Real-time genomic surveillance of pathogen evolution and spread
Trevor Bedford (@trvrb)
5 Jun 2017
Global Infectious Disease Seminar
Center for Infectious Disease Research
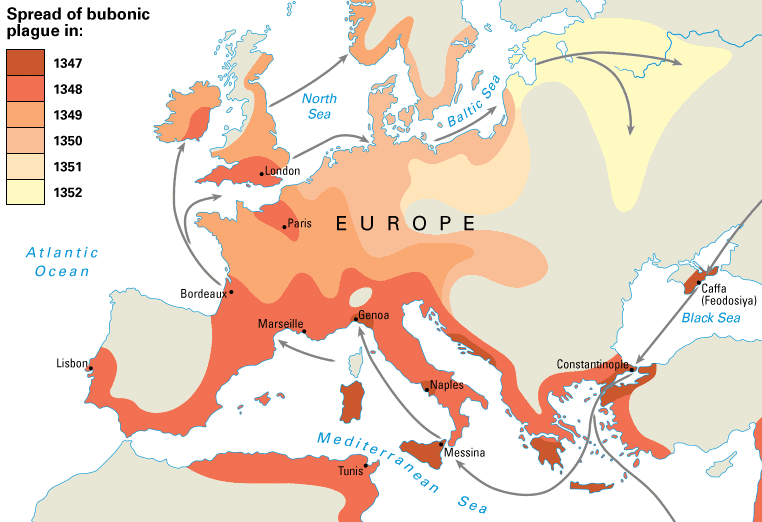
Spread of plague in 14th century
Spread of swine flu in 2009
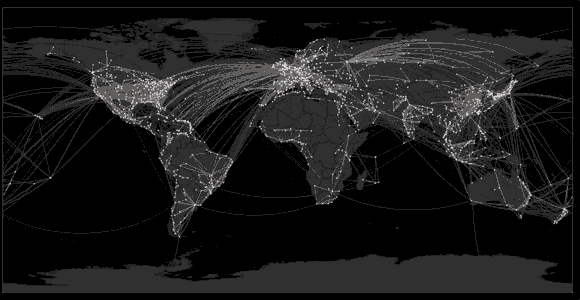