Real-time genomic surveillance of pathogen evolution and spread
Trevor Bedford (@trvrb)
27 Aug 2018
ICEID 2018
Atlanta, GA
Sequencing to reconstruct pathogen spread
Epidemic process
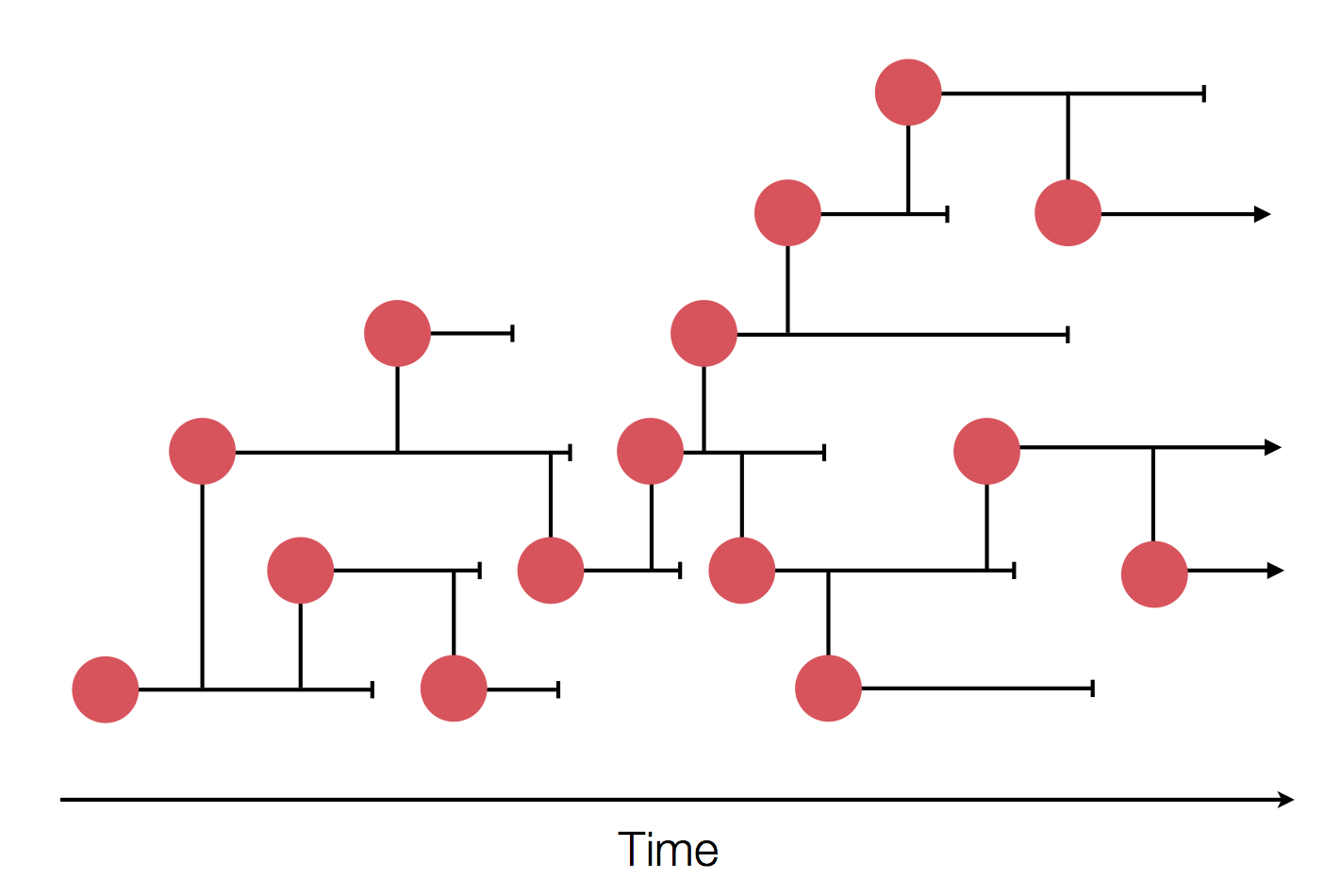
Sample some individuals
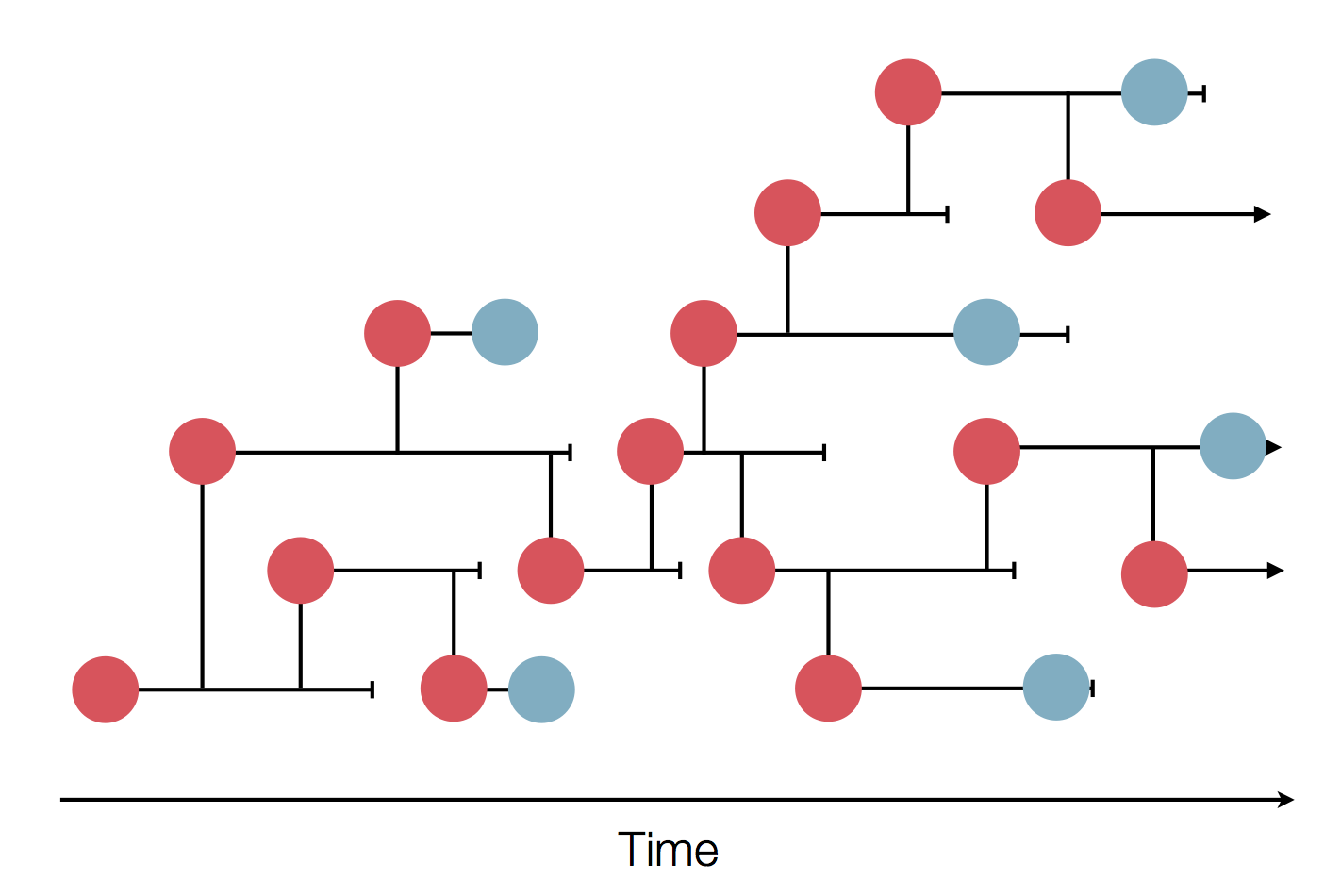
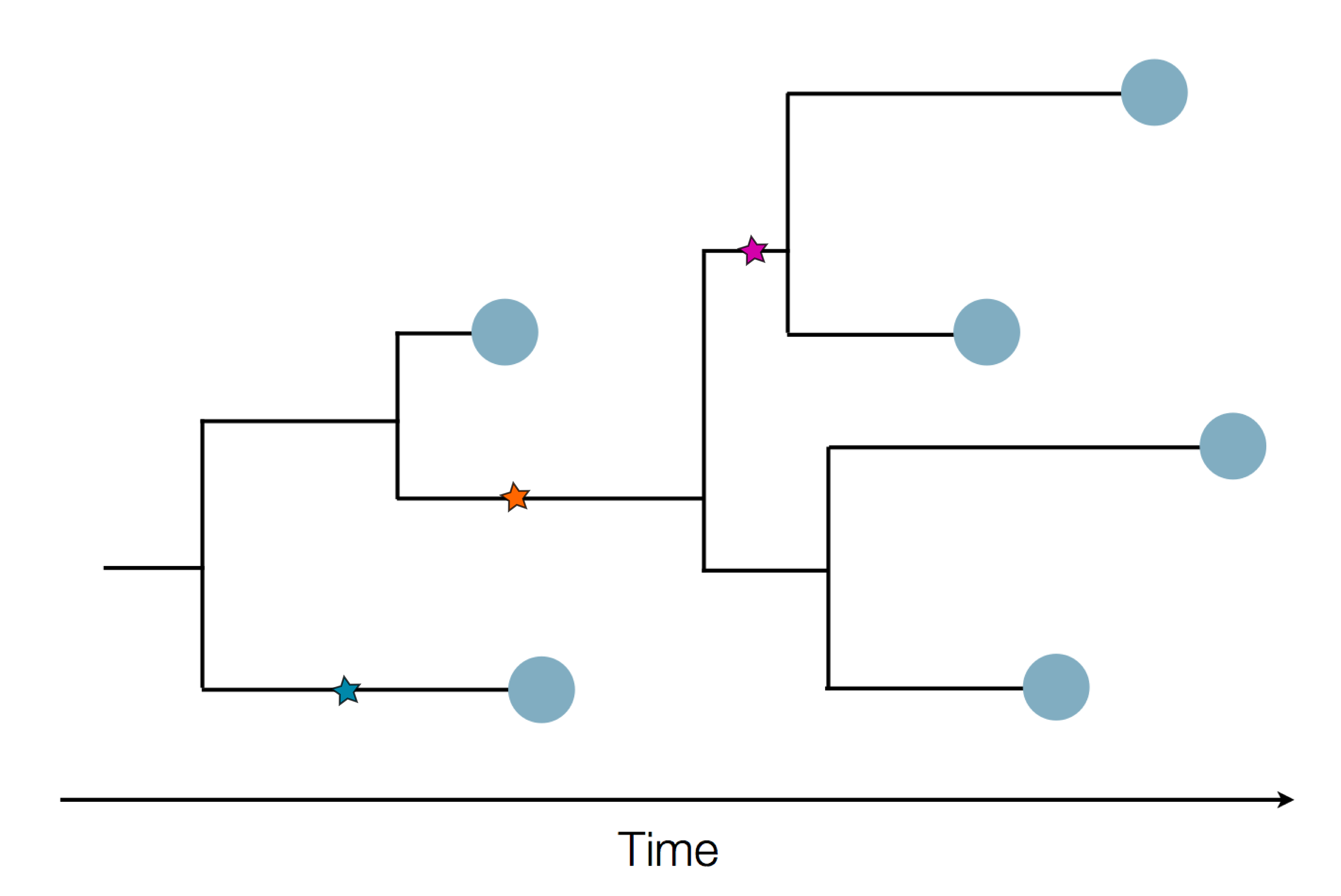
Sequence and determine phylogeny
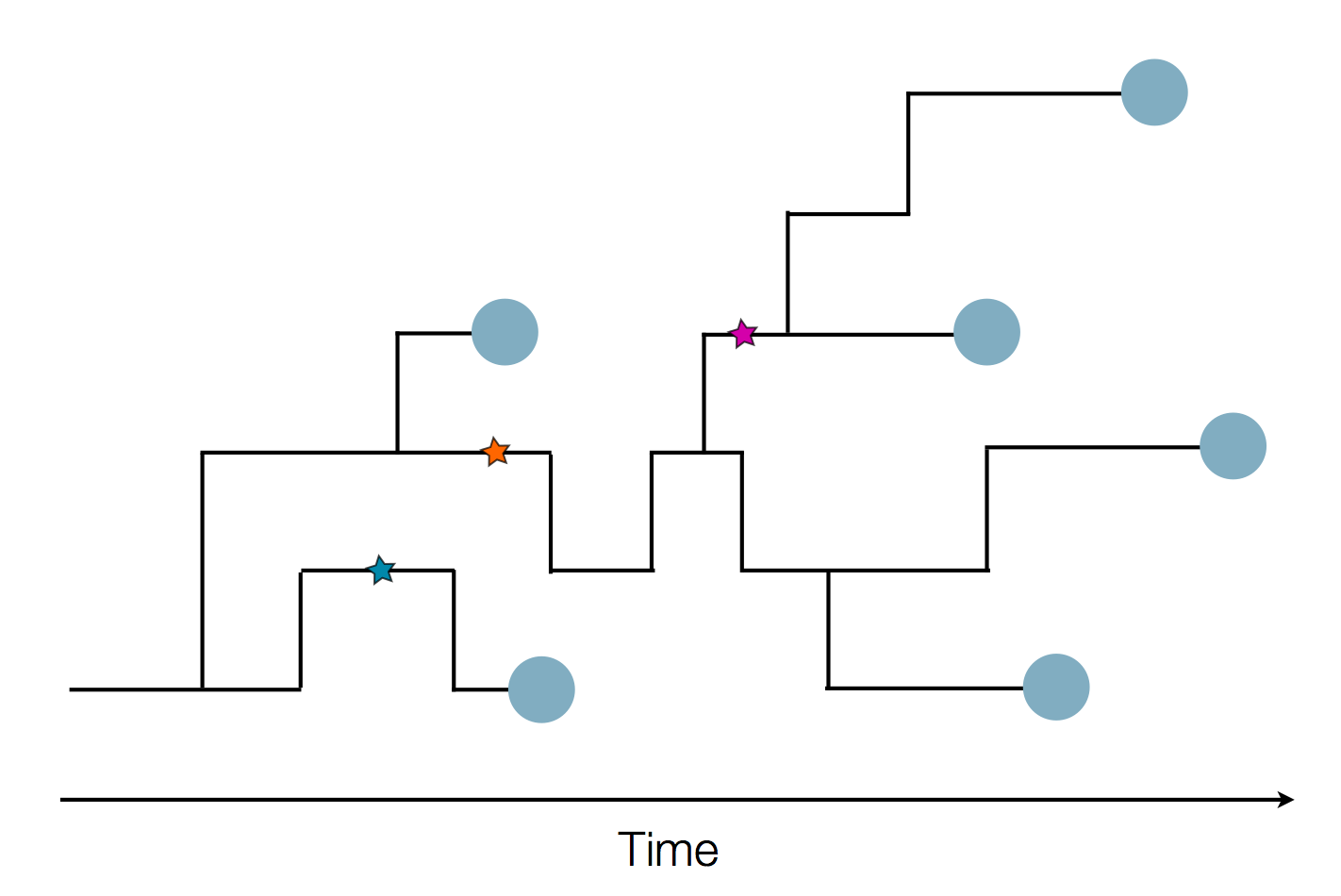
Sequence and determine phylogeny
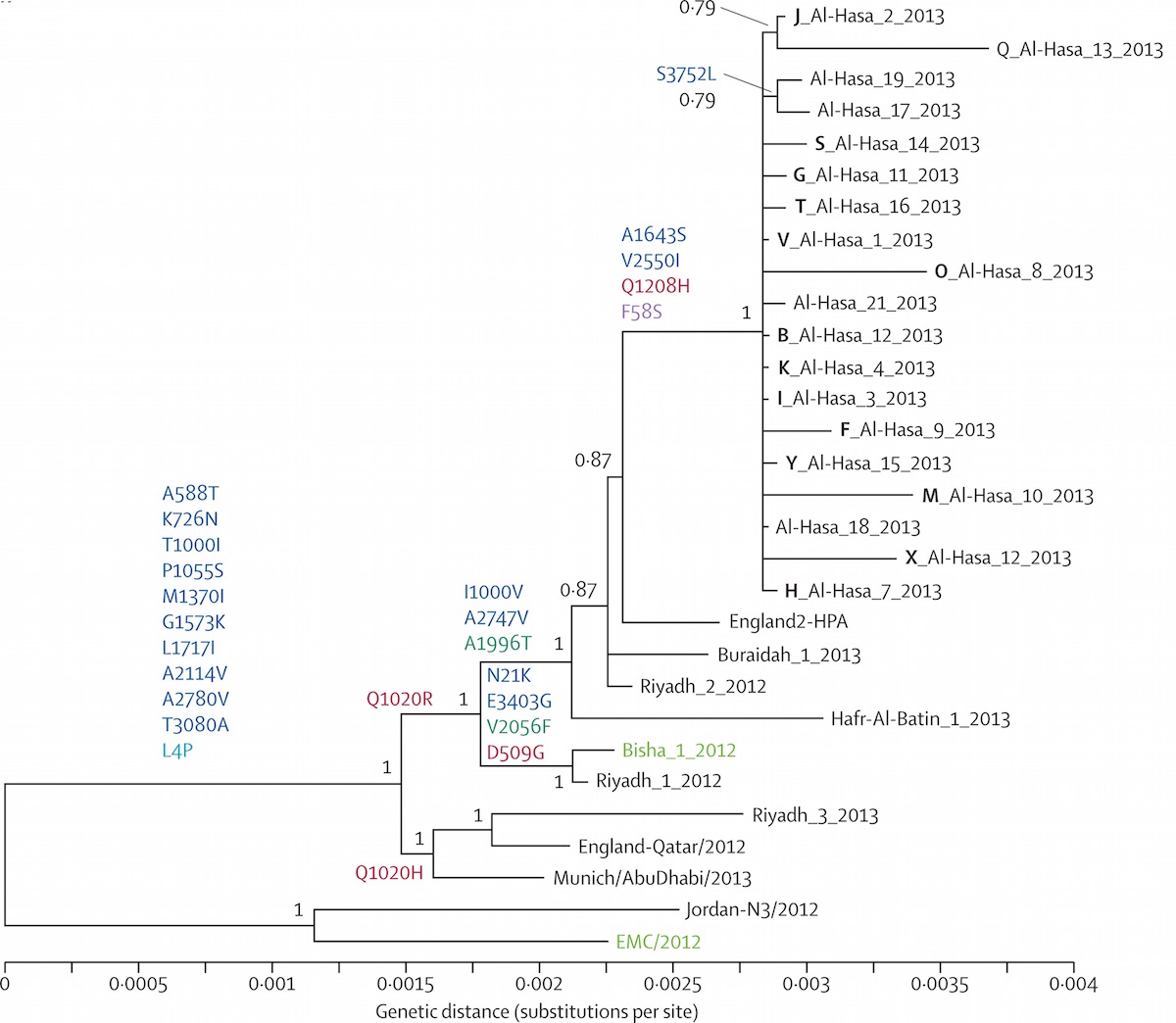
Localized Middle Eastern MERS-CoV phylogeny
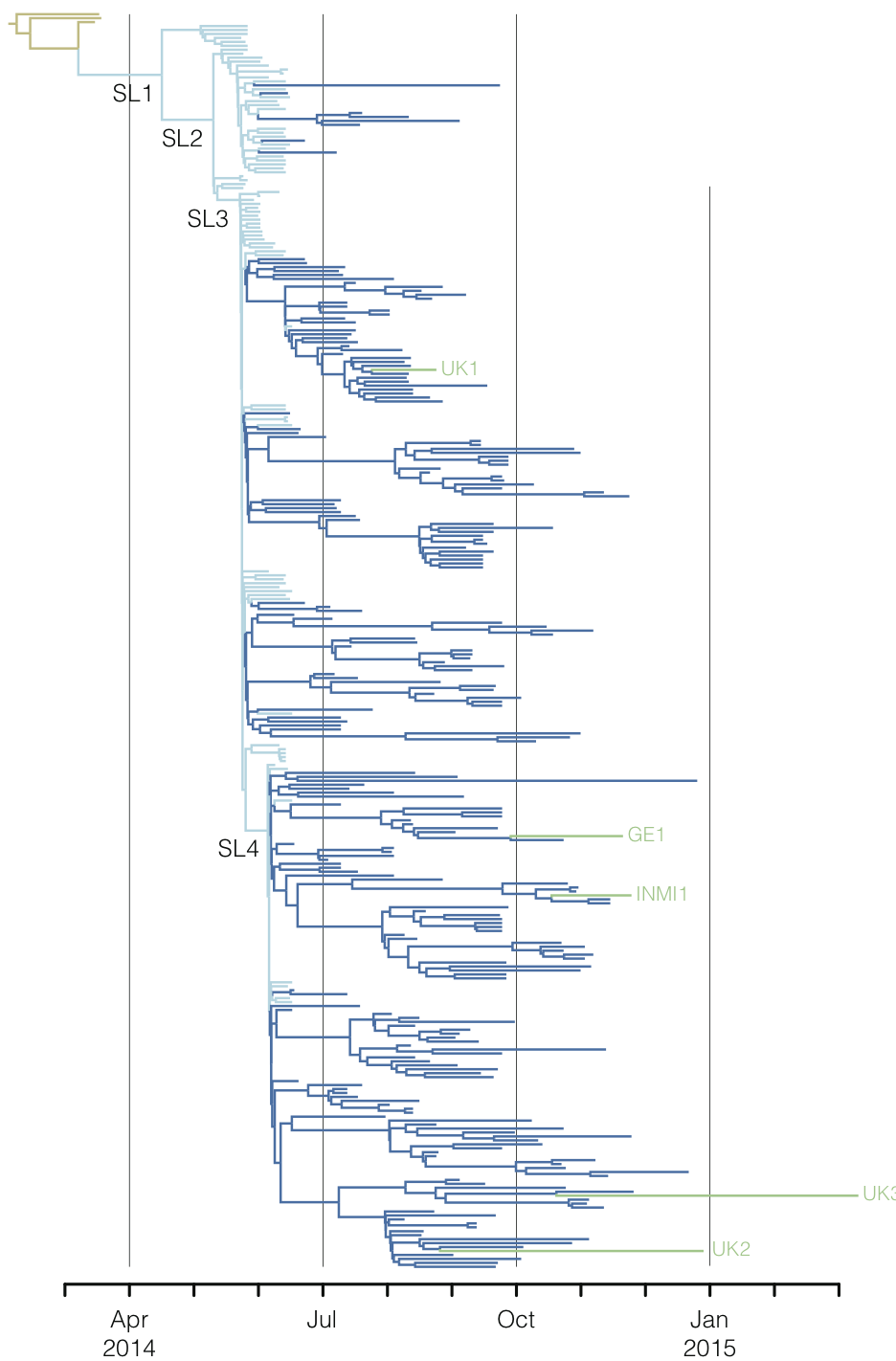
Regional West African Ebola phylogeny
Global influenza phylogeny
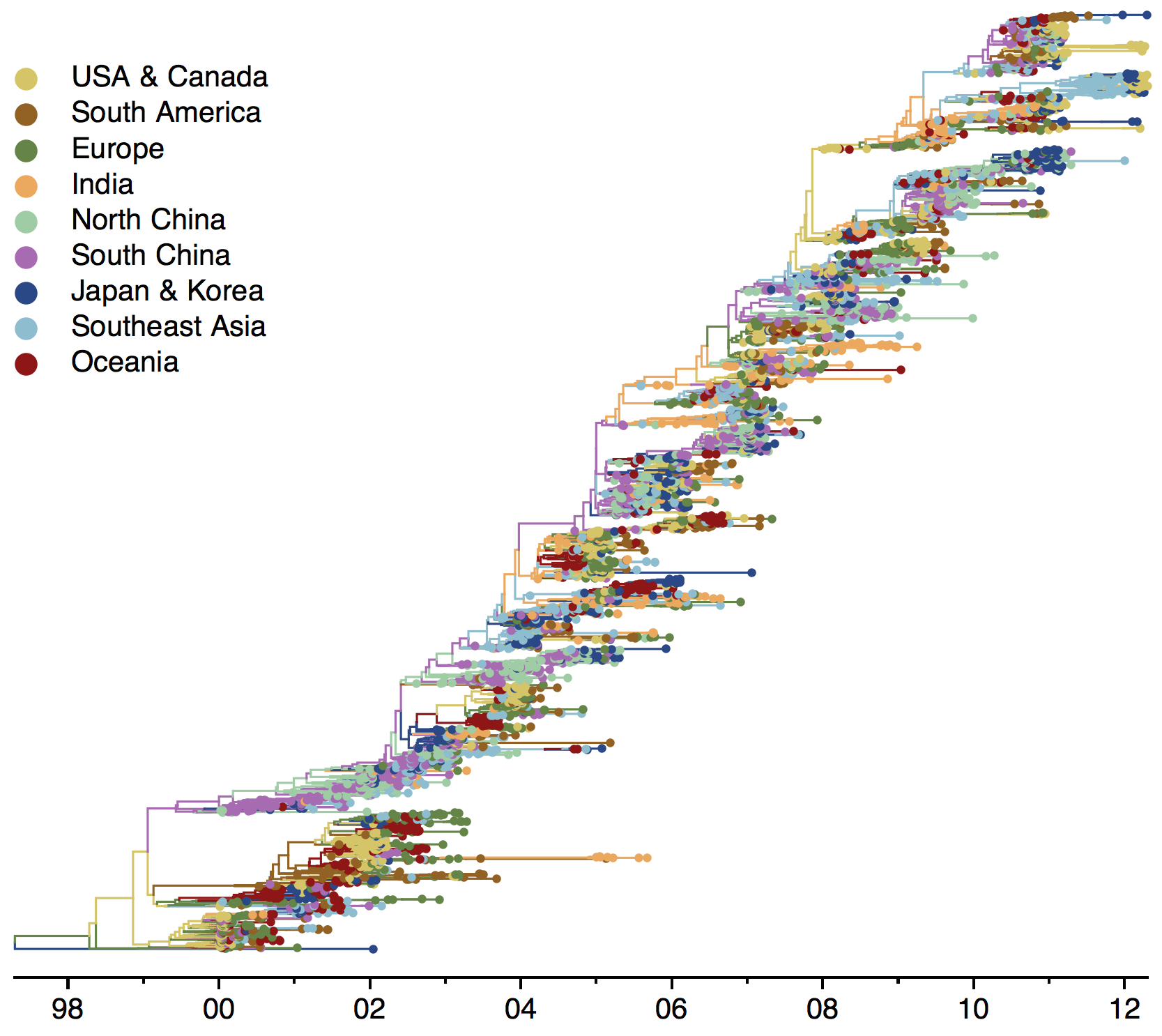
Phylogenetic tracking has the capacity to revolutionize epidemiology
Stuttering chains and animal-to-human spillover
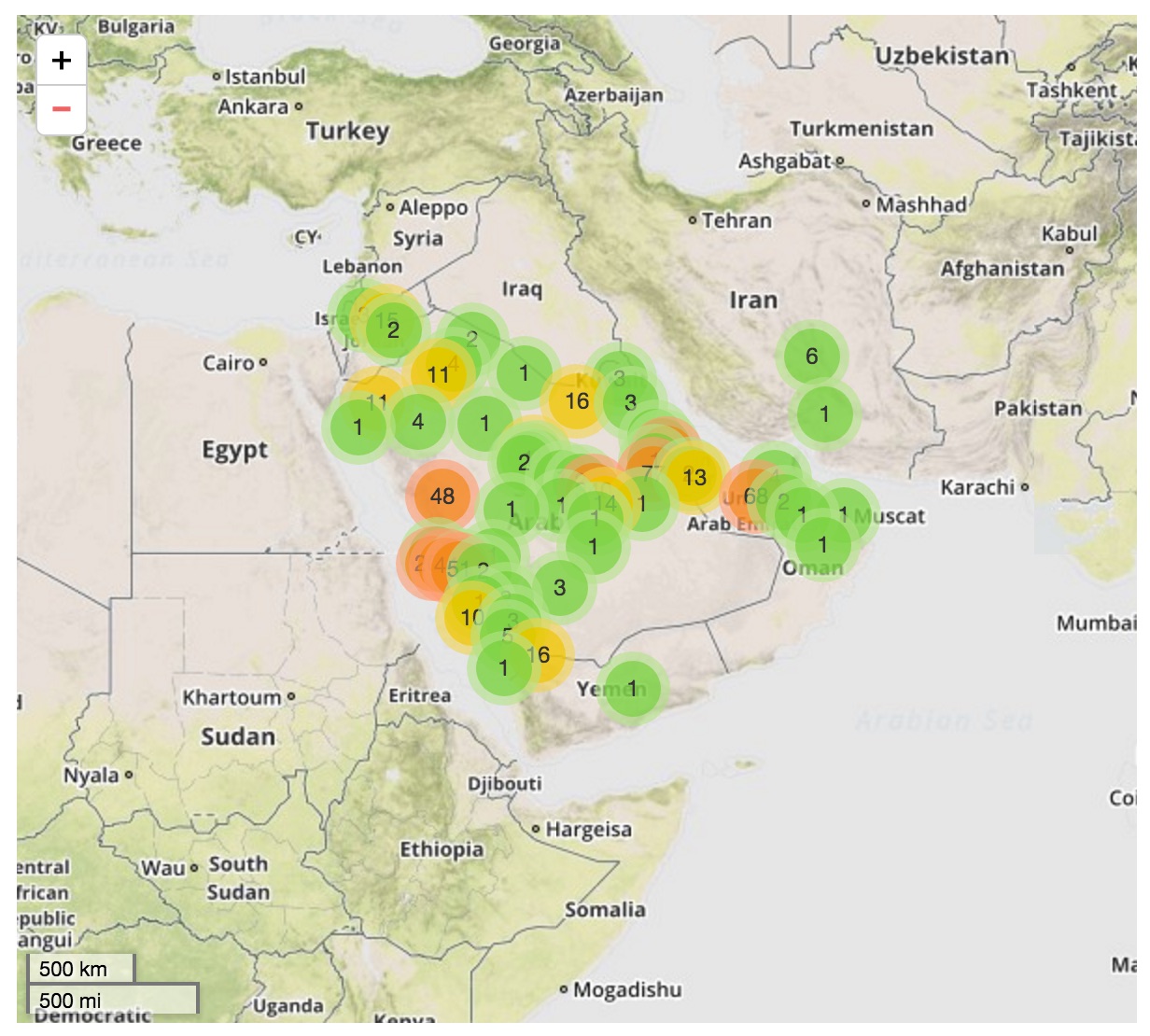
MERS spillover in the Arabian Peninsula
Epidemic growth and human-to-human transmission
Ebola spread in West Africa
Zika spread in the Americas
New methods for rapid phylogenetics and visualization
MERS-CoV
Middle East respiratory syndrome coronavirus (MERS-CoV)
- First identified in Saudi Arabia in 2012
