Real-time genomic surveillance for public health response
Trevor Bedford (@trvrb)
Fred Hutchinson Cancer Center / Howard Hughes Medical Institute
13 Apr 2022
Africa PGI Webinar Series
Africa CDC
Slides at: bedford.io/talks
Sequencing to reconstruct pathogen spread
Epidemic process
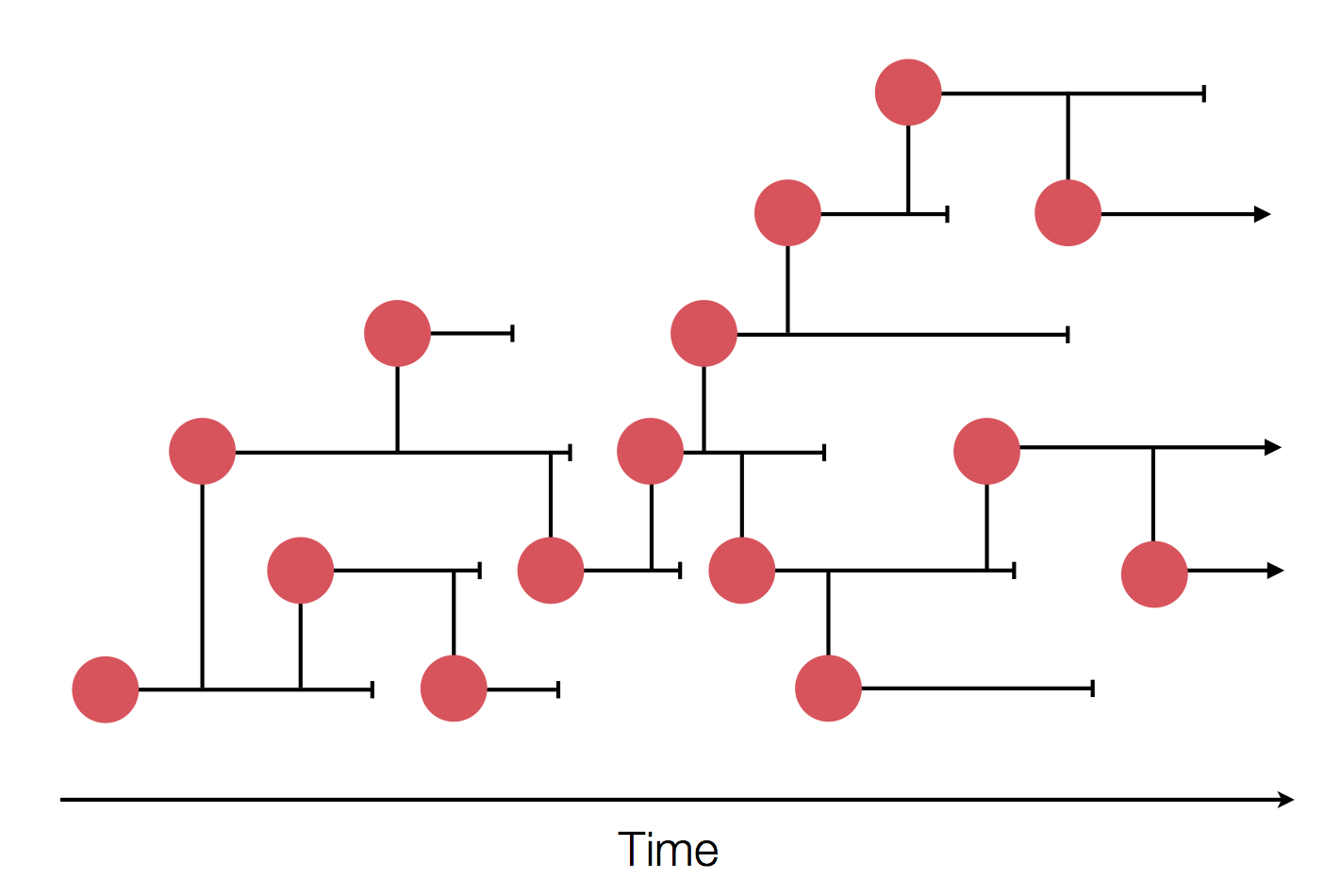
Sample some individuals
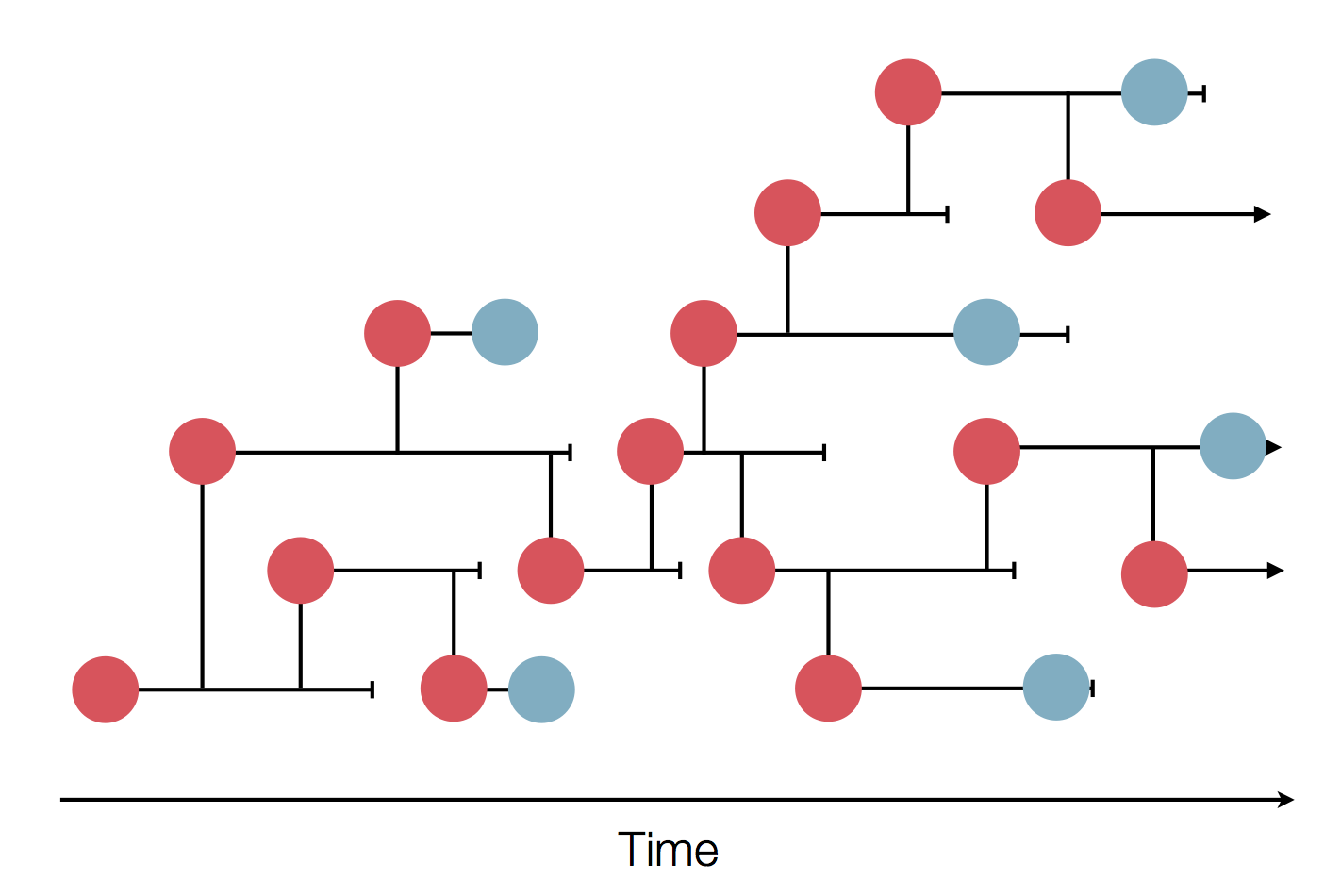
Sequence and determine phylogeny
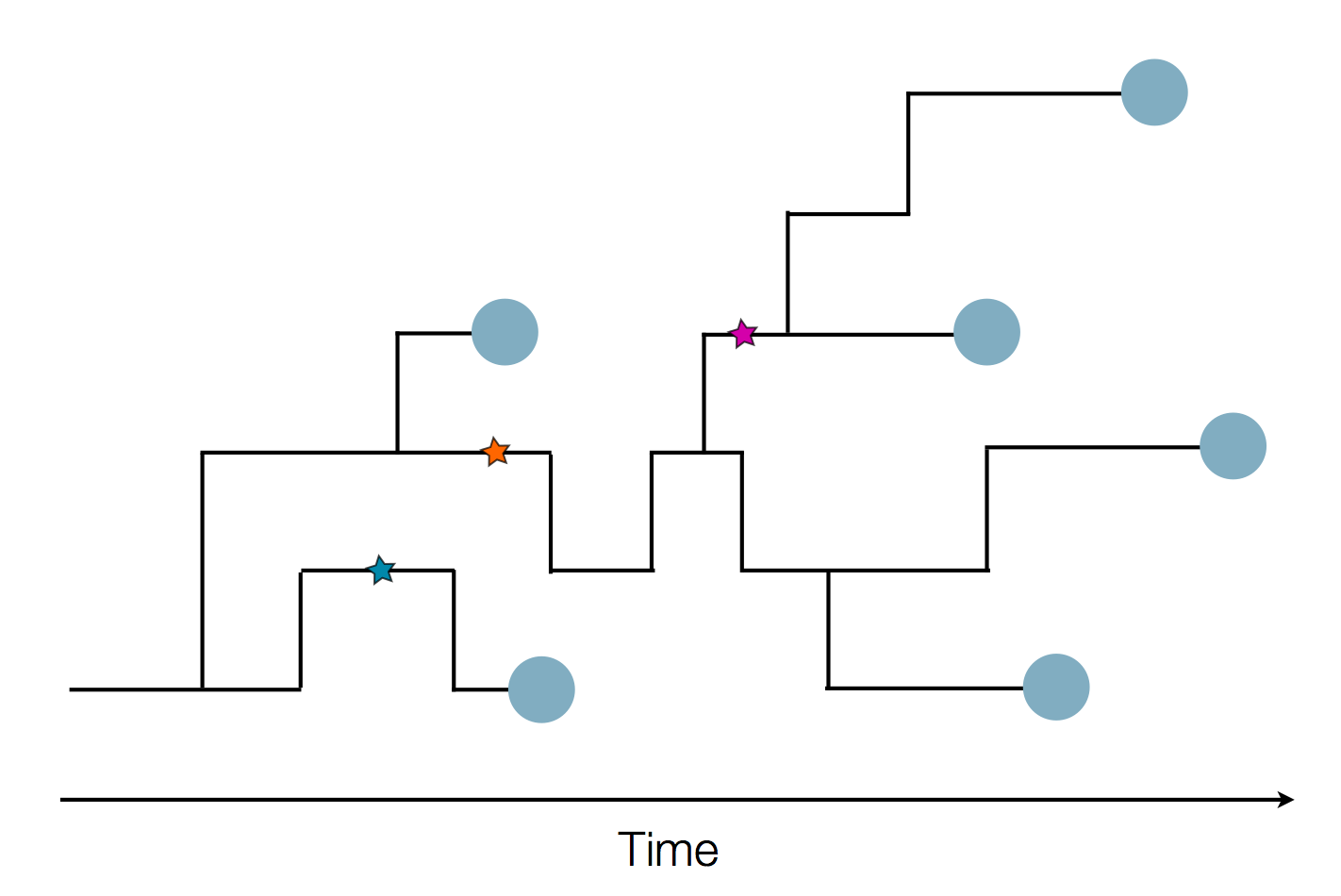
Sequence and determine phylogeny
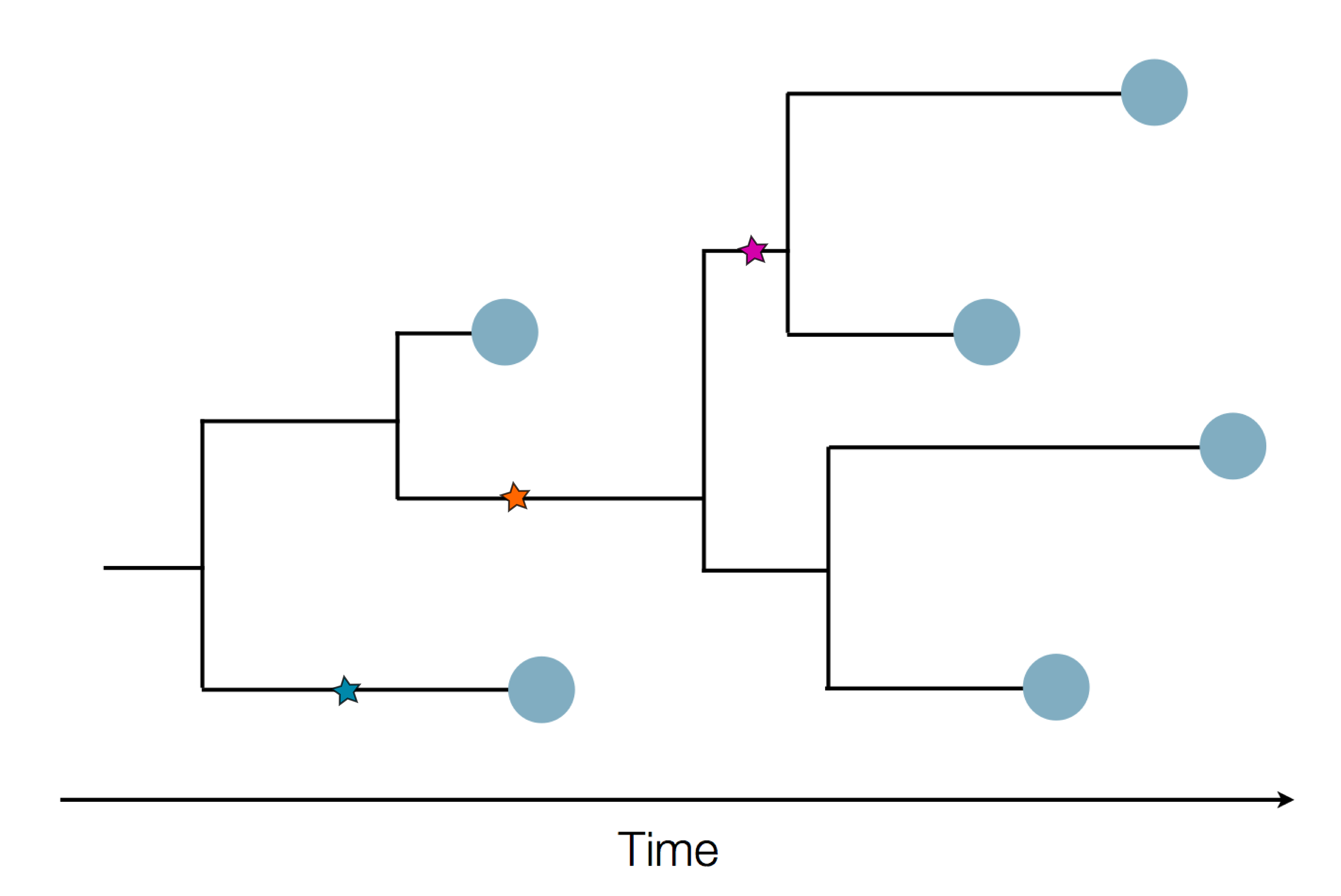
Pathogen genomes may reveal
- Evolution of new adaptive variants
- Epidemic origins
- Patterns of geographic spread
