Real-time tracking of virus evolution
Trevor Bedford (@trvrb)
7 Mar 2017
Pew Annual Meeting
Santa Barbara, CA
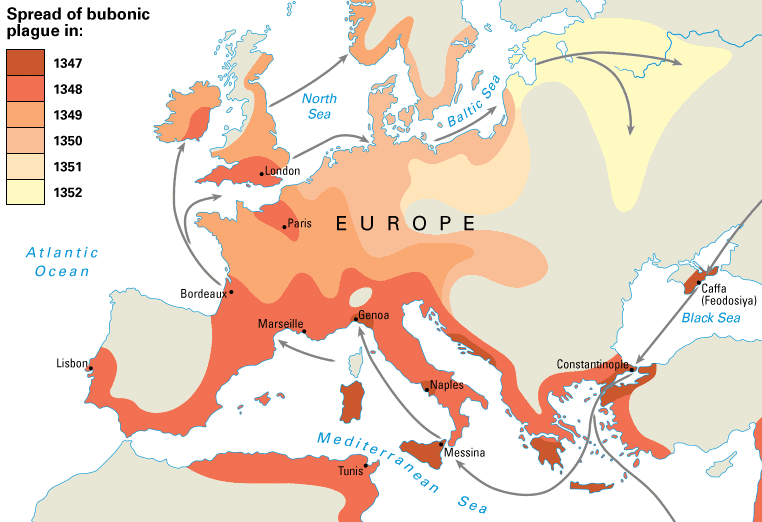
Spread of plague in 14th century
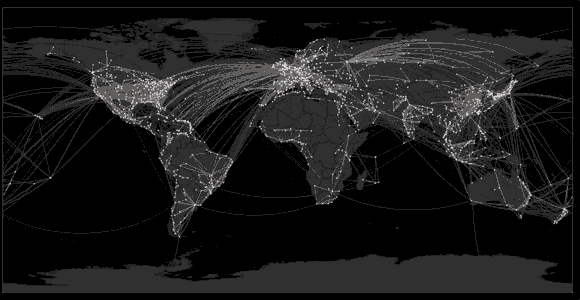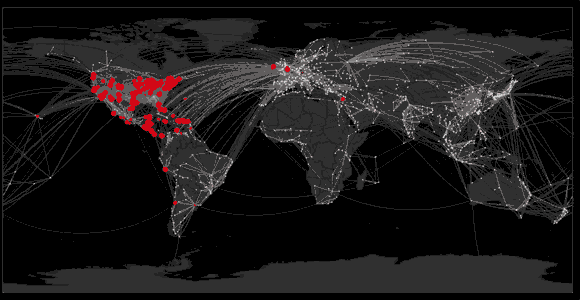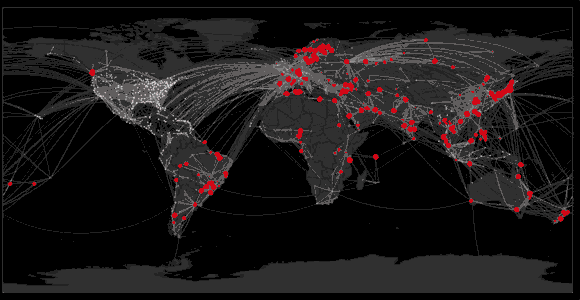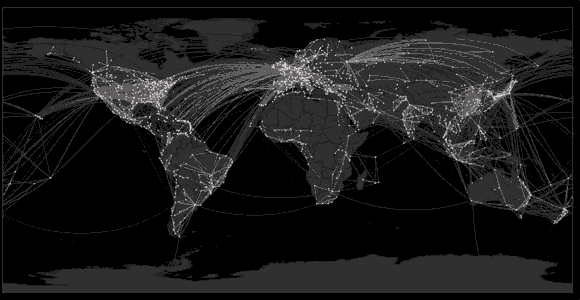
