Real-time tracking of virus evolution
Trevor Bedford (@trvrb)
7 May 2019
Northwest Data Science Summit
University of Washington
Slides at: bedford.io/talks
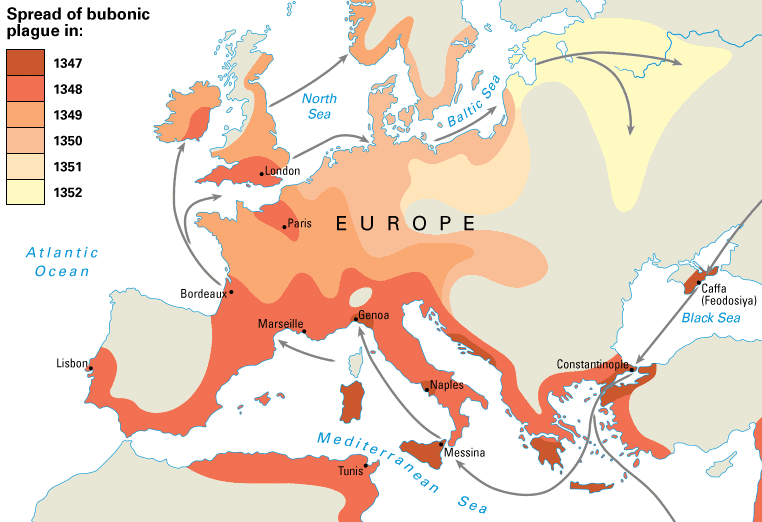
Spread of plague in 14th century
Spread of swine flu in 2009
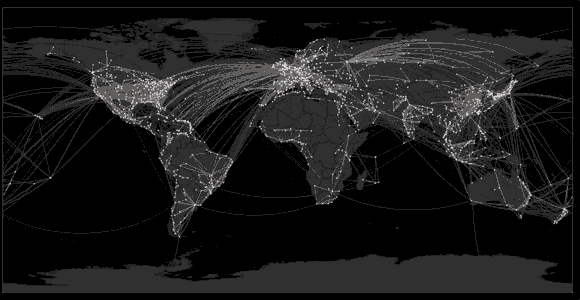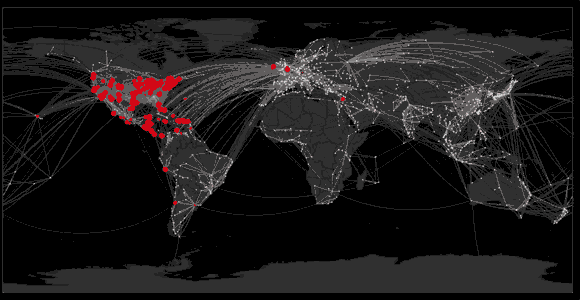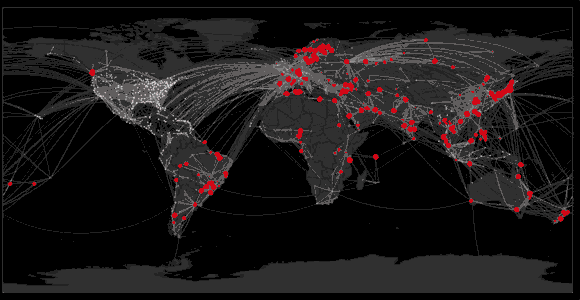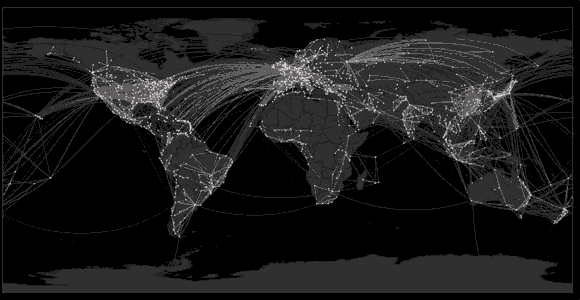
Sequencing to reconstruct pathogen spread
Epidemic process
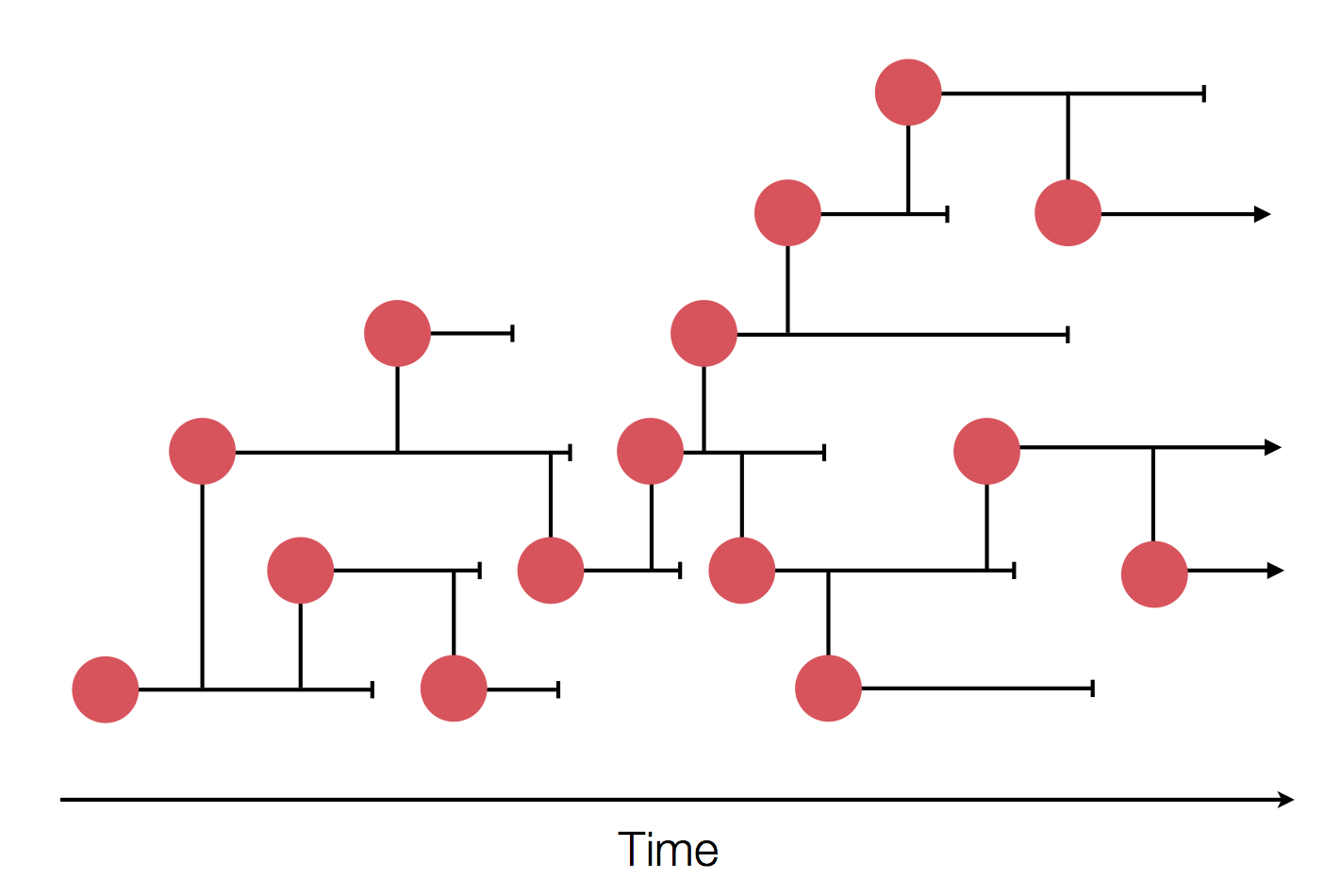
Sample some individuals
