Real-time genomic surveillance of pathogen evolution and spread
Trevor Bedford (@trvrb)
29 Nov 2017
Epidemics6
Sitges, Spain
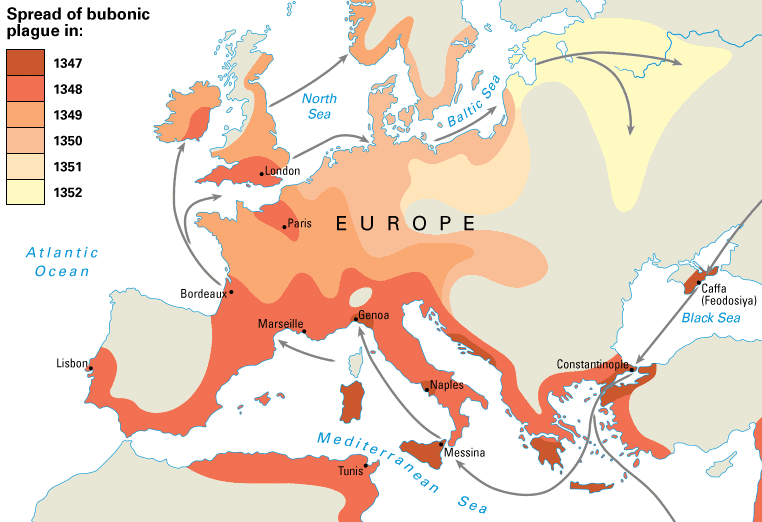
Spread of plague in 14th century
Spread of swine flu in 2009
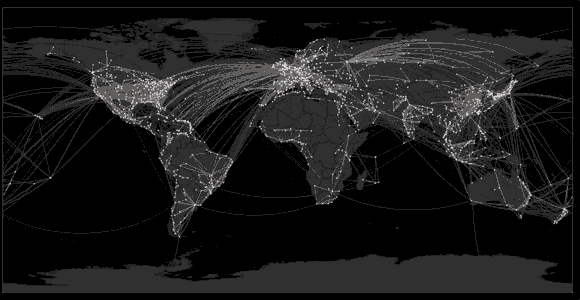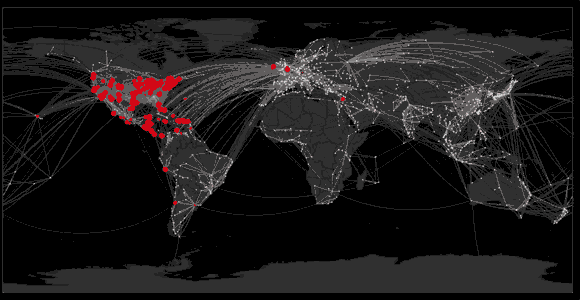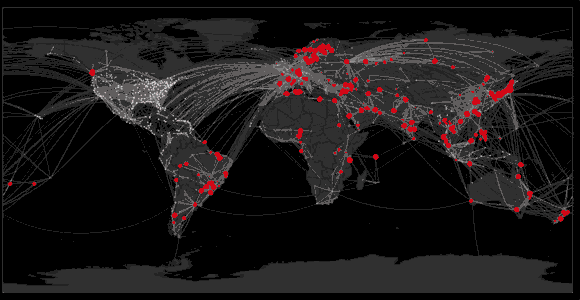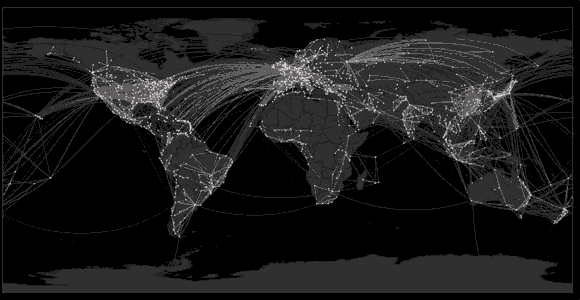
Sequencing to reconstruct pathogen spread