- 2229 confirmed cases to date and 791 deaths
- Camels thought to be the intermediate host
- 30% of common colds due to endemic human coronaviruses
Ongoing incidence, but lack of epidemic growth
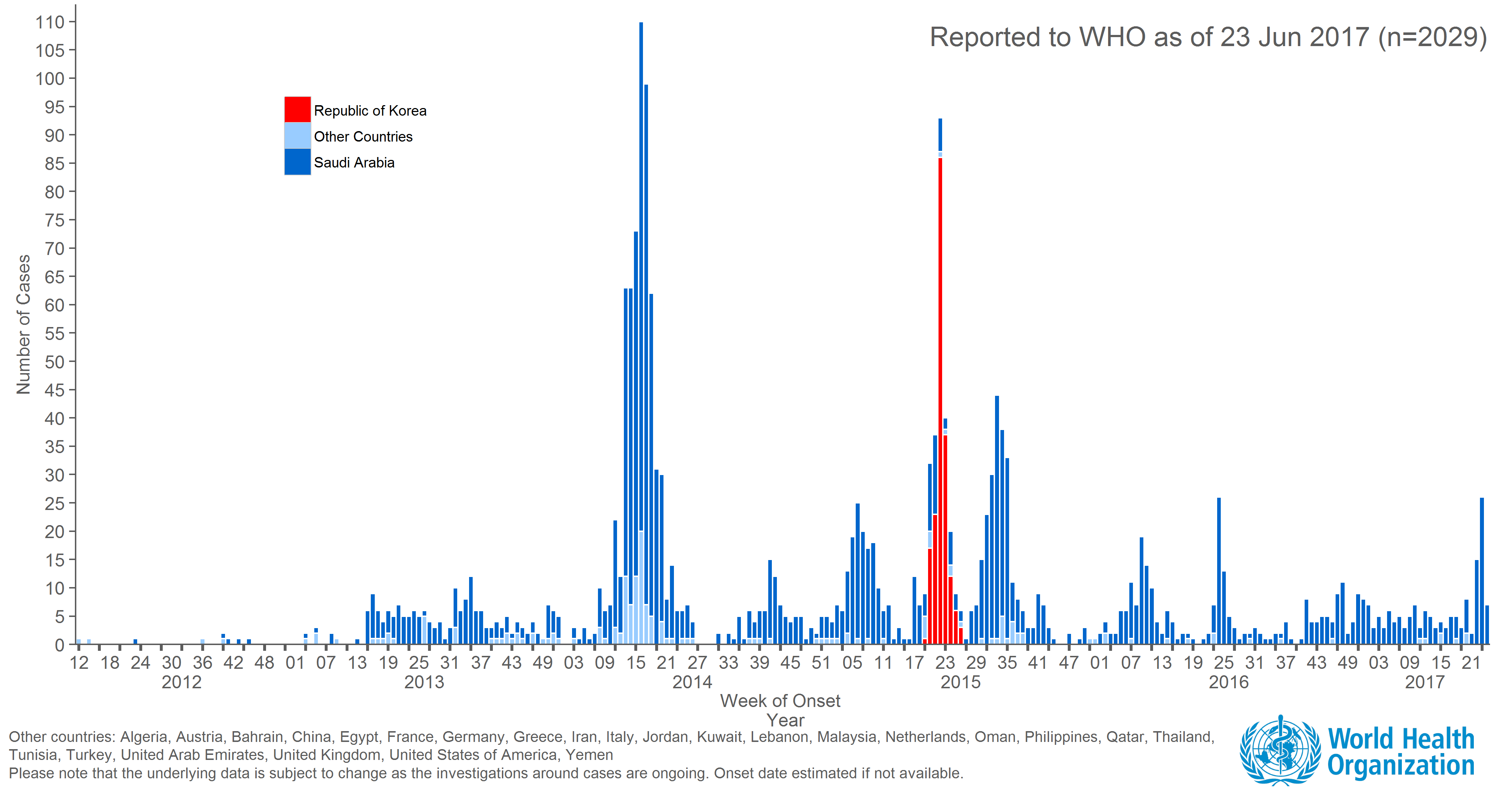
Cases localized to the Arabian Peninsula
Hypotheses for MERS transmission
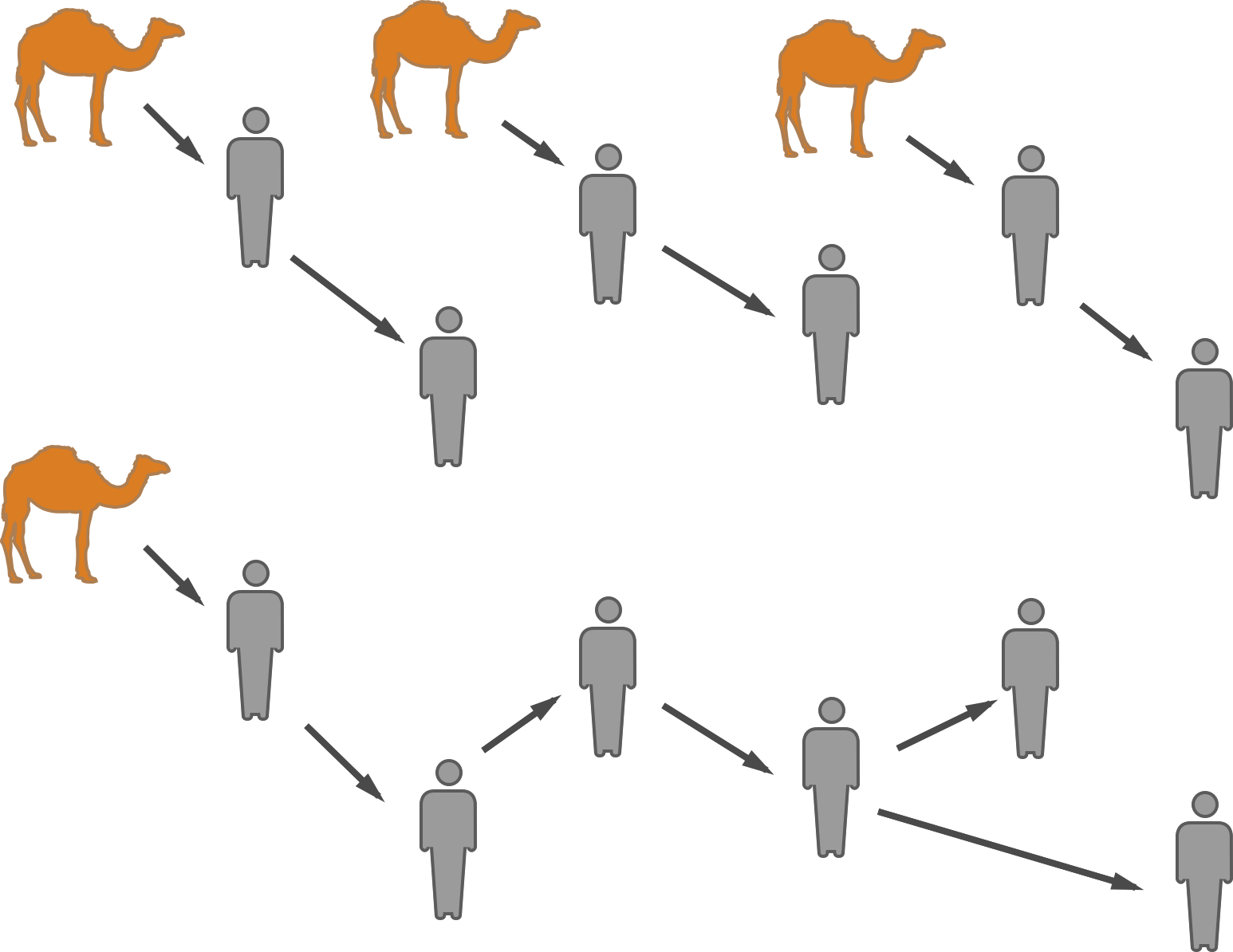
MERS-CoV spillover at the camel-human interface
with ![]() Gytis Dudas, Luiz Carvalho and Andrew Rambaut
Gytis Dudas, Luiz Carvalho and Andrew Rambaut
Genomic dataset
- 174 virus genomes from human infections
- 100 virus genomes from camel infections
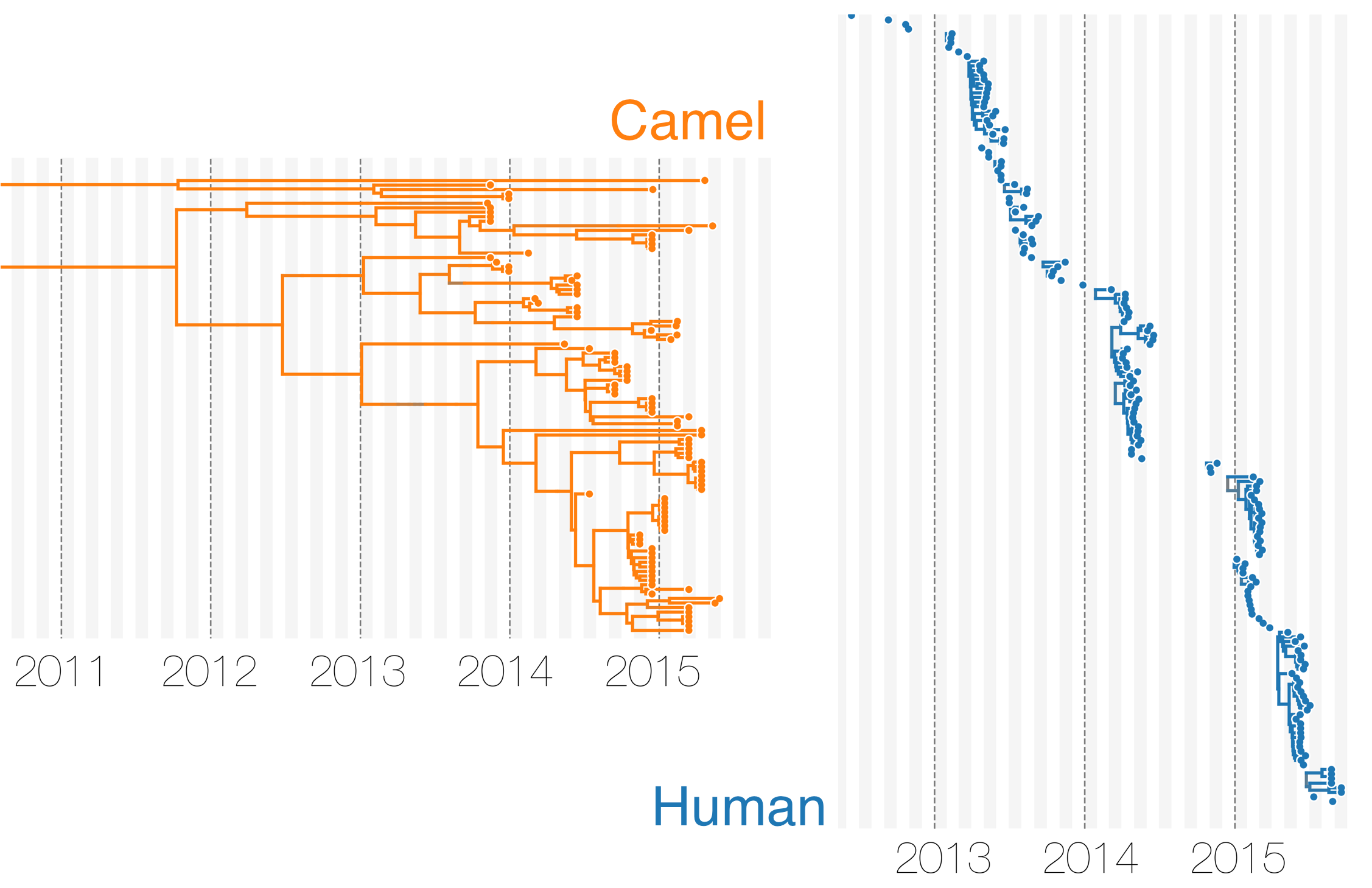
MERS tree with host state
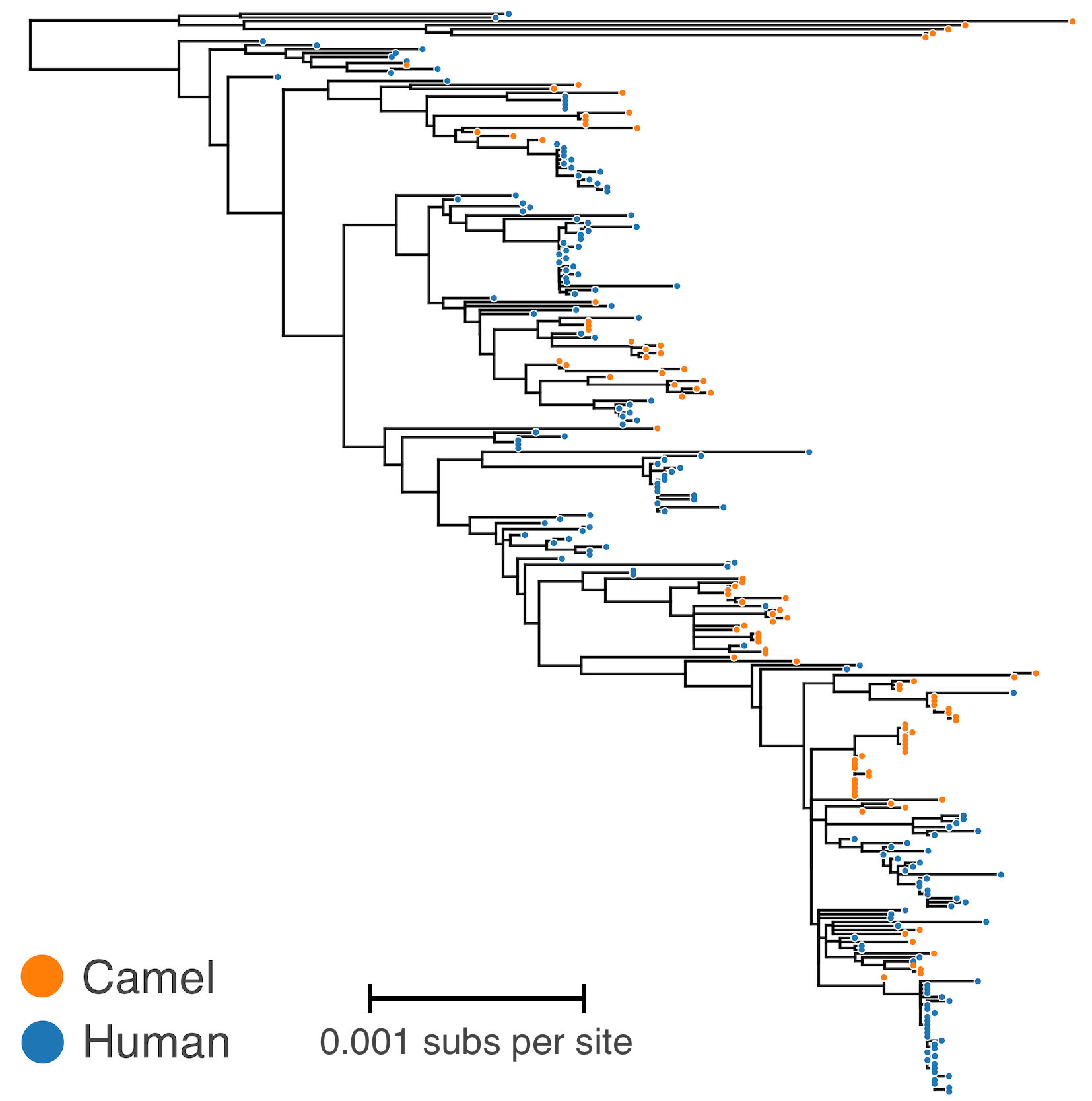
Phylodynamic reconstruction of host state
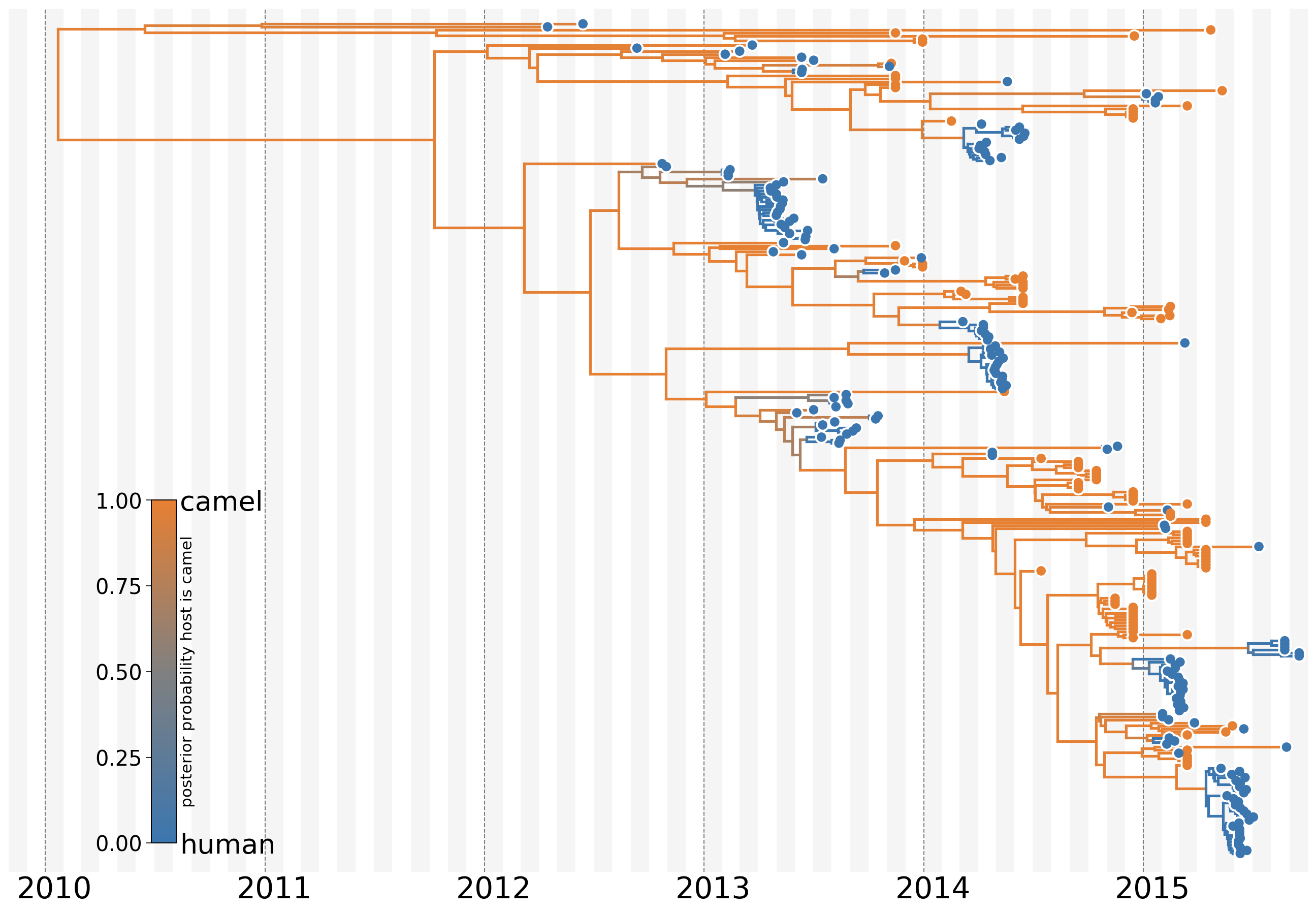
Humans are transient hosts
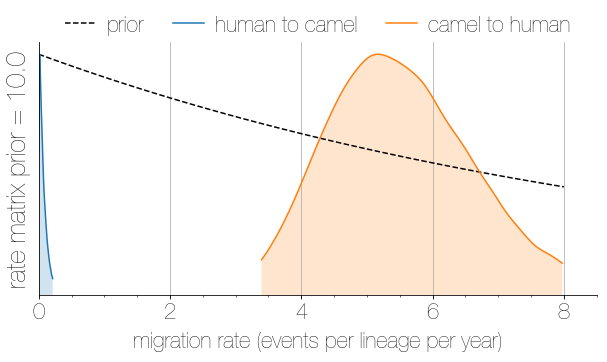
Asymmetric migration rates
- 56 (48–63) camel-to-human transmission events resulting in 174 sequenced human infections
- 3 (0-12) human-to-camel transmission events
Introduction events tend to occur between April and July
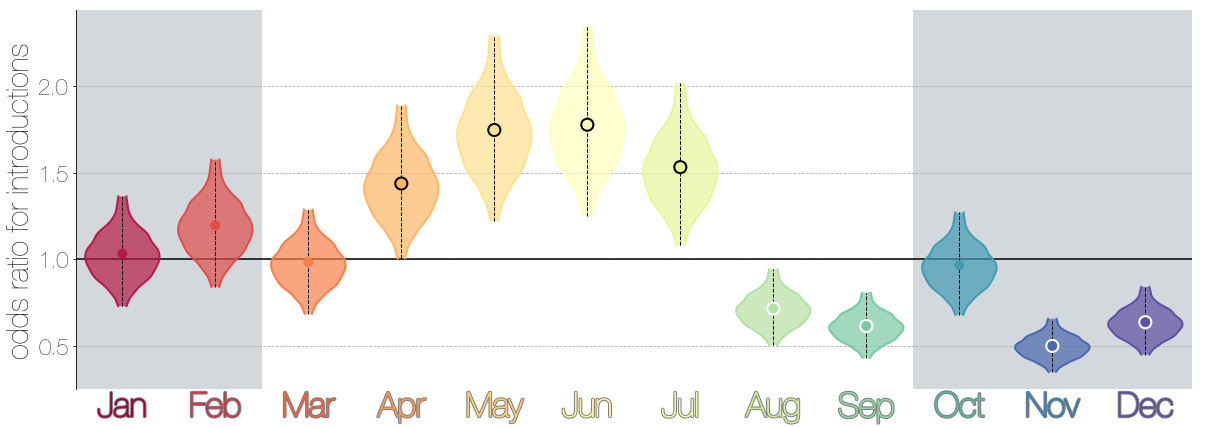