- Animal-to-human spillover
- Transmission chains
Influenza: Forecasting spread of new variants for vaccine strain selection
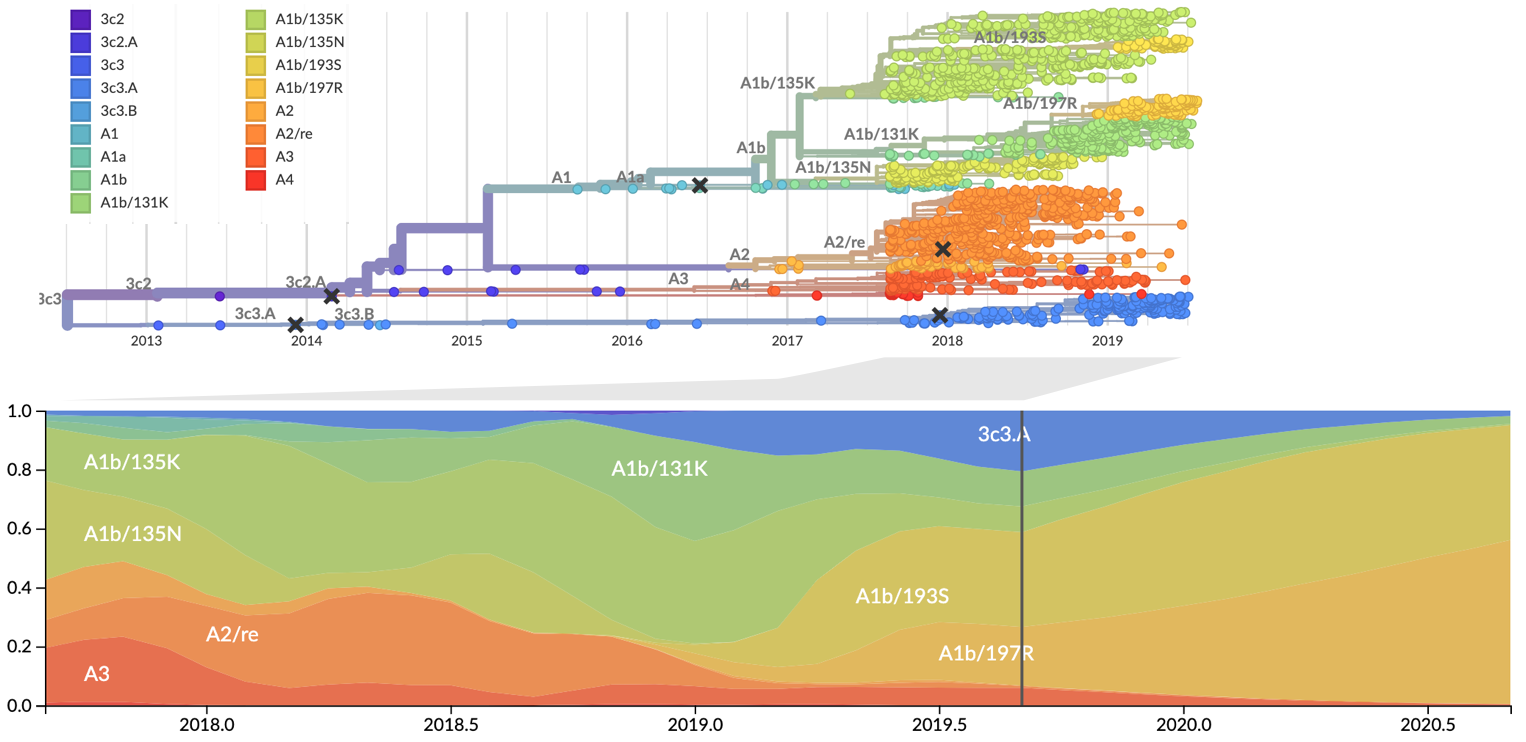
Zika: Uncovering origins of the epidemic in the Americas
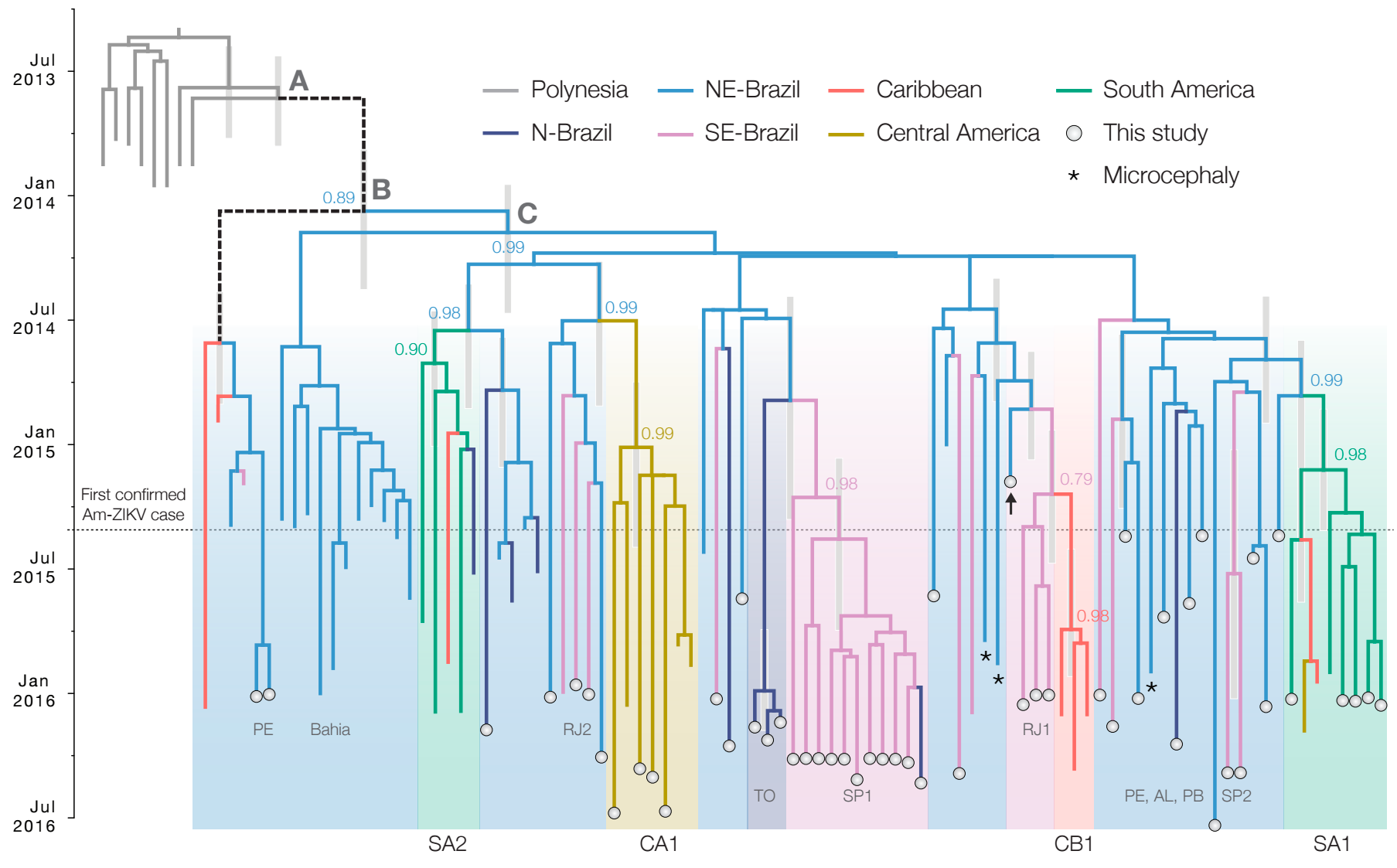
Ebola: Revealing spatial spread and persistence in West Africa
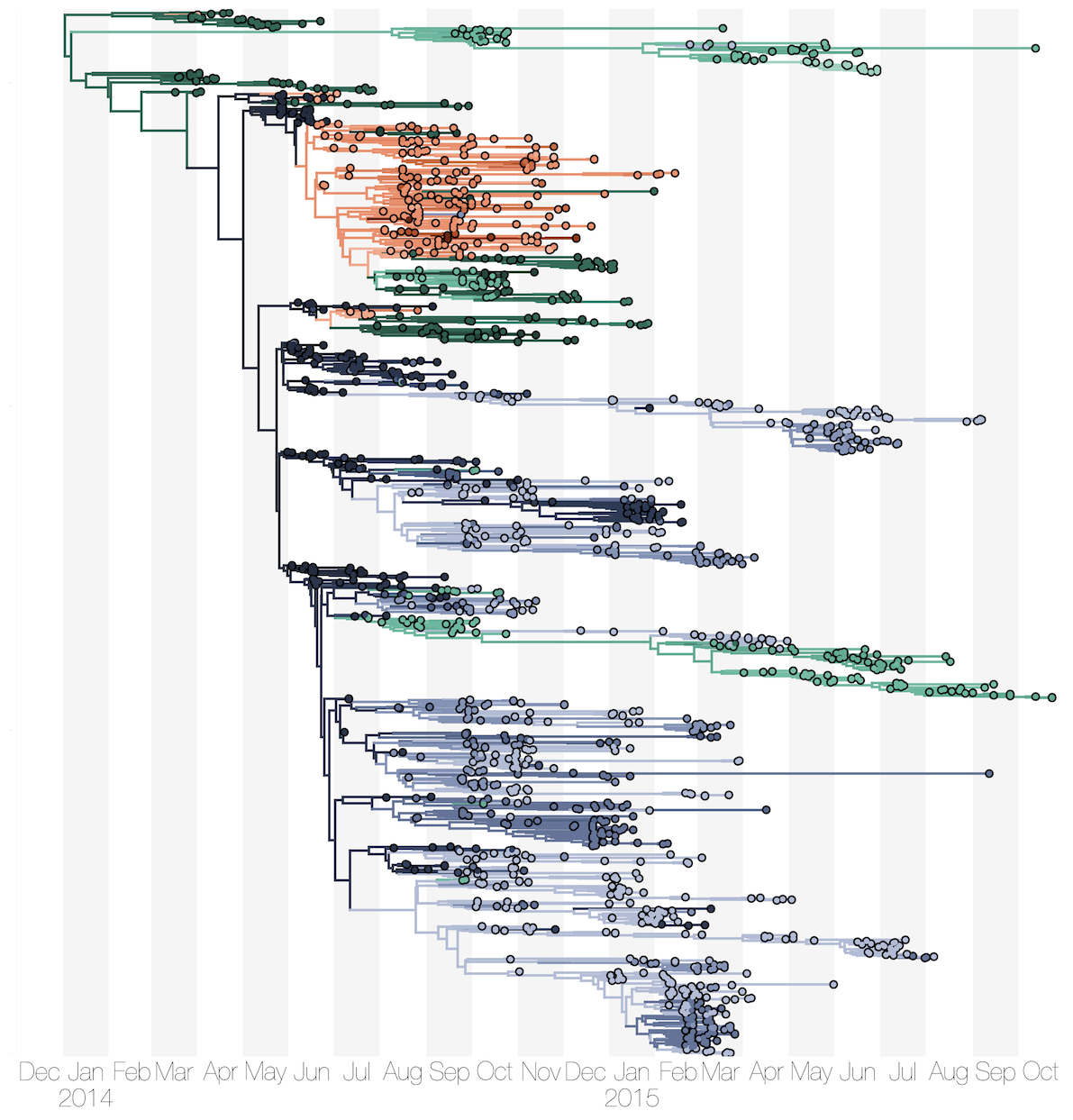
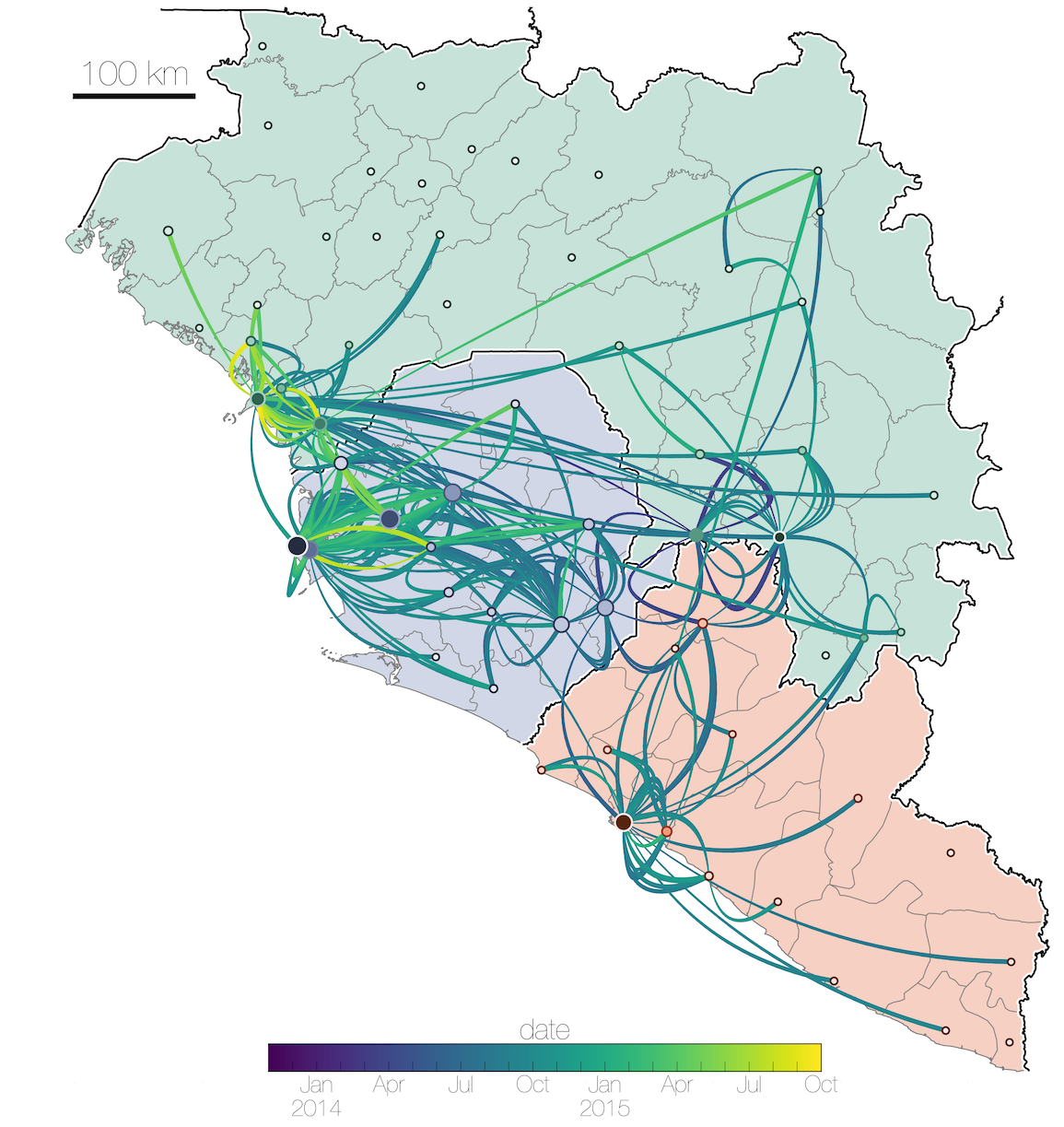
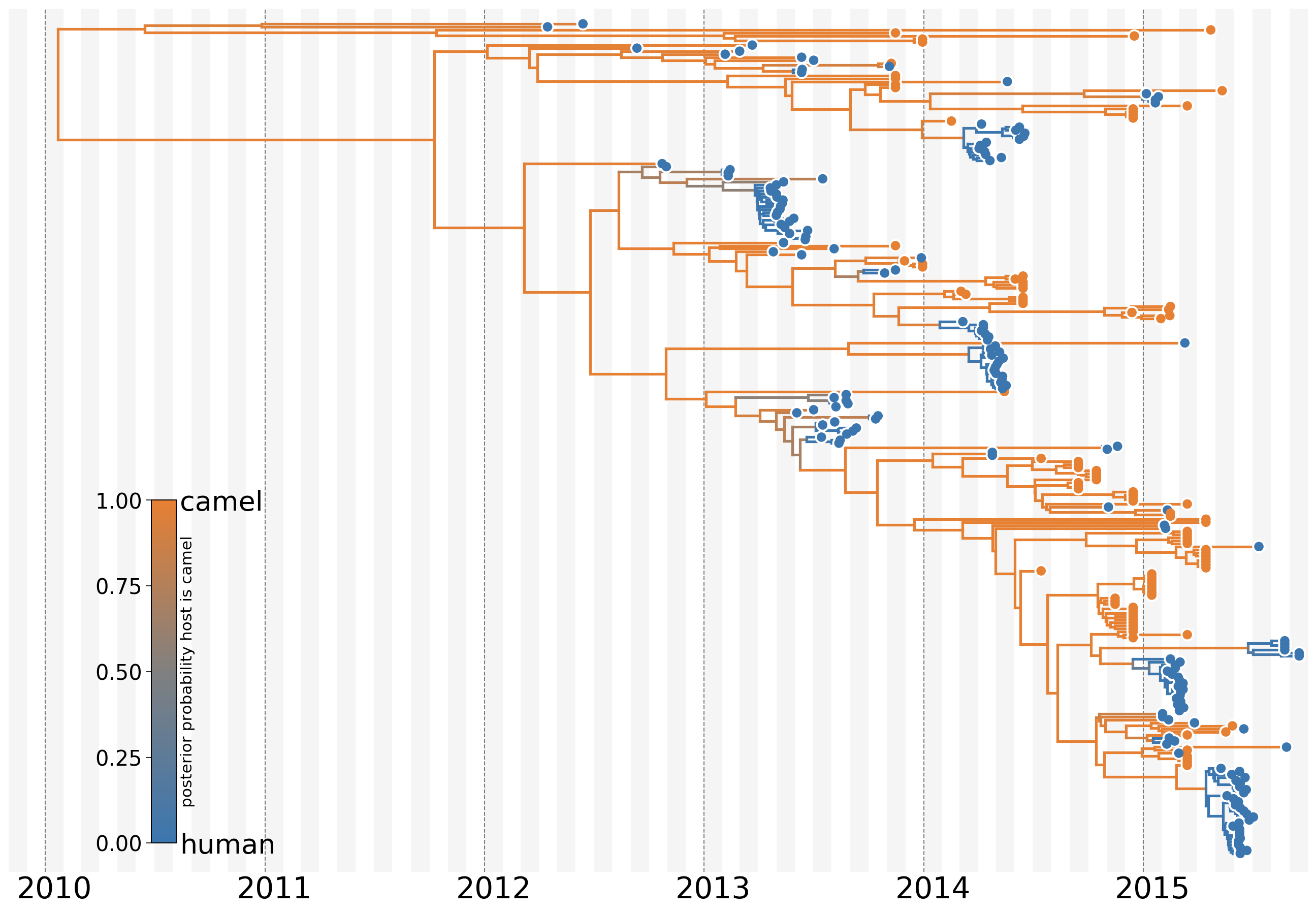
MERS: Repeated spillover into the human population from camel reservoir
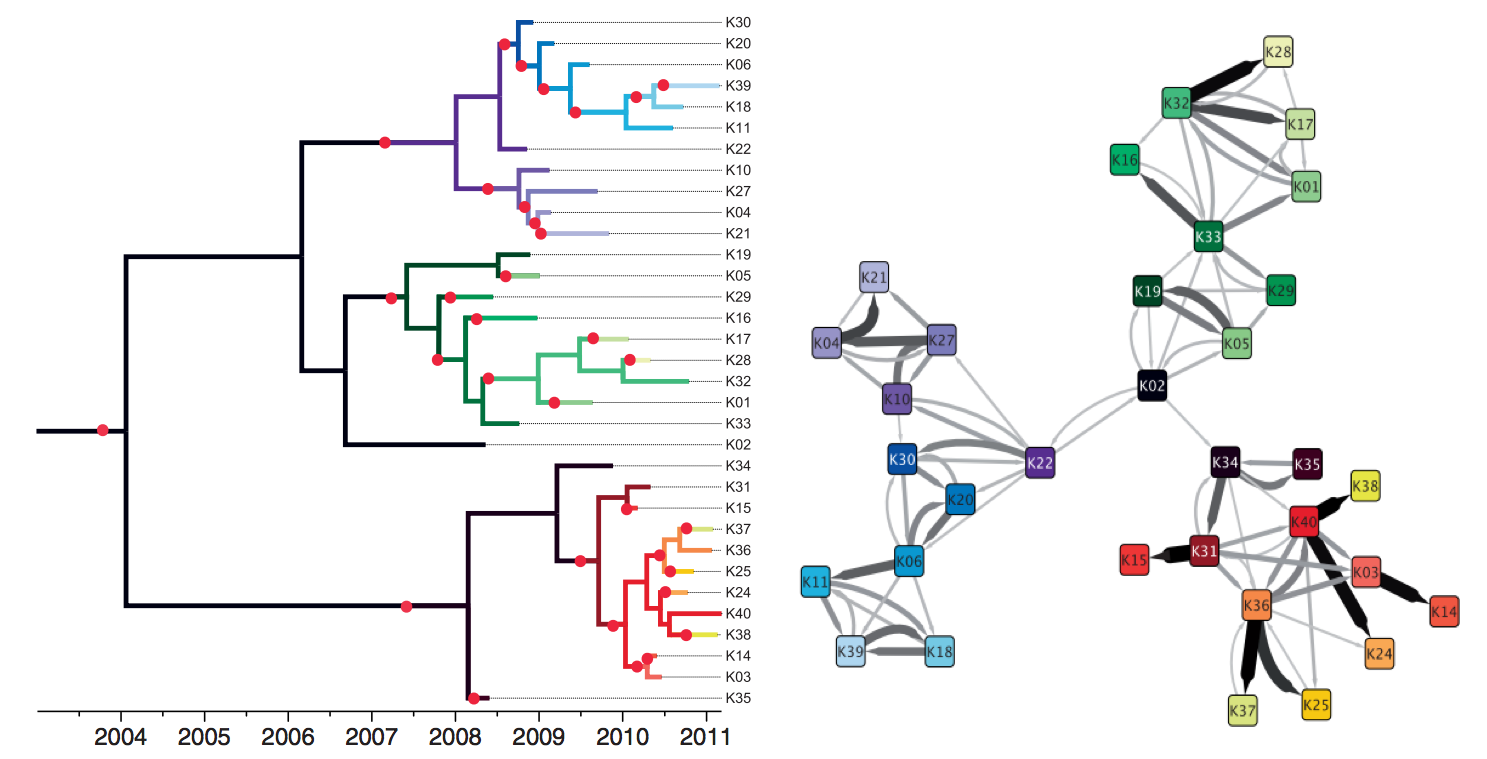
TB: Tracking individual transmission chains
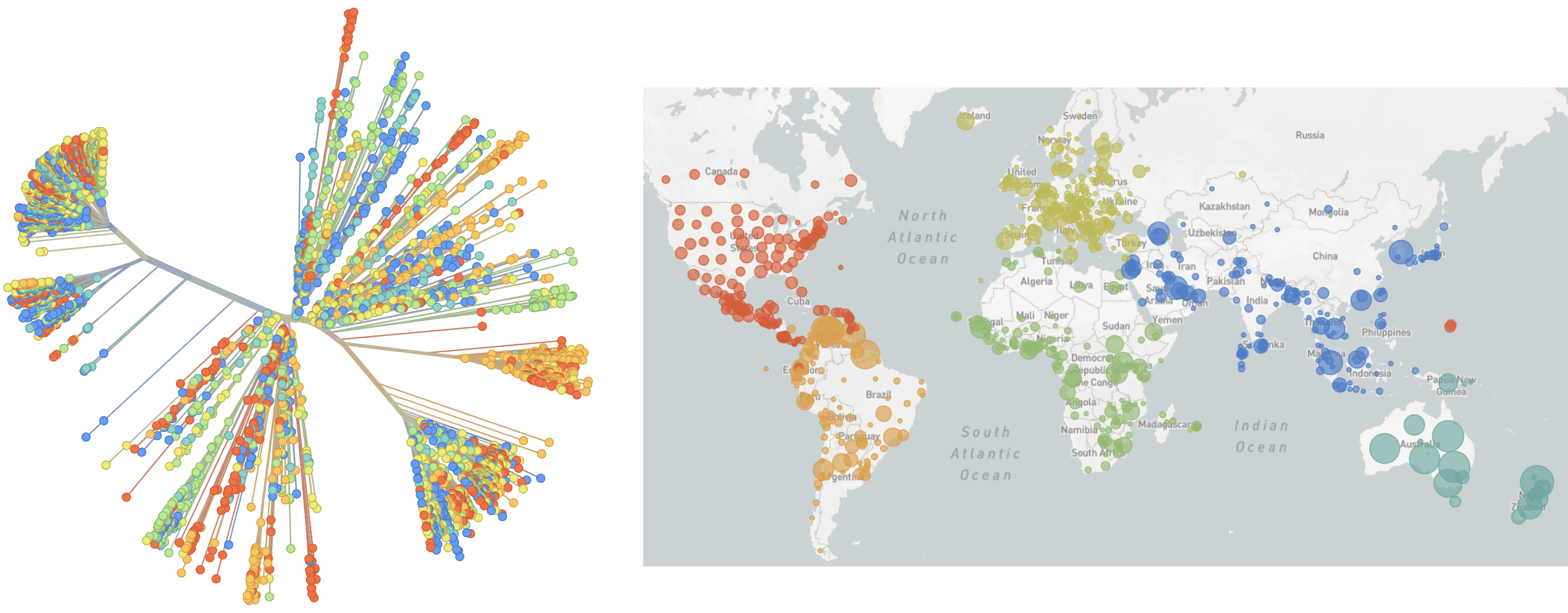