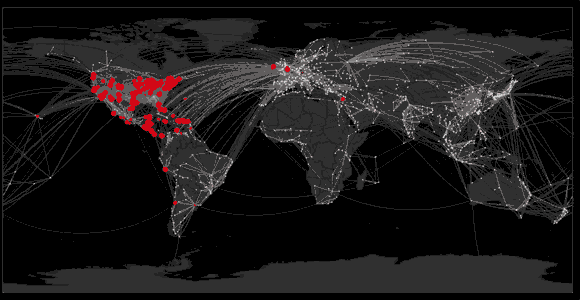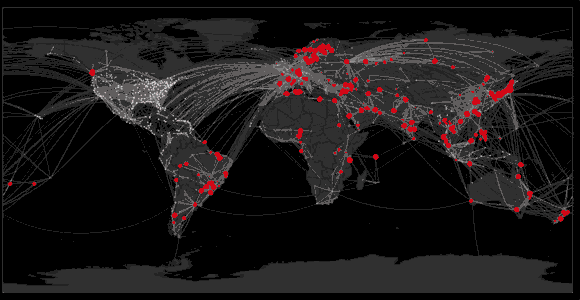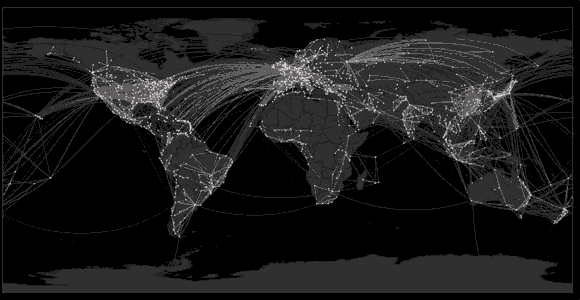
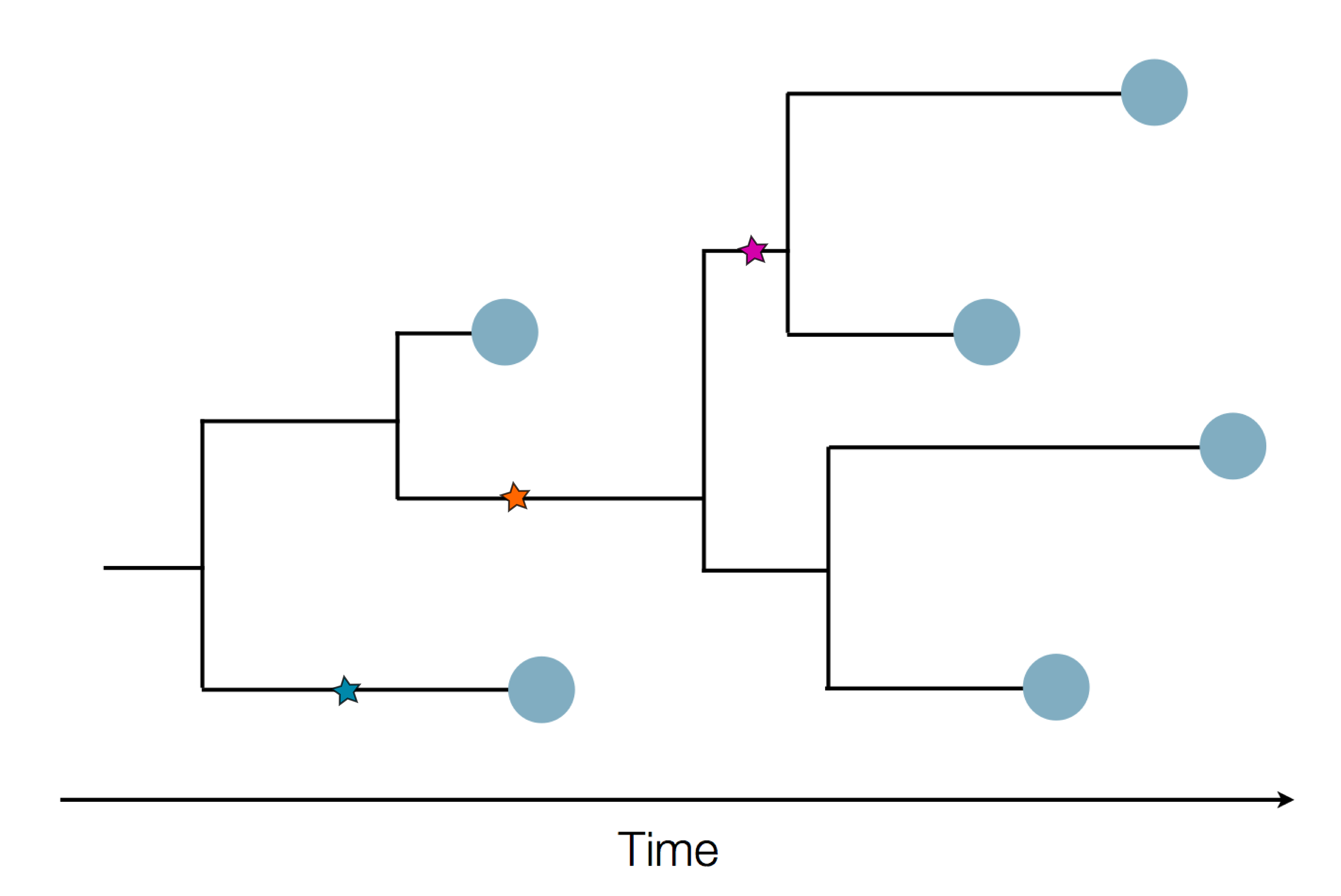