Dromedary camel calving occurs between Nov and Feb
Monte Carlo simulation
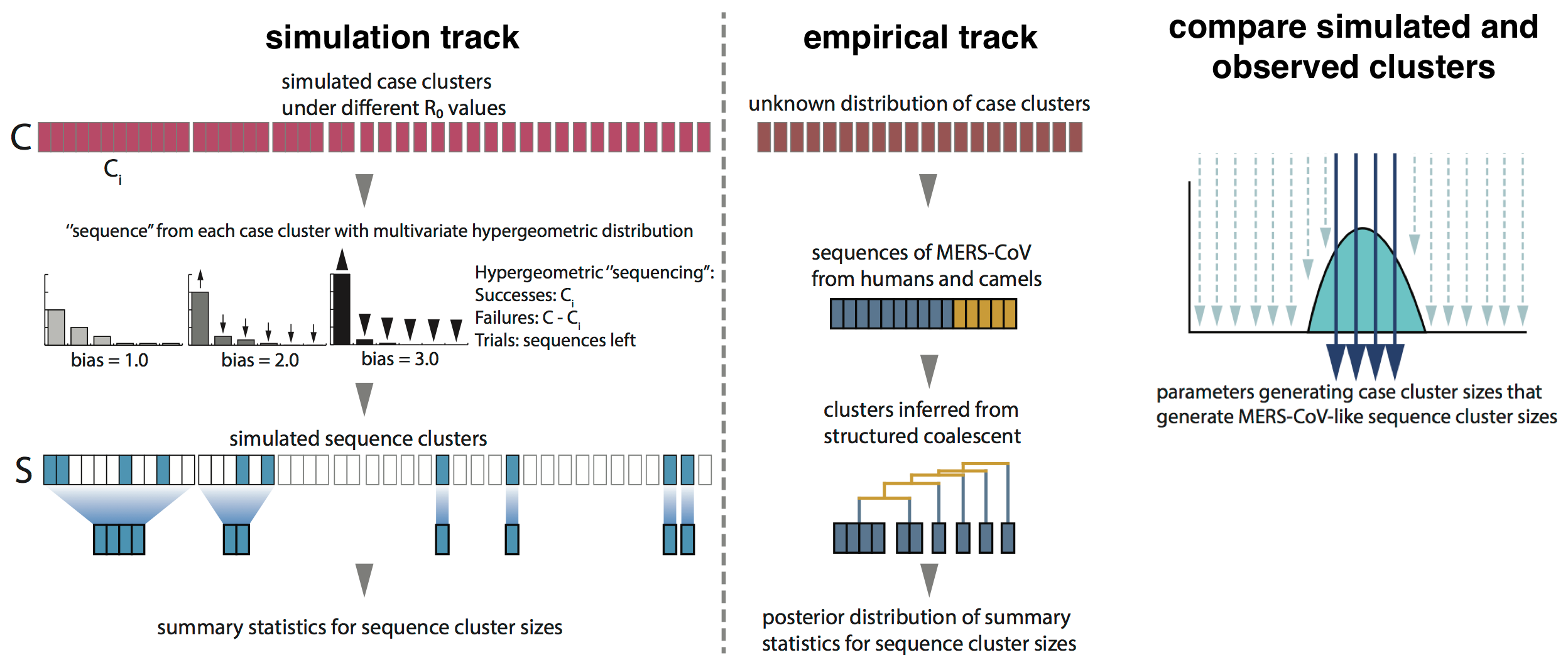
Phylogenetic clustering suggests $R_0$ below 1.0 and ~2000 human cases driven by ~600 introduction events
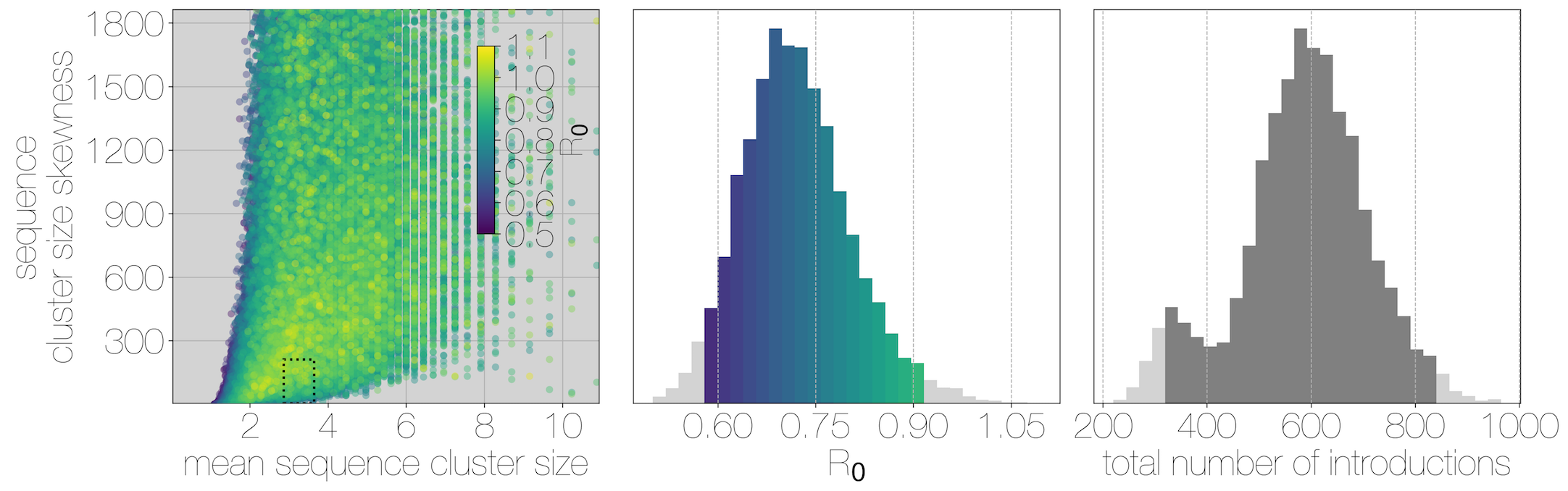
Critically, no evidence of increasing cluster sizes through time
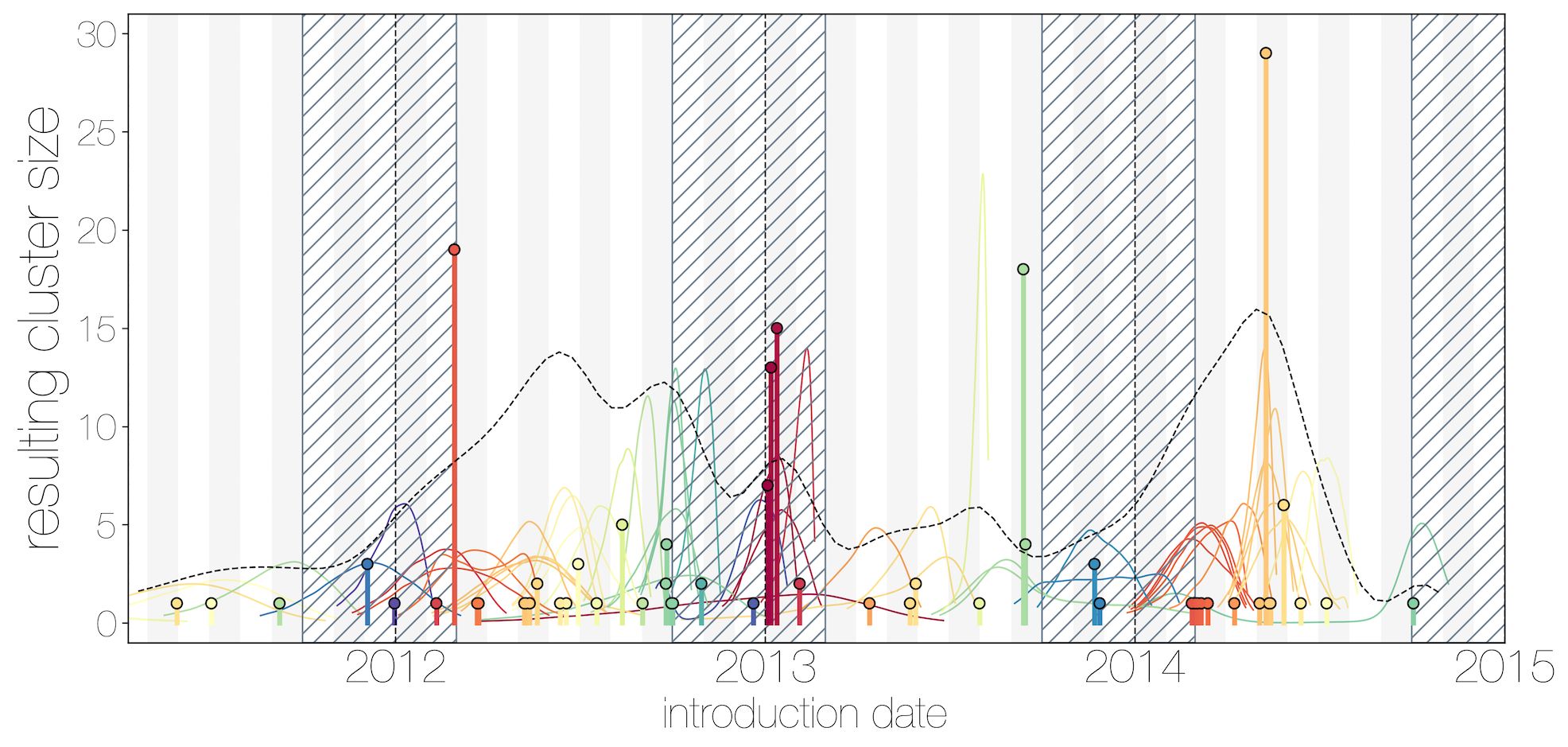
Many other viruses that exhibit stuttering chains of human infection
- Nipah virus (fruit bats / pigs, Southeast Asia)
- Lassa virus (rodents, West Africa)
- Avian influenza (birds, mainland China)
Sylvatic introductions of yellow fever virus show similar dynamics
Ebola
Ebola epidemic of 2014-2016 was unprecedented in scope
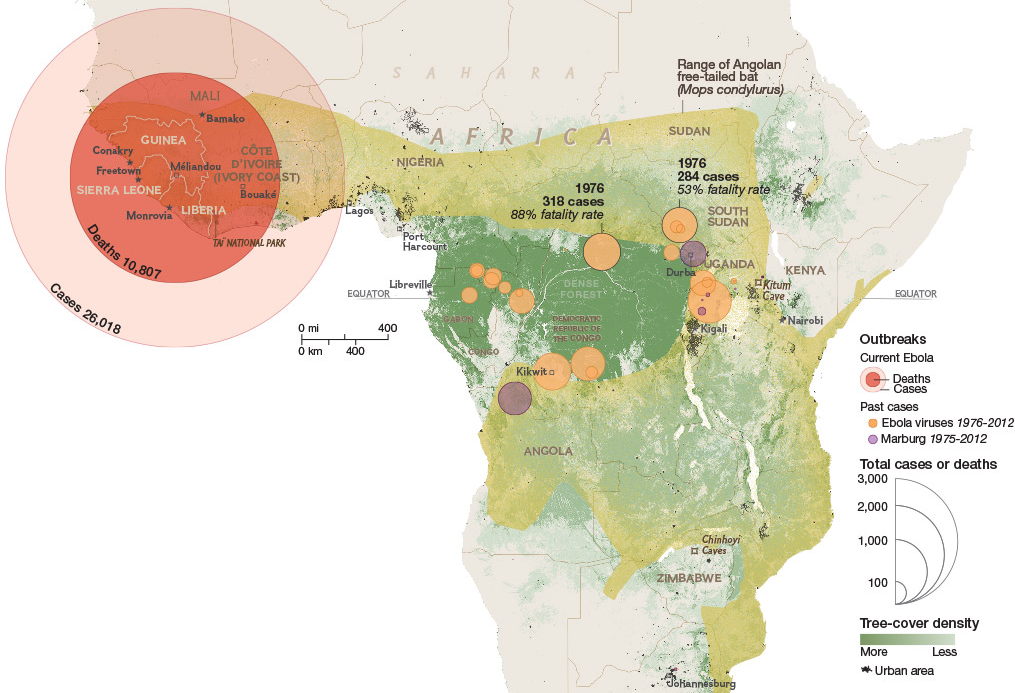
Ebola epidemic in West Africa
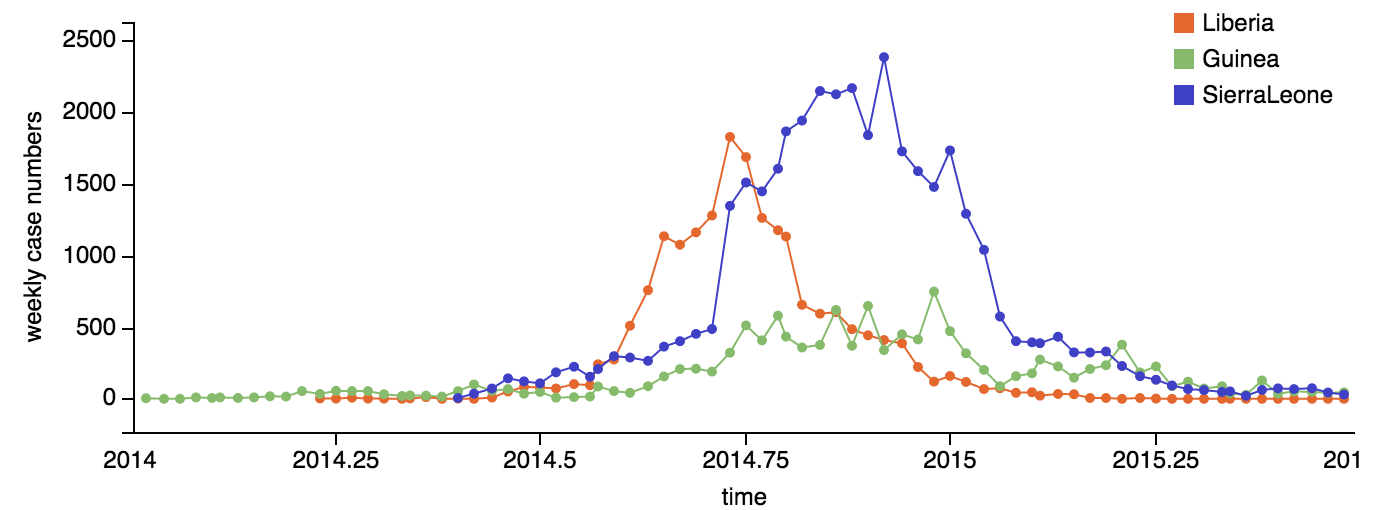
Ebola epidemic within Sierra Leone
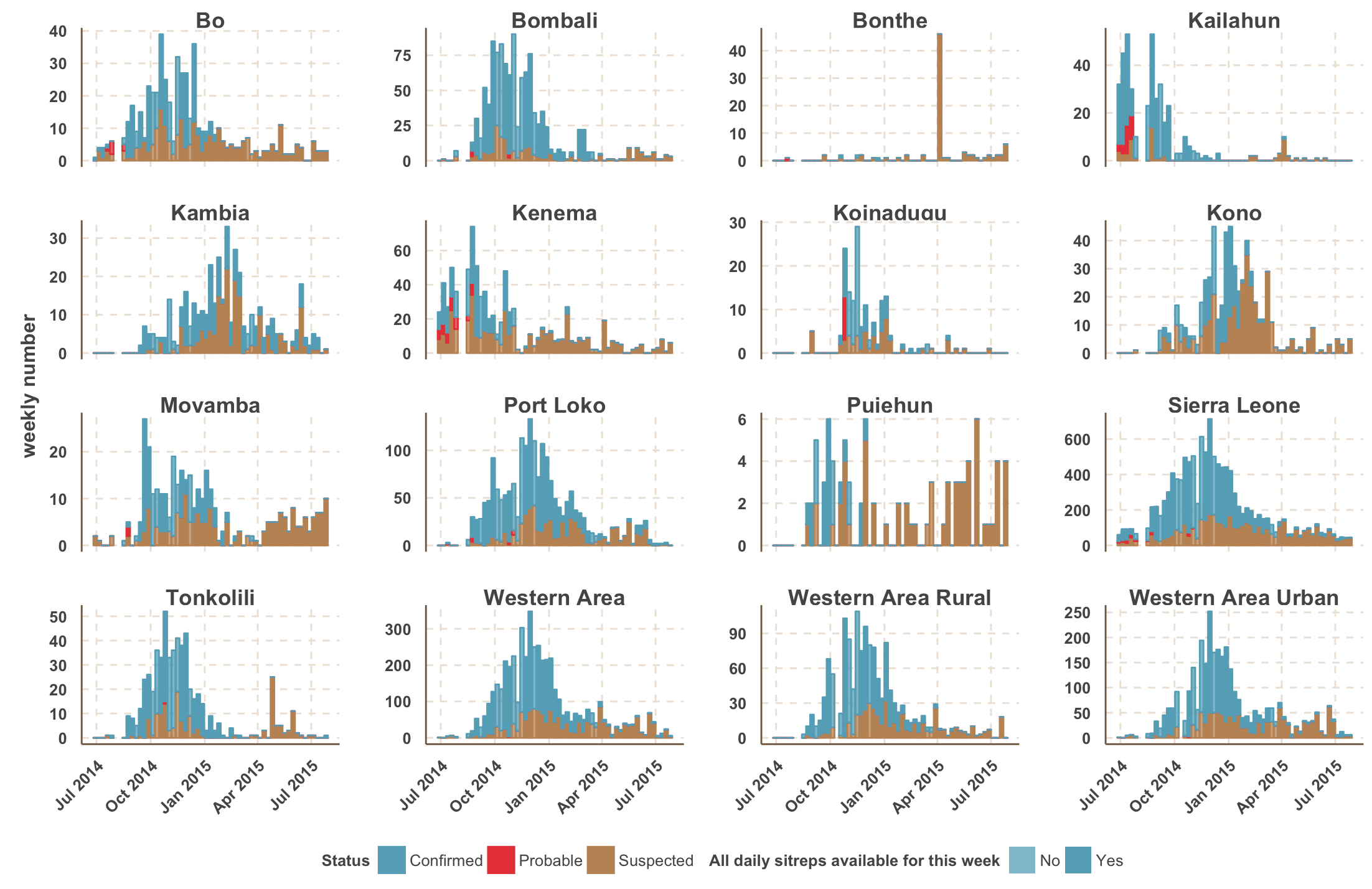
Virus genomes reveal factors that spread and sustained the Ebola epidemic
with ![]() Gytis Dudas, Andrew Rambaut, Luiz Carvalho, Marc Suchard, Philippe Lemey,
Gytis Dudas, Andrew Rambaut, Luiz Carvalho, Marc Suchard, Philippe Lemey,
and many others
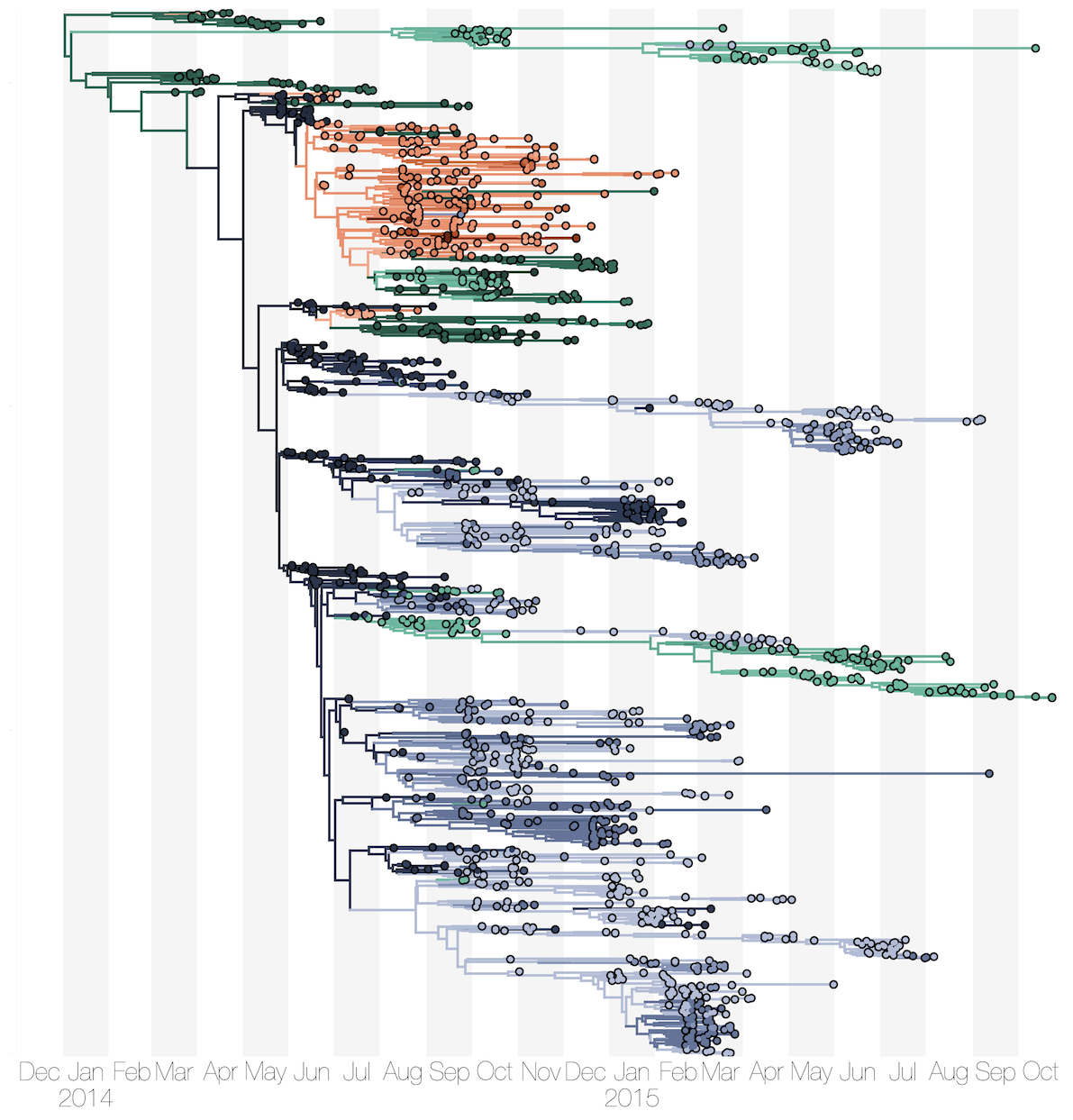