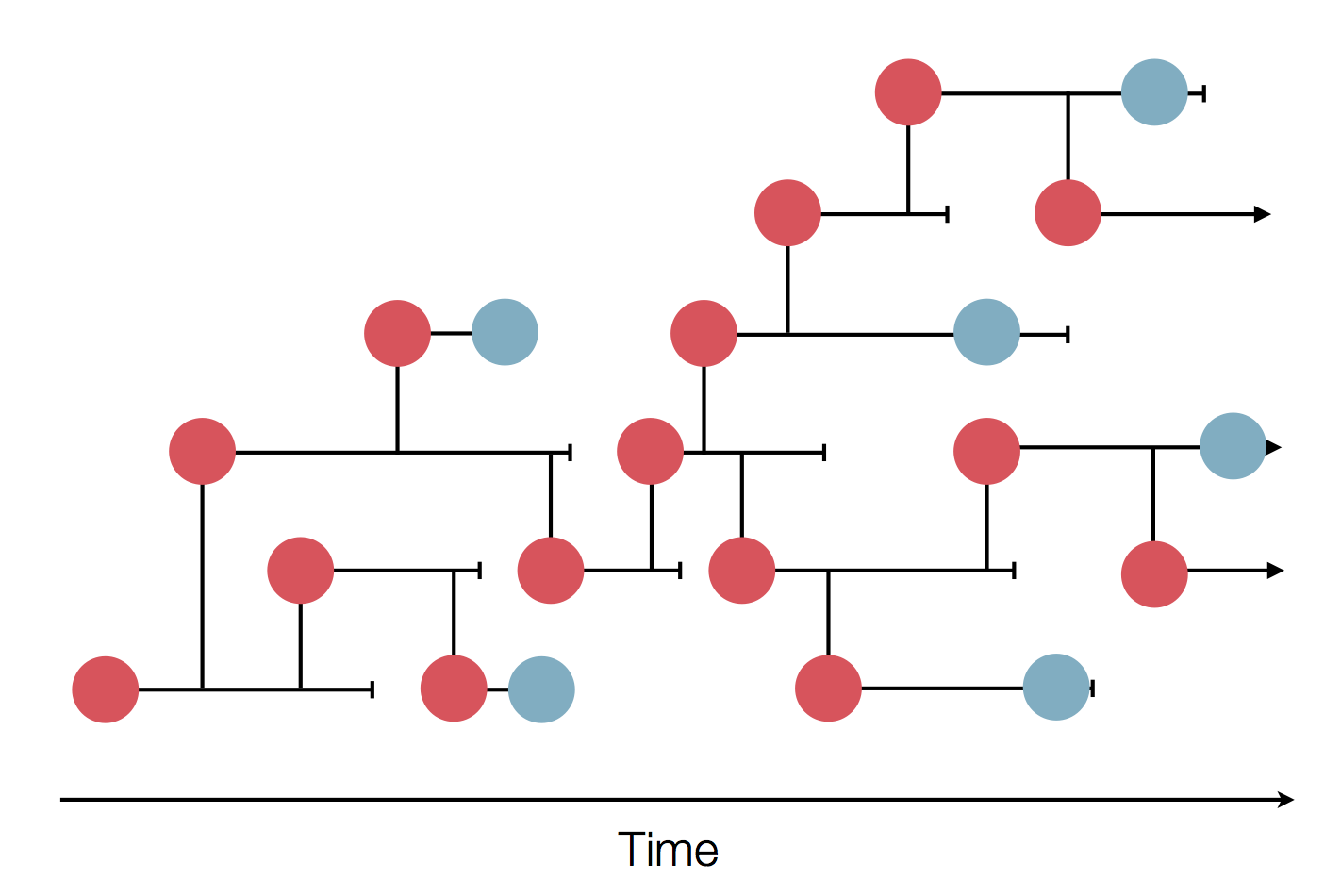
Sequence and determine phylogeny
Sequence and determine phylogeny
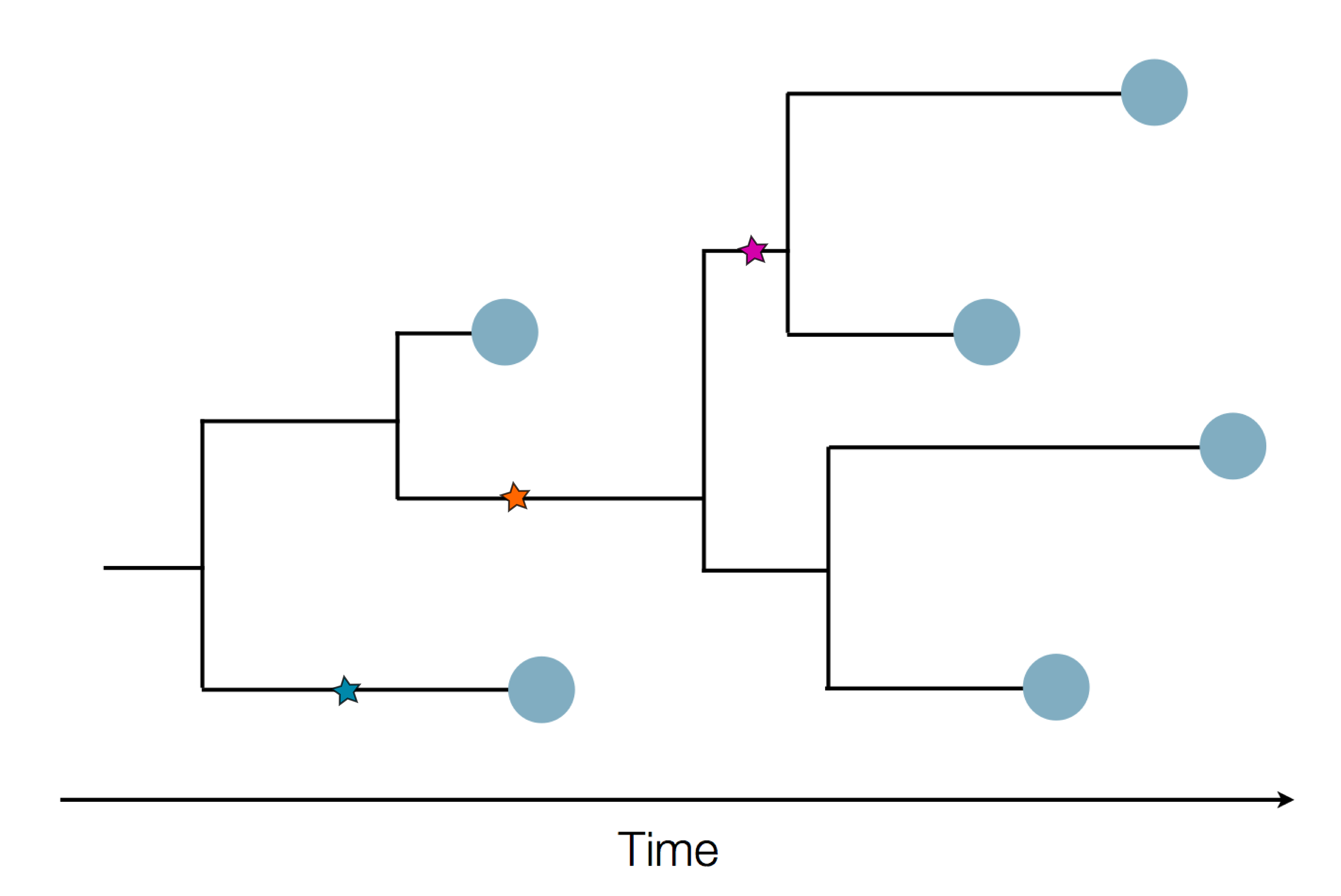
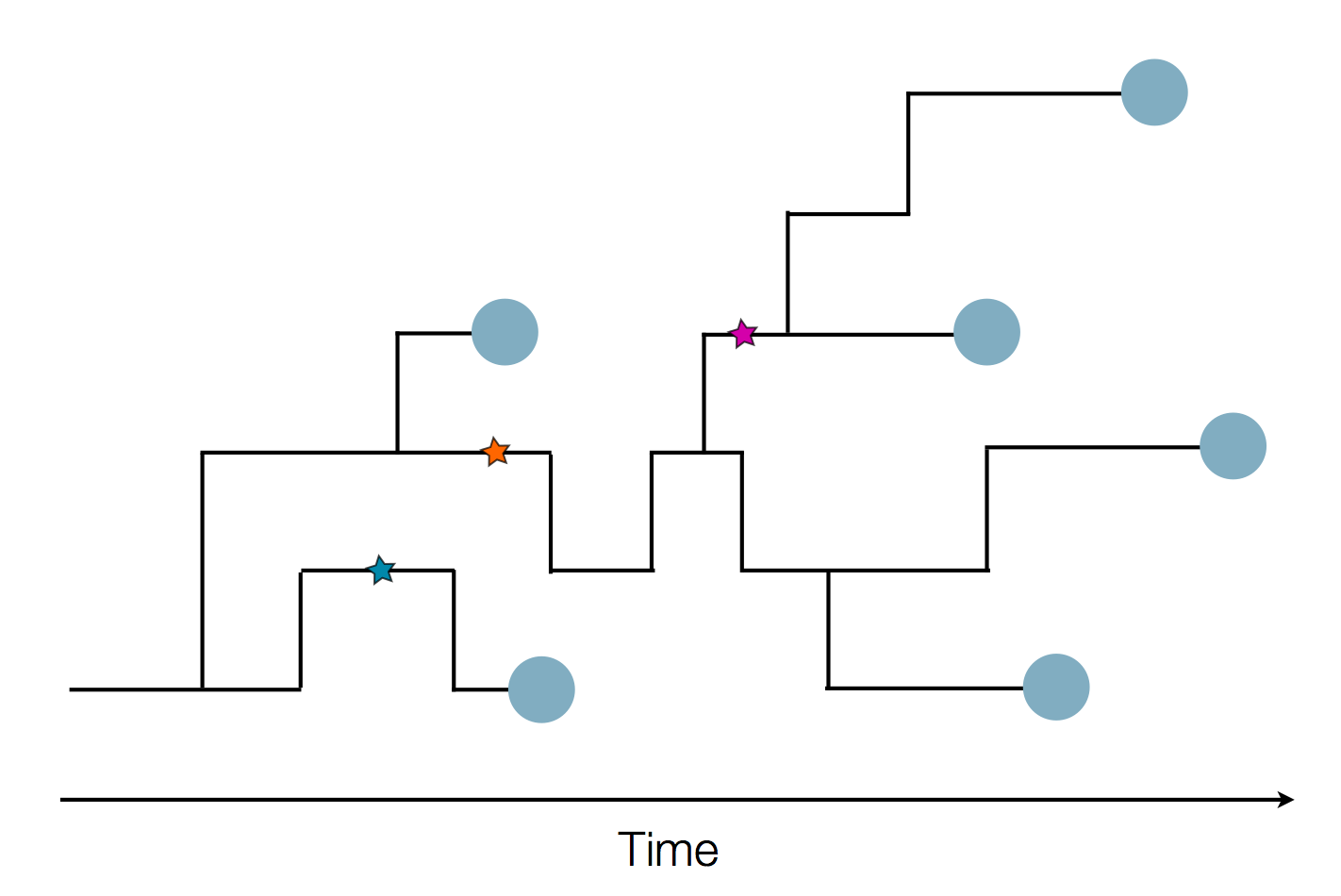
Phylogenetic tracking has the capacity to revolutionize epidemiology
Pathogen genomes may reveal:
- Evolution of new adaptive variants
- Epidemic origins
- Patterns of geographic spread
- Animal-to-human spillover
- Transmission chains
Influenza: Forecasting spread of new variants for vaccine strain selection
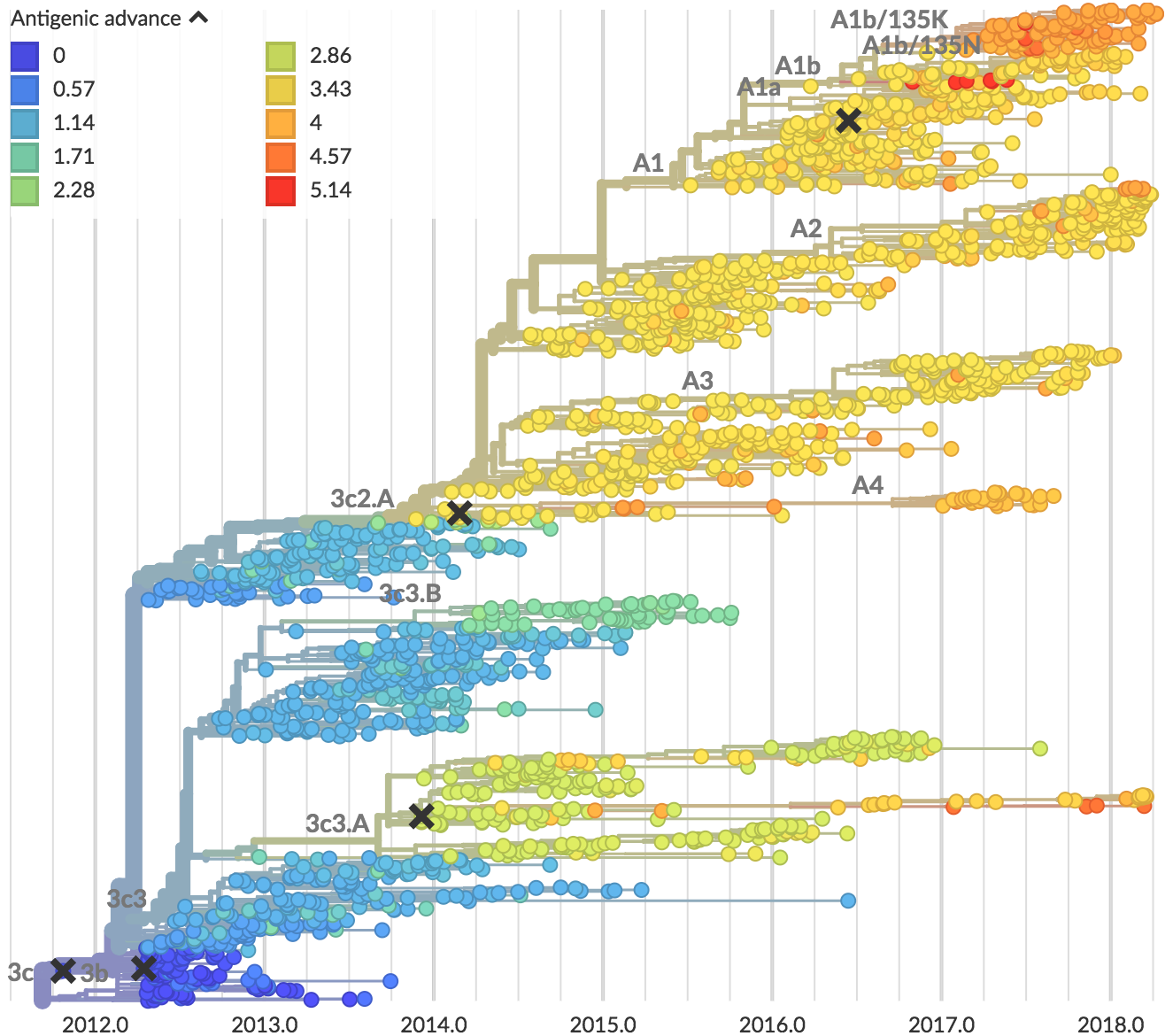