Epidemic process
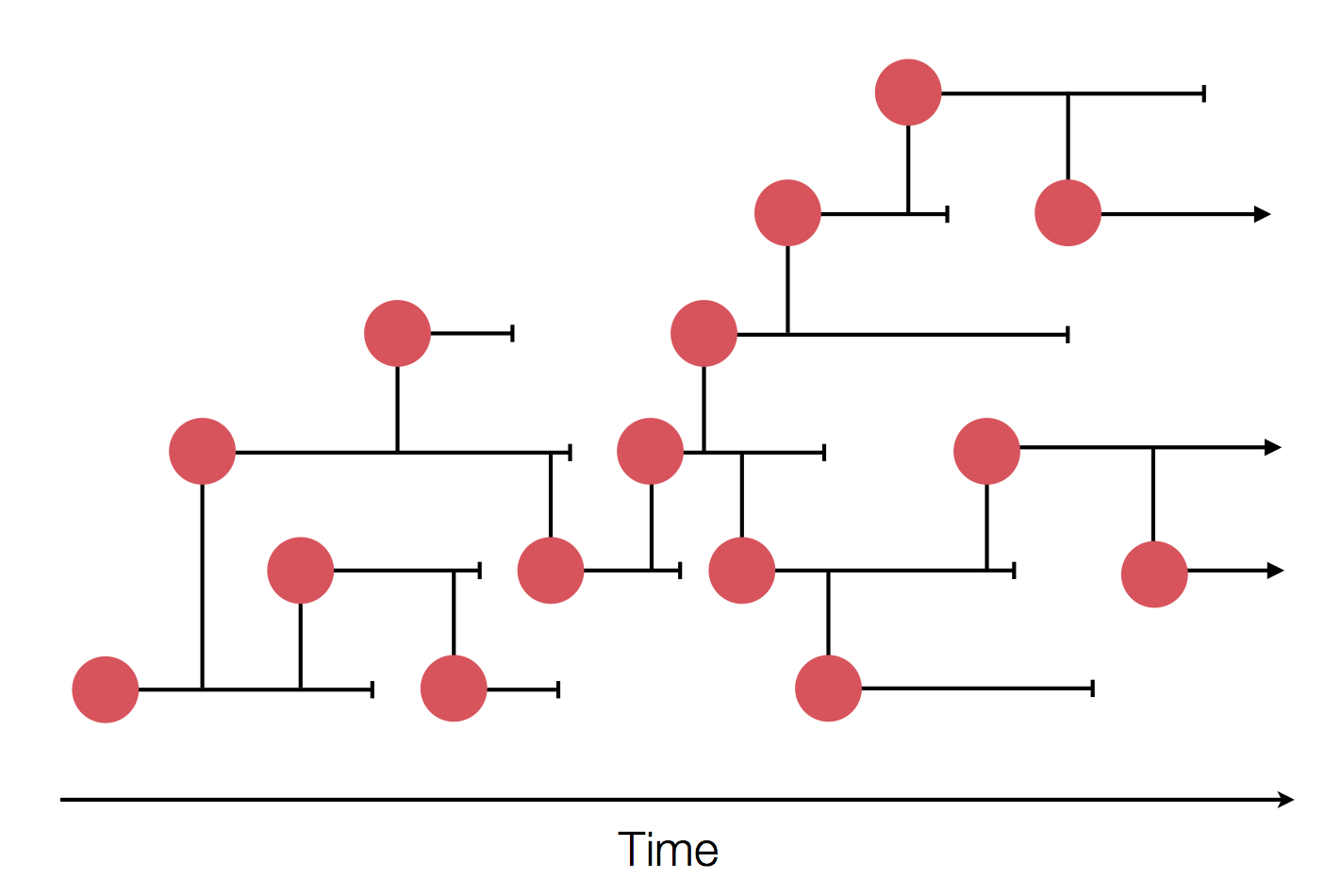
Sample some individuals
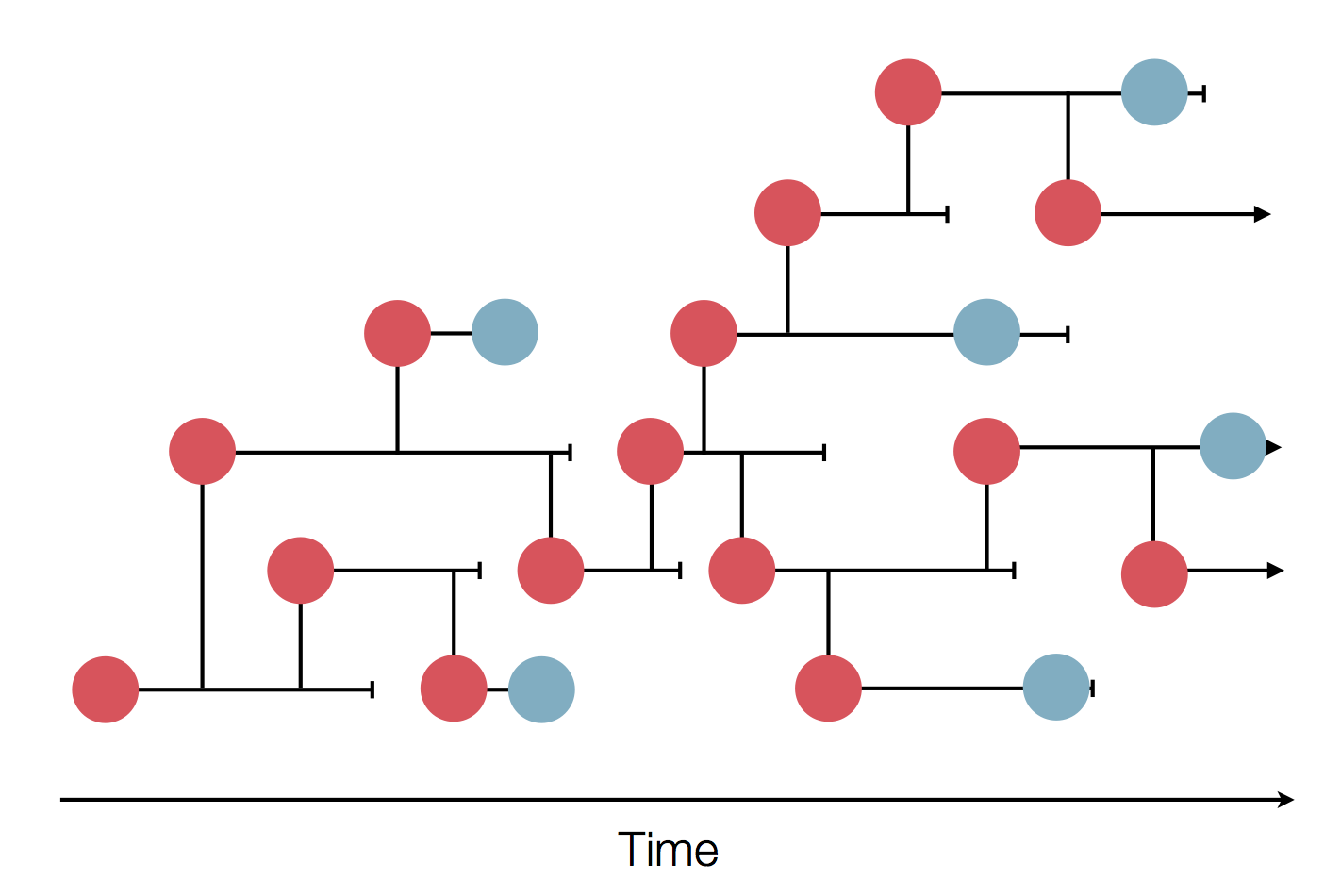
Sequence and determine phylogeny
Sequence and determine phylogeny
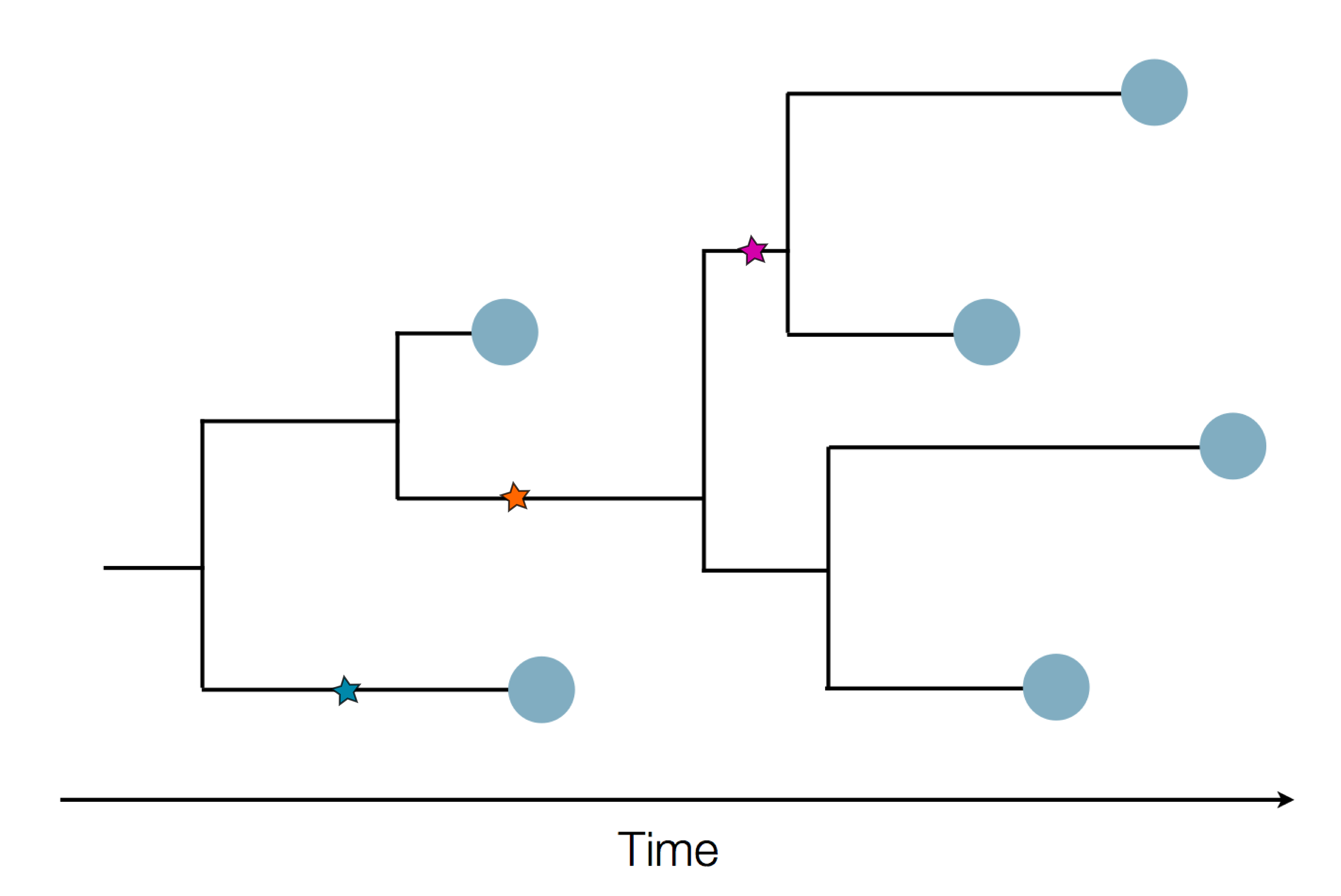
Phylogenetic tracking has the capacity to revolutionize epidemiology
Virus genomes reveal factors that spread and sustained the Ebola epidemic
with ![]() Gytis Dudas, Andrew Rambaut, Luiz Carvalho, Marc Suchard, Philippe Lemey,