Sequencing of 1610 Ebola virus genomes collected during the 2013-2016 West African epidemic
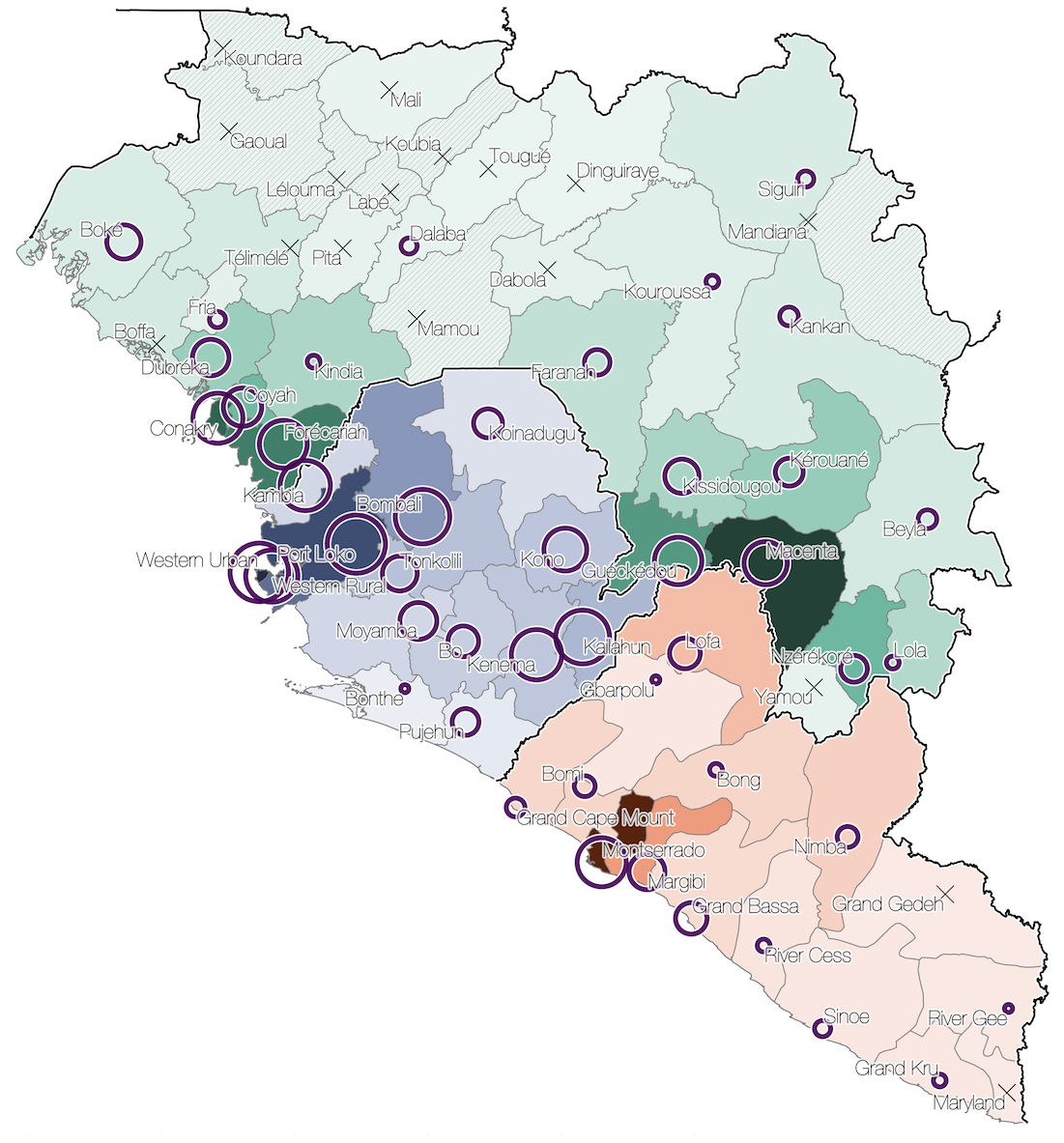
Sequenced genomes were representative of spatiotemporal diversity
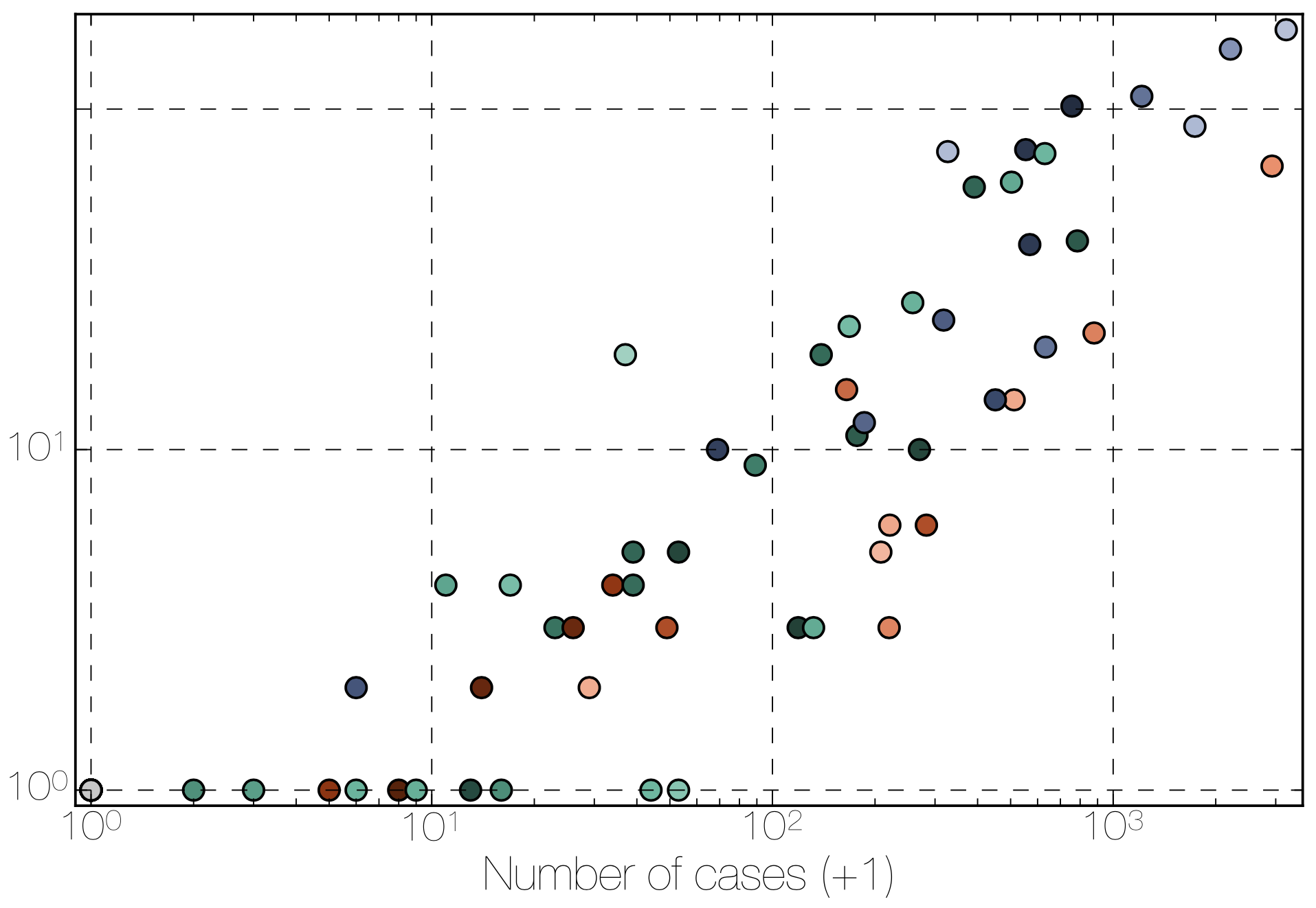
Phylogenetic reconstruction of epidemic
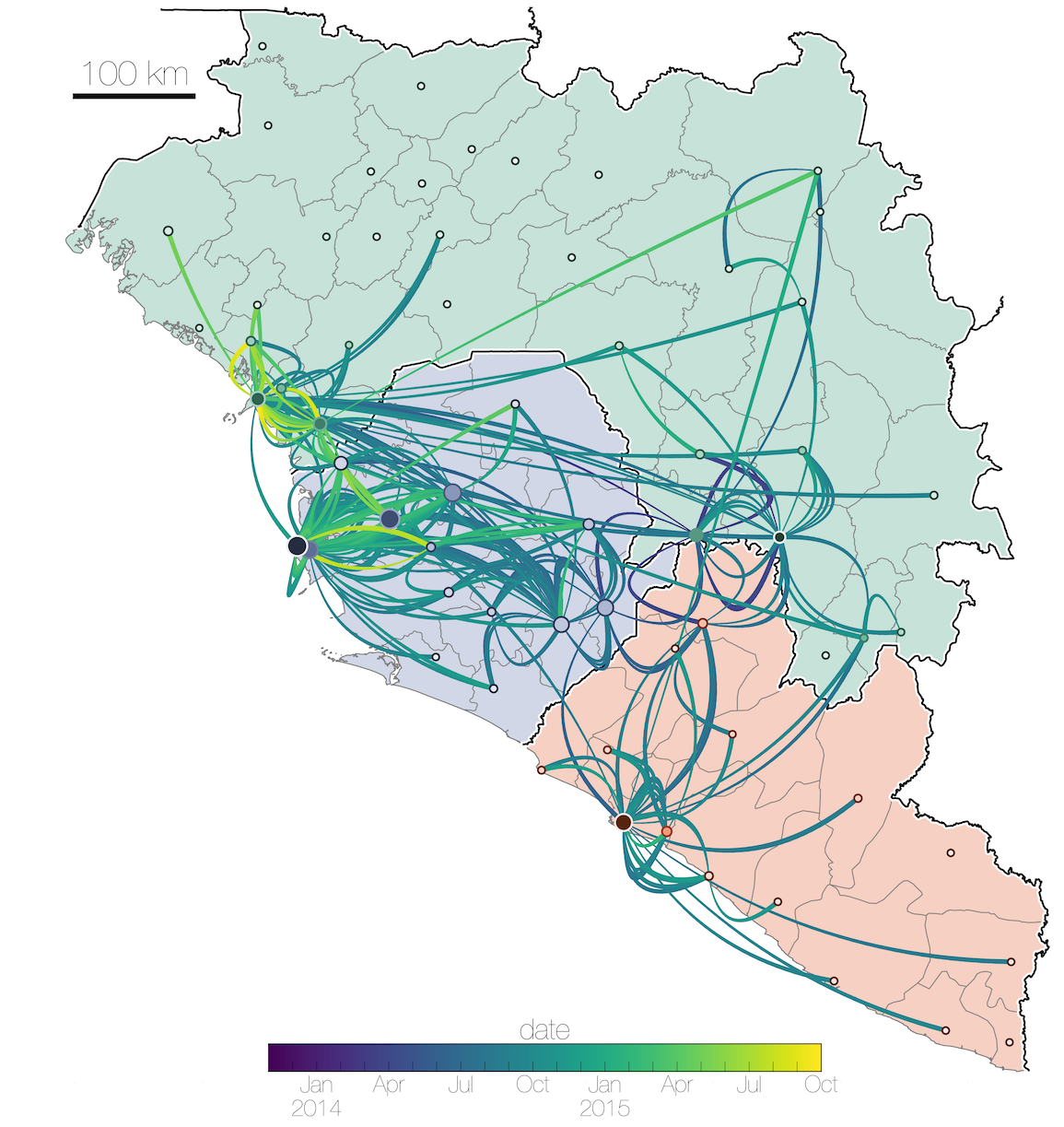
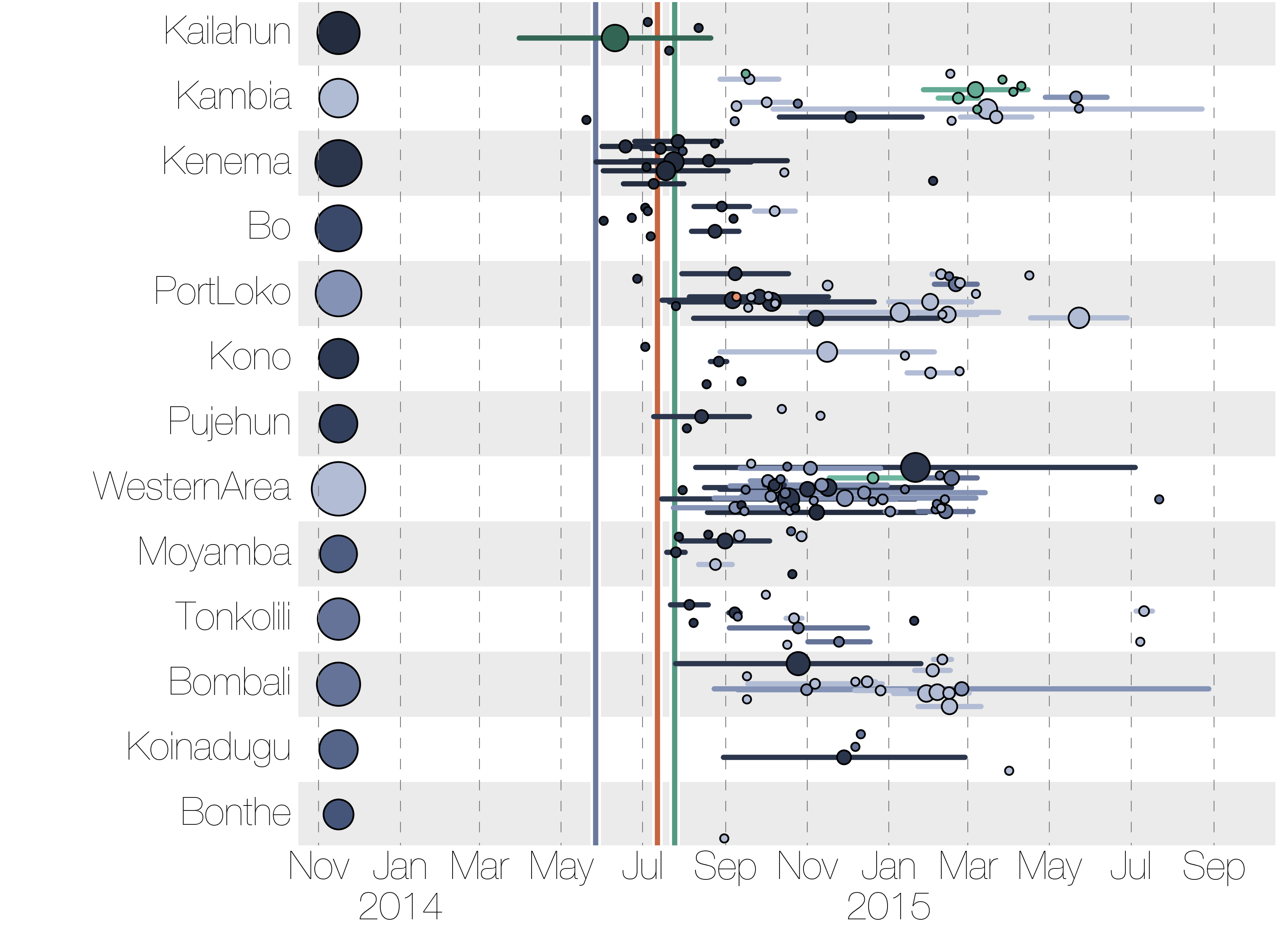
Tracking migration events
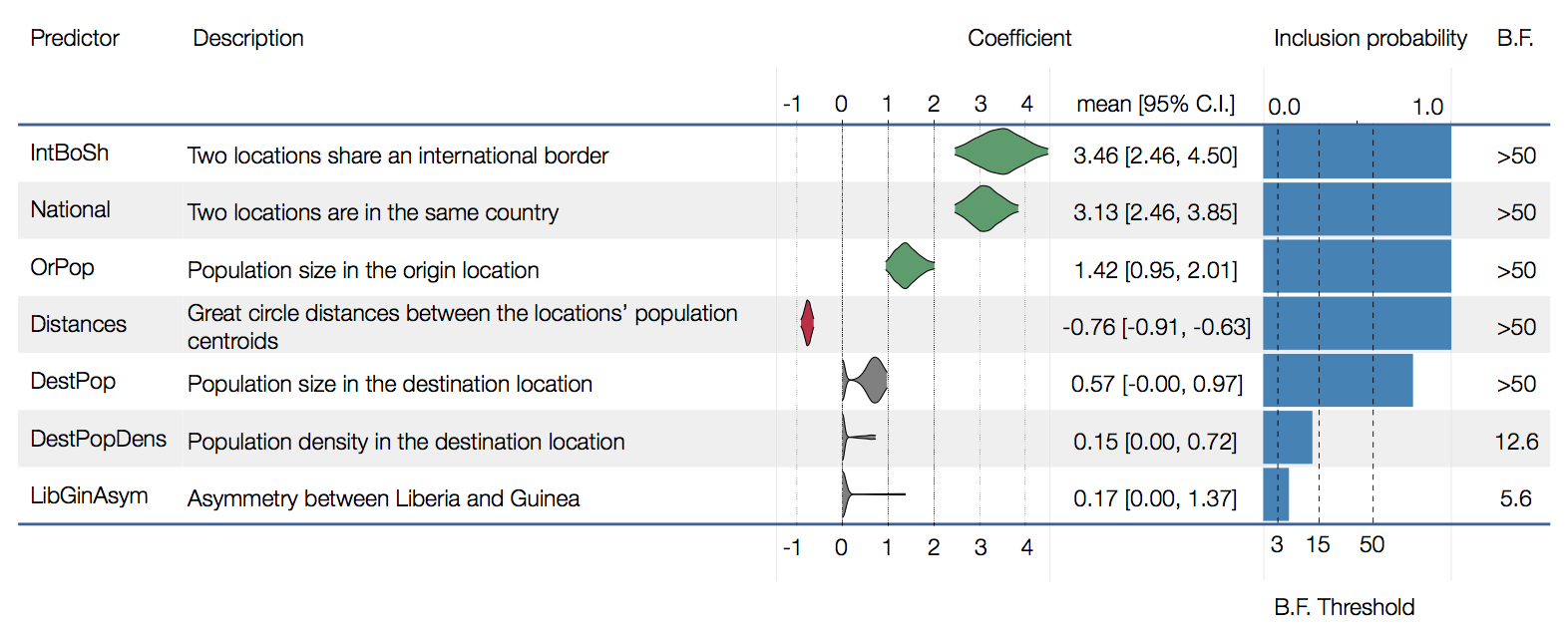
Factors influencing migration rates
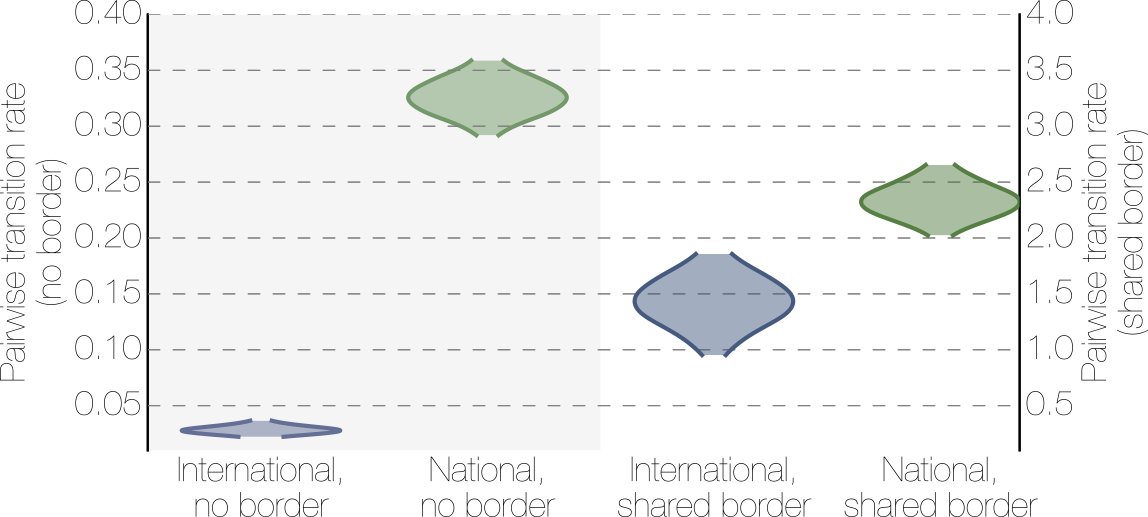
Effect of borders on migration rates
Spatial structure at the country level
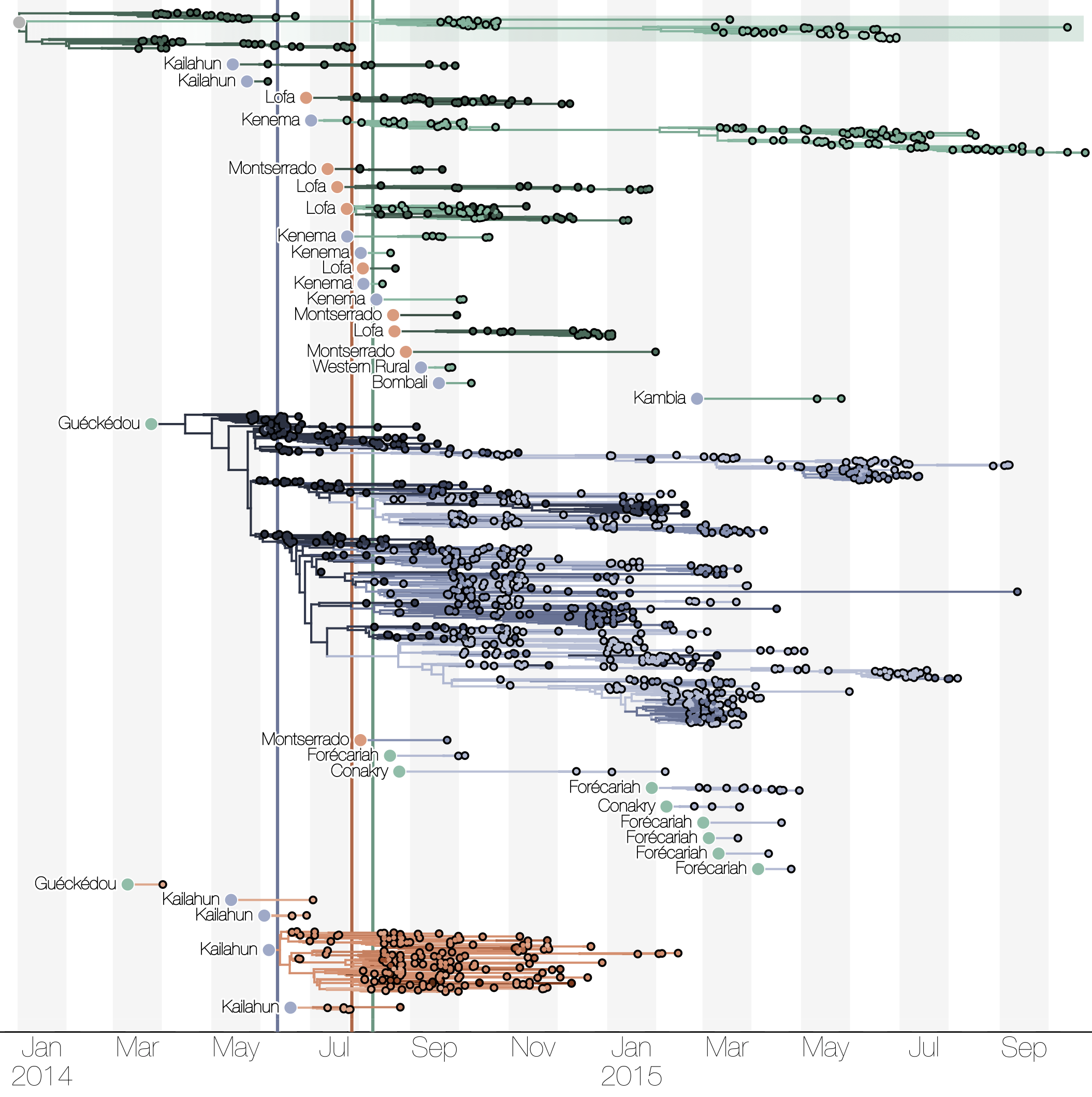
Substantial mixing at the regional level
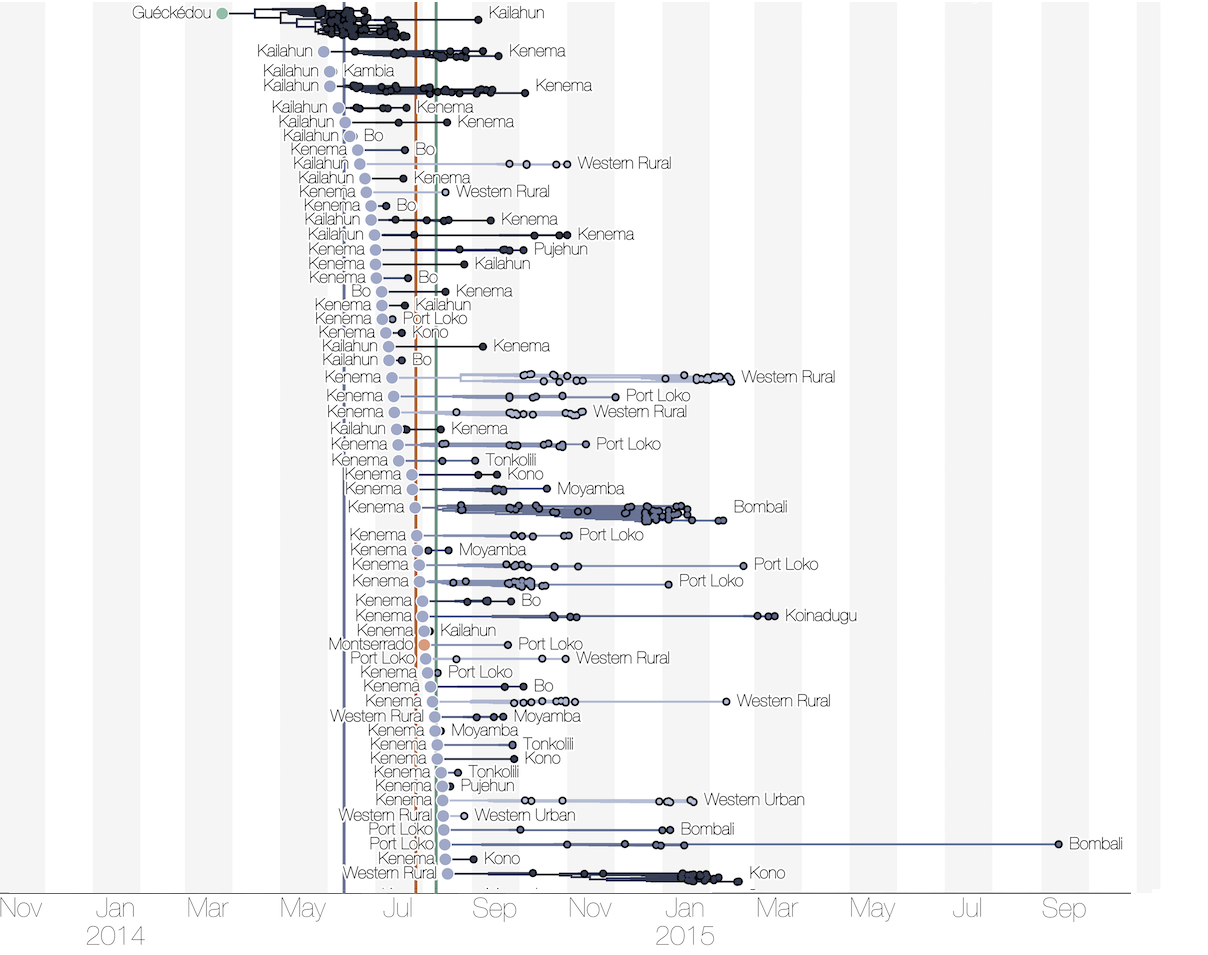
Each introduction results in a minor outbreak
Regional outbreaks due to multiple introductions
Regional outbreaks due to multiple introductions
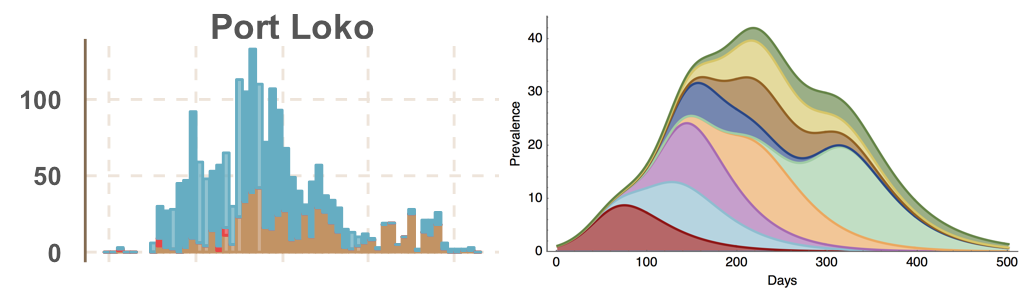
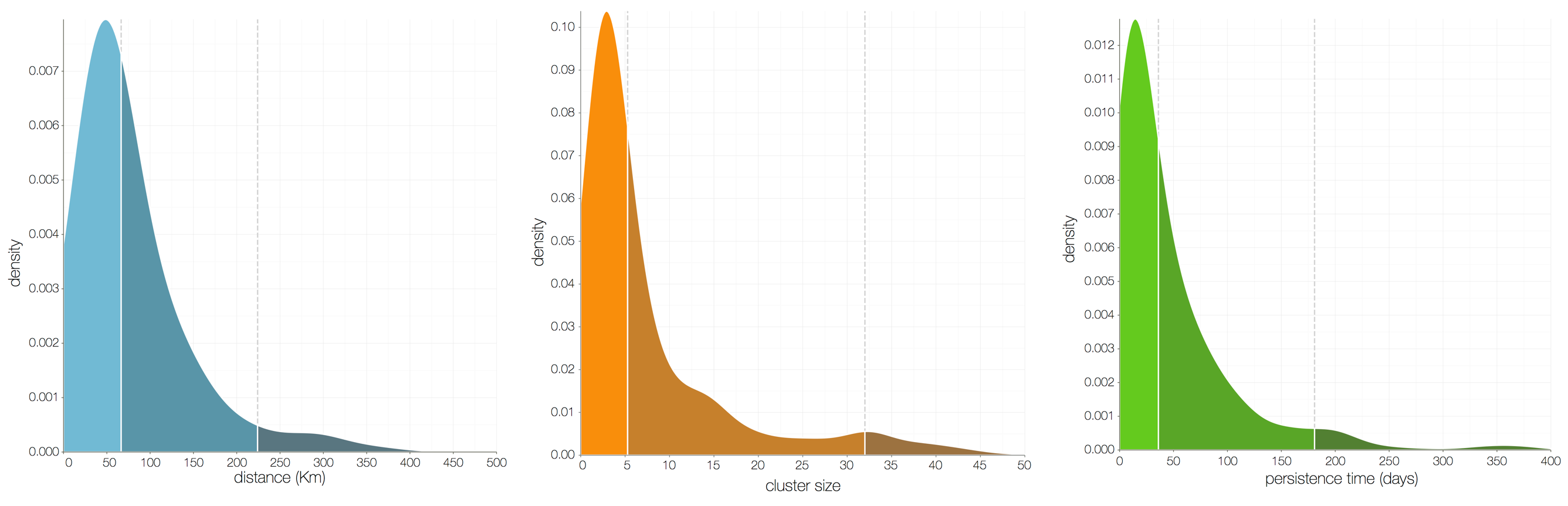
Ebola spread in West Africa followed a gravity model with moderate slowing by international borders, in which spread is driven by short-lived migratory clusters
Zika
Zika's arrival and spread in the Americas