Zika: Uncovering origins of the epidemic in the Americas
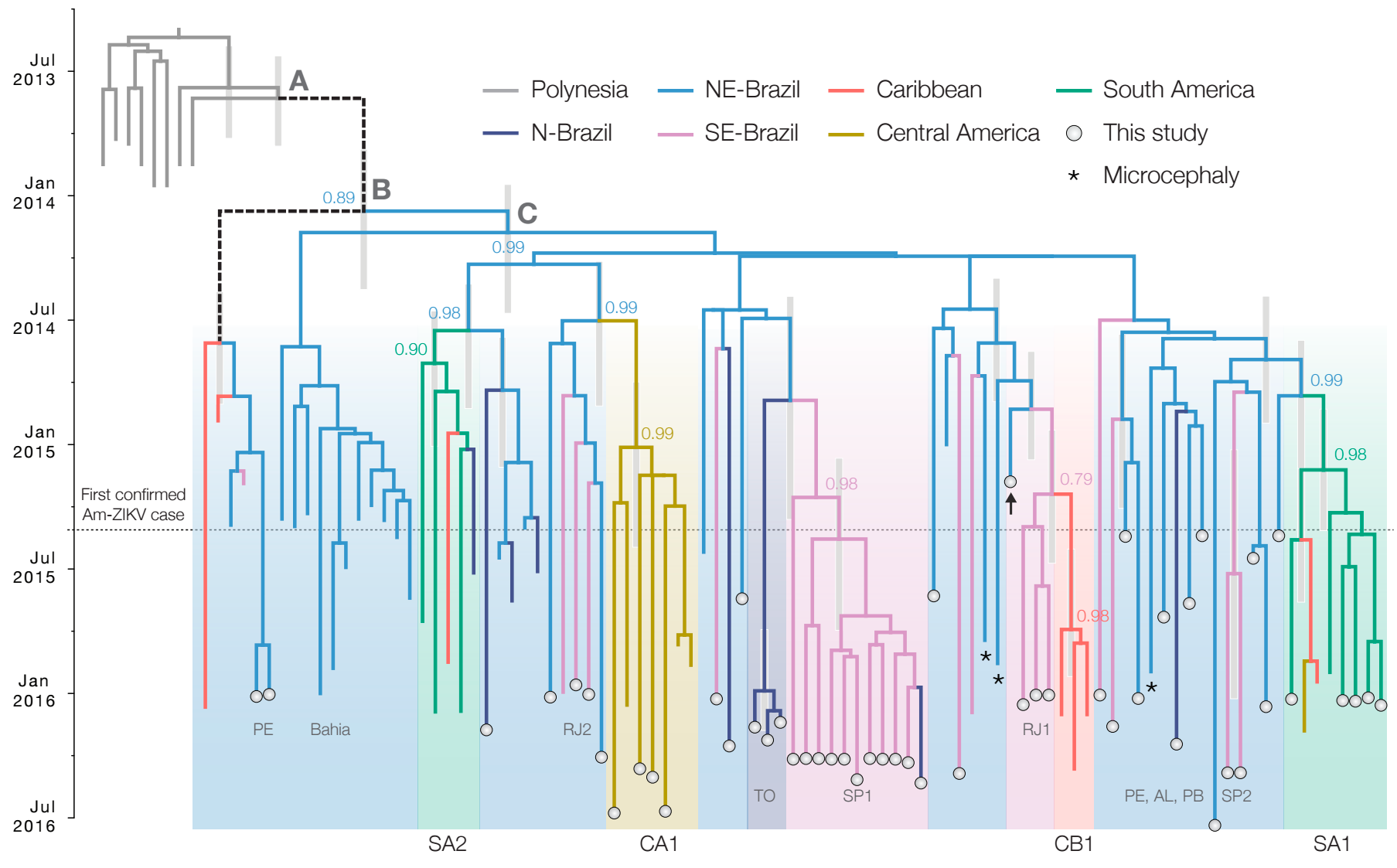
Ebola: Revealing spatial spread and persistence in West Africa
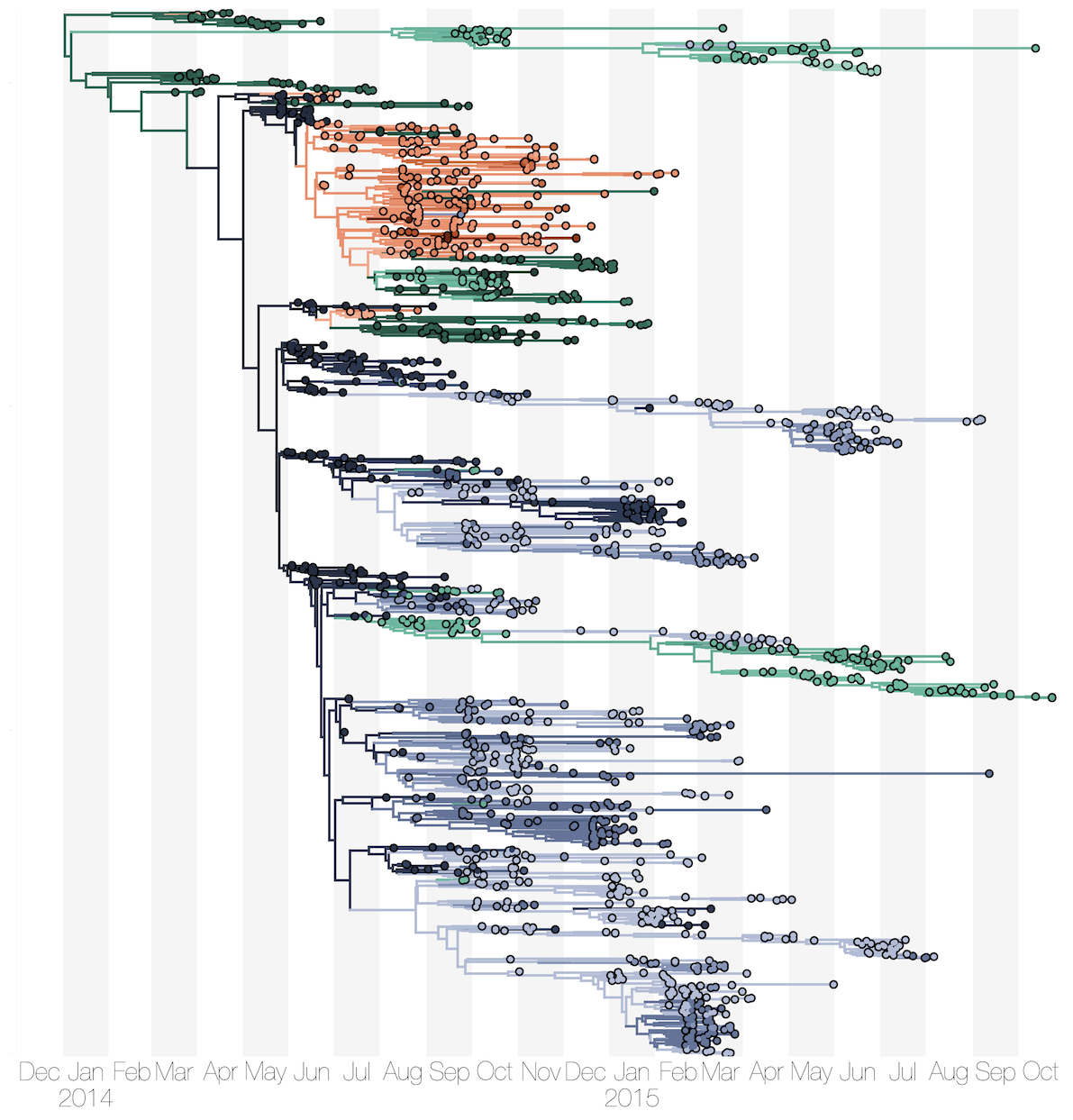
Ebola: Revealing spatial spread and persistence in West Africa
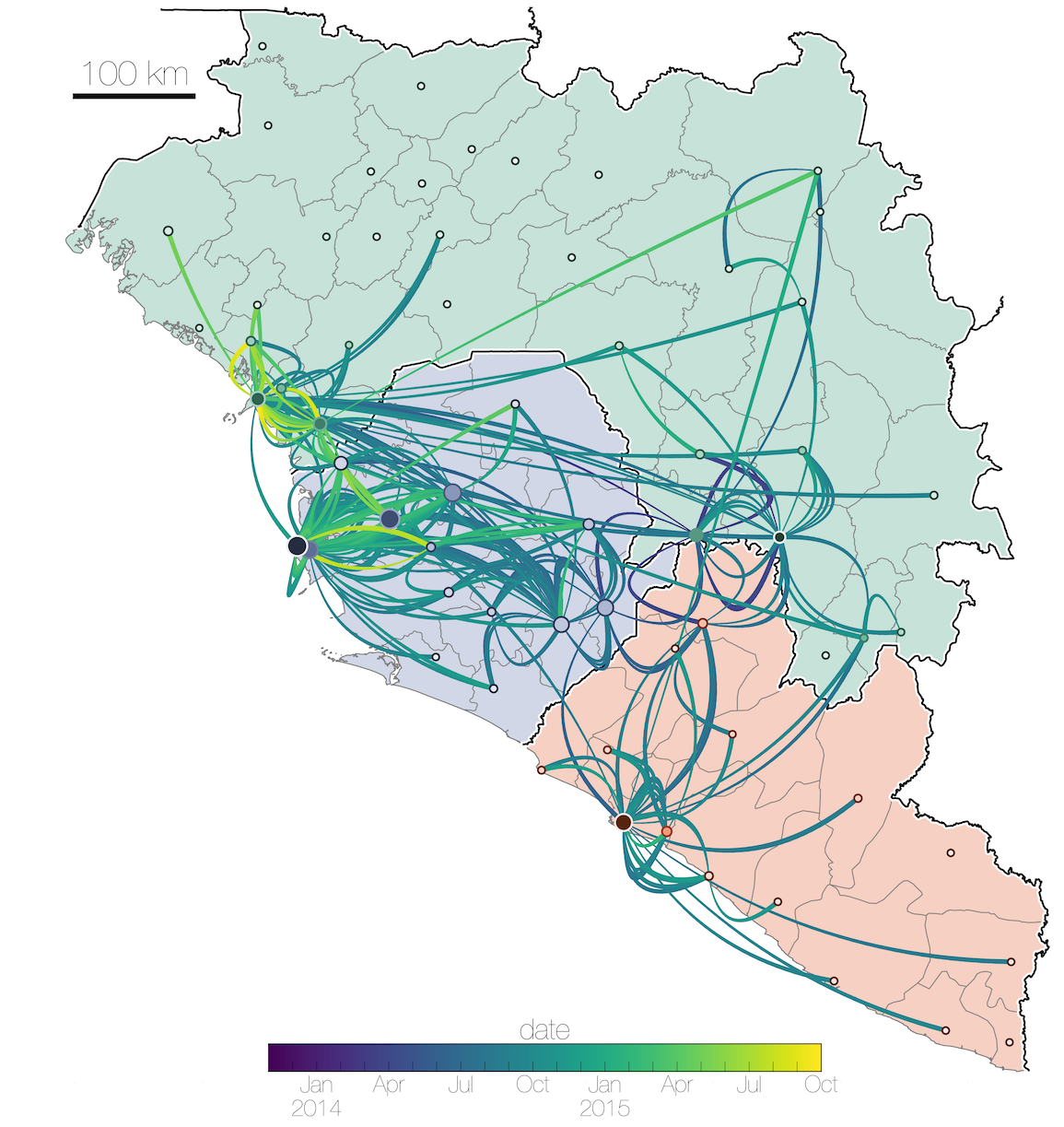
Ebola: Revealing spatial spread and persistence in West Africa
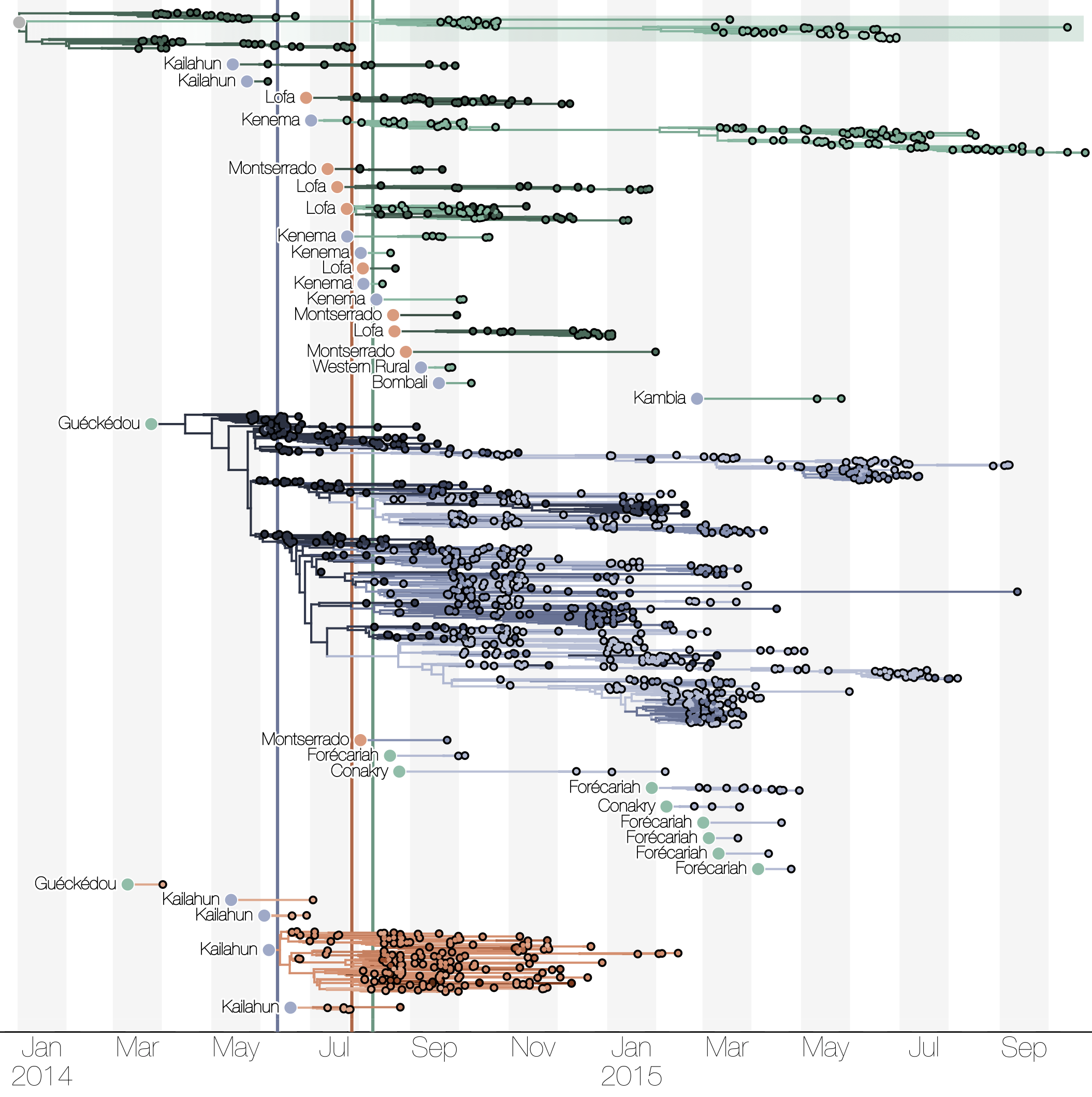
Actionable inferences
Genomic analyses were mostly done in a retrospective manner
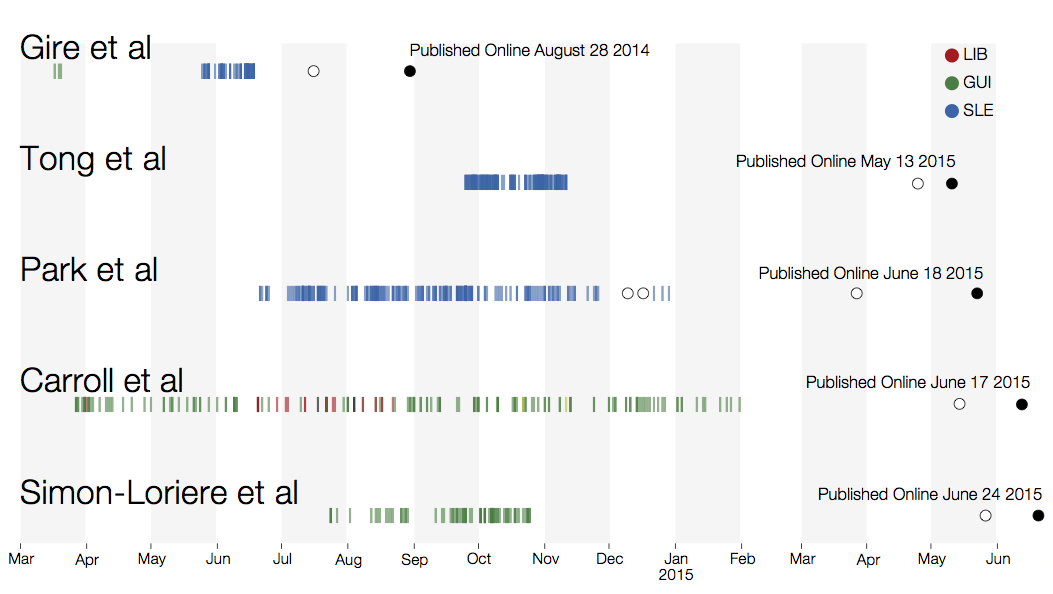
