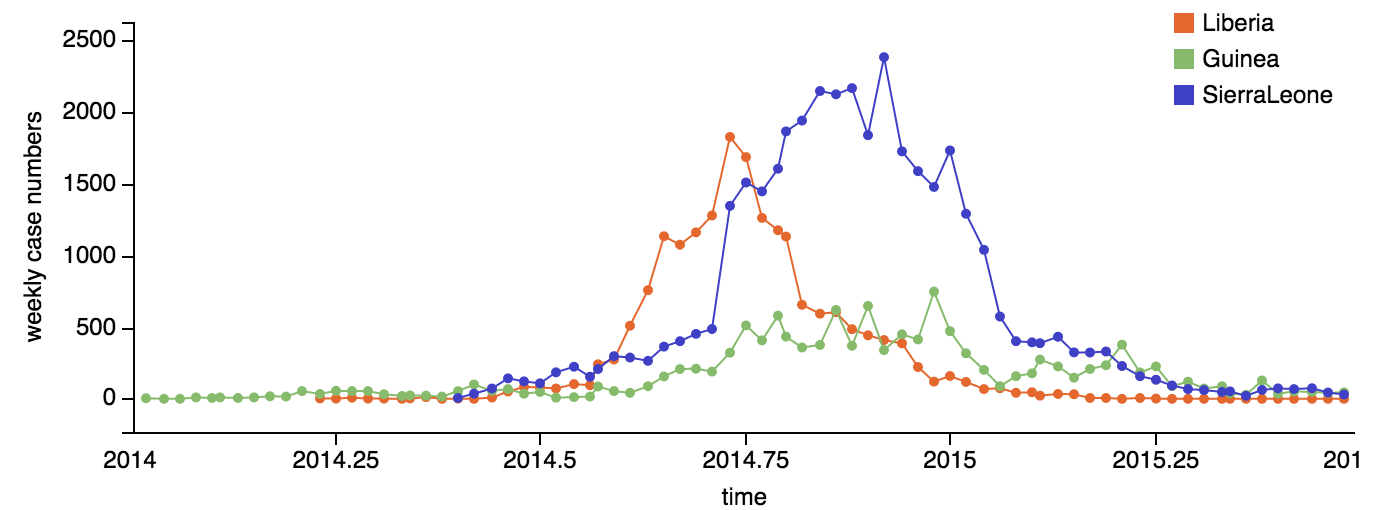
Continued spread through Dec 2014
At epidemic height, geographic spread of particular interest
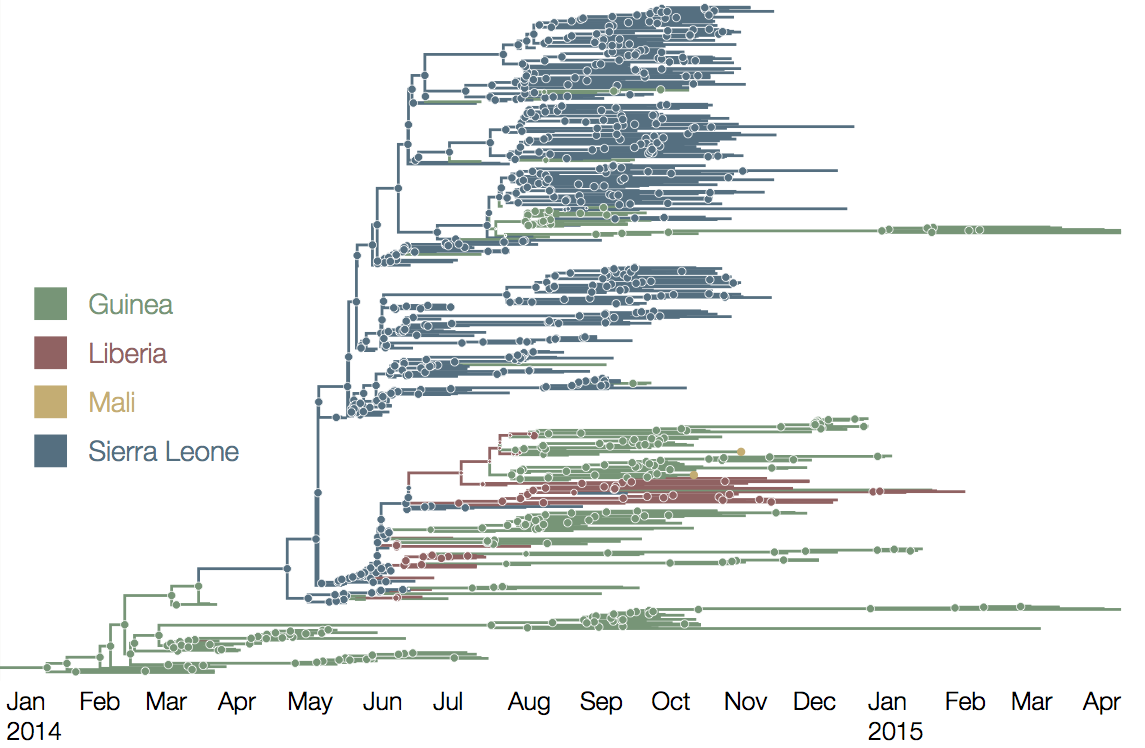
Later on, tracking transmission clusters of primary importance
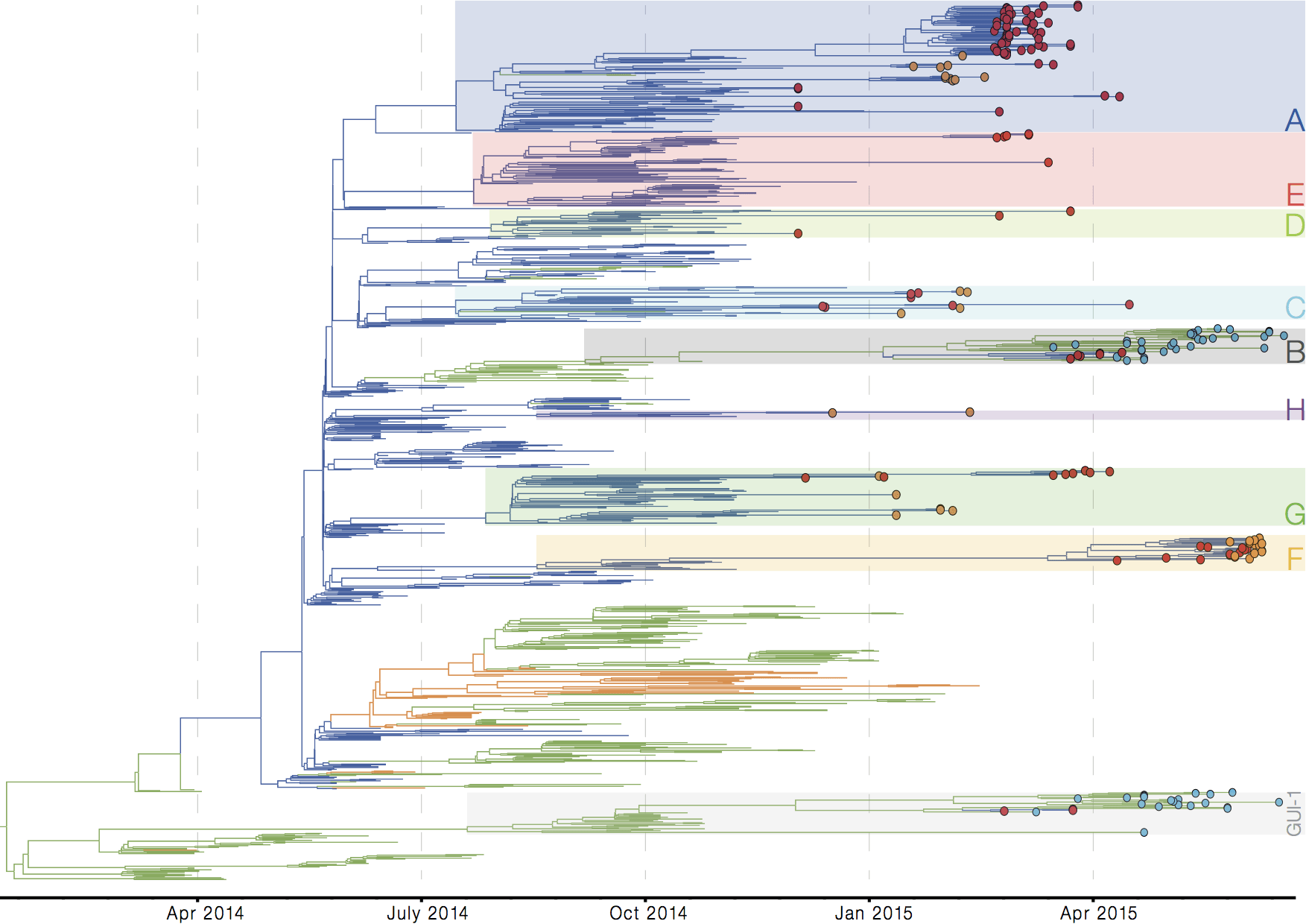
ebola.nextflu.org
Middle East respiratory syndrome coronavirus (MERS-CoV)
Cases concentrated in the Arabian Peninsula with occasional exports
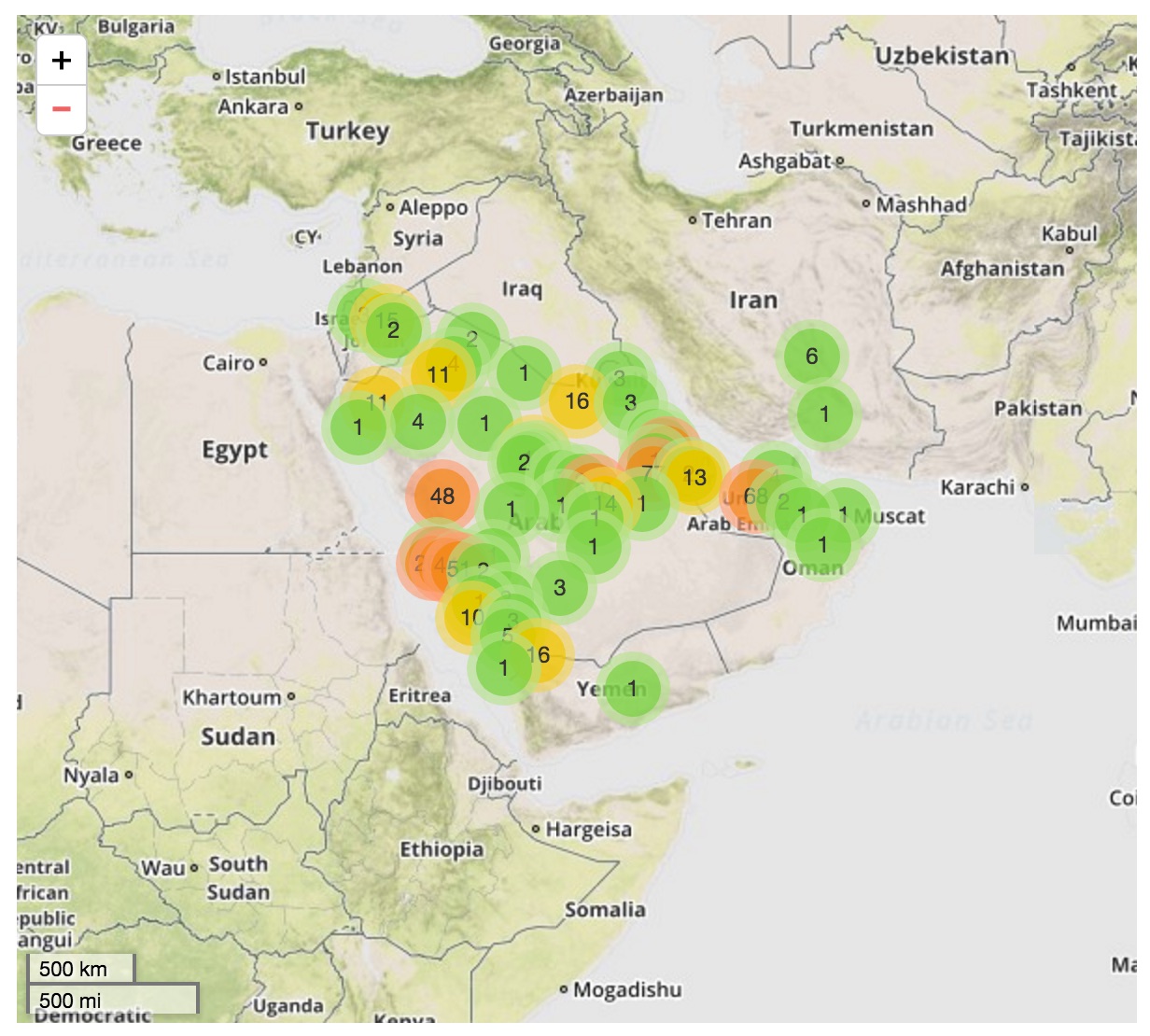
No evidence of epidemic growth, spill-over transmission clusters

Bats ➞ Camels ➞ Humans
mers.nextflu.org
Moving forward, genetically-informed outbreak response requires:
- Rapid sharing of sequence data, genetic context critical
- Technologies to rapidly conduct phylogenetic inference
- Technologies to explore genetic relationships and inform epidemiological investigation
Acknowledgements
Richard Neher (Max Planck Tübingen), Andrew Rambaut (University of Edinburgh), Colin Russell (Cambridge University), Michael Lässig (University of Cologne), Marta Łuksza (Institute for Advanced Study), Gytis Dudas (University of Edinburgh), Pardis Sabeti (Harvard University), Danny Park (Harvard University), Nick Loman (University of Birmingham) Matthew Cotten (Sanger Institute), Paul Kellam (Sanger Institute), WHO Global Influenza Surveillance Network, GISAID



Contact
- Website: bedford.io
- Twitter: @trvrb
- Slides: bedford.io/talks/real-time-tracking-uw/