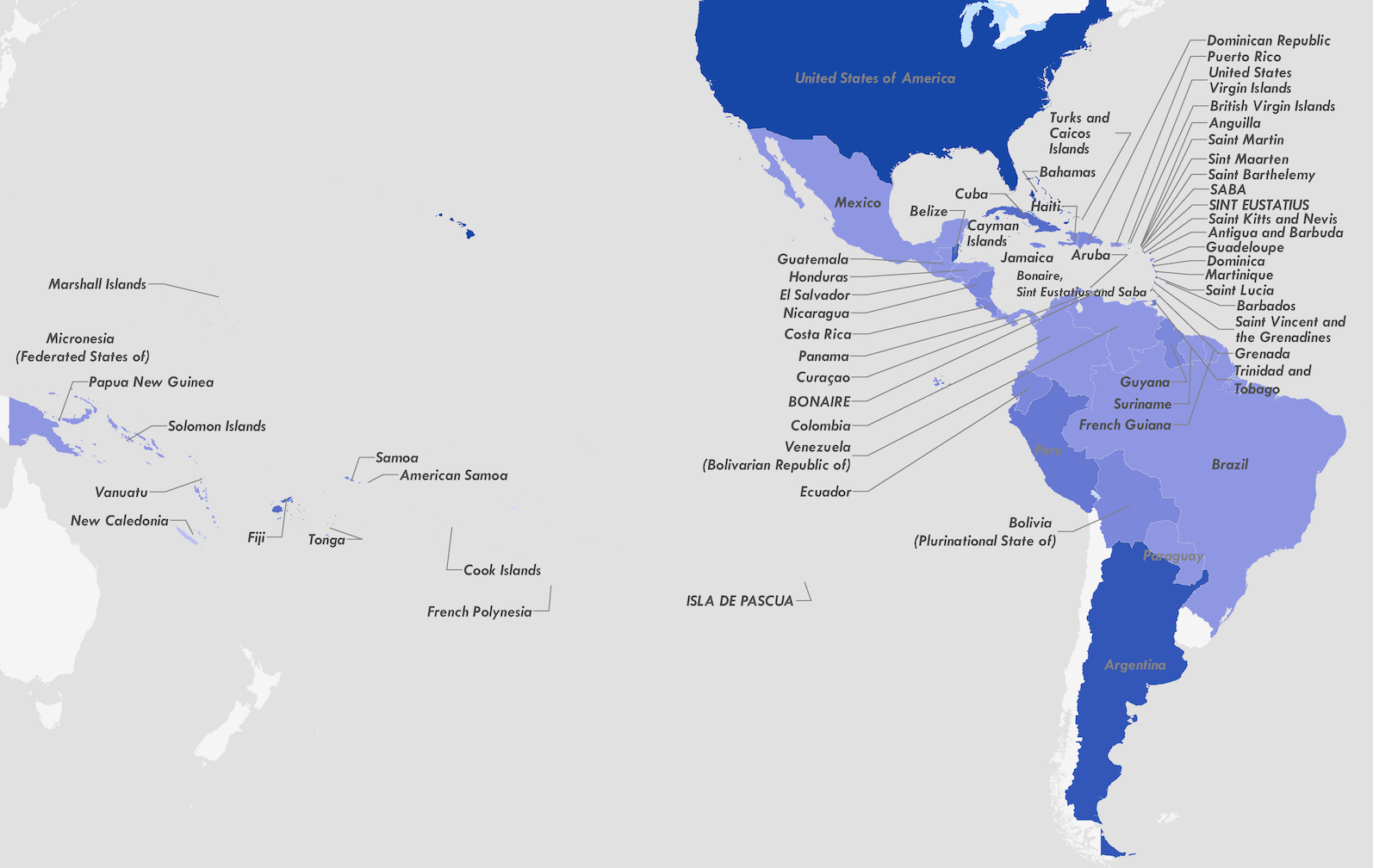
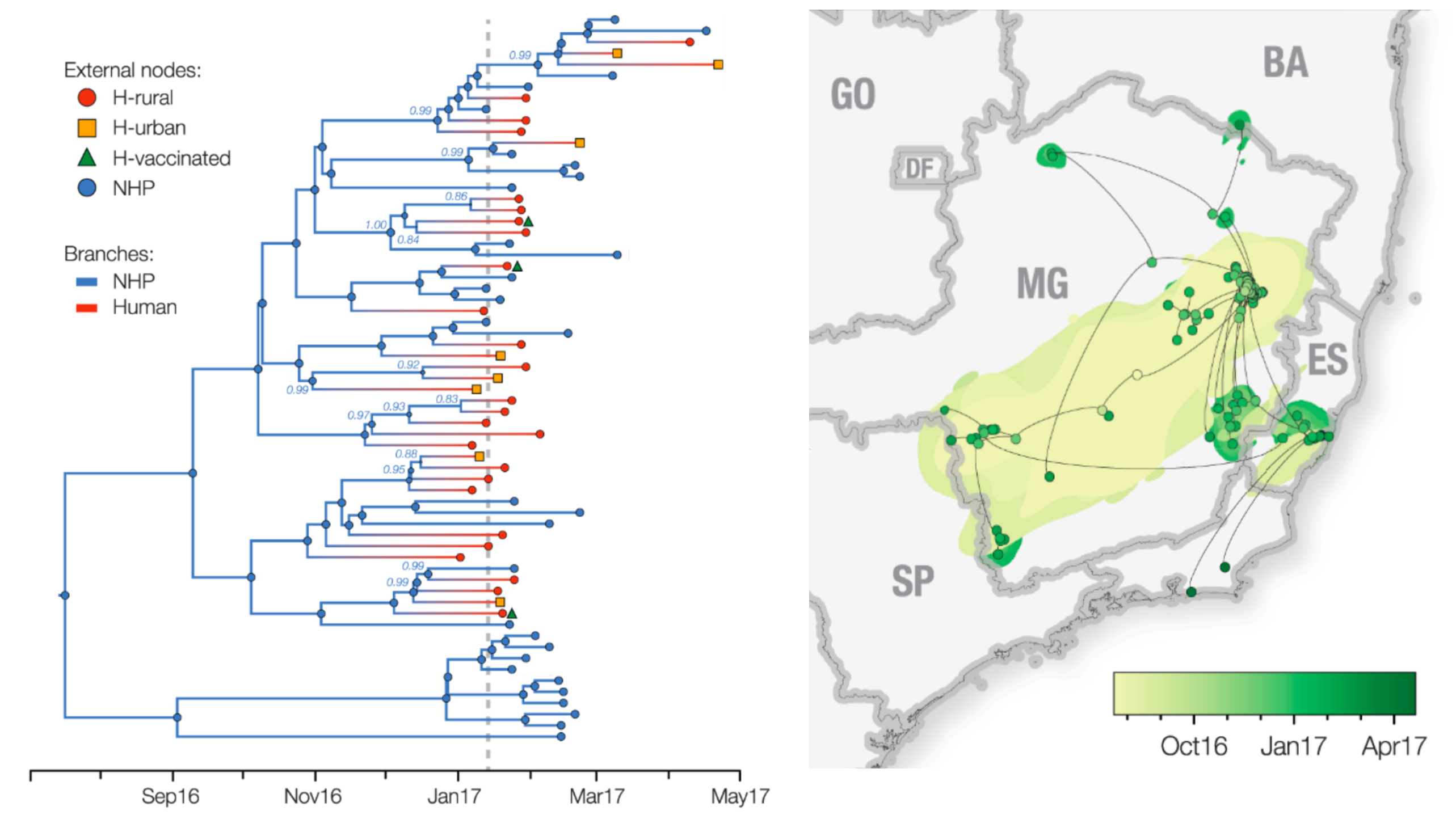
Establishment and cryptic transmission of Zika virus in Brazil and the Americas
with Nuno Faria, Nick Loman, Oli Pybus, Luiz Alcantara, Ester Sabino, Josh Quick,
![]() Alli Black,
Ingra Morales, Julien Thézé, Marcio Nunes, Jacqueline de Jesus,
Alli Black,
Ingra Morales, Julien Thézé, Marcio Nunes, Jacqueline de Jesus,
Marta Giovanetti, Moritz Kraemer, Sarah Hill and many others

Road trip through northeast Brazil to collect samples and sequence
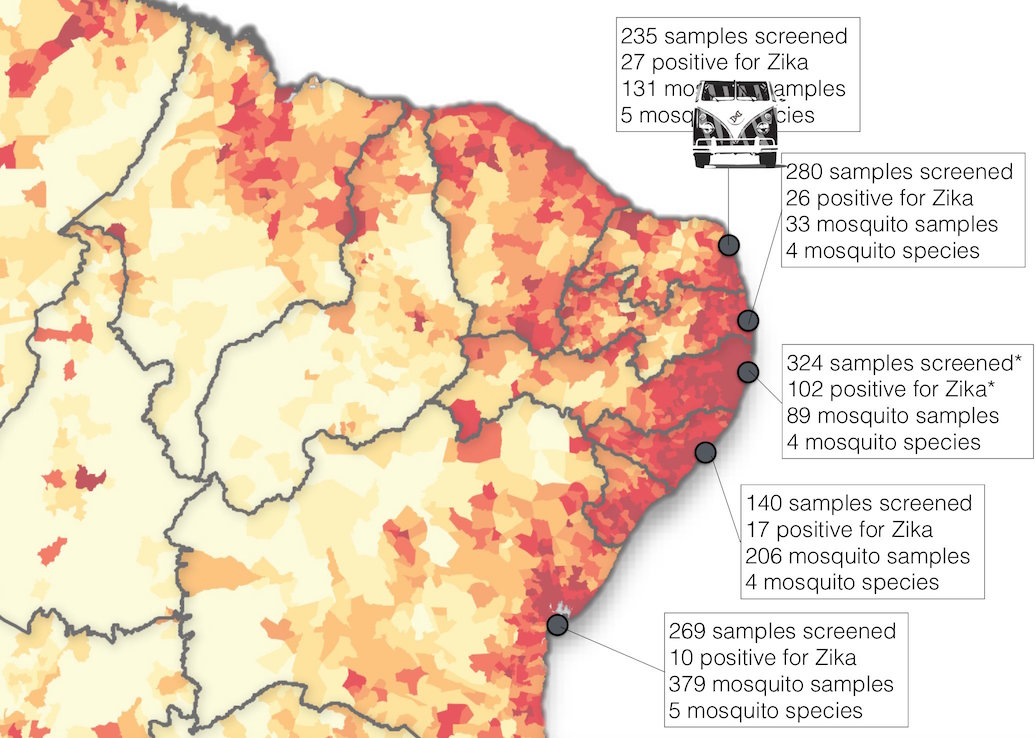
Case reports and diagnostics suggest initiation in northeast Brazil
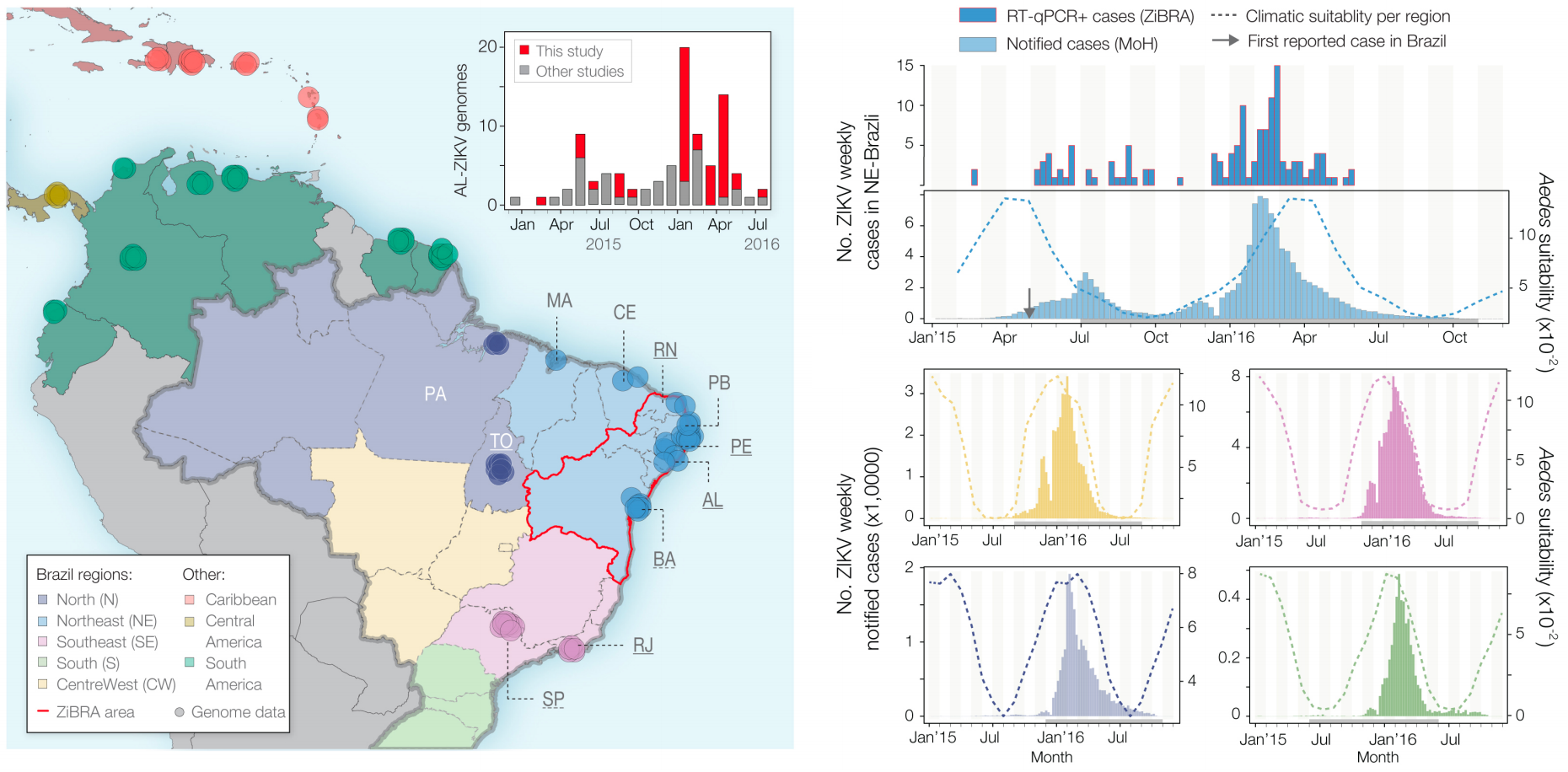
Phylogeny infers an origin in northeast Brazil
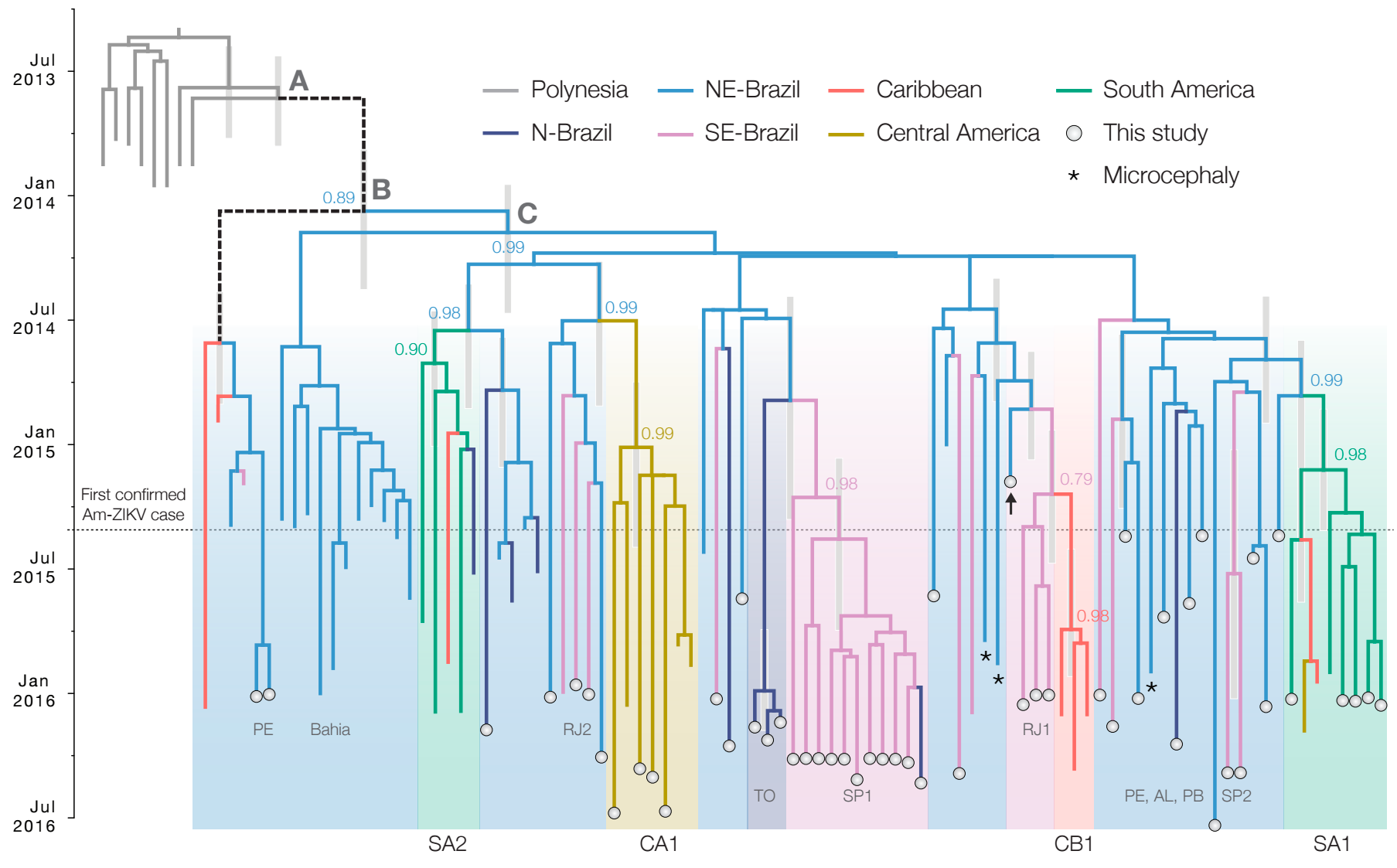
Actionable inferences
Genomic analyses were mostly done in a retrospective manner
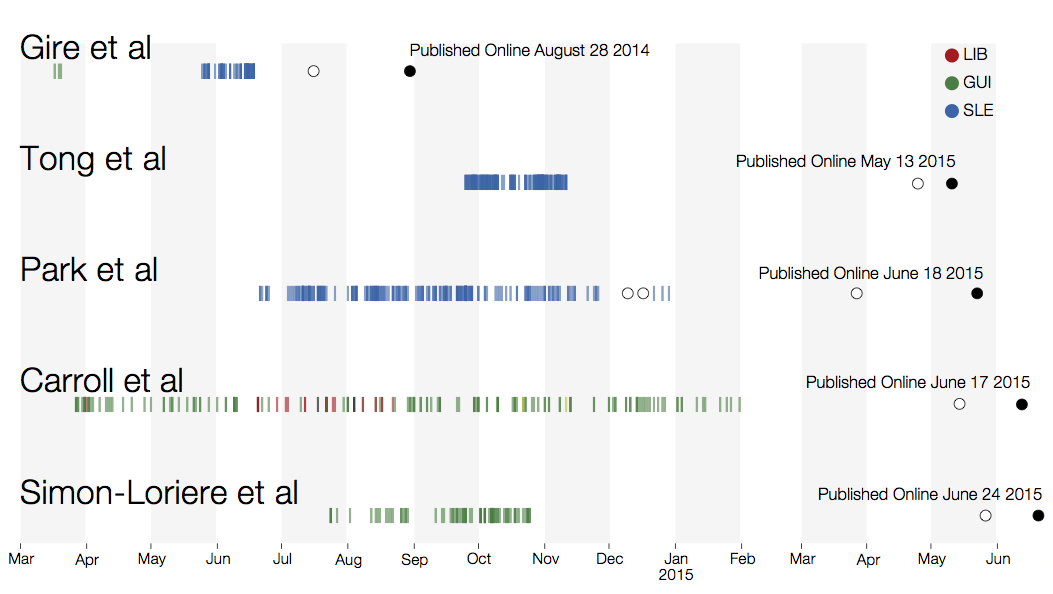
Key challenges to making genomic epidemiology actionable
- Timely analysis and sharing of results critical
- Dissemination must be scalable
- Integrate many data sources
- Results must be easily interpretable and queryable