Key challenges to making genomic epidemiology actionable
- Timely analysis and sharing of results critical
- Dissemination must be scalable
- Integrate many data sources
- Results must be easily interpretable and queryable
Nextstrain
Project to conduct real-time molecular epidemiology and evolutionary analysis of emerging epidemics
with
![]() Richard Neher,
Richard Neher,
![]() James Hadfield,
James Hadfield,
![]() Emma Hodcroft,
Emma Hodcroft,
![]() Thomas Sibley,
Thomas Sibley,
![]() John Huddleston,
John Huddleston,
![]() Colin Megill,
Colin Megill,
![]() Sidney Bell,
Sidney Bell,
![]() Barney Potter,
Barney Potter,
![]() Charlton Callender
Charlton Callender
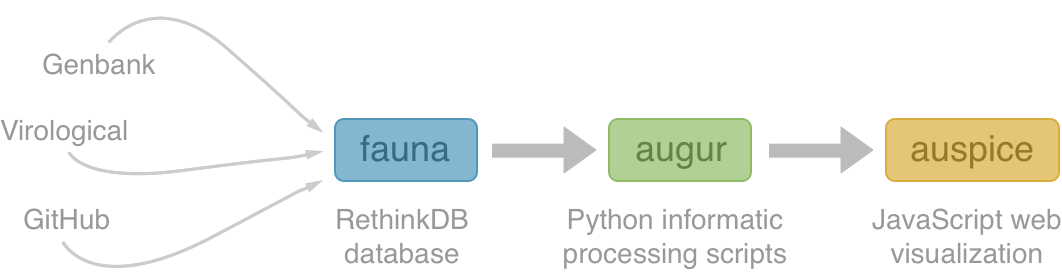
Nextstrain architecture
All code open source at github.com/nextstrain
Two central aims: (1) rapid and flexible phylodynamic analysis and
(2) interactive visualization
Rapid build pipeline for 1600 Ebola genomes
- Align with MAFFT (34 min)
- Build ML tree with RAxML (54 min)
- Temporally resolve tree and geographic ancestry with TreeTime (16 min)
- Total pipeline (1 hr 46 min)