Sequencing to reconstruct pathogen spread
Epidemic process
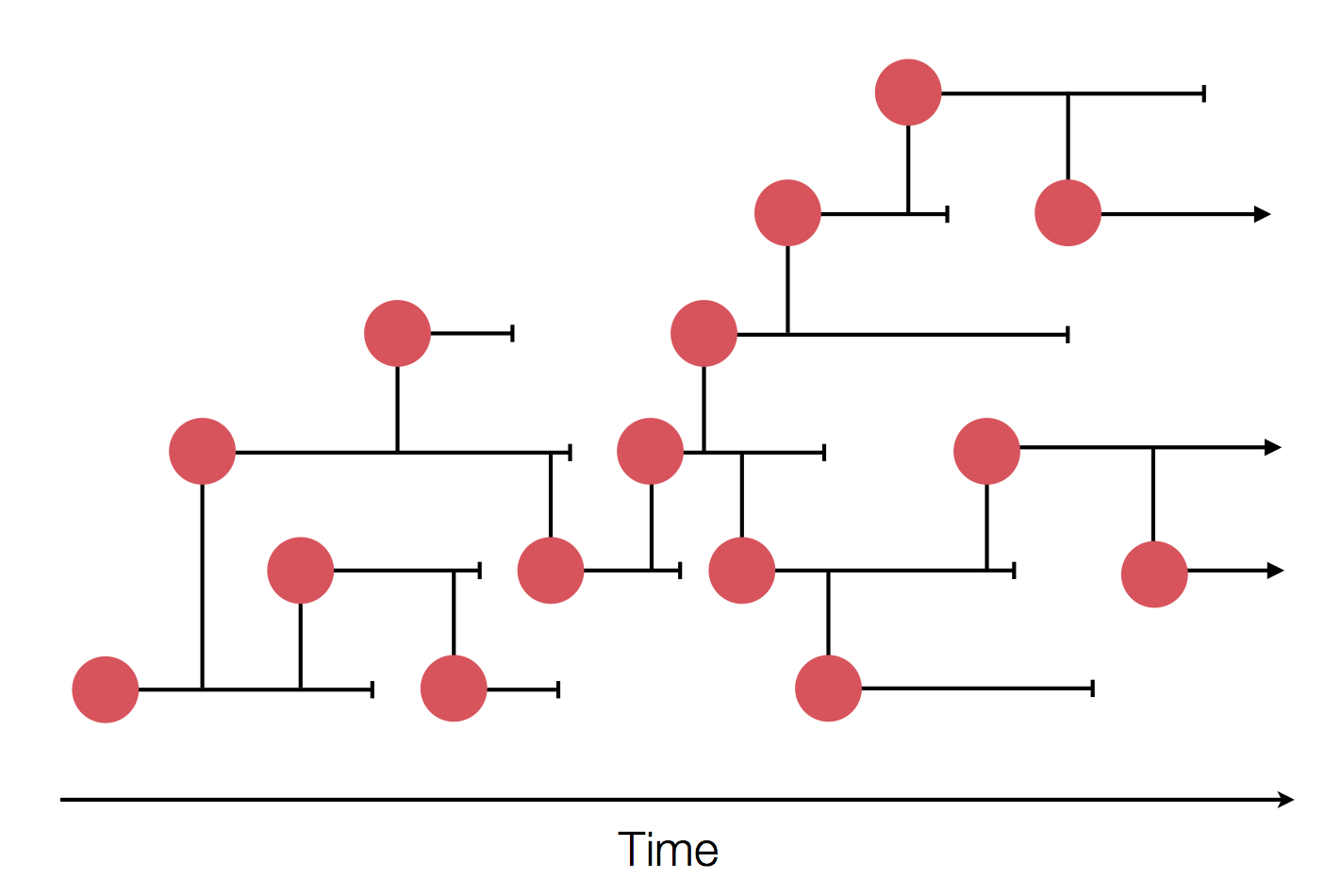
Sample some individuals
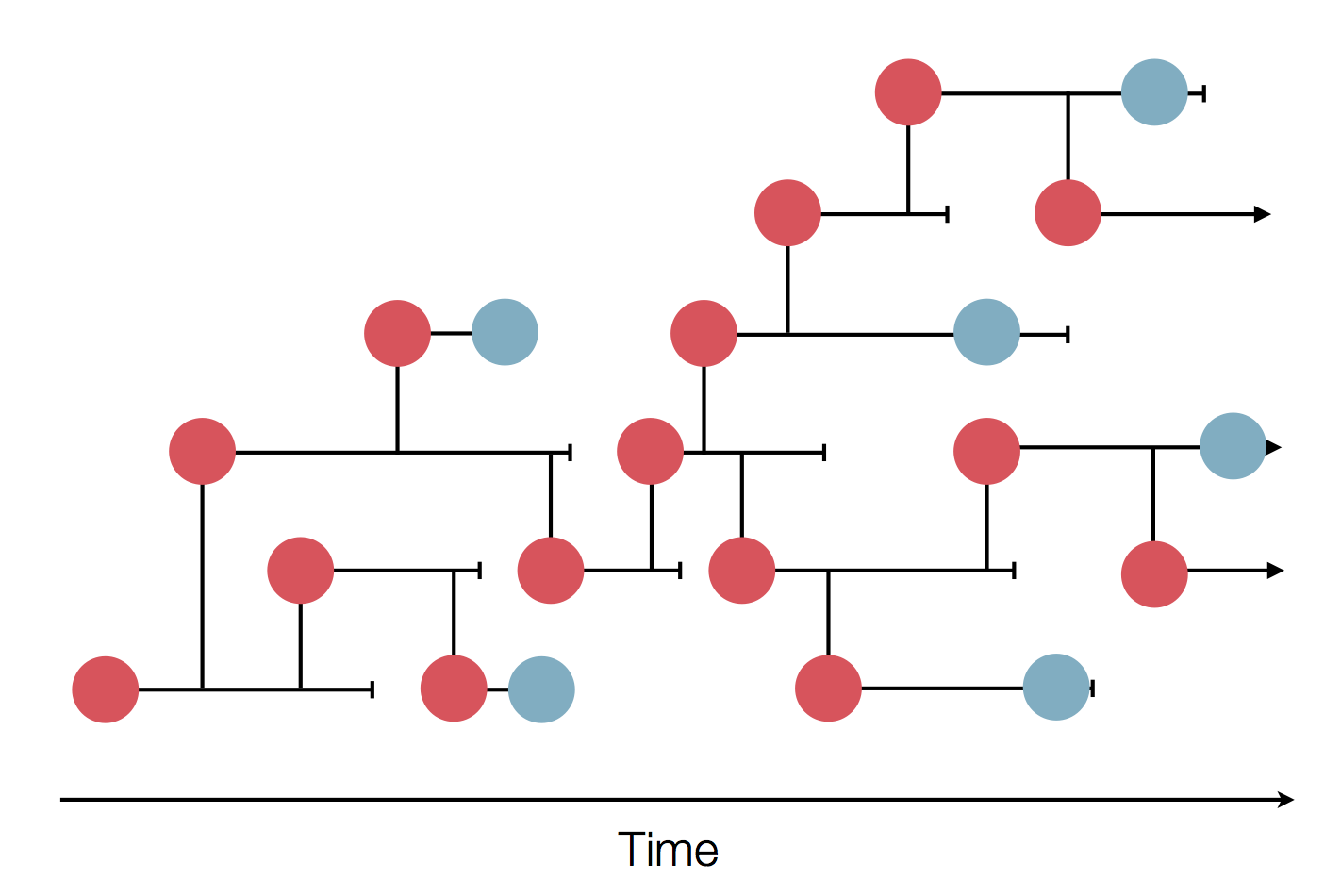