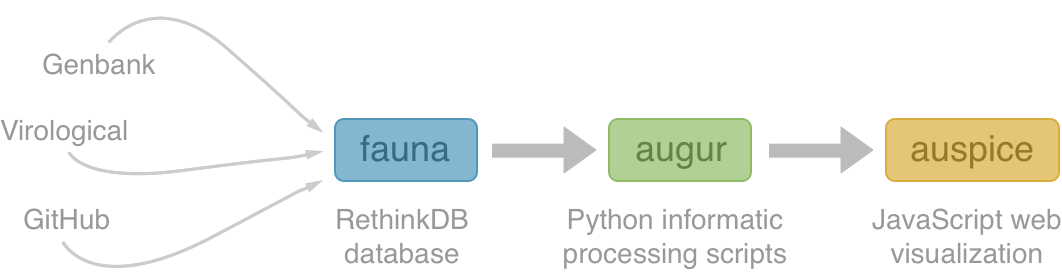
Nextstrain
Project to conduct real-time molecular epidemiology and evolutionary analysis of emerging epidemics
with
![]() Richard Neher,
Richard Neher,
![]() James Hadfield,
James Hadfield,
![]() Emma Hodcroft,
Emma Hodcroft,
![]() Tom Sibley,
Tom Sibley,
![]() John Huddleston,
John Huddleston,
![]() Colin Megill,
Colin Megill,
![]() Sidney Bell,
Sidney Bell,
![]() Barney Potter,
Barney Potter,
![]() Charlton Callender
Charlton Callender
Nextstrain architecture
All code open source at github.com/nextstrain
Two central aims: (1) rapid and flexible phylodynamic analysis and
(2) interactive visualization
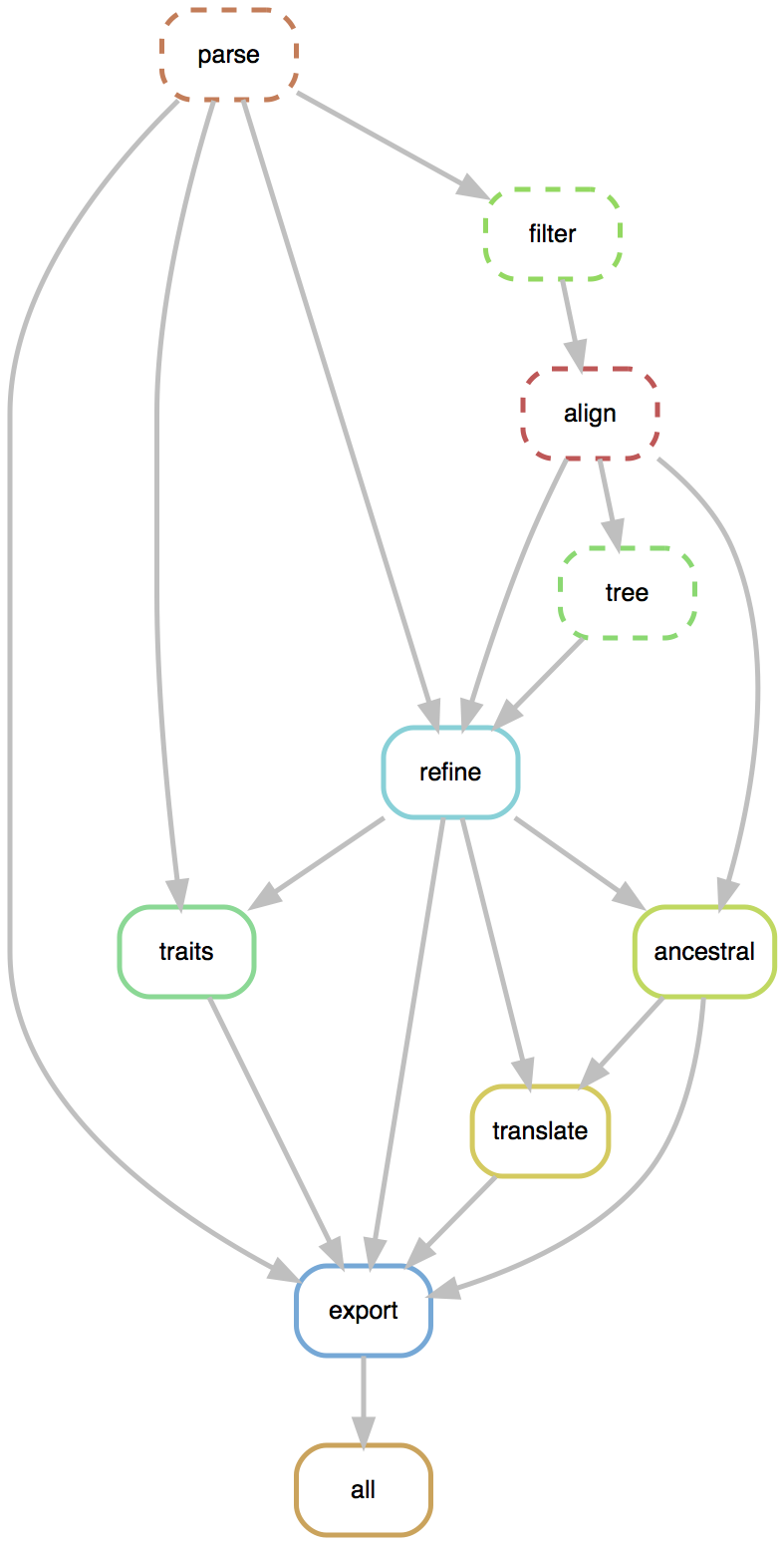
Rapid build pipeline for 1600 Ebola genomes
- Align with MAFFT (34 min)
- Build ML tree with RAxML (54 min)
- Temporally resolve tree and geographic ancestry with TreeTime (16 min)
- Total pipeline (1 hr 46 min)
Flexible pipelines constructed through command line modules
- Modules called via
augur filter,augur tree,augur traits, etc... - Designed to be composable across pathogen builds
- Defined pipeline, making steps obvious
- Provides dependency graph for fast recomputation