Spread of swine flu in 2009
Sequencing to reconstruct epidemic spread
Epidemic process
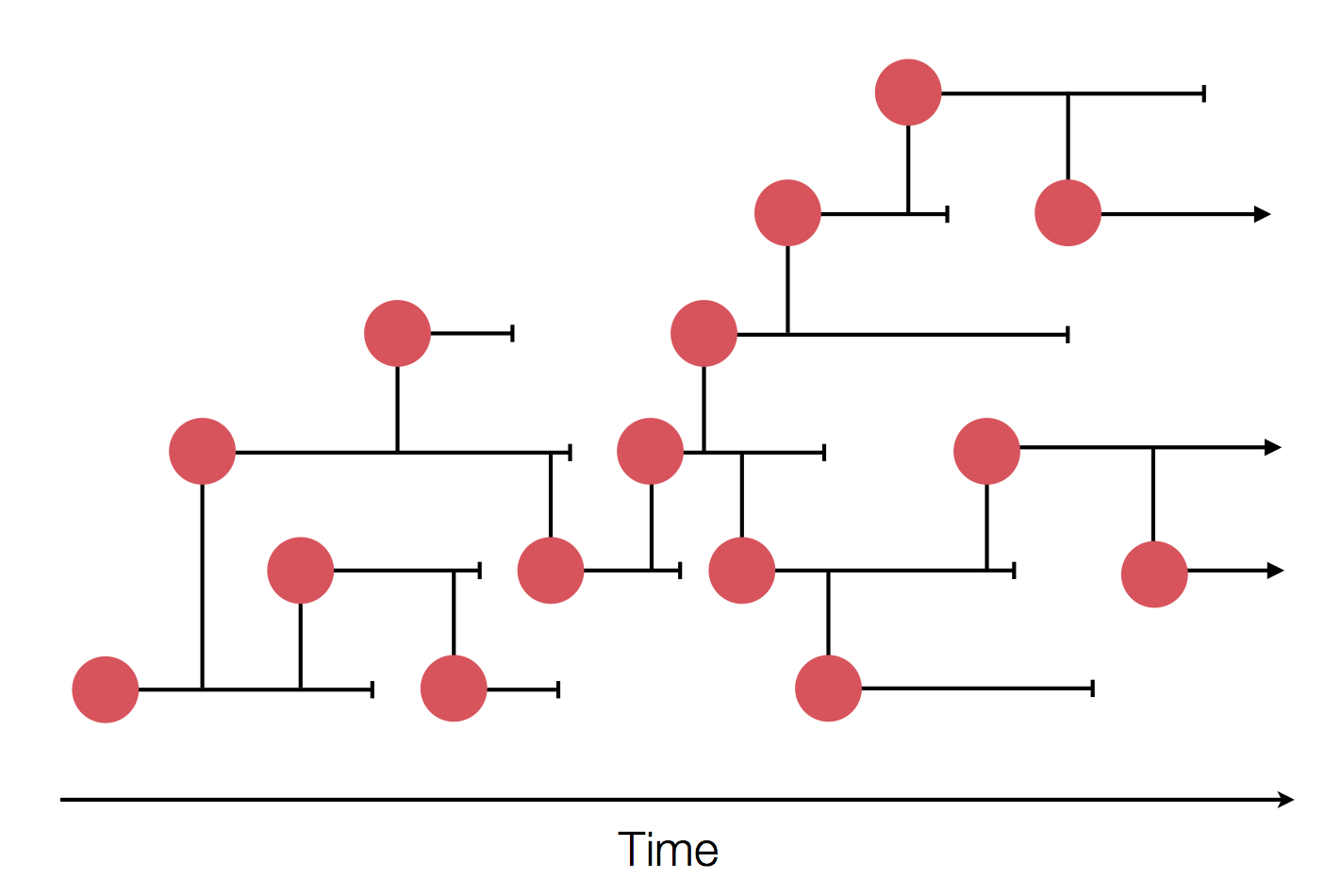
Sample some individuals
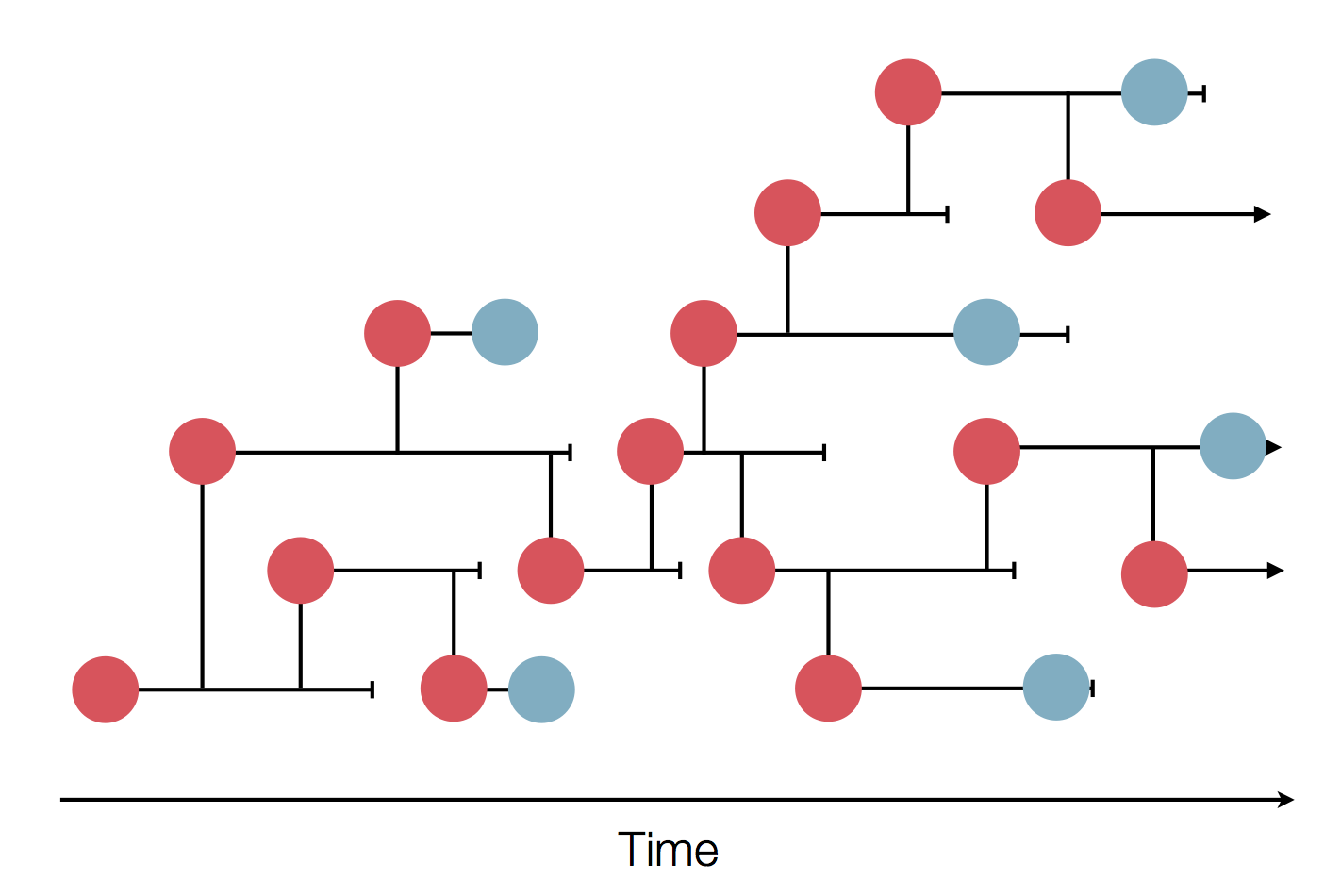
Sequence and determine phylogeny
Sequence and determine phylogeny
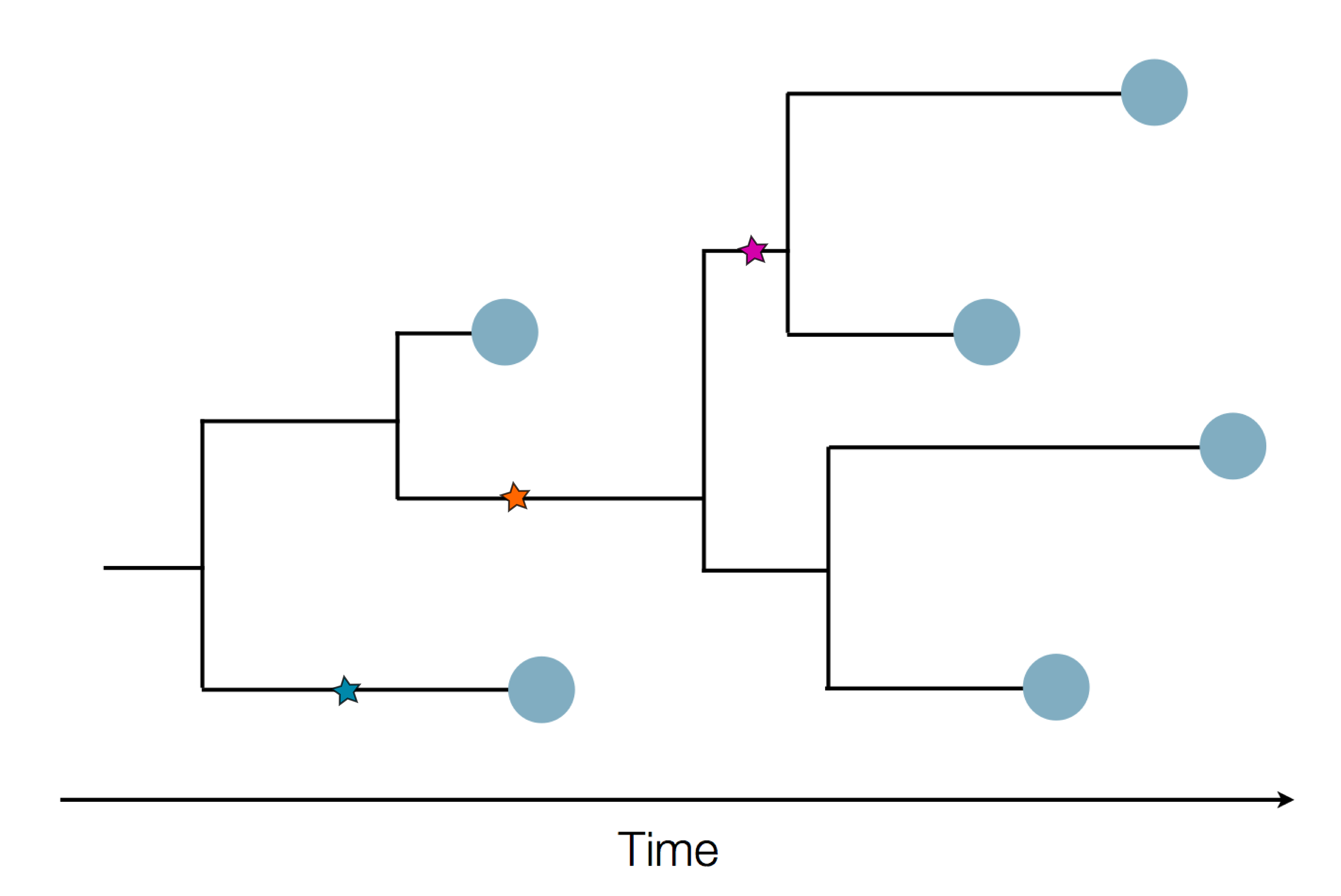