Sequence and determine phylogeny
Sequence and determine phylogeny
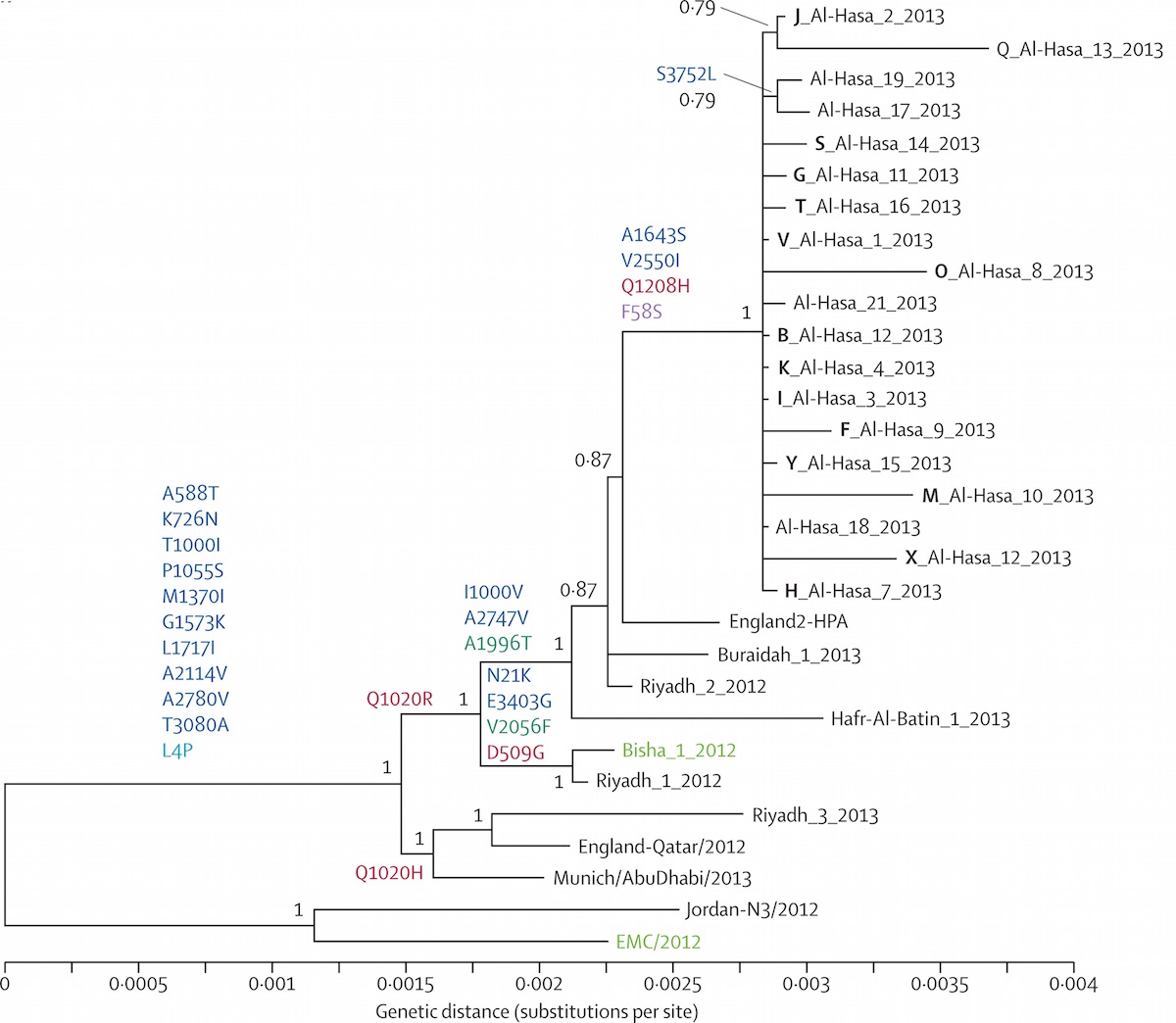
Localized Middle Eastern MERS-CoV phylogeny