- Pathogen-specific repos give users an obvious foundation to build from
Nextstrain is two things
- a bioinformatics toolkit and visualization app, which can be used for a broad range of datasets
- a collection of real-time pathogen analyses kept up-to-date on the website nextstrain.org
nextstrain.org
Rapid on-the-ground sequencing in Makeni, Sierra Leone
Newly released features
- Bacteria build pipelines using VCF rather than FASTA
- "Community" builds to promote frictionless sharing of results
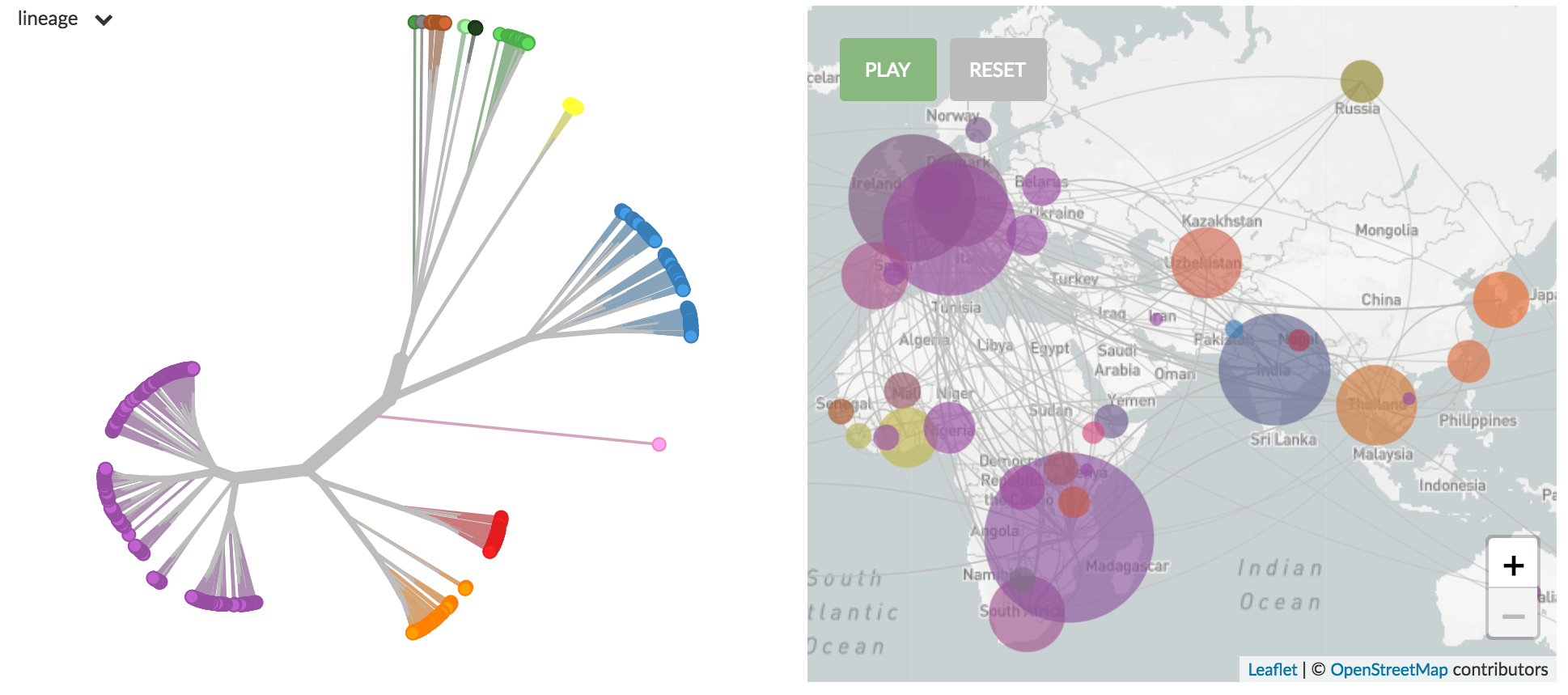
Global distribution of TB lineages
Tuberculosis builds courtesy of Emma Hodcroft
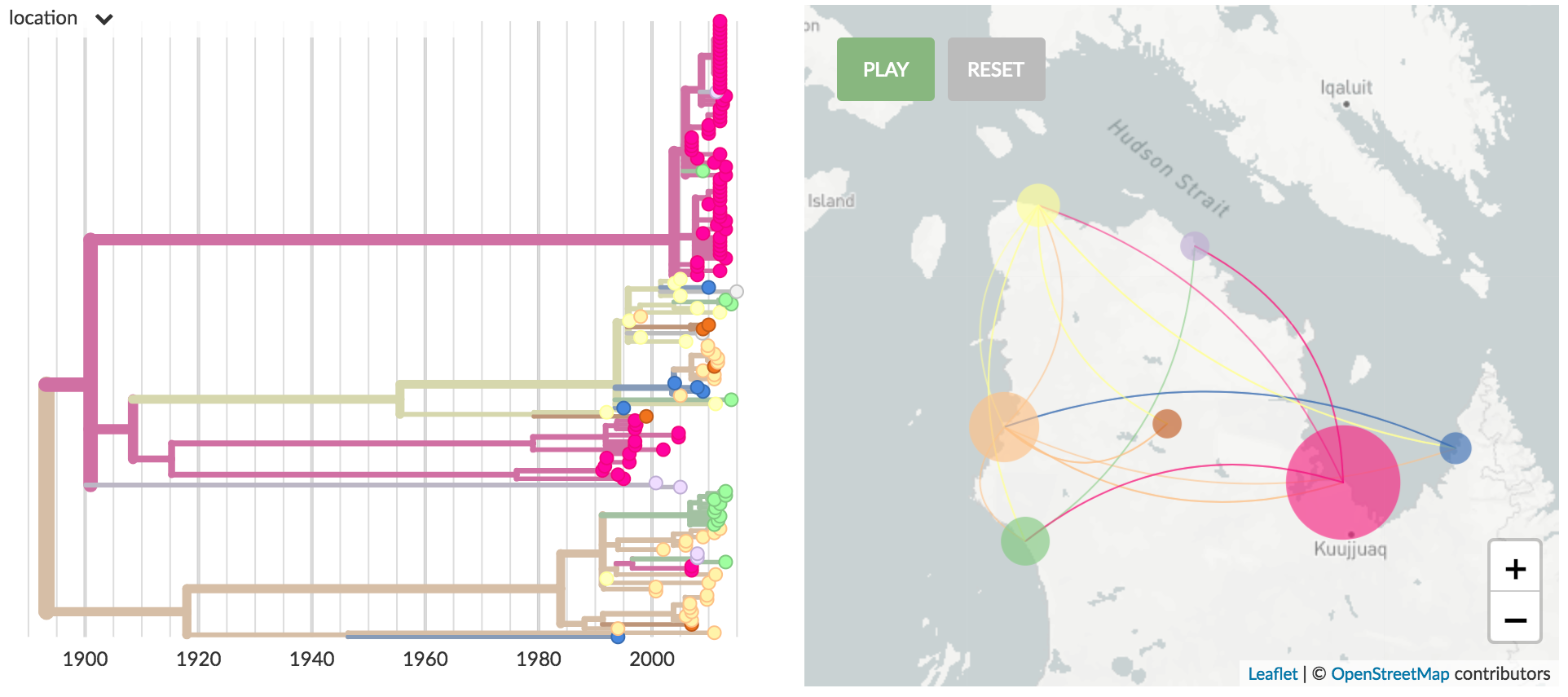
Detailed spread of TB in Nunavik
Tuberculosis builds courtesy of Emma Hodcroft
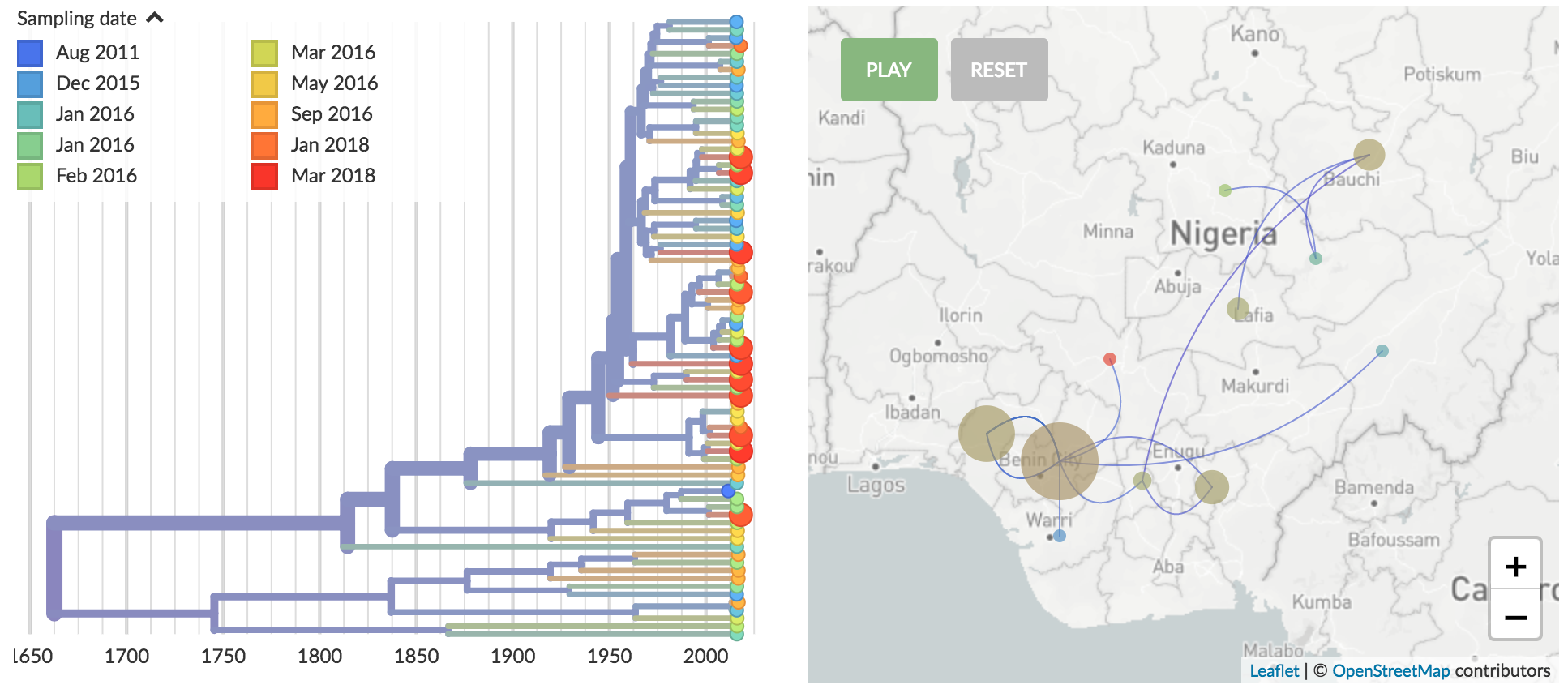
Genomic epidemiology of Lassa virus in Nigeria
Data and builds courtesy of Paul Oluniyi, Christian Happi and ACEGID at Redeemer's University
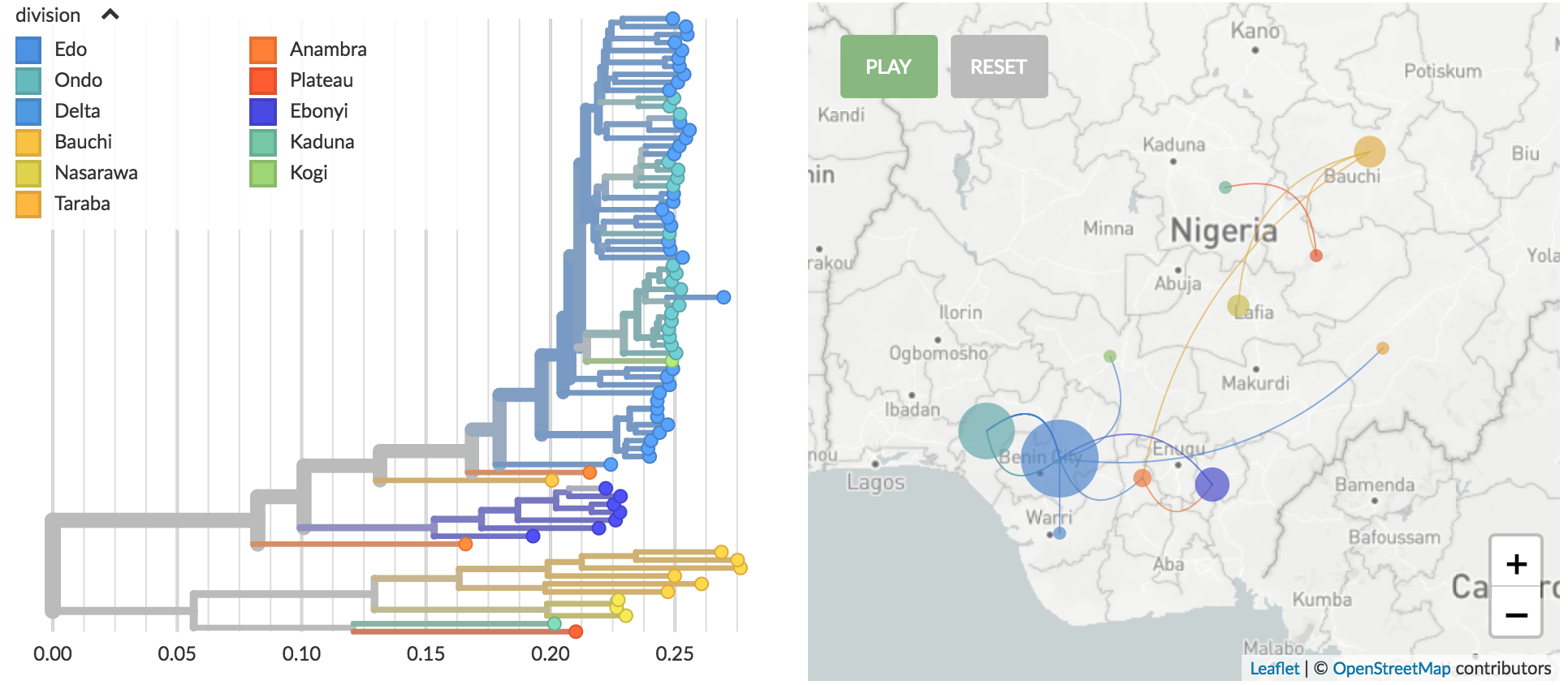
Stable clustering by geography
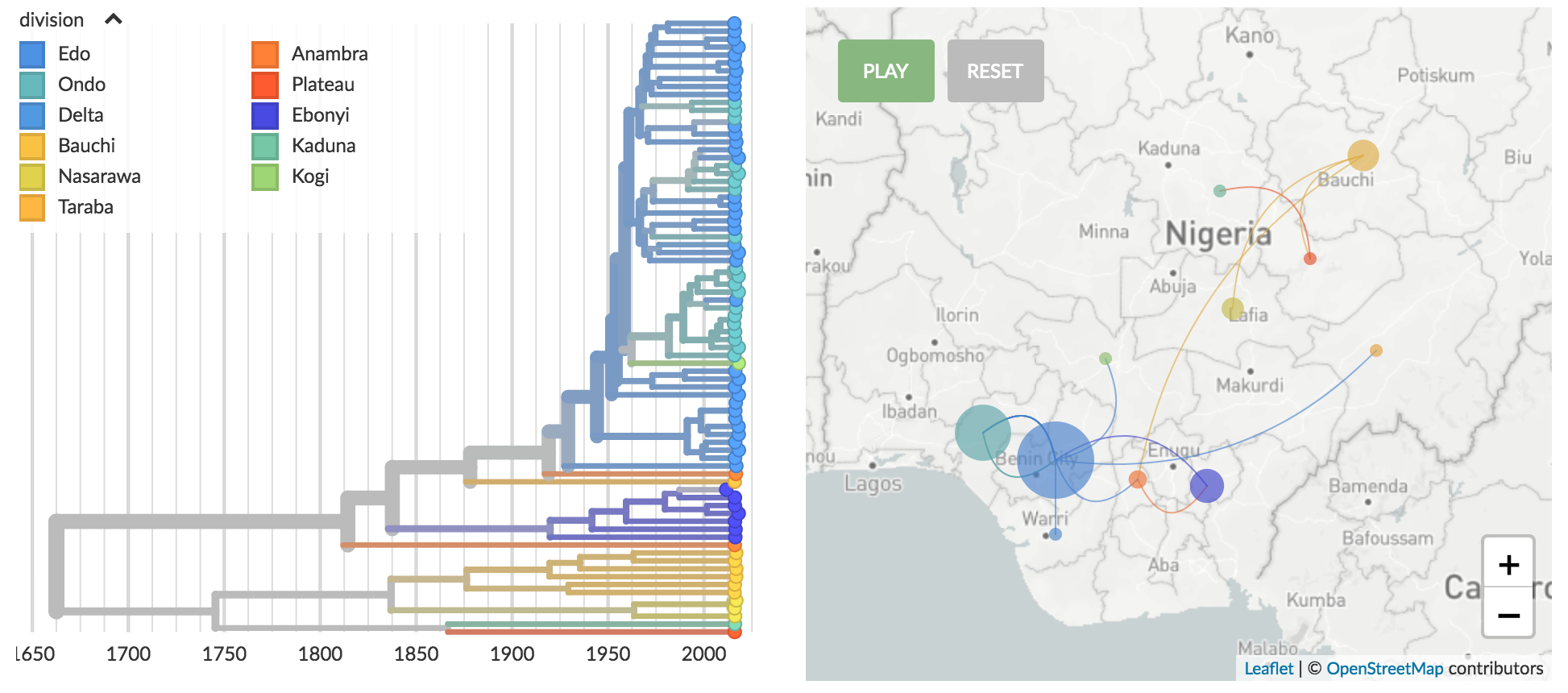
Recent "outbreak" originates from a diverse group of viruses