Nextstrain is two things
- a bioinformatics toolkit and visualization app, which can be used for a broad range of datasets
- a collection of real-time pathogen analyses kept up-to-date on the website nextstrain.org
nextstrain.org
Rapid on-the-ground sequencing in Makeni, Sierra Leone
Newly released features
- Bacteria build pipelines using VCF rather than FASTA
- "Community" builds to promote frictionless sharing of results
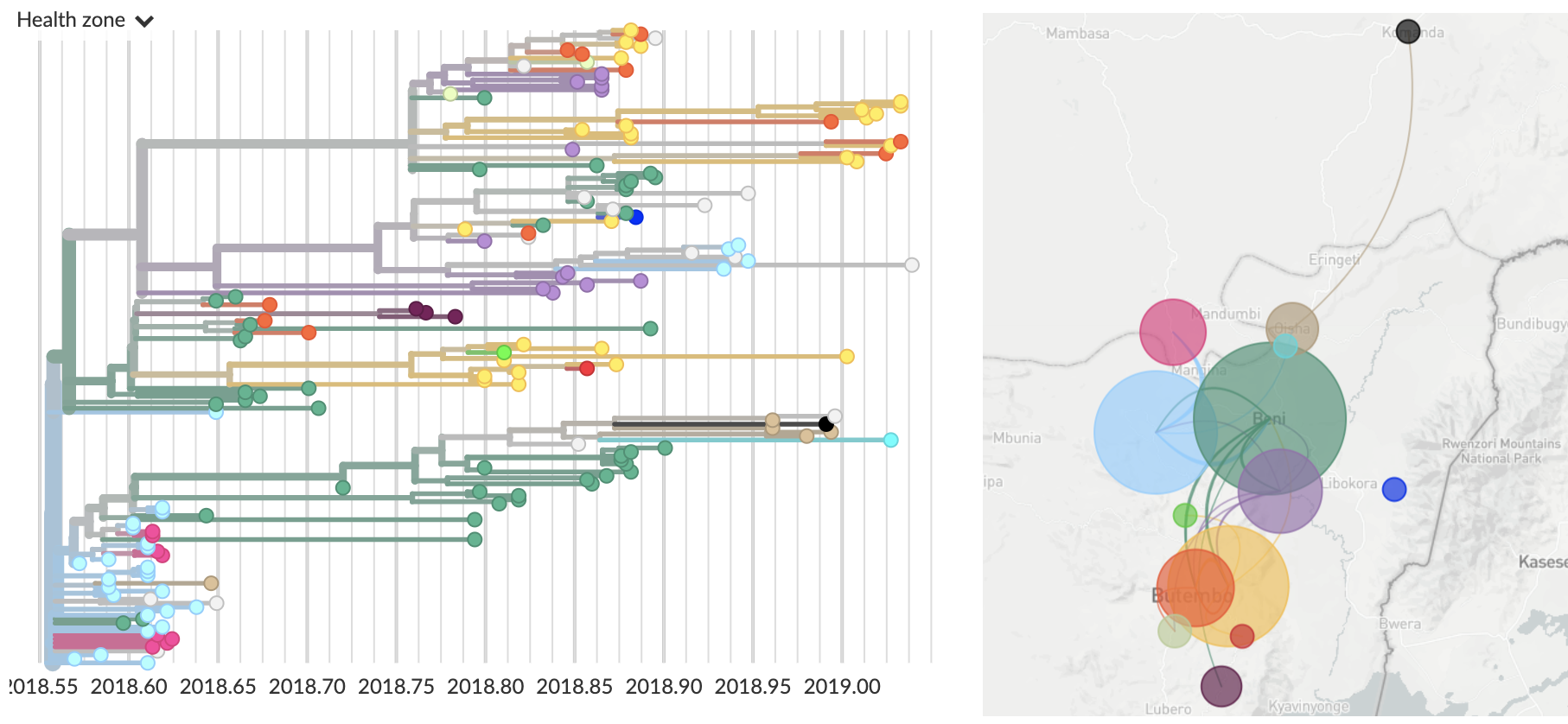
Genomic epidemiology of the ongoing DRC Ebola epidemic






Acknowledgements