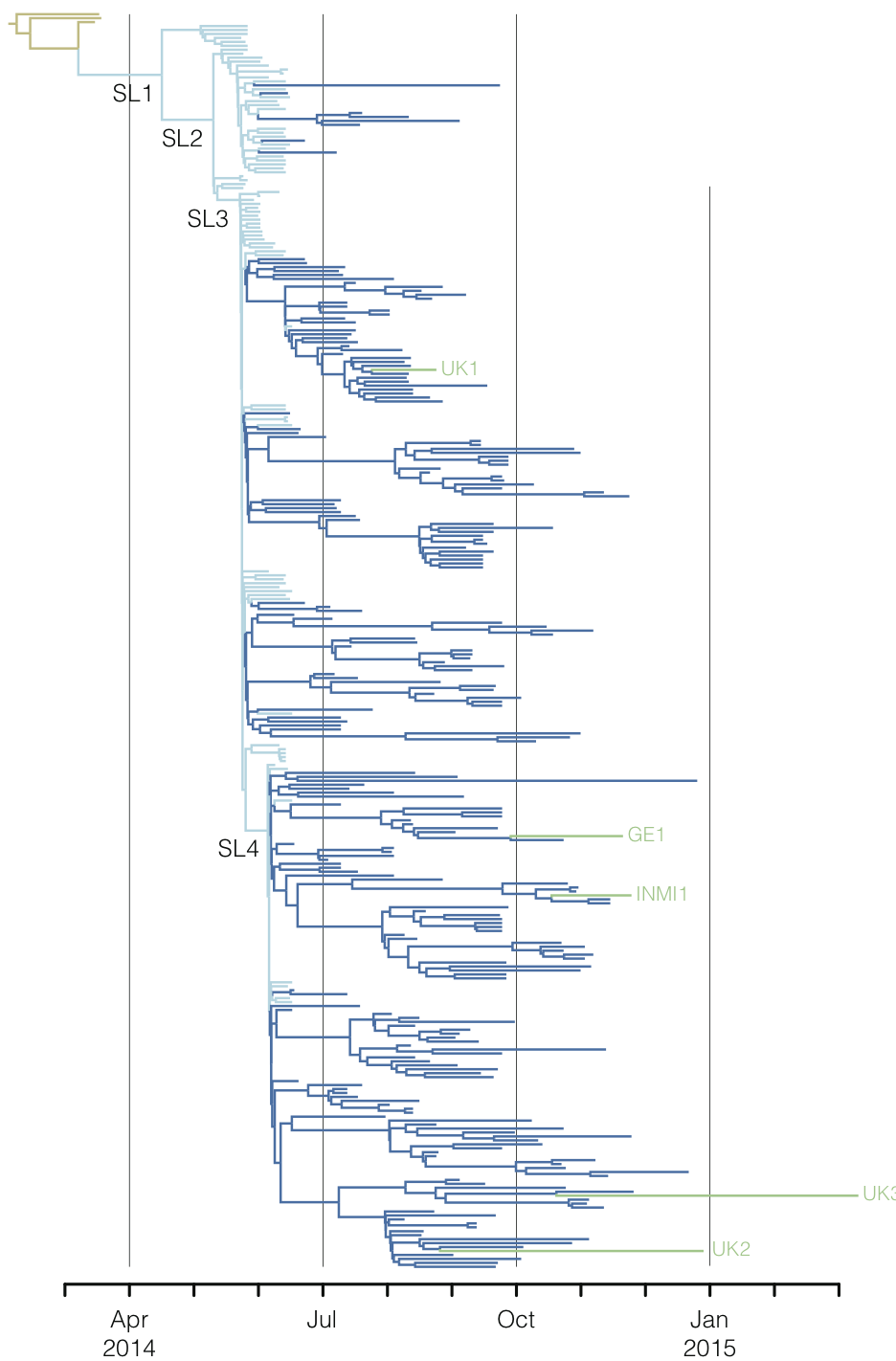
Regional West African Ebola phylogeny
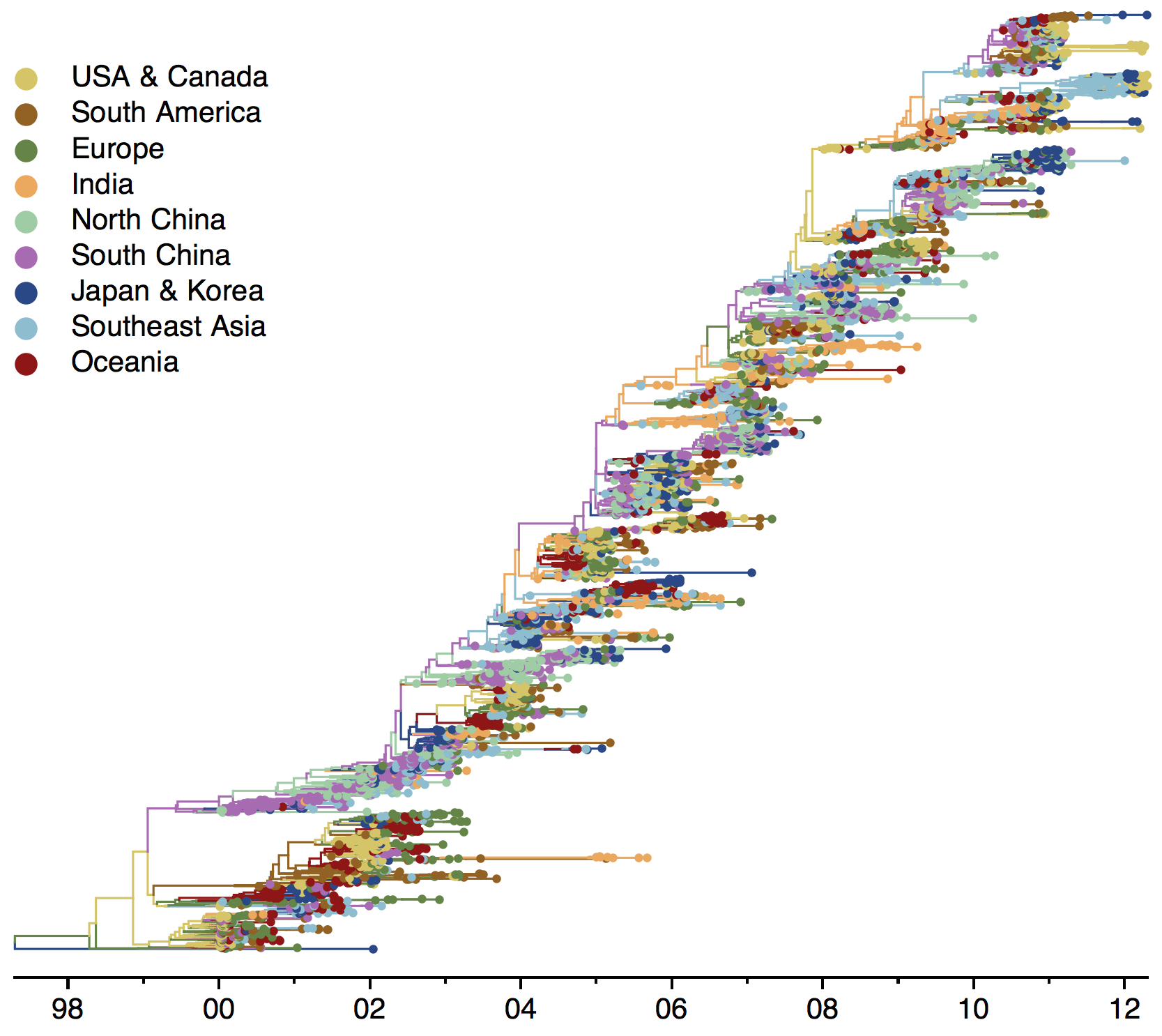
Global influenza phylogeny