Genomic epidemiology during the COVID-19 pandemic
Over 10M SARS-CoV-2 genomes shared to GISAID and evolution tracked in real-time at nextstrain.org
![]() Richard Neher,
Richard Neher,
![]() Emma Hodcroft,
Emma Hodcroft,
![]() James Hadfield,
James Hadfield,
![]() Thomas Sibley,
Thomas Sibley,
![]() John Huddleston,
John Huddleston,
![]() Ivan Aksamentov,
Ivan Aksamentov,
![]() Moira Zuber,
Moira Zuber,
![]() Jover Lee,
Jover Lee,
![]() Cassia Wagner,
Cassia Wagner,
![]() Denisse Sequeira,
Denisse Sequeira,
![]() Cornelius Roemer,
Cornelius Roemer,
![]() Victor Lin,
Victor Lin,
![]() Jennifer Chang
Jennifer Chang
Three key insights that genomic epi provided during pandemic
- Rapid human-to-human spread in Wuhan beyond initial market outbreak
- Extensive local transmission while testing was rare
- Identification of variants of concern and mapping of increased transmission
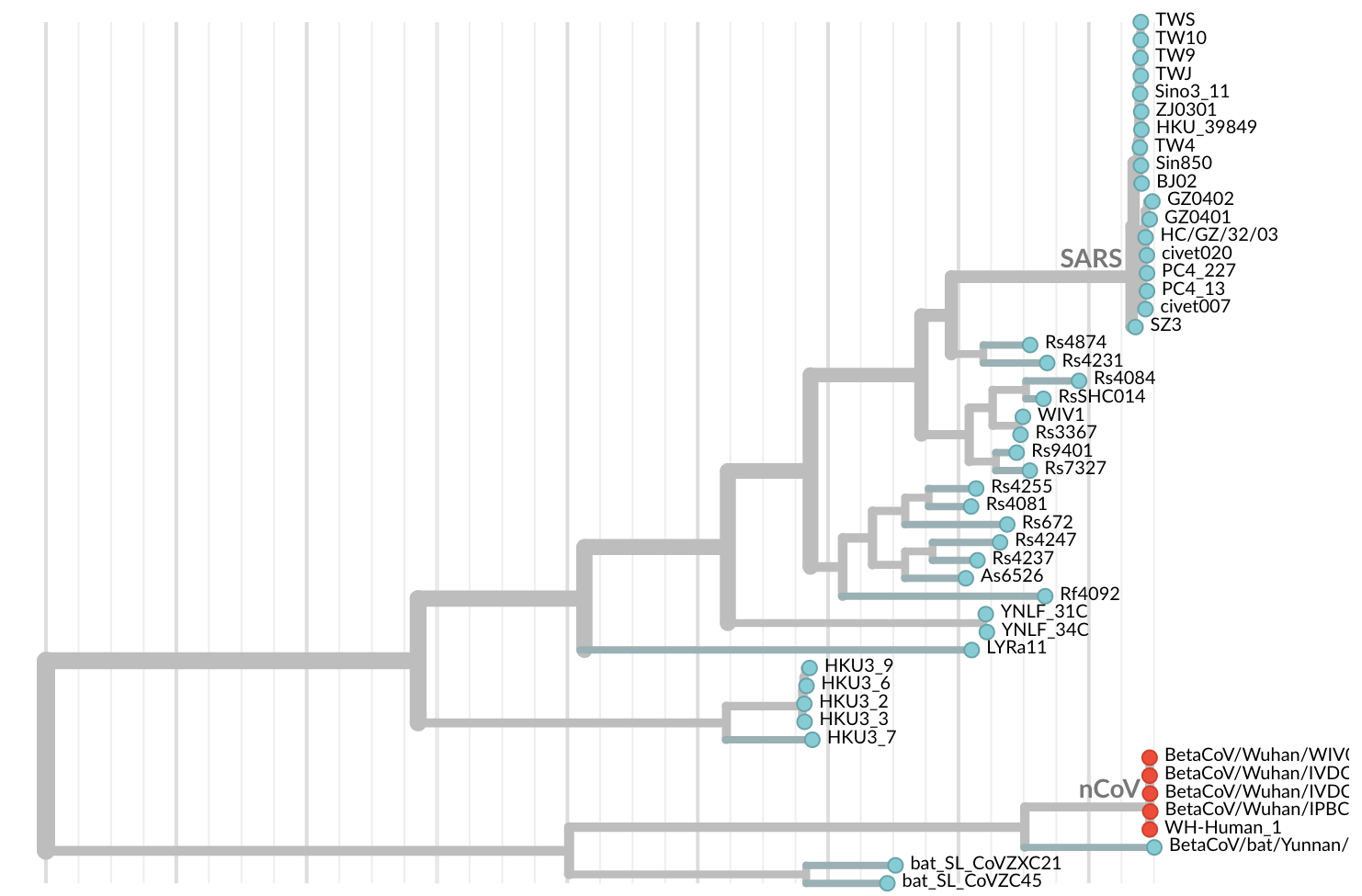
Jan 11: First five genomes from Wuhan showed a novel SARS-like coronavirus
Jan 19: First 12 genomes from Wuhan and Bangkok showed lack of genetic diversity
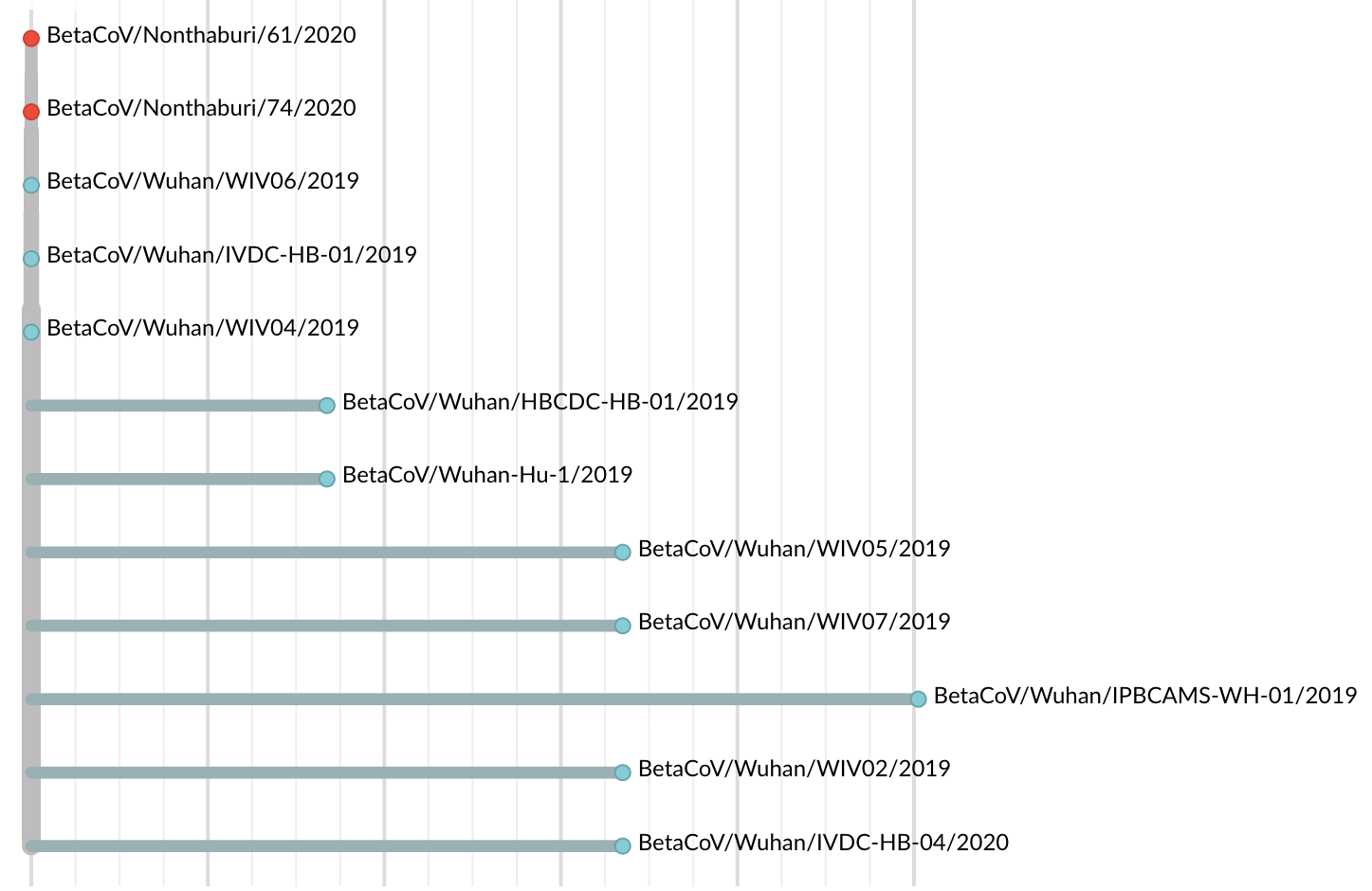
Single introduction into the human population between Nov 15 and Dec 15 and subsequent rapid human-to-human spread
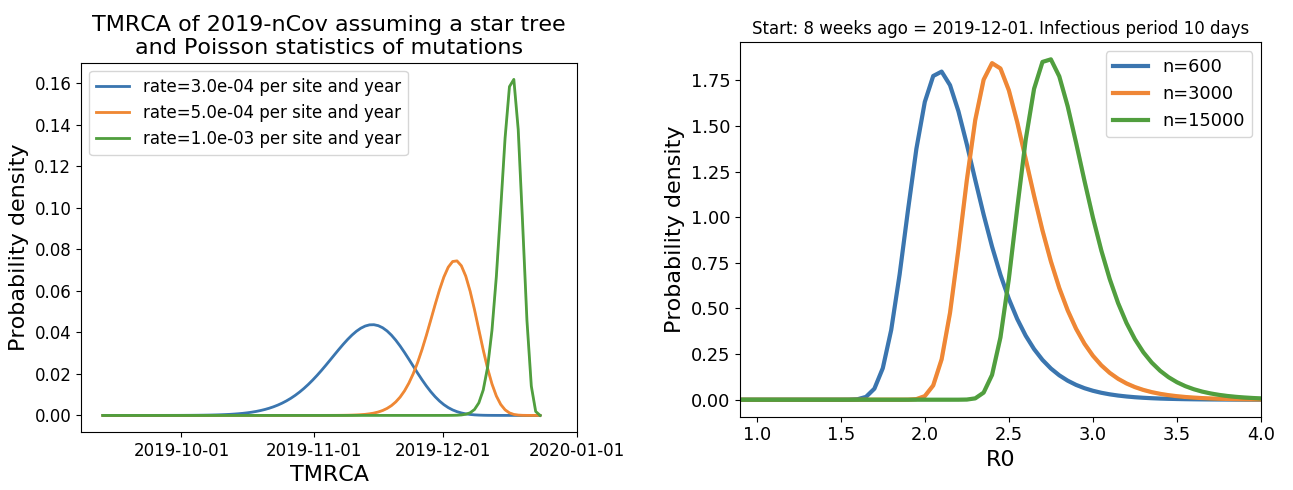
Rapid global epidemic spread from China
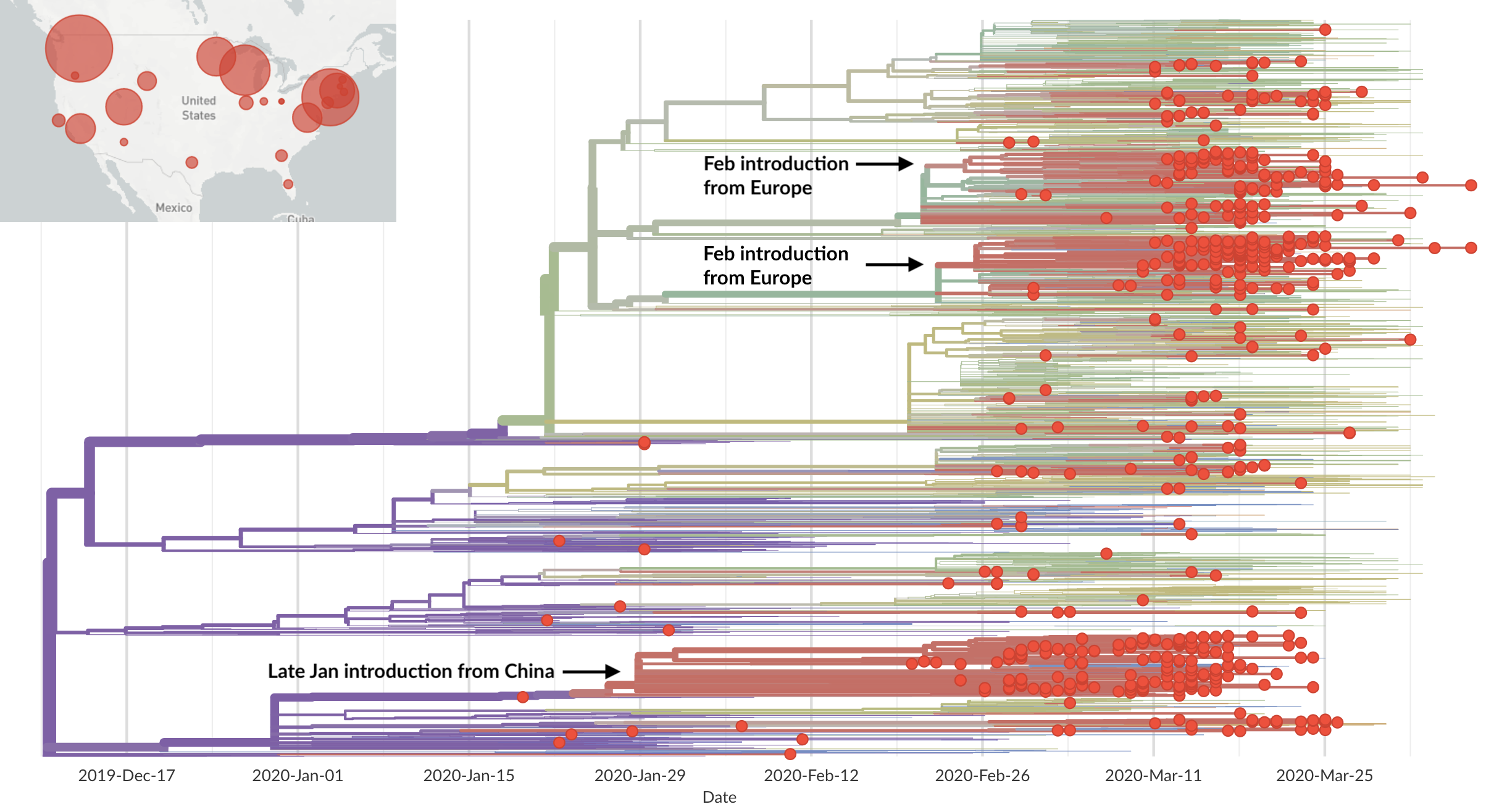