Gytis Dudas, Andrew Rambaut, Luiz Carvalho, Marc Suchard, Philippe Lemey,
and many others
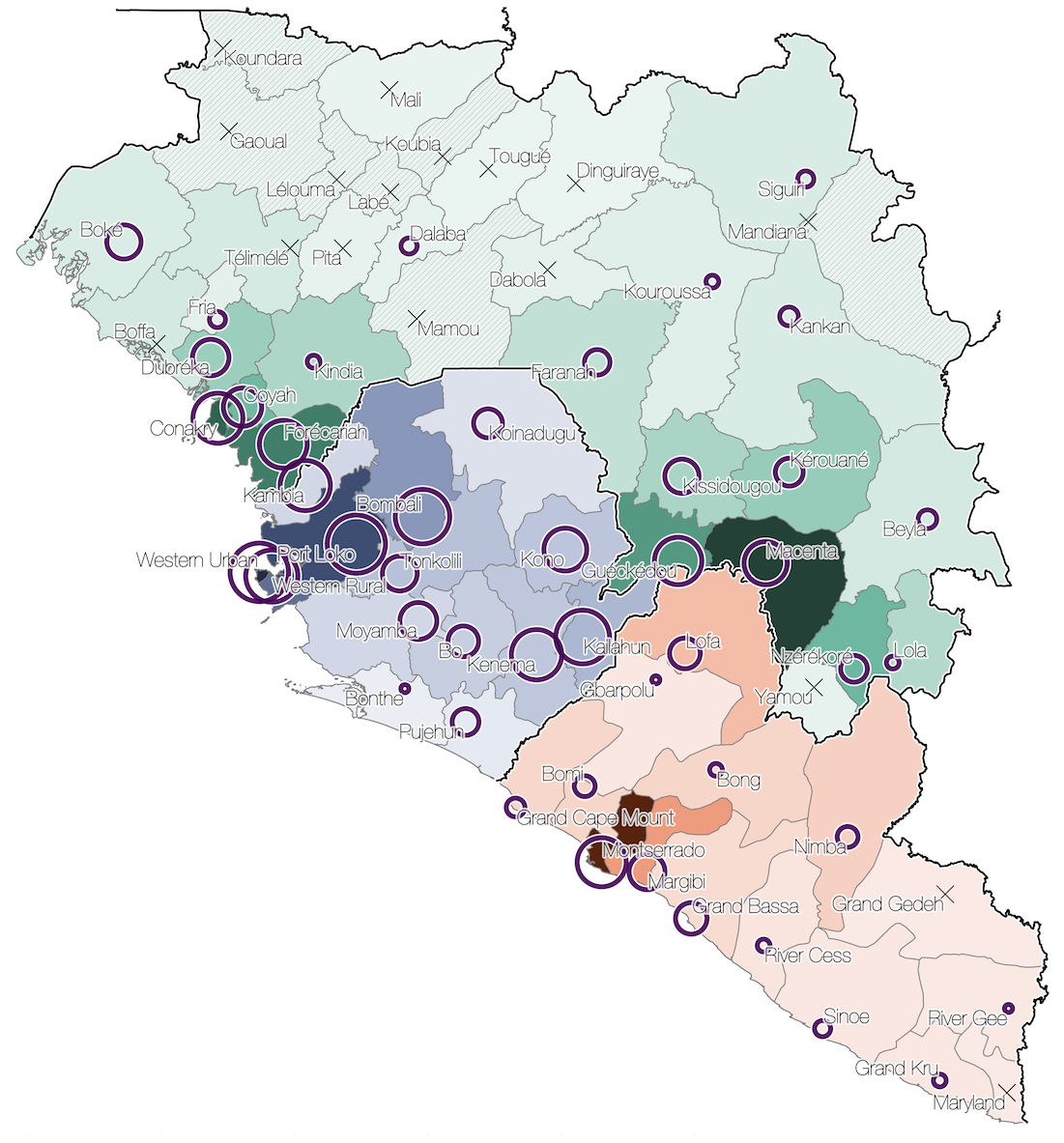
Sequencing of 1610 Ebola virus genomes collected during the 2013-2016 West African epidemic
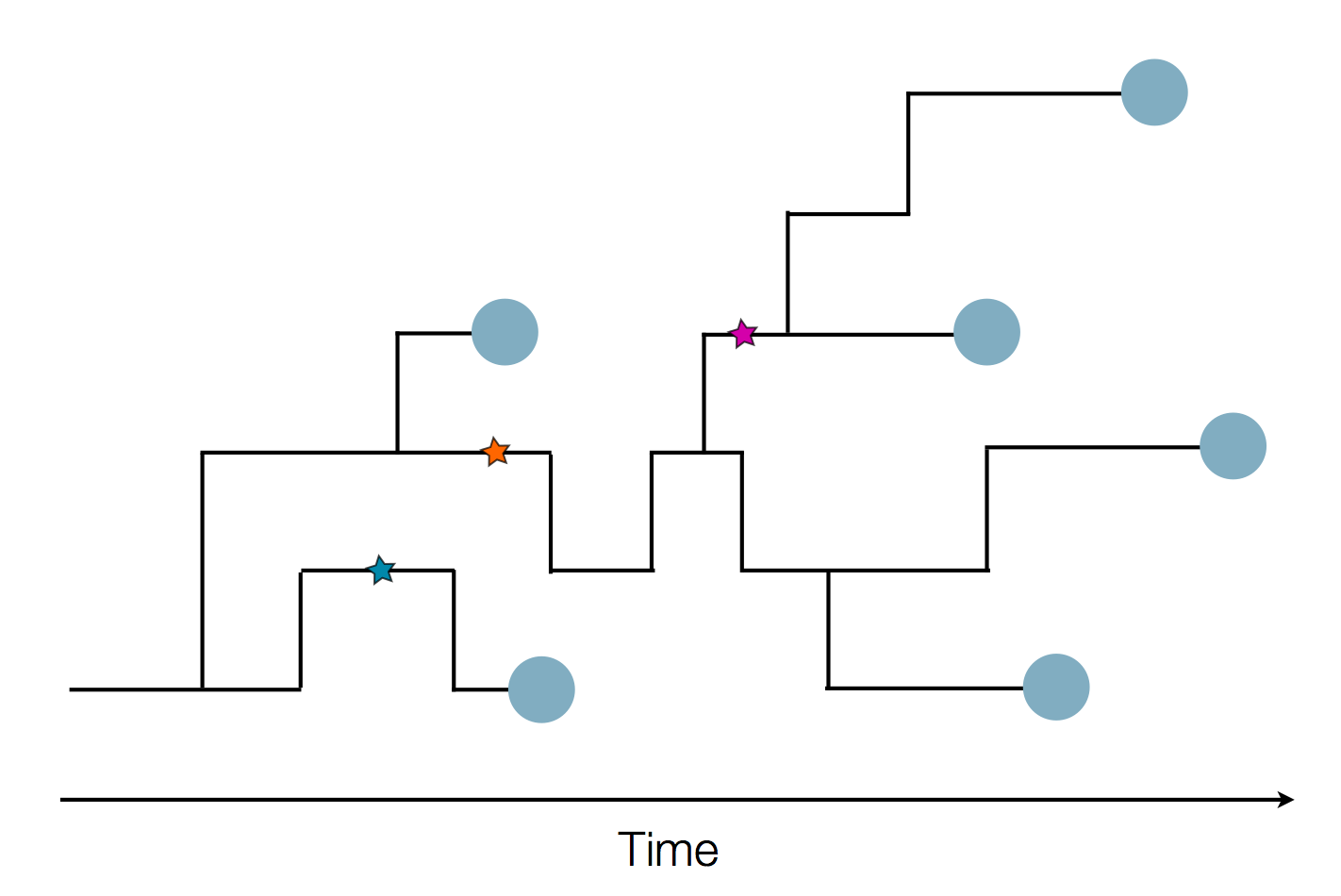
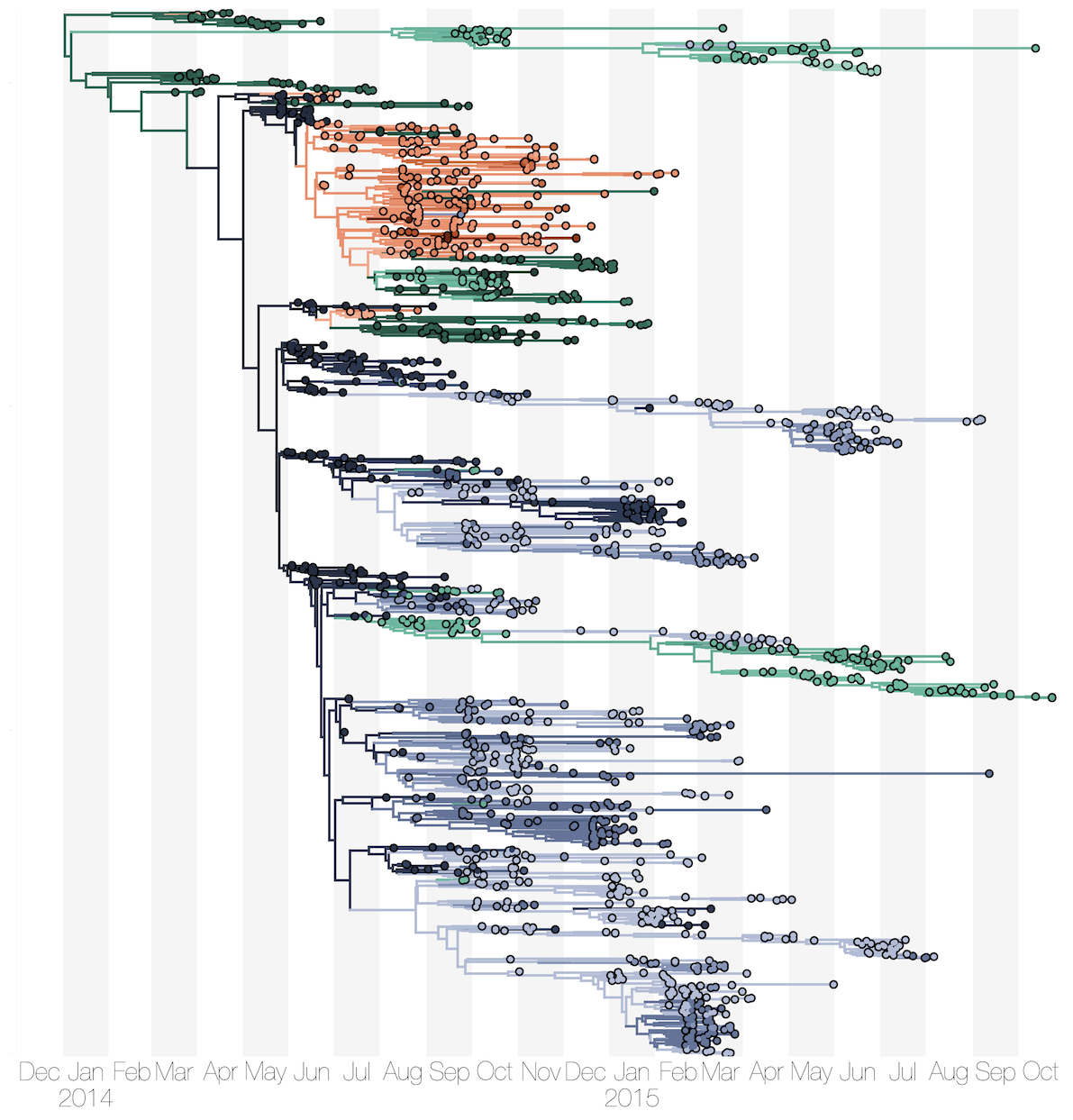
Phylogenetic reconstruction of evolution and spread
Tracking migration events