Phylogenetic tracking has the capacity to revolutionize epidemiology
Outline
- Influenza evolution and forecasting
- Ebola spread in West Africa
- Zika spread in the Americas
- "Real-time" analyses
Influenza
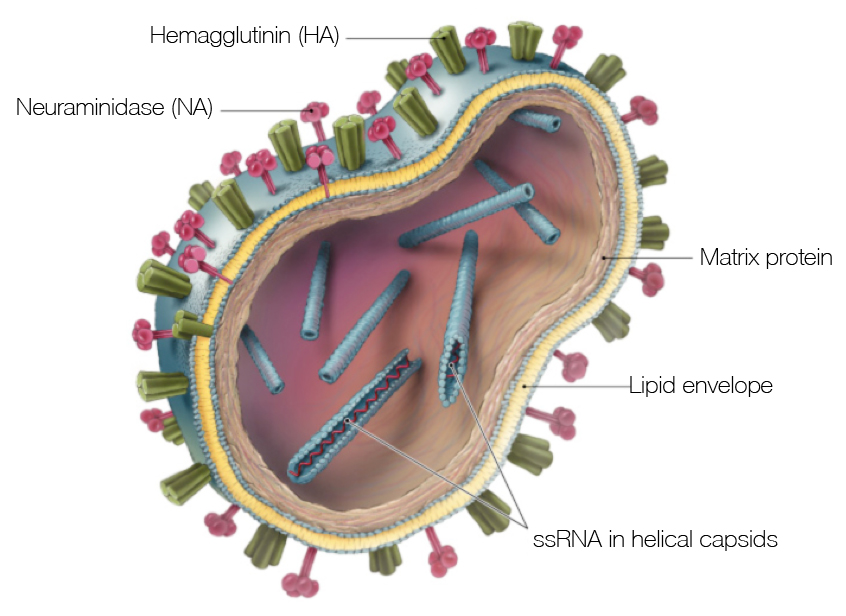
Influenza virion
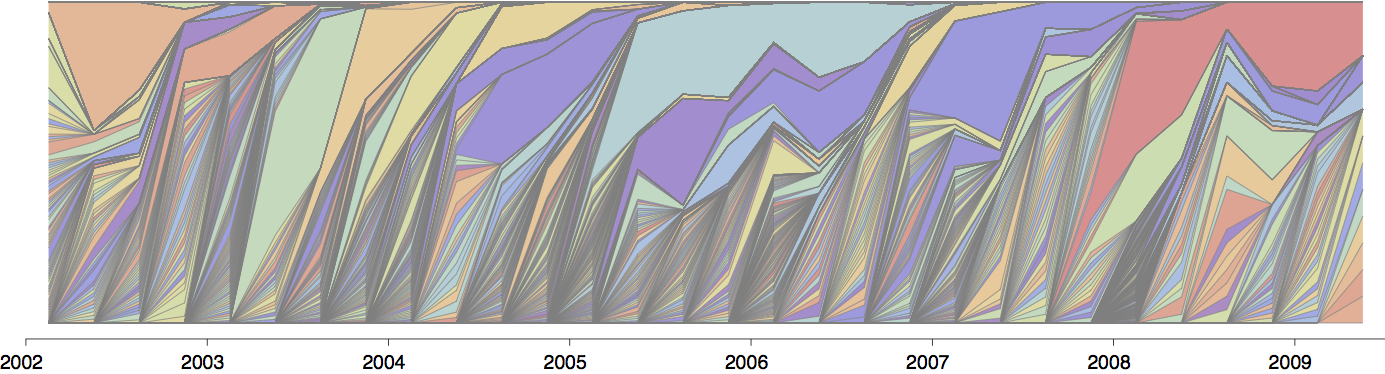