Epidemic in the USA was introduced from China in late Jan and from Europe during Feb
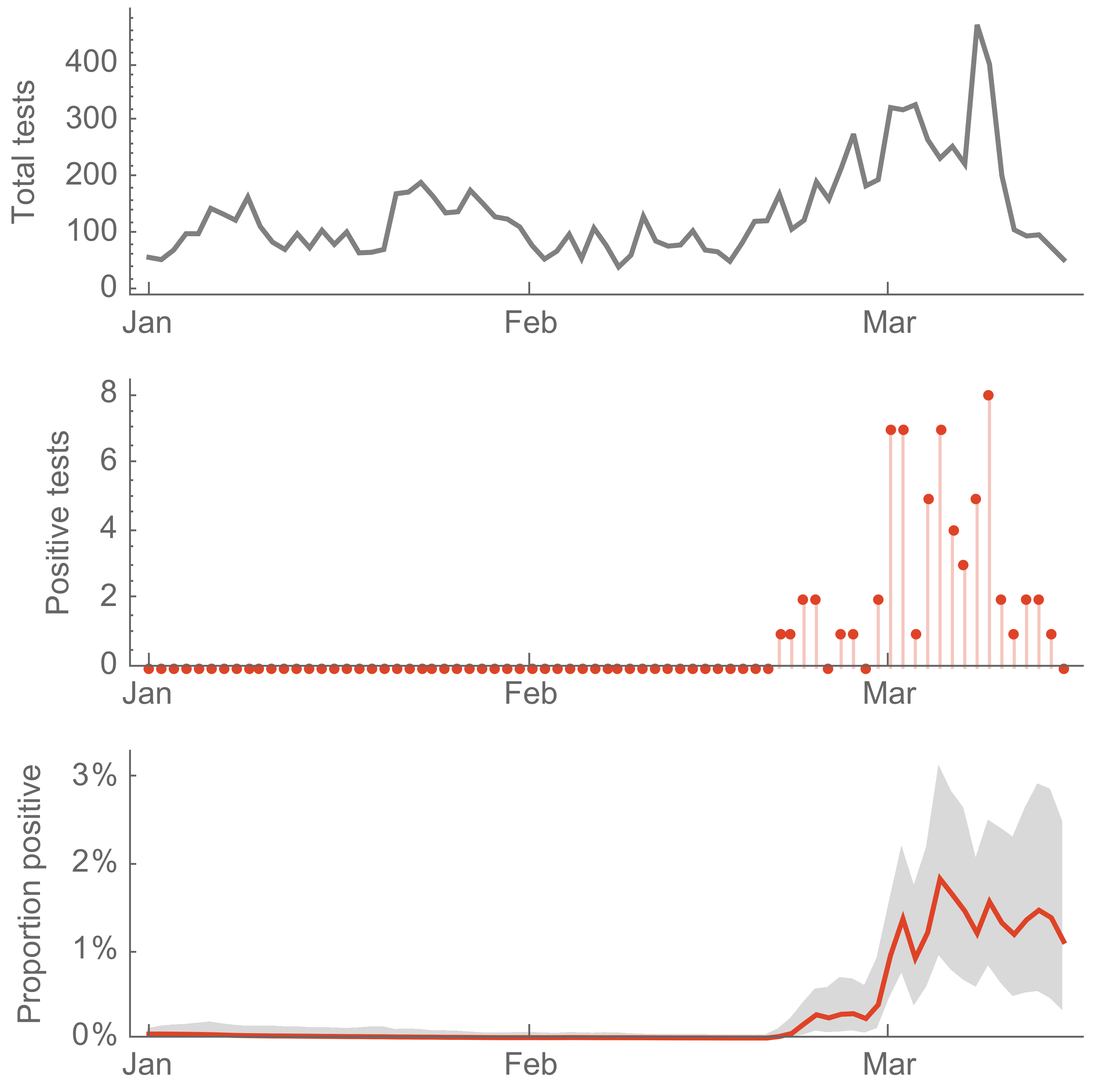
Seattle Flu Study detects local circulation and charts early epidemic in Washington State
Direct introduction from China ~Feb 1 responsible for the majority of the epidemic
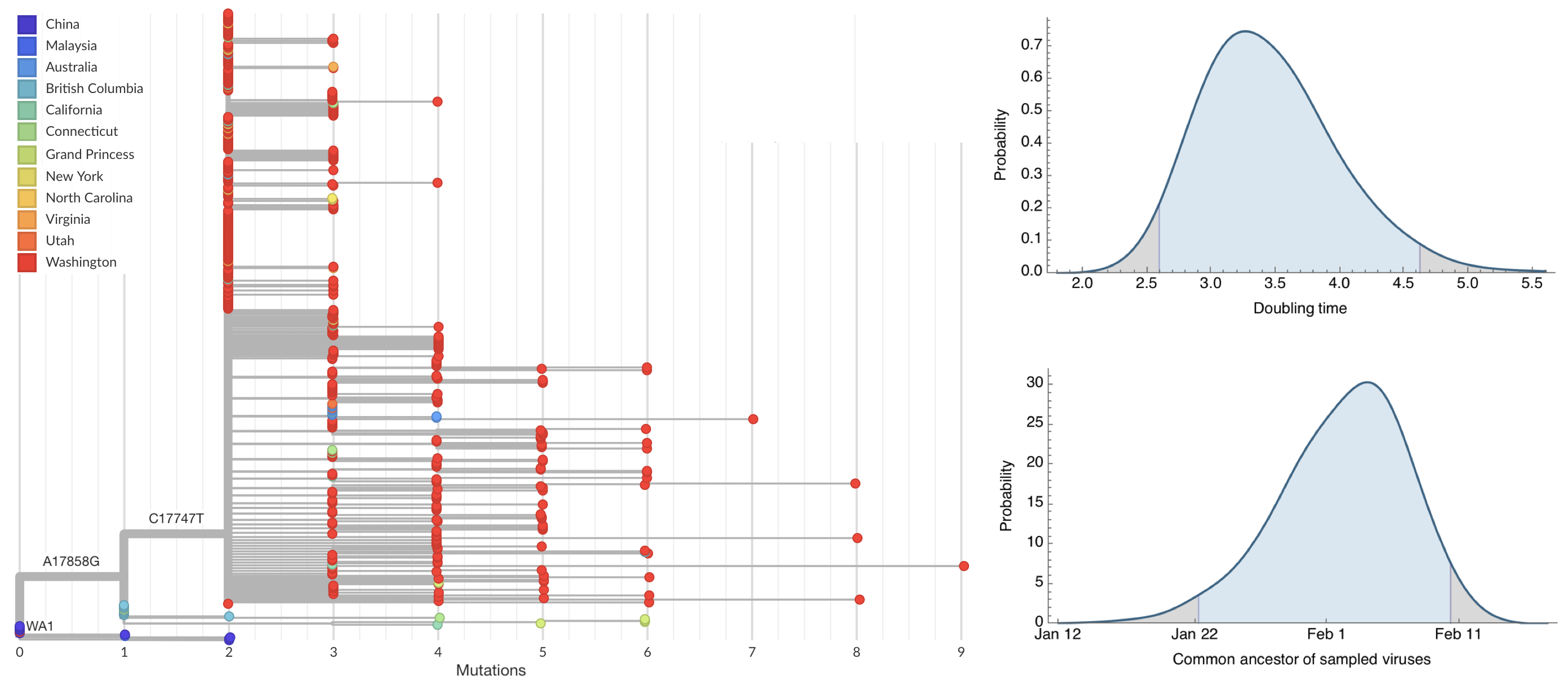


Mitigation efforts visible in virus genetic diversity
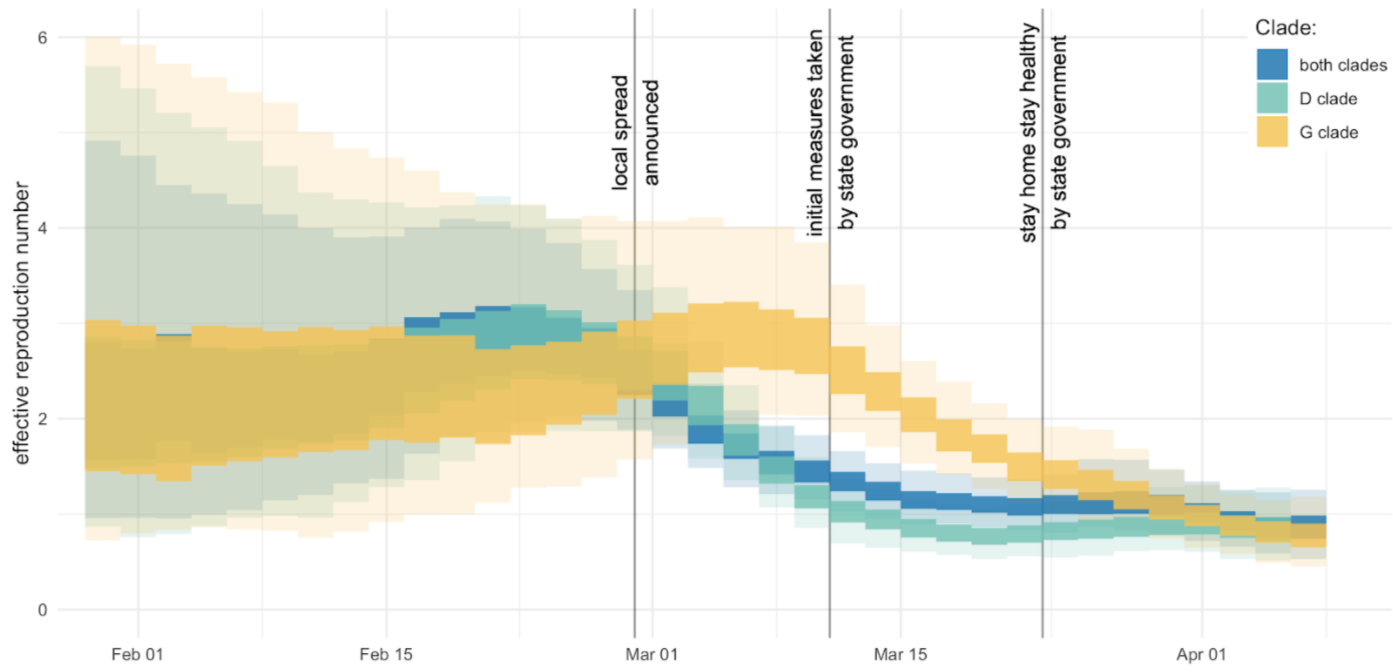
After initial wave, with mitigation
efforts and decreased travel,
regional clades emerge
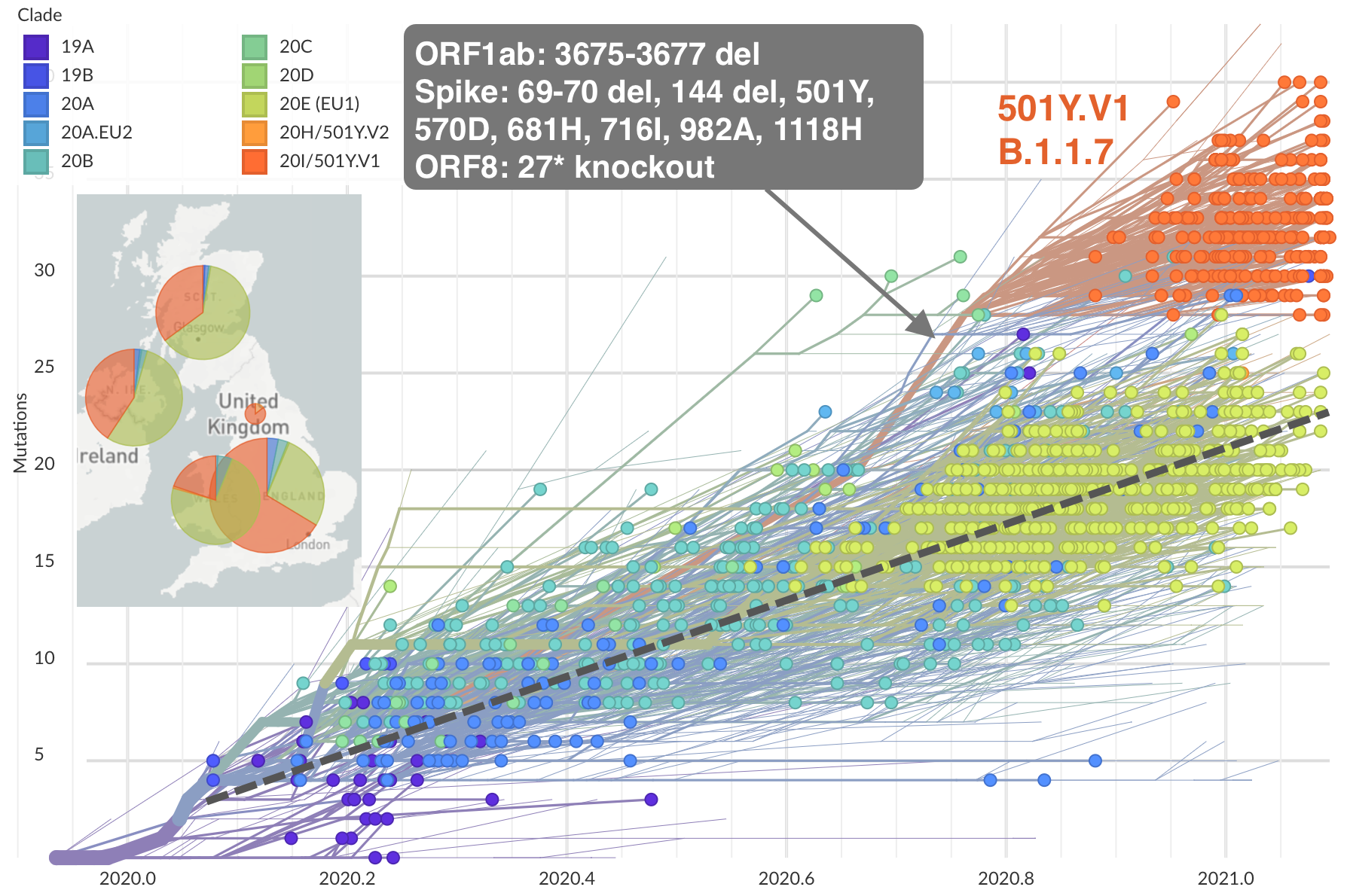
Repeated emergence of variant of concern viruses
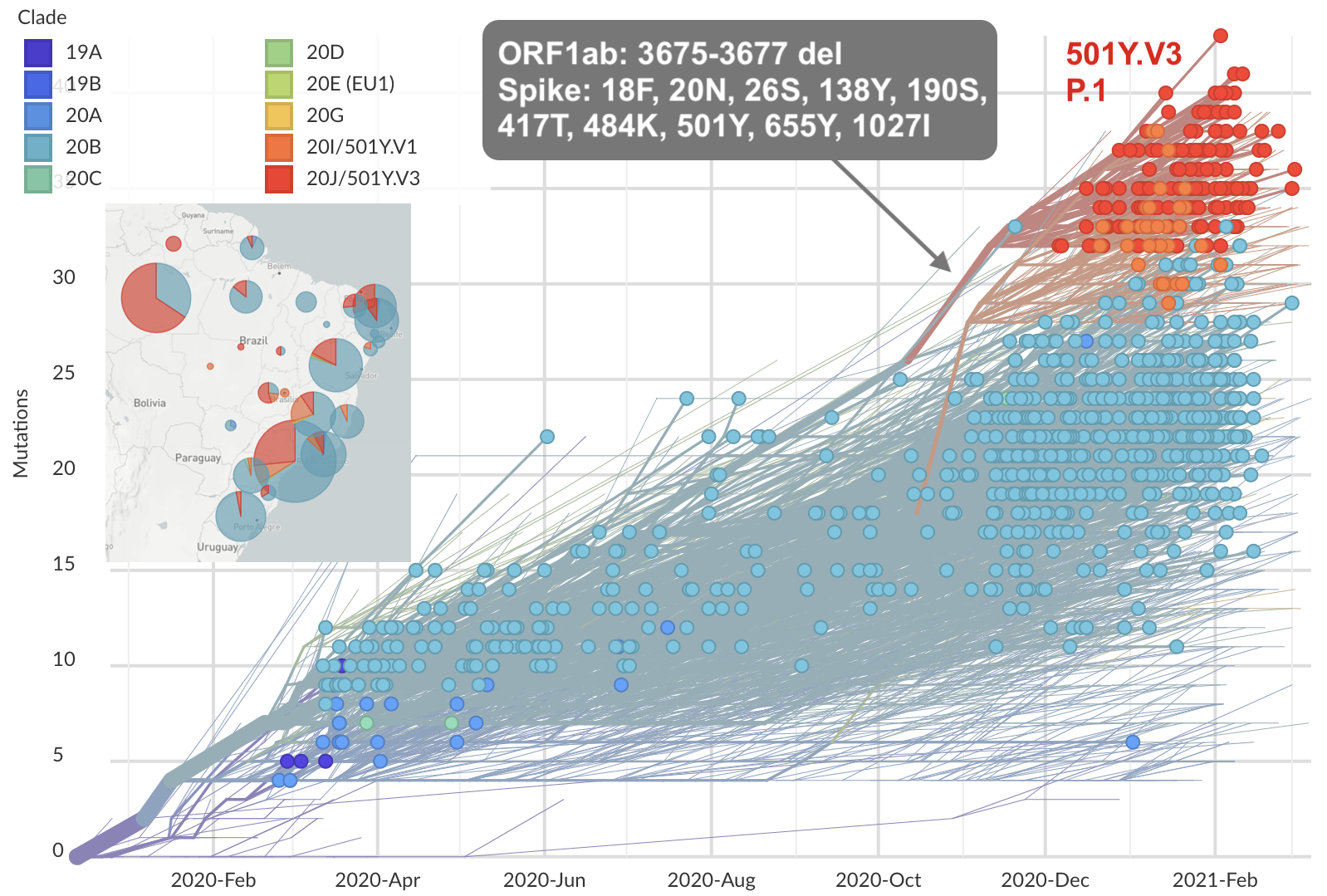