Bedford Lab:
![]() Alli Black,
Alli Black,
![]() John Huddleston,
John Huddleston,
![]() Barney Potter,
Barney Potter,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Louise Moncla,
Louise Moncla,
![]() Maya Lewinsohn,
Maya Lewinsohn,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Kairsten Fay,
Kairsten Fay,
![]() Misja Ilcisin
Misja Ilcisin
Genomic epi: Richard Neher, Gytis Dudas, Andrew Rambaut, Luiz Carvalho, Nick Loman, Nuno Faria, Oli Pybus, Josh Quick, Matt Cotten, Ian Goodfellow, Alli Black, Louise Moncla, John Huddleston
Nextstrain: Richard Neher, James Hadfield, Emma Hodcroft, Thomas Sibley, John Huddleston, Sidney Bell, Barney Potter, Colin Megill, Charlton Callender
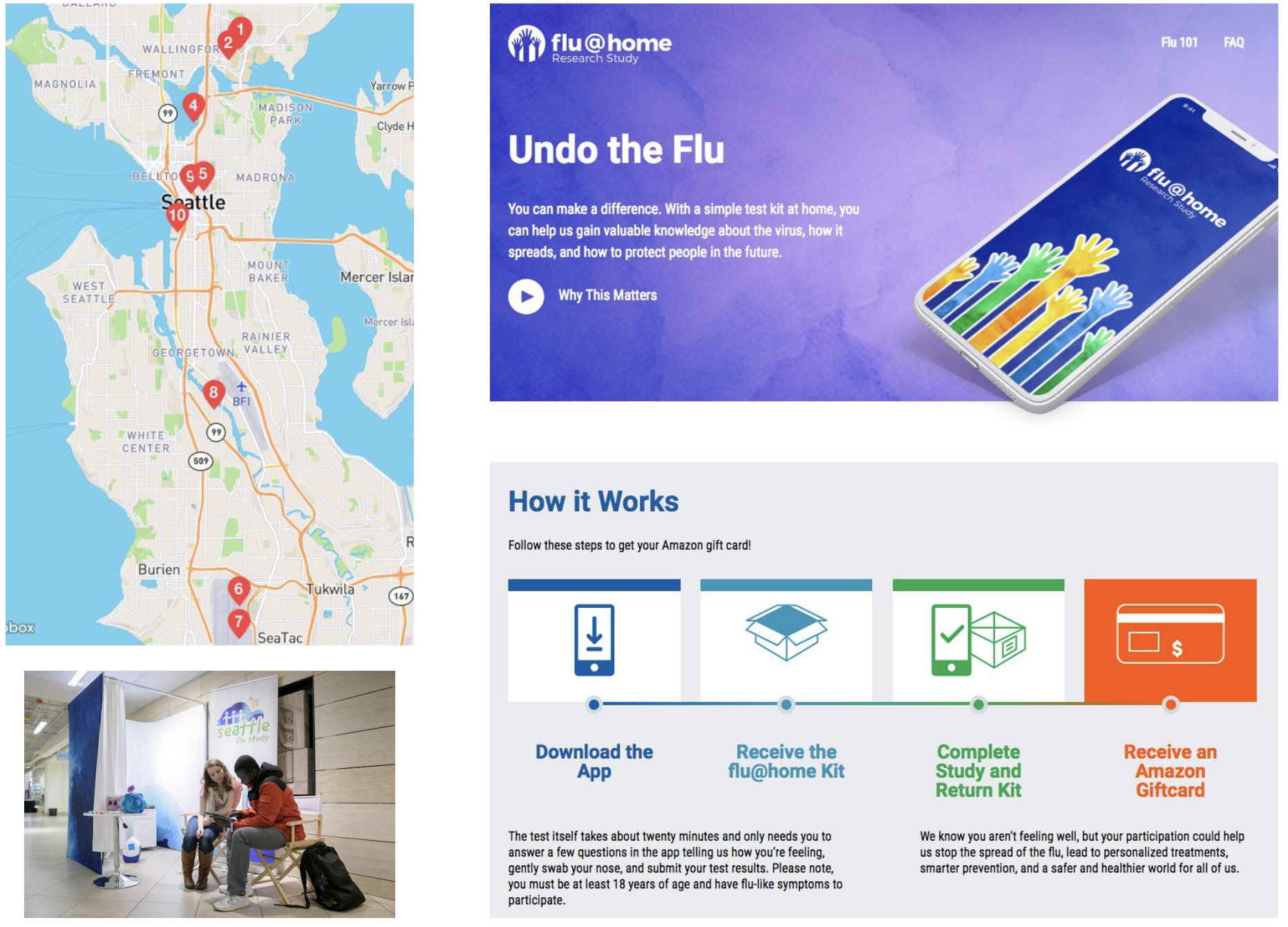
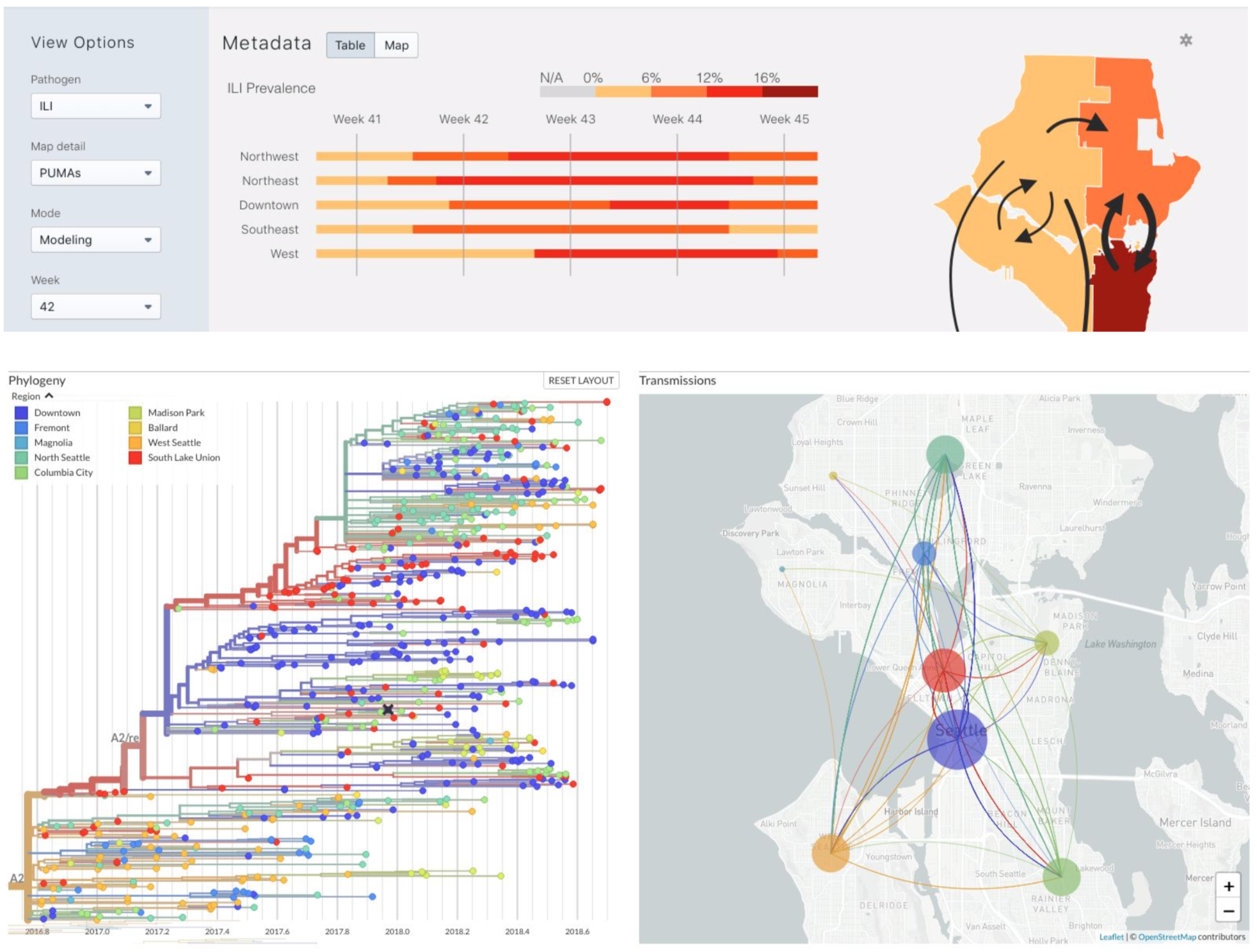
Seattle Flu Study: Helen Chu, Michael Boeckh, Janet Englund, Michael Famulare, Barry Lutz, Debbie Nickerson, Mark Rieder, Lea Starita, Matthew Thompson, Jay Shendure, Jeris Bosua, Thomas Sibley, Louise Moncla, Barney Potter, Jover Lee, Kairsten Fay, Misja Ilcisin, James Hadfield, Antonio Solano