Phylogenetic tracking has the capacity to revolutionize epidemiology
Tracking geographic spread of the Ebola epidemic
with Gytis Dudas, Luiz Carvalho, Marc Suchard, Philippe Lemey, Andrew Rambaut
and many others
Sequencing of 1610 Ebola virus genomes collected during the 2013-2016 West African epidemic
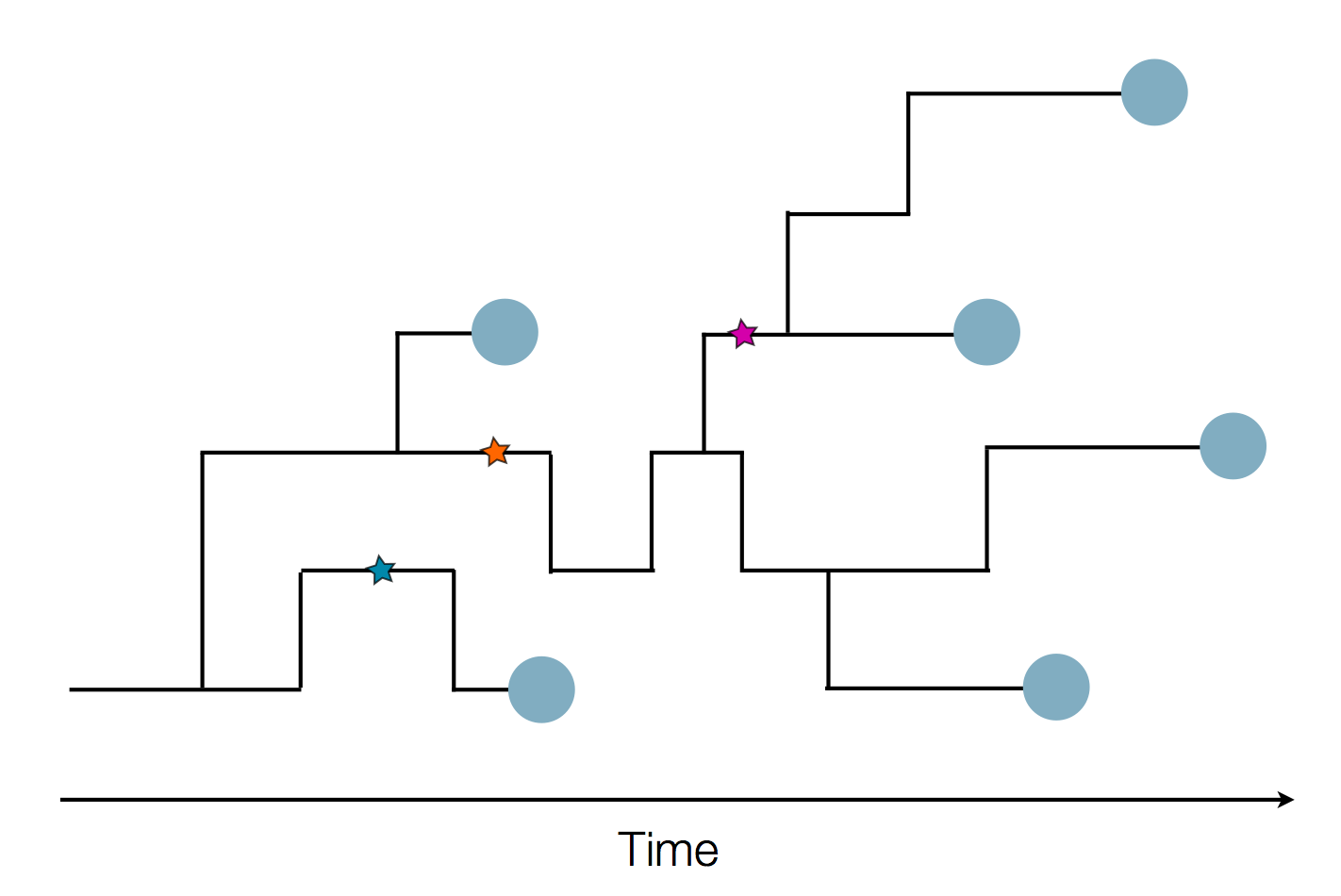