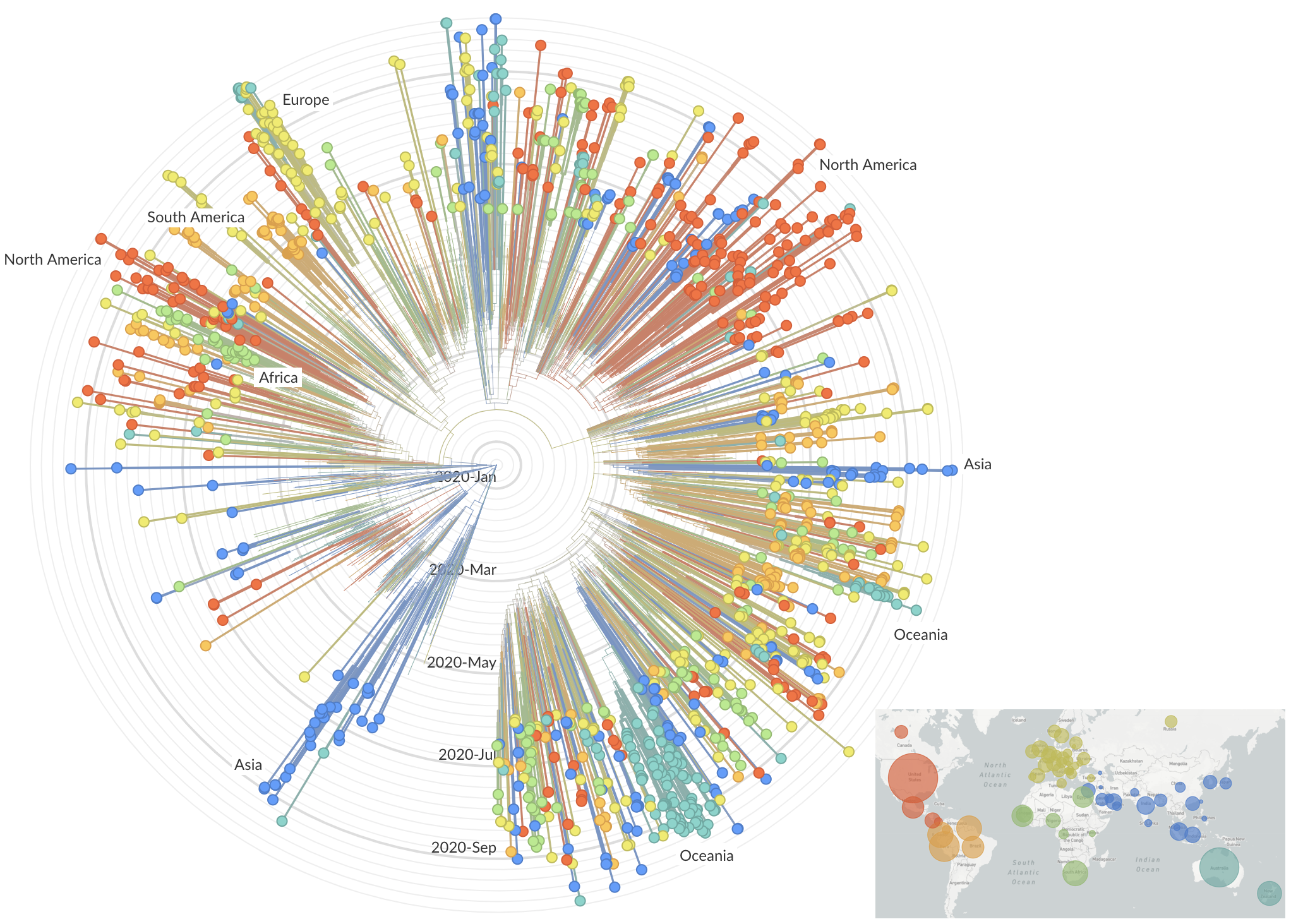
Genetic relationships of globally sampled SARS-CoV-2 to present
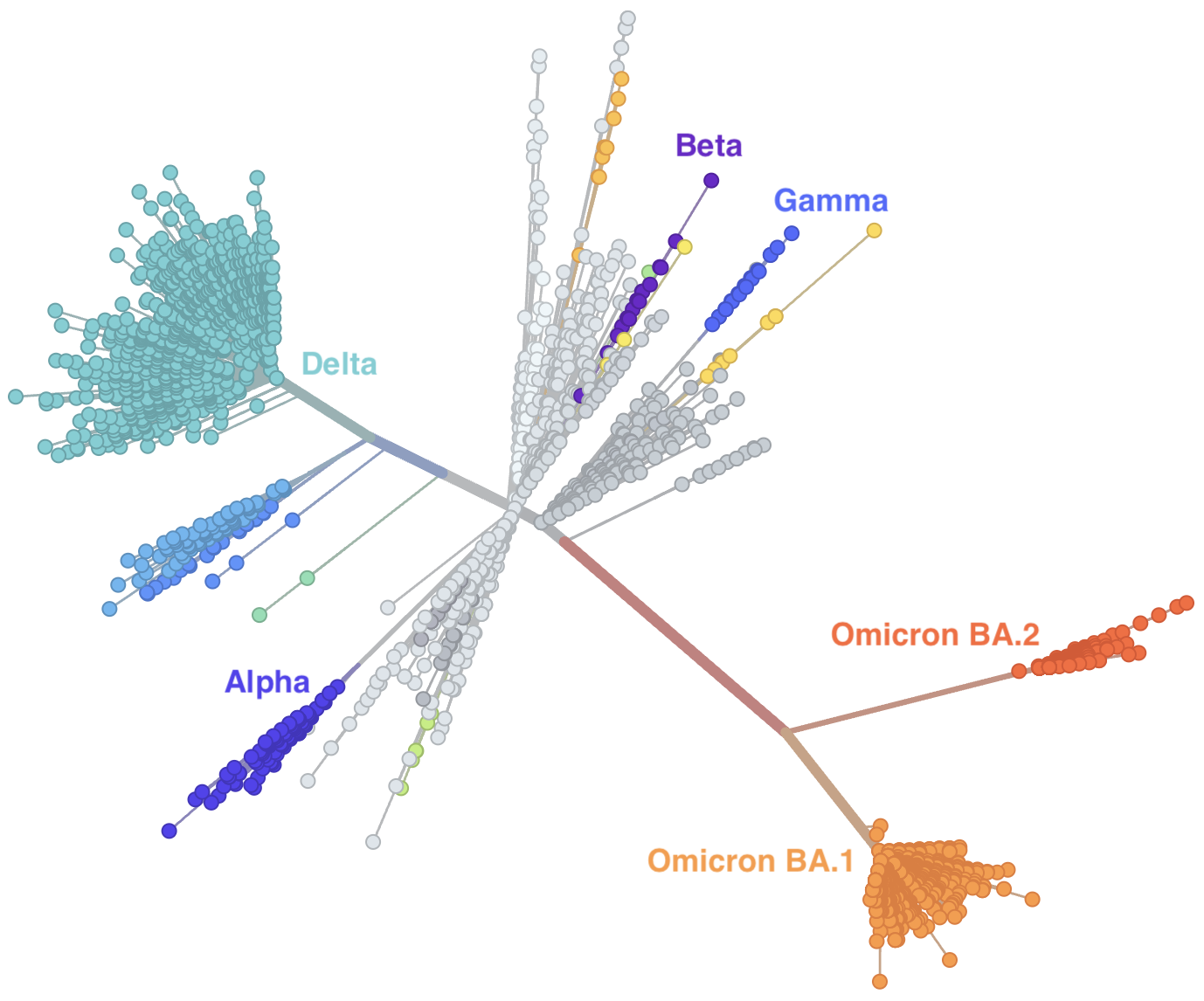
Rapid displacement of existing diversity by emerging variants
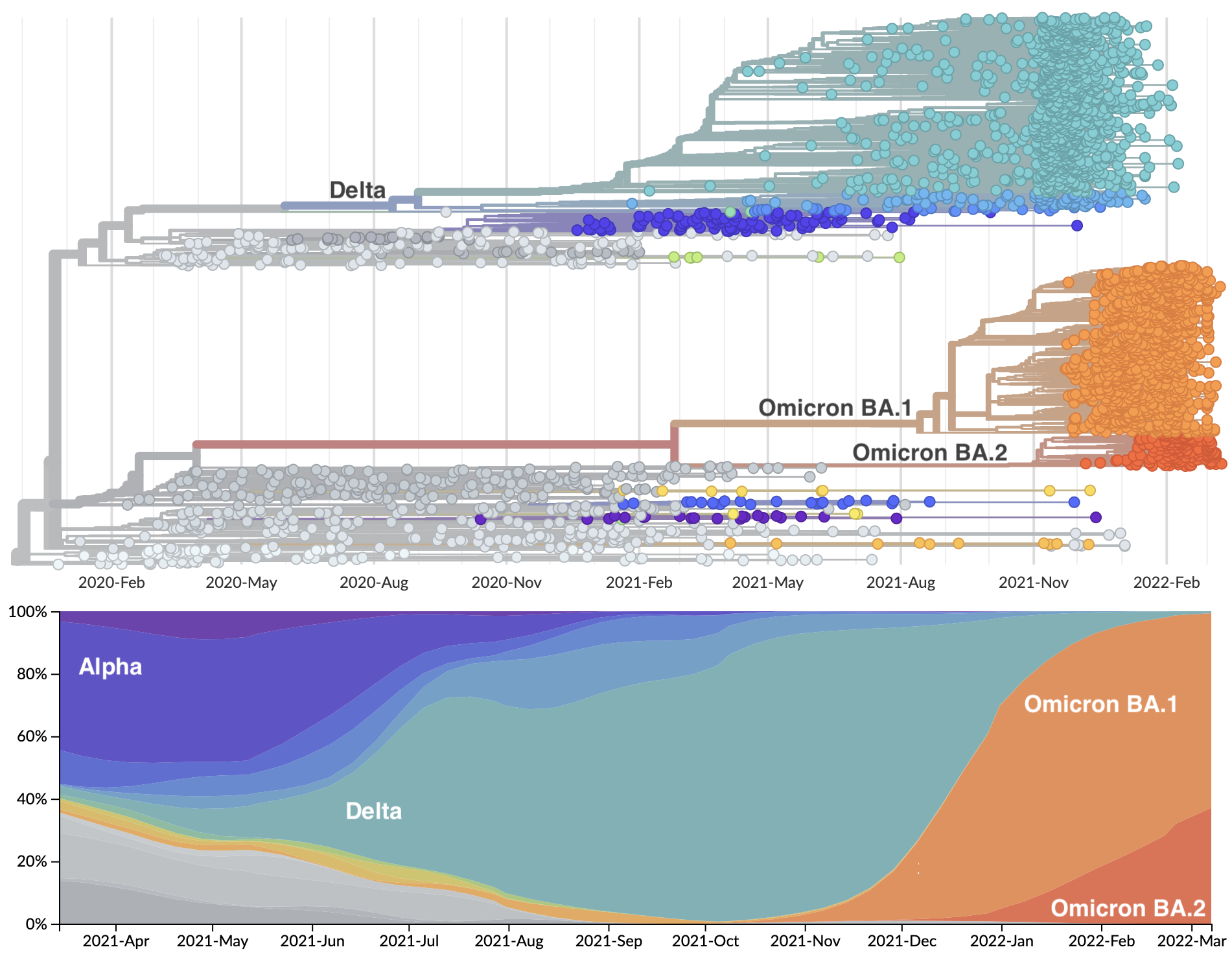
Rapid displacement of existing diversity by emerging variants

Genomic surveillance
Sequences generated and shared at an unprecedented pace with >1M genomes shared in Mar 2022 alone

My favorite metric is number of sequences available from samples collected in the past 30 days
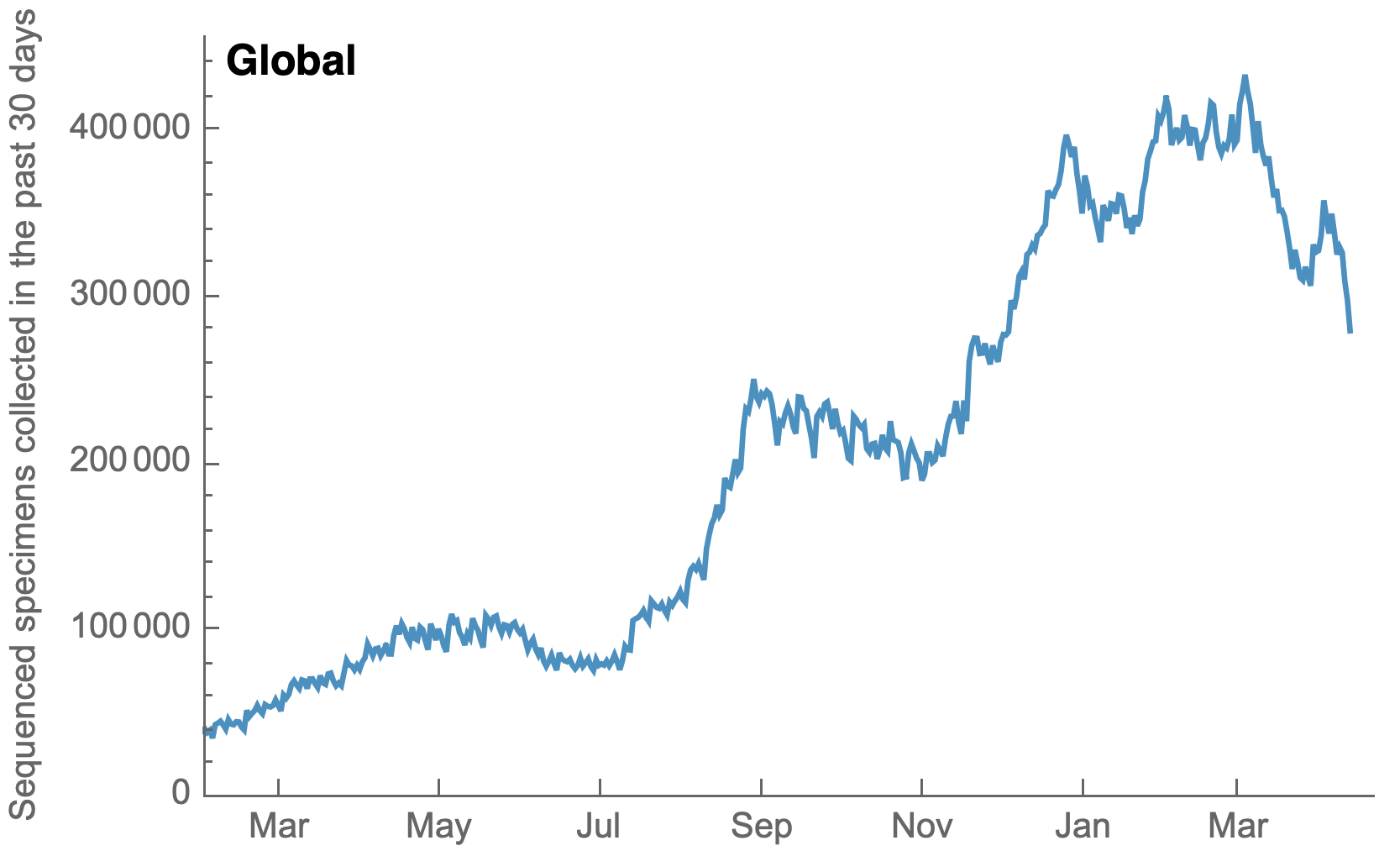
US, UK and Europe all have stunning genomic surveillance programs