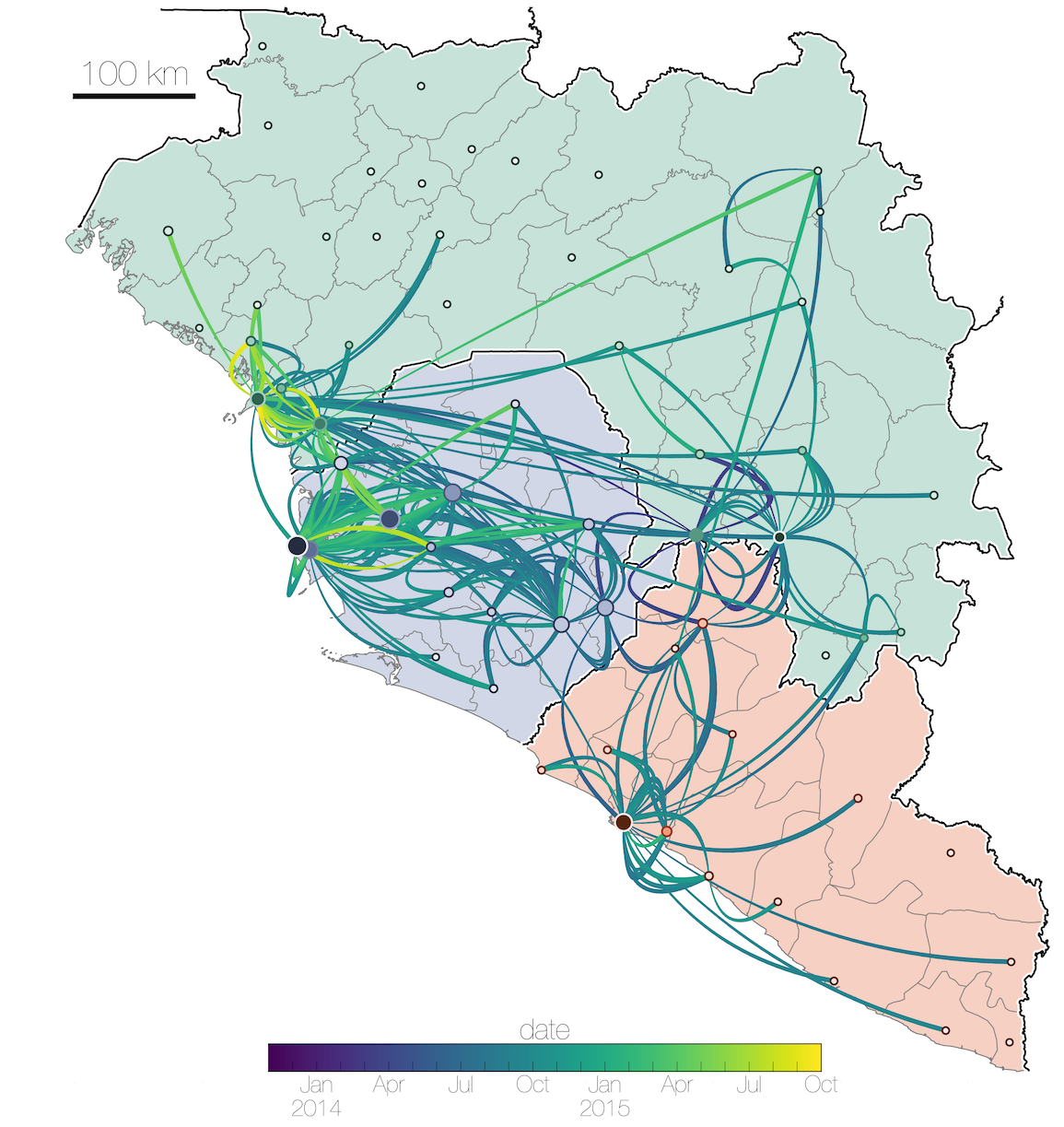
Factors influencing migration rates
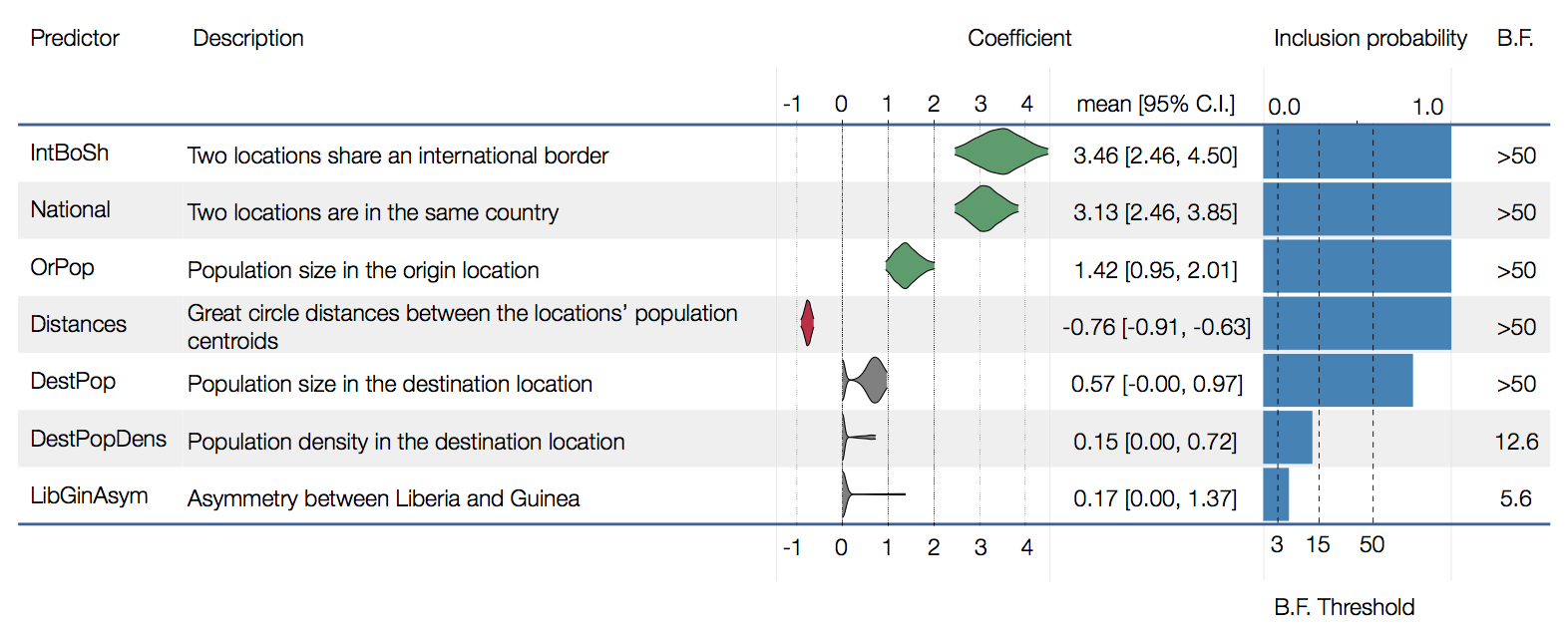
Spatial structure at the country level
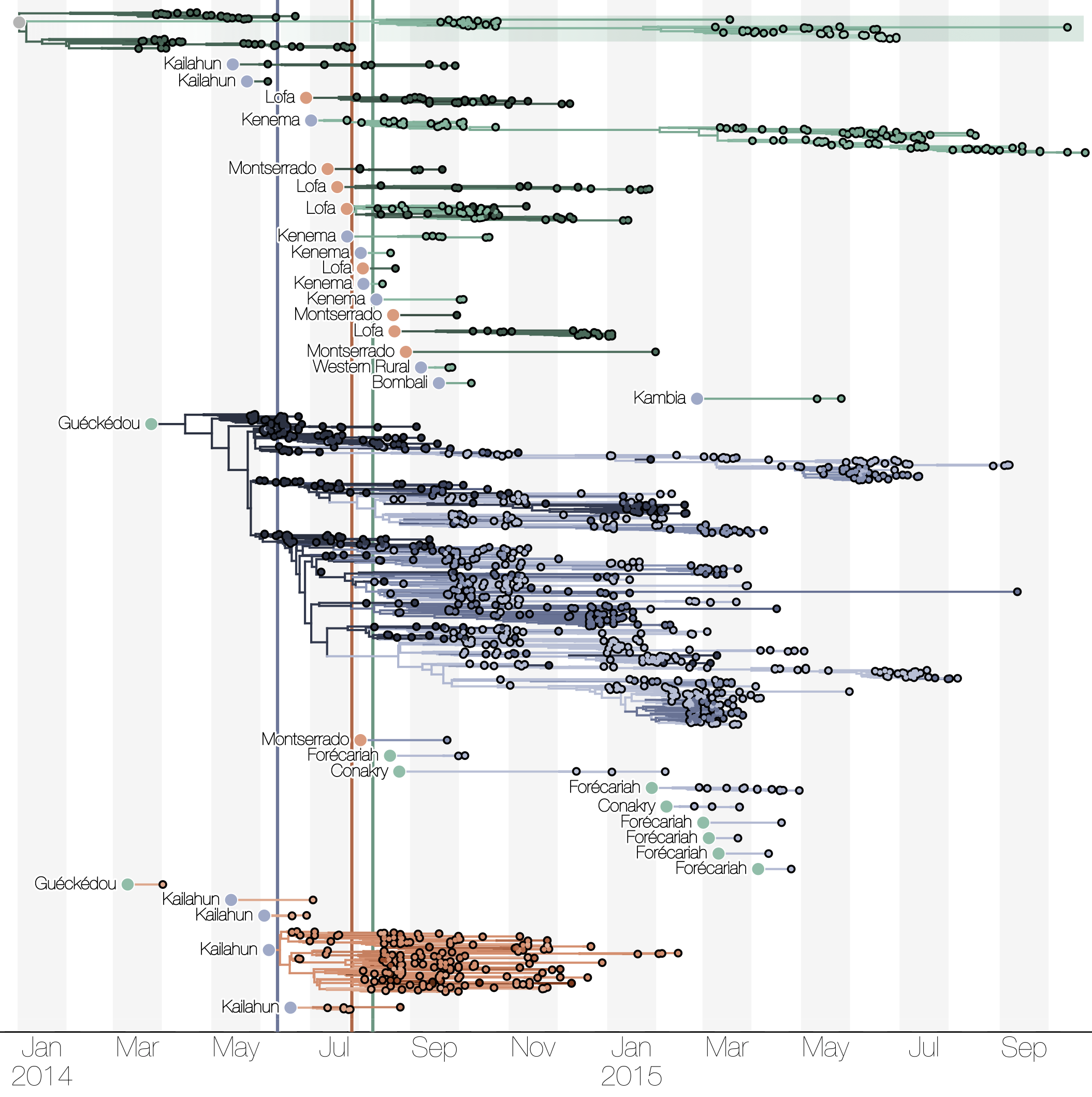
Substantial mixing at the regional level
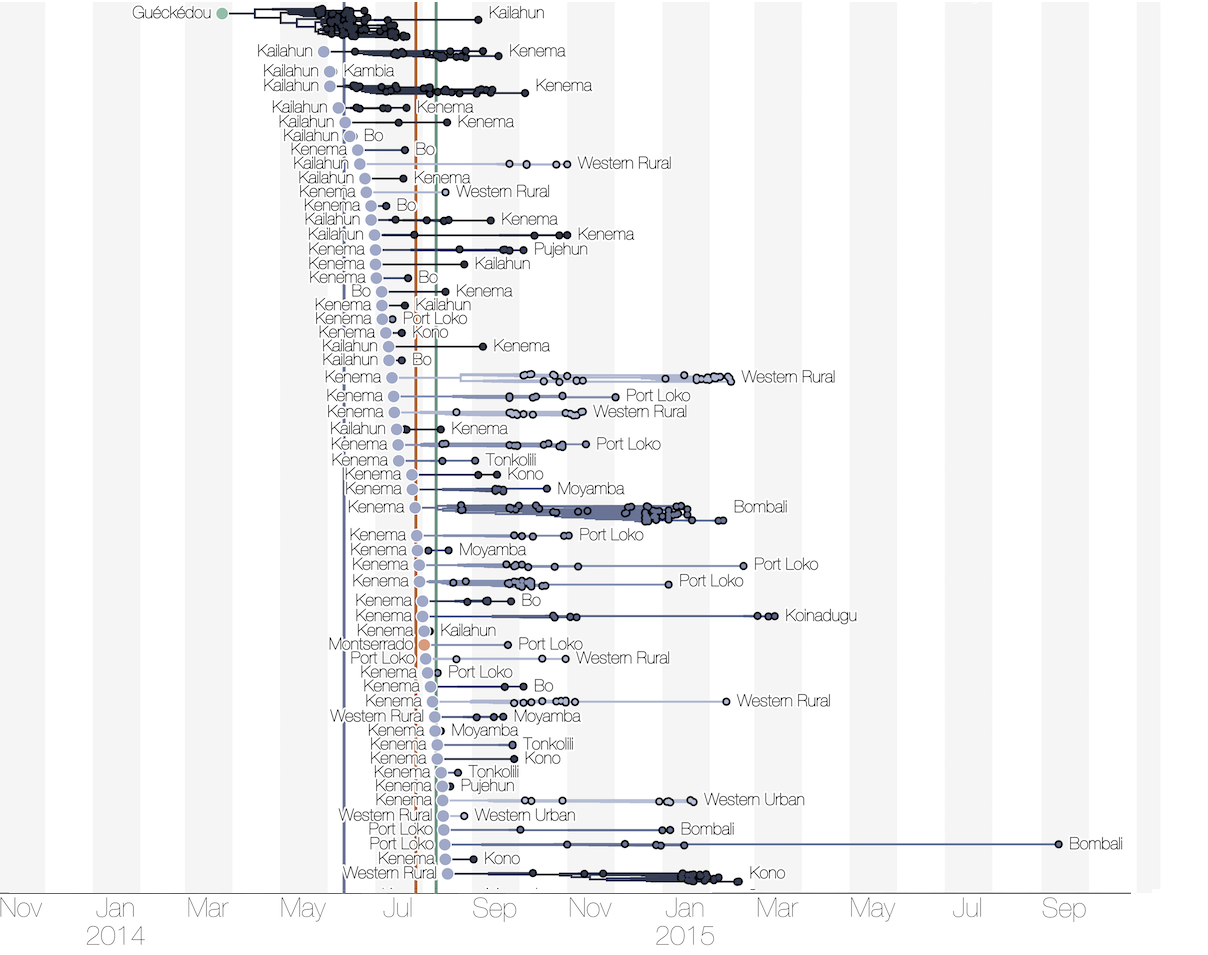
Regional outbreaks due to multiple introductions
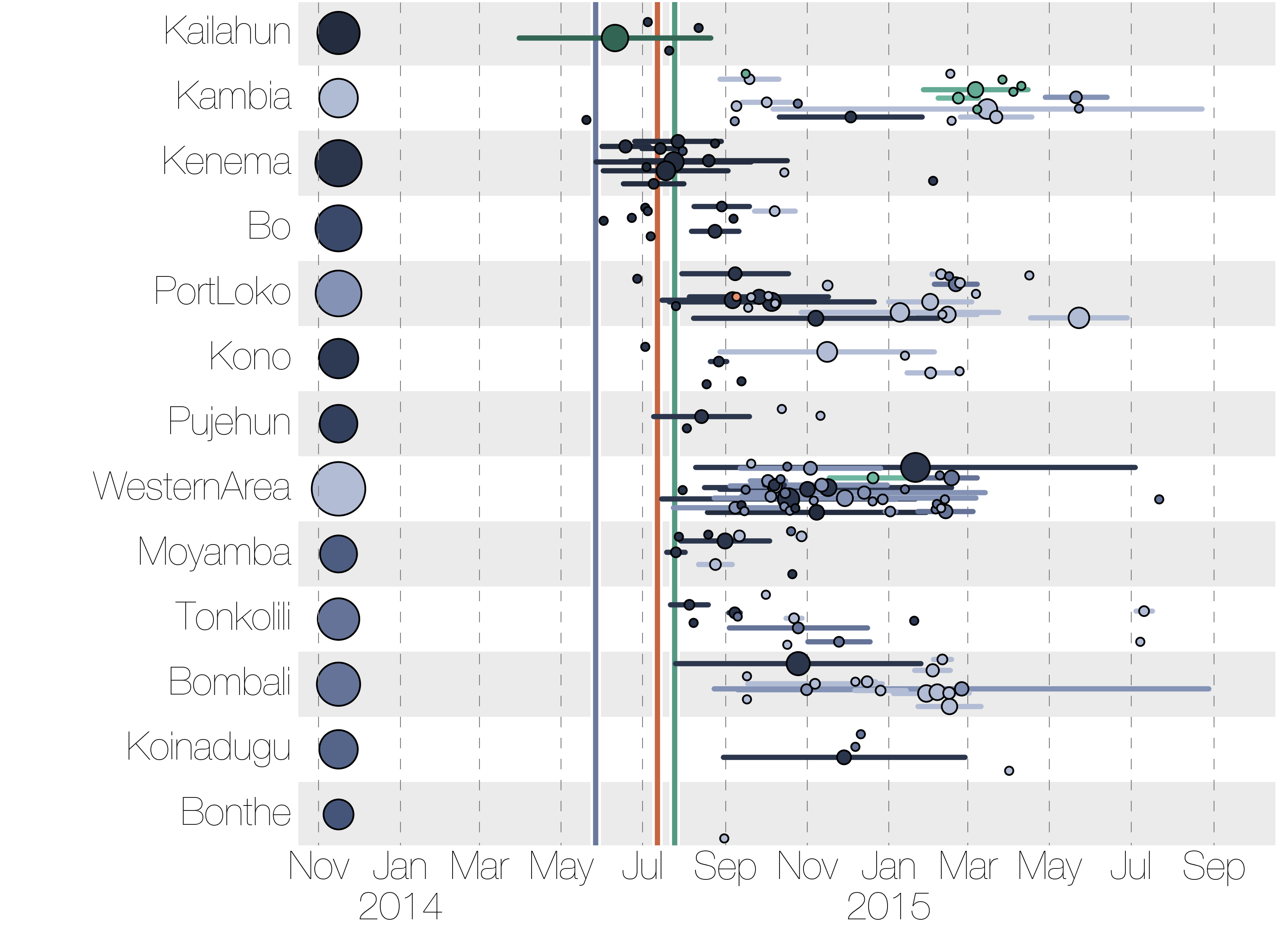
Genomic analyses were mostly done in a retrospective manner