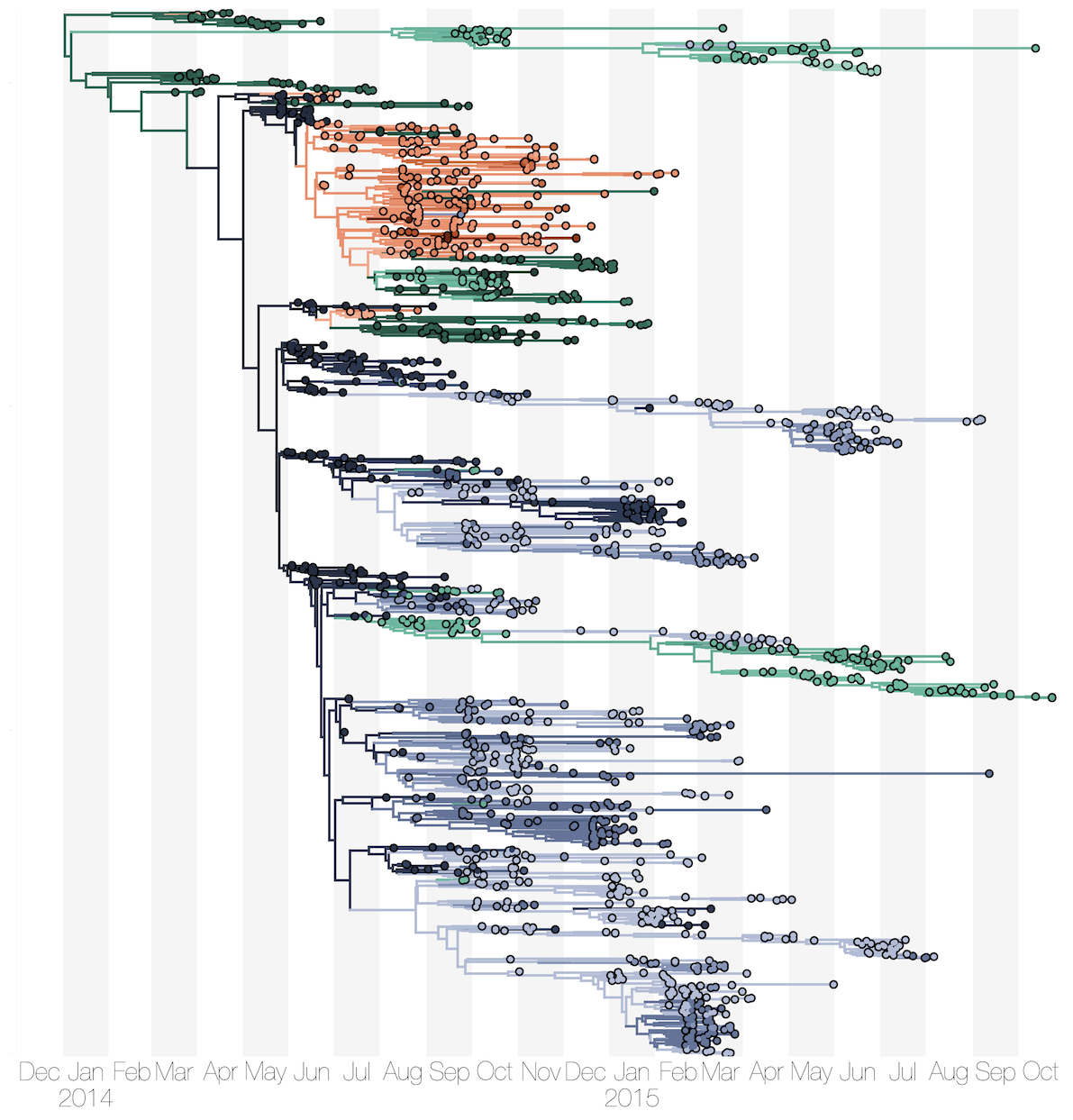
Phylogenetic reconstruction of evolution and spread
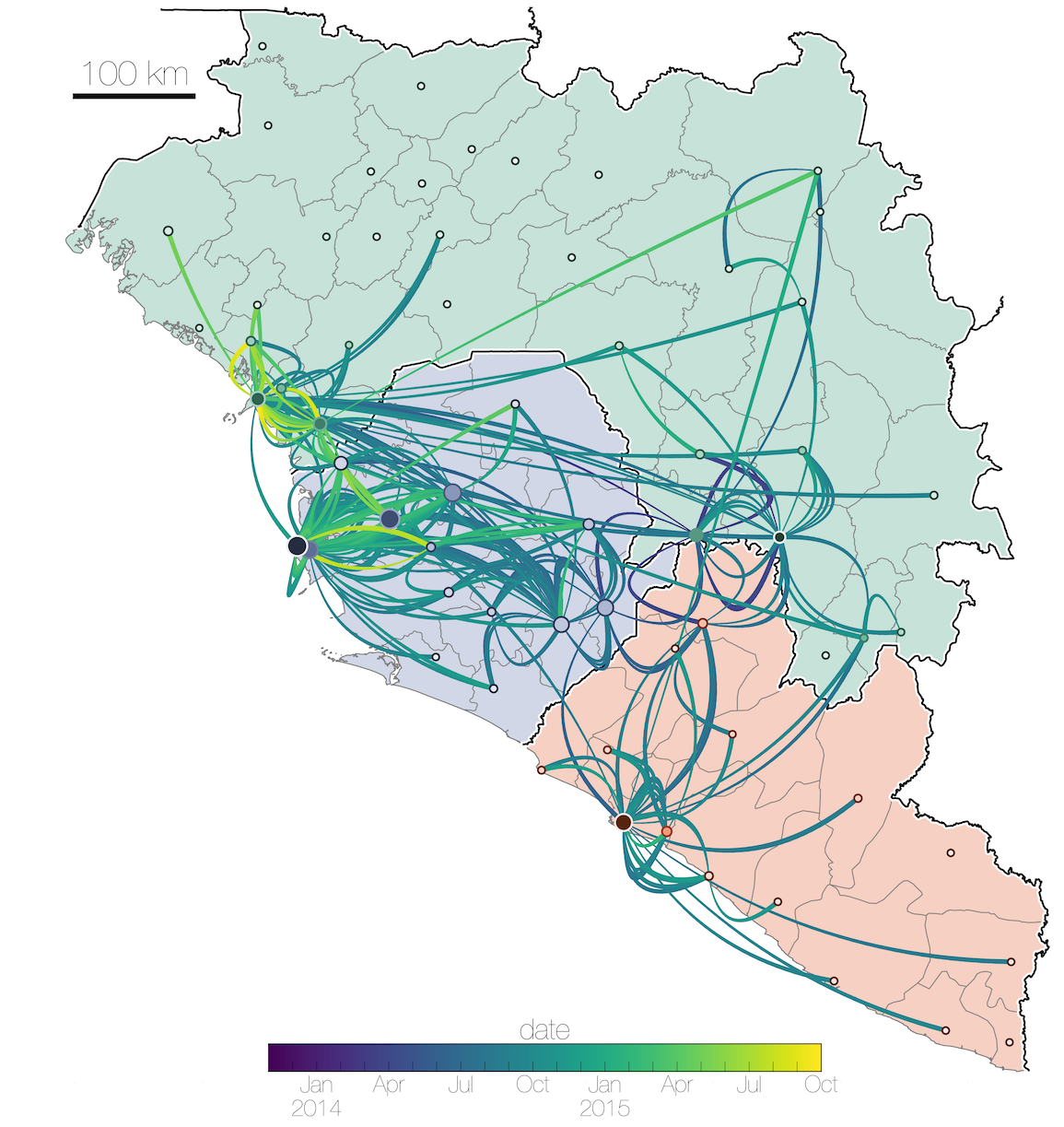
Tracking migration events
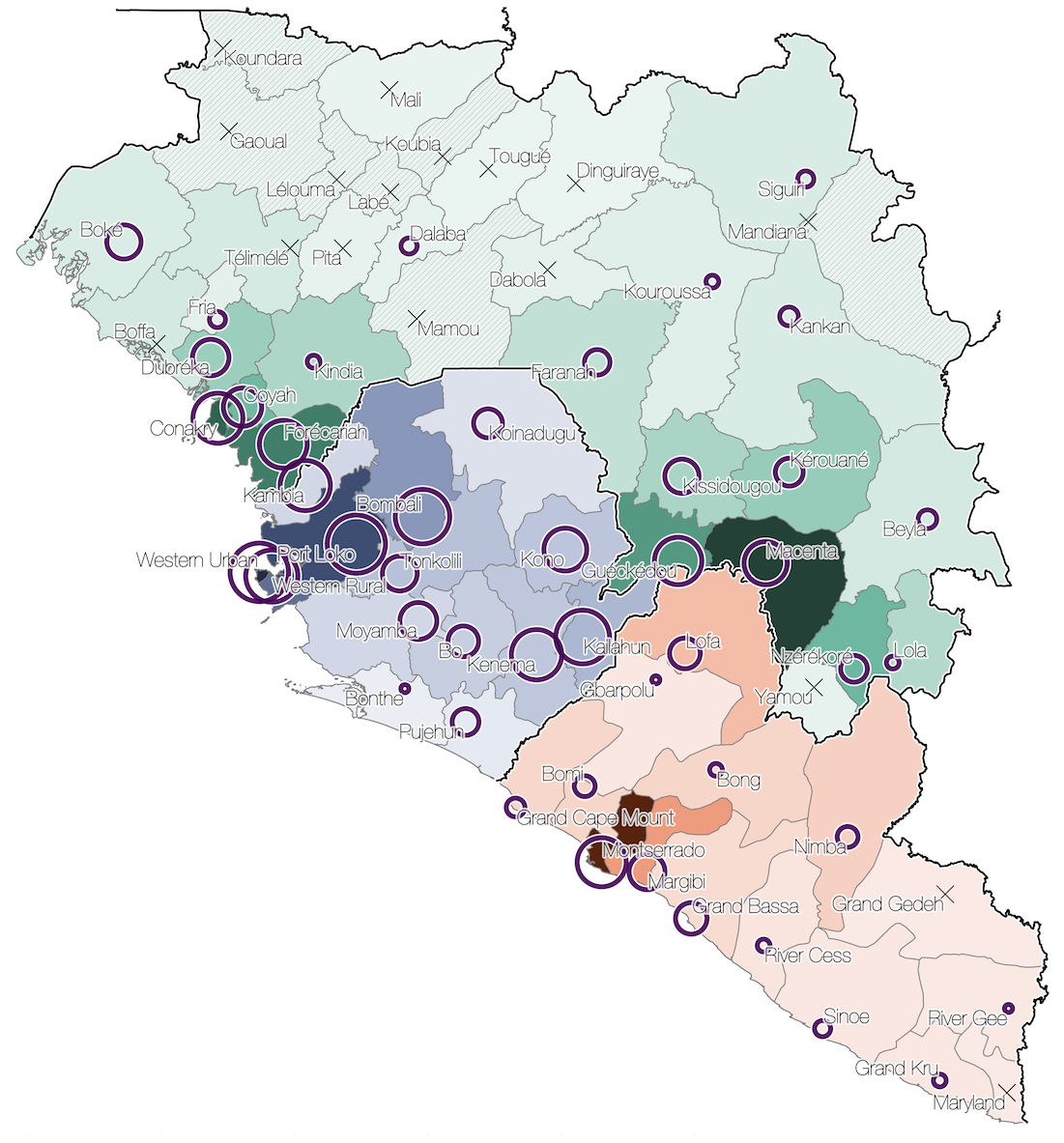
Spatial structure at the country level
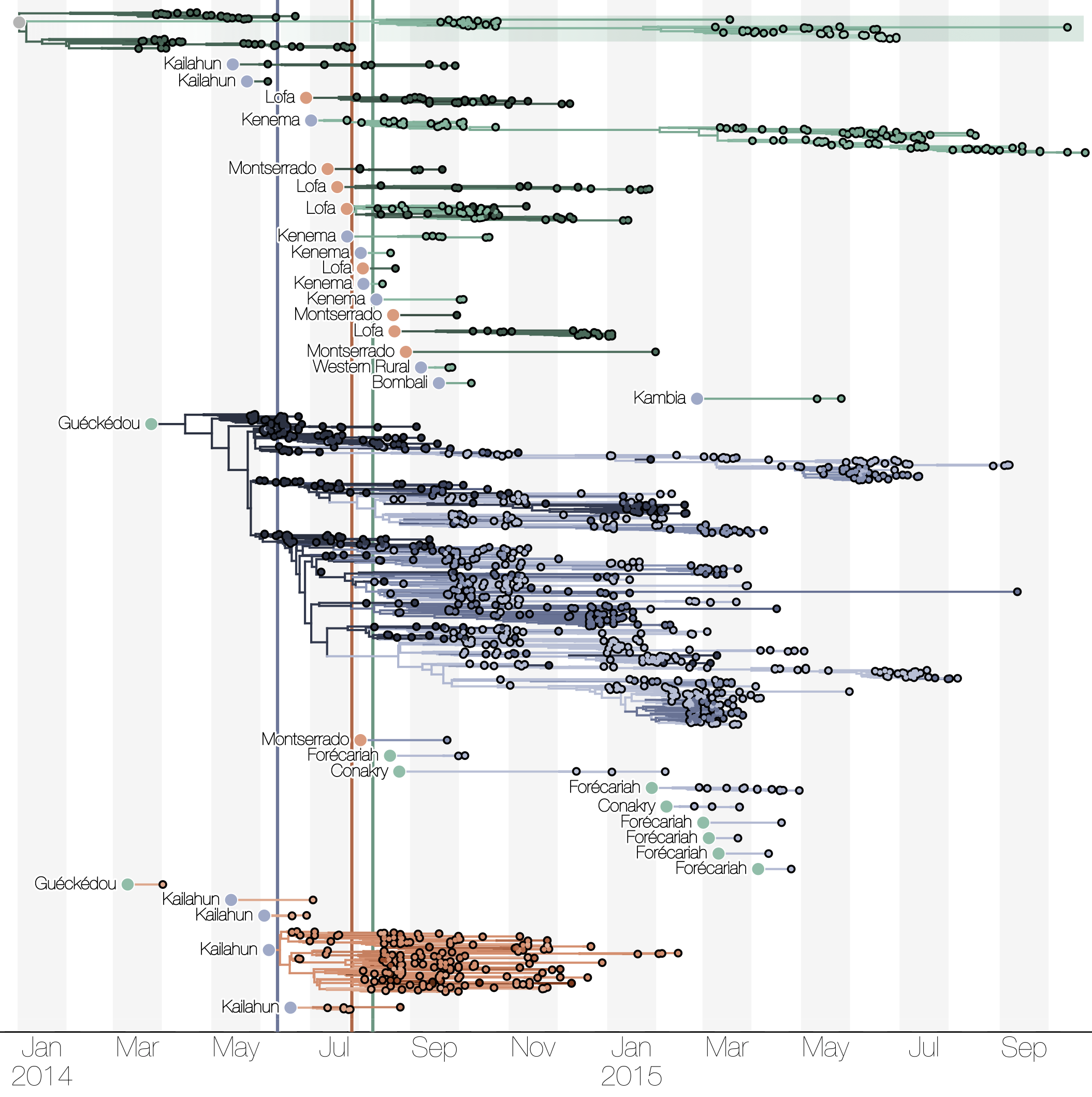
Substantial mixing at the regional level
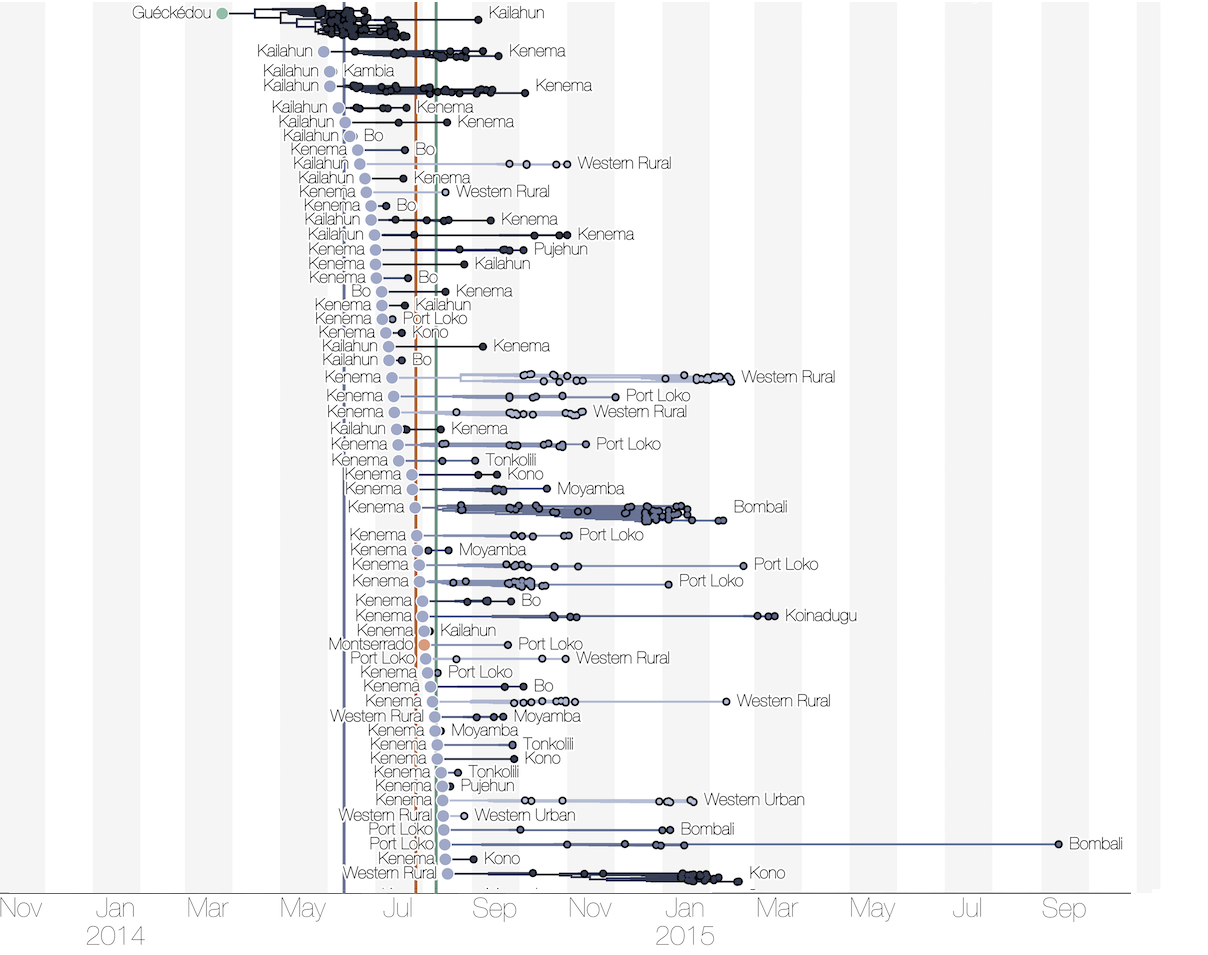