Page live at nextstrain.org/community/pauloluniyi/lassa/s
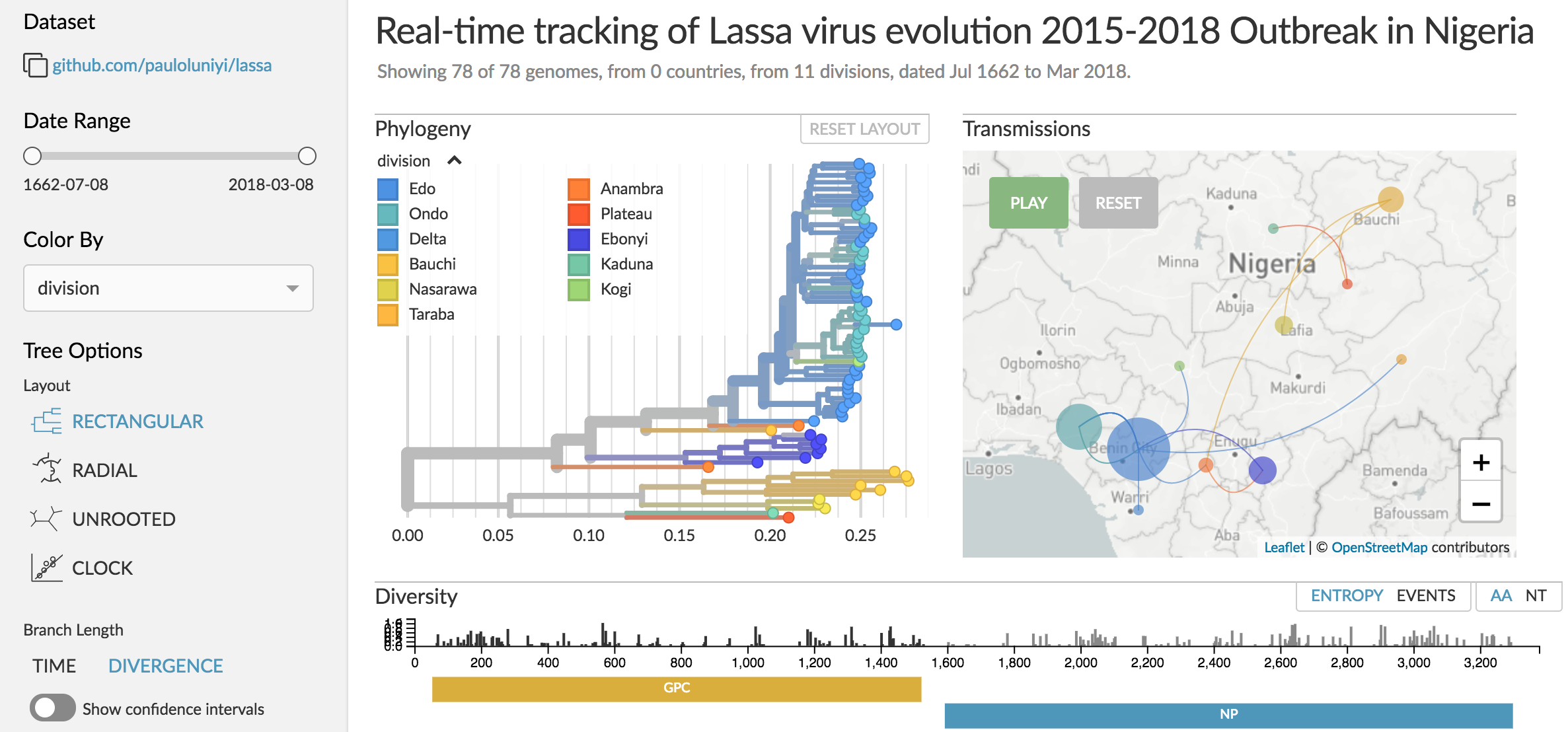
Results sourced from Paul's GitHub page
Recent headway towards "actionable" genomic epidemiology
- affordable and portable full genome sequencing (ONT MinION)
- rapid phylodynamic methods (TreeTime, treedater, etc...)
- rapid distribution of results (nextstrain.org, microreact.org, virological.org)
Acknowledgements
Bedford Lab:
 Alli Black,
Alli Black,
 Sidney Bell,
Sidney Bell,
 Gytis Dudas,
Gytis Dudas,
 John Huddleston,
John Huddleston,
 Barney Potter,
Barney Potter,
 James Hadfield,
James Hadfield,
 Louise Moncla,
Louise Moncla,
 Tom Sibley
Tom Sibley
MERS: Gytis Dudas, Andrew Rambaut, Luiz Carvalho
Ebola: Gytis Dudas, Andrew Rambaut, Luiz Carvalho, Philippe Lemey, Marc Suchard, Andrew Tatem
Zika: Nick Loman, Nuno Faria, Oli Pybus, Josh Quick, Ingra Claro, Julien Thézé, Jaquilene de Jesus, Marta Giovanetti, Moritz Kraemer, Sarah Hill, Allison Black, Ester Sabino, Luiz Alcantara
Nextstrain: Richard Neher, James Hadfield, Emma Hodcroft, Tom Sibley, Paul Oluniyi, John Huddleston, Sidney Bell, Barney Potter, Colin Megill, Charlton Callender