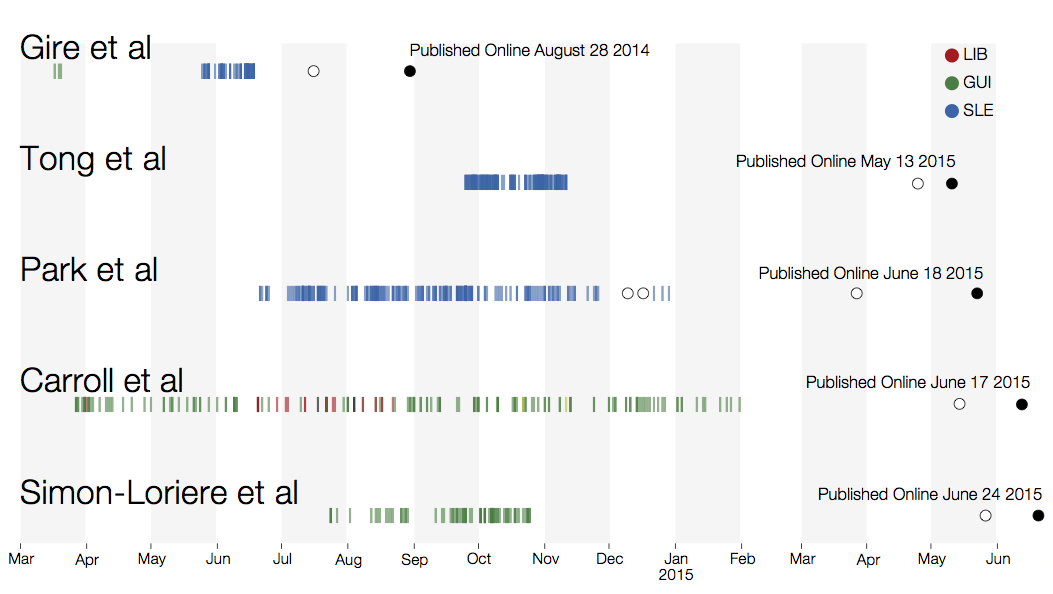
Key challenges to making genomic epidemiology actionable
- Timely analysis and sharing of results critical
- Dissemination must be scalable
- Integrate many data sources
- Results must be easily interpretable and queryable
nextstrain
Project to conduct real-time molecular epidemiology and evolutionary analysis of emerging epidemics
with
![]() Richard Neher,
Richard Neher,
![]() James Hadfield,