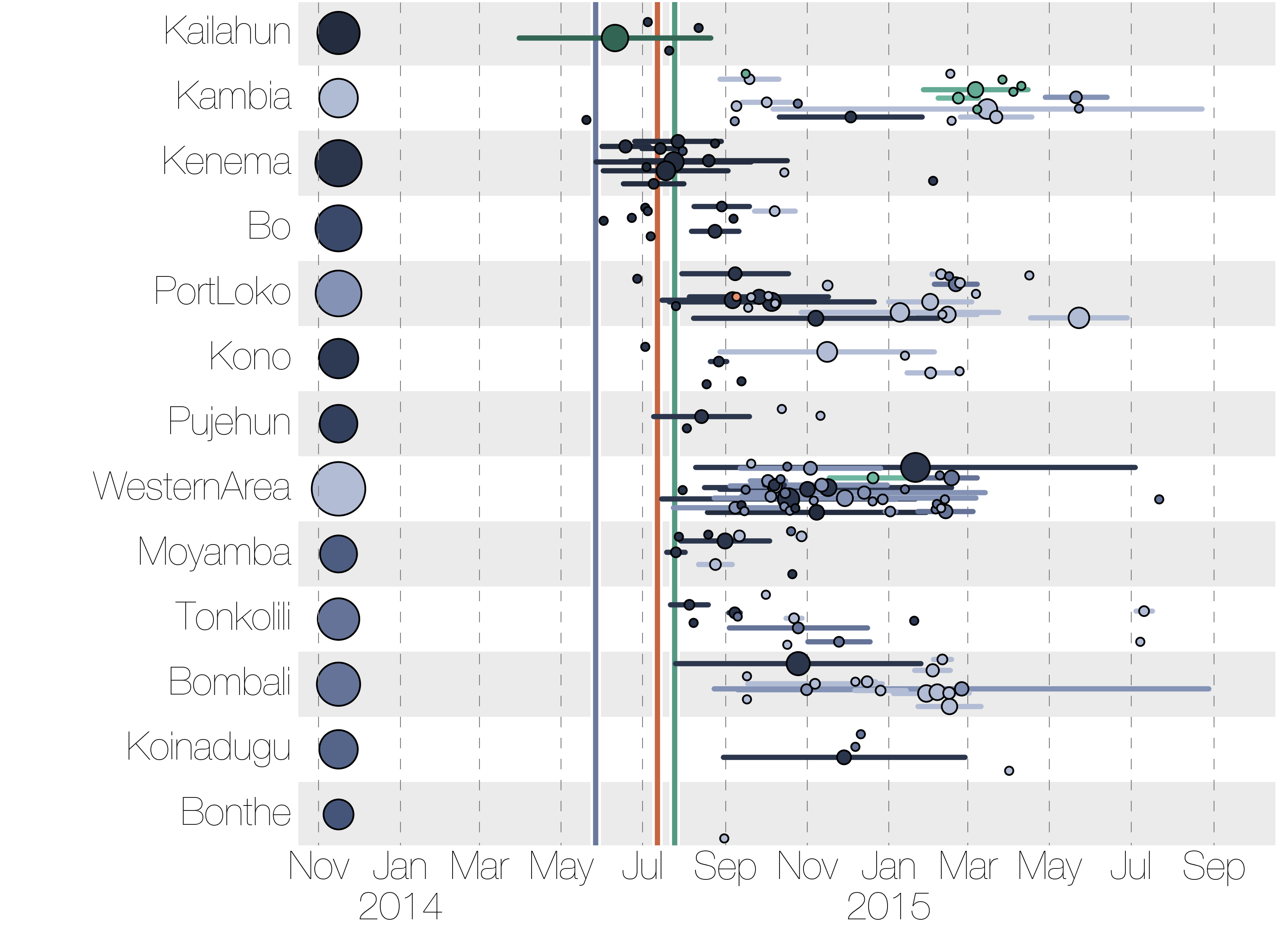
Regional outbreaks due to multiple introductions
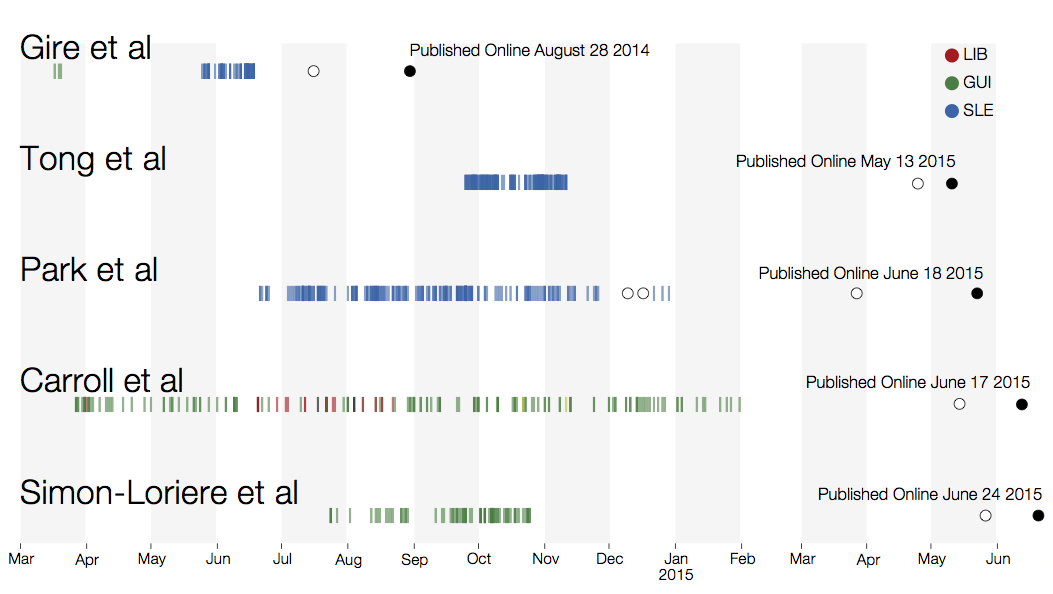
However, this has mostly been done in a retrospective manner
(with some notable exceptions, ala Kristian's recent Zika work)
nextstrain
Project to conduct real-time molecular epidemiology and evolutionary analysis of emerging epidemics
 Richard Neher,
Richard Neher,
 Colin Megill,
Colin Megill,
 James Hadfield,
James Hadfield,
 Charlton Callender,
Charlton Callender,
 Sidney Bell,
Sidney Bell,
 Barney Potter,
Barney Potter,
 Sarah Murata,
Sarah Murata,
nextstrain.org
Rapid on-the-ground sequencing by Ian Goodfellow, Matt Cotten and colleagues
Now with the Zika epidemic, we have the possibility of doing this in a closer to real-time fashion
Zika's arrival and spread in the Americas
Acknowledgements
Ebola: data producers, Gytis Dudas, Andrew Rambaut, Luiz Carvalho, Philippe Lemey, Marc Suchard, Andrew Tatem, Nick Loman, Ian Goodfellow, Matt Cotten, Paul Kellam, Kristian Andersen, Pardis Sabeti, many others
Zika: data producers, Kristian Andersen, Nathan Grubaugh, Nick Loman, Nuno Faria, Oliver Pybus, Josh Quick, Allison Black, Gytis Dudas, many others
Nextstrain: Richard Neher, Colin Megill, James Hadfield, Charlton Callender, Sarah Murata, Sidney Bell, Barney Potter