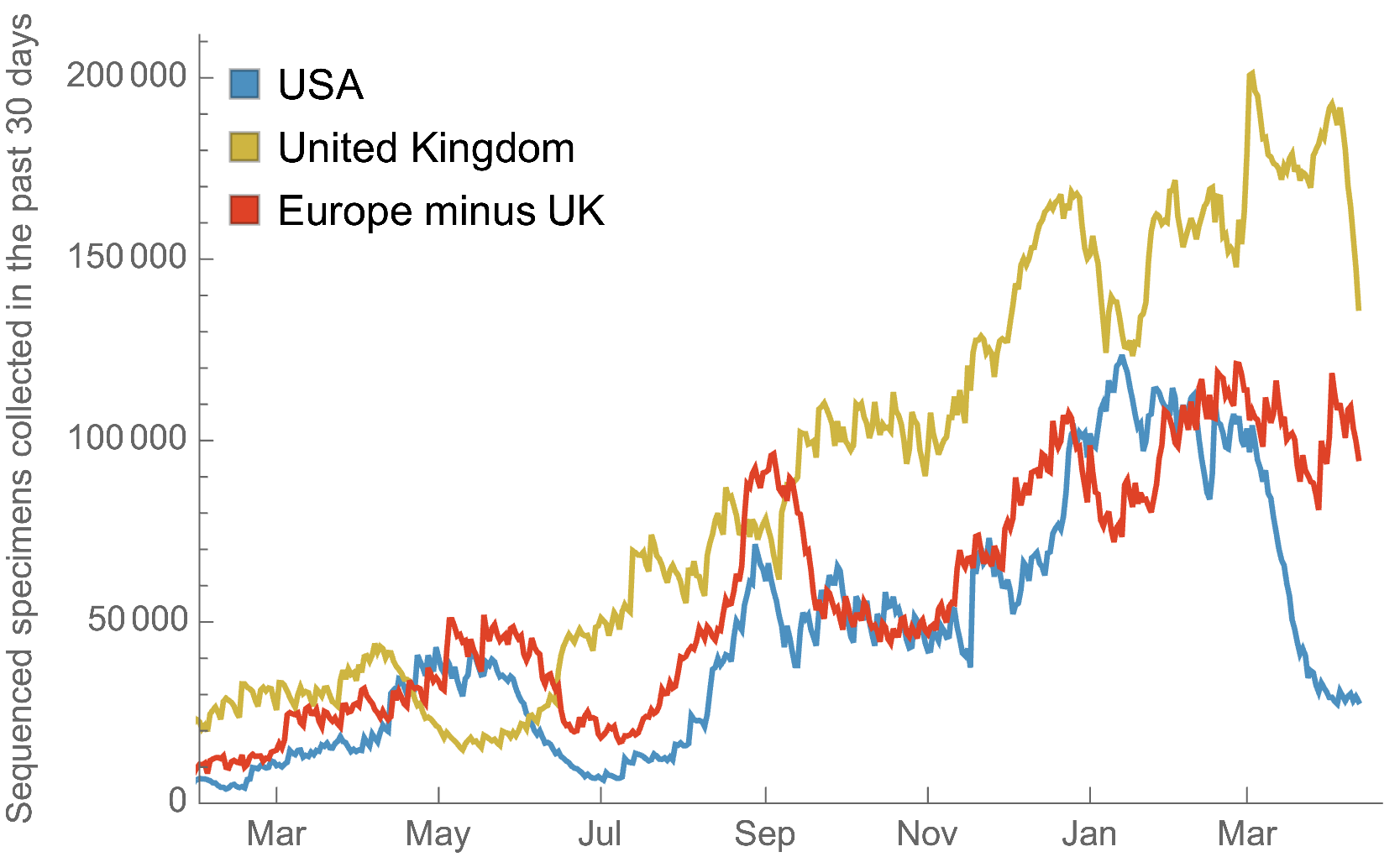
Africa and South America have picked up dramatically but there are still geographic gaps
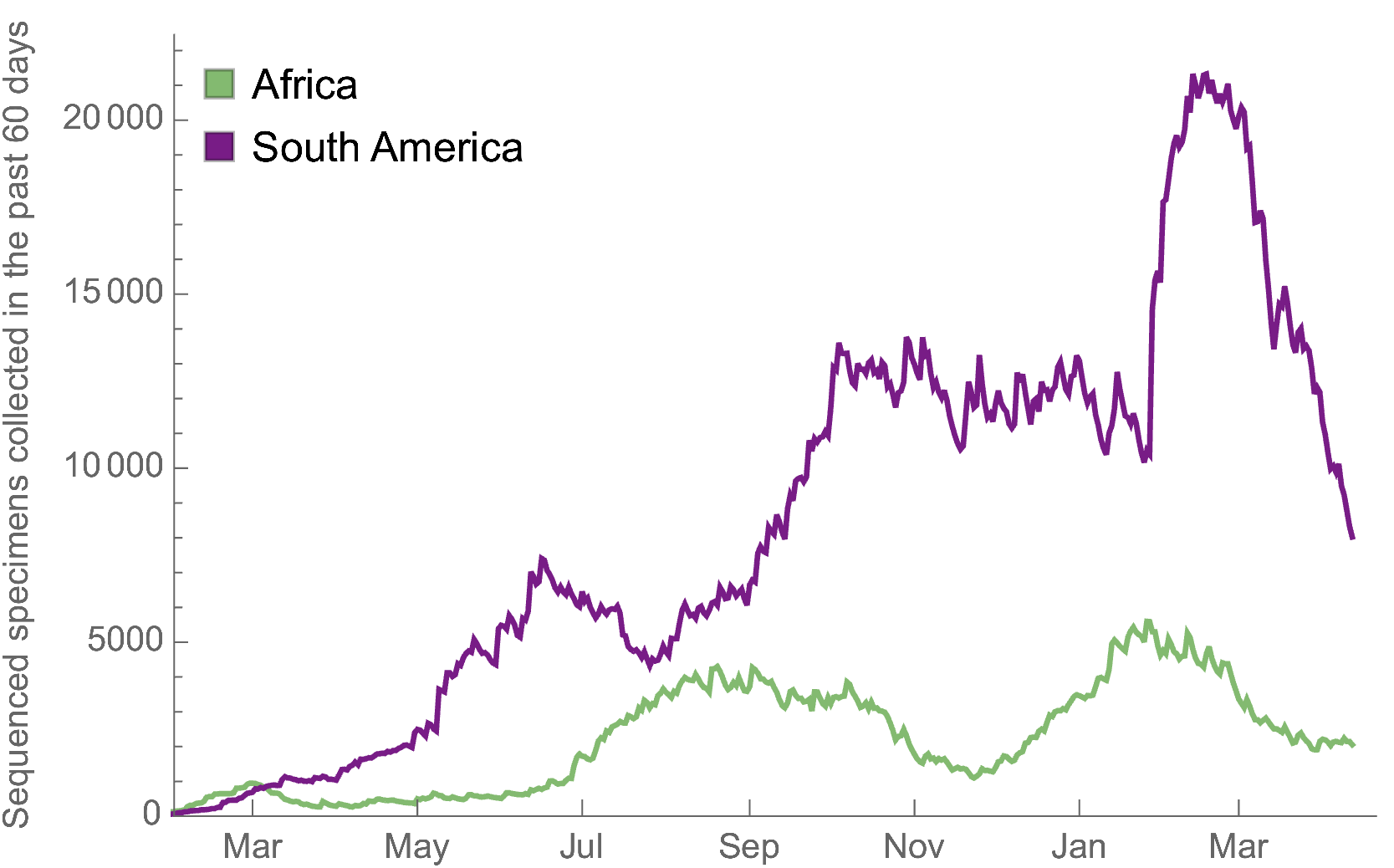
Global genomic surveillance aimed at rapid turnaround times important to catch new variants early, additional sequences from Africa or South Amerca would be far more valuable than additional sequences from the US or Europe
In depth genomic surveillance tied to data systems allows investigation of variant severity, secondary attack rate, etc...
Future perspectives
Response to the pandemic has spurred sequencing capacity across the world. This sequencing capacity should shift to non-SARS-CoV-2 pathogens of local public health concern as acute impacts of pandemic dissipate.
Acknowledgements
SARS-CoV-2 genomic epi: Data producers from all over the world, GISAID and the Nextstrain team
Bedford Lab:
![]() John Huddleston,
John Huddleston,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Louise Moncla,
Louise Moncla,
![]() Maya Lewinsohn,
Maya Lewinsohn,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Cassia Wagner,
Cassia Wagner,
![]() Miguel Paredes,
Miguel Paredes,
![]() Nicola Müller,
Nicola Müller,
![]() Marlin Figgins,
Marlin Figgins,
![]() Denisse Sequeira,
Denisse Sequeira,
![]() Victor Lin,
Victor Lin,
![]() Jennifer Chang
Jennifer Chang