James Hadfield,
![]() Colin Megill,
Colin Megill,
![]() Sidney Bell,
Sidney Bell,
![]() Charlton Callender,
Charlton Callender,
![]() Barney Potter,
Barney Potter,
and ![]() John Huddleston
John Huddleston
Nextstrain architecture
All code open source at github.com/nextstrain
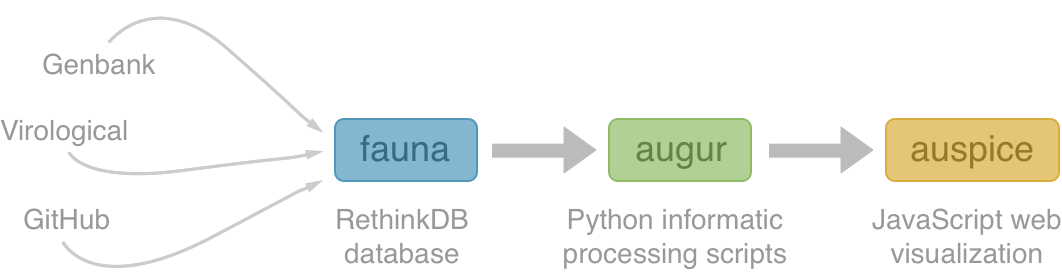
Example augur pipeline for 1600 Ebola genomes
- Align with MAFFT (34 min)
- Build ML tree with RAxML (54 min)
- Temporally resolve tree and geographic ancestry with TreeTime (16 min)
- Total pipeline (1 hr 46 min)
nextstrain.org
Rapid on-the-ground sequencing by Ian Goodfellow, Matt Cotten and colleagues
Desired analytics are pathogen specific and tied to response measures
Build out pipelines for different pathogens, improve databasing and lower
bioinformatics bar
Acknowledgements
Bedford Lab:
![]() Alli Black,
Alli Black,
![]() Sidney Bell,
Sidney Bell,
![]() Gytis Dudas,
Gytis Dudas,
![]() John Huddleston,
John Huddleston,
![]() Barney Potter,
Barney Potter,
![]() James Hadfield,
James Hadfield,
![]() Louise Moncla
Louise Moncla
Ebola: Gytis Dudas, Andrew Rambaut, Luiz Carvalho, Philippe Lemey, Marc Suchard, Andrew Tatem
Nextstrain: Richard Neher, James Hadfield, Colin Megill, Sidney Bell, Charlton Callender, Barney Potter, John Huddleston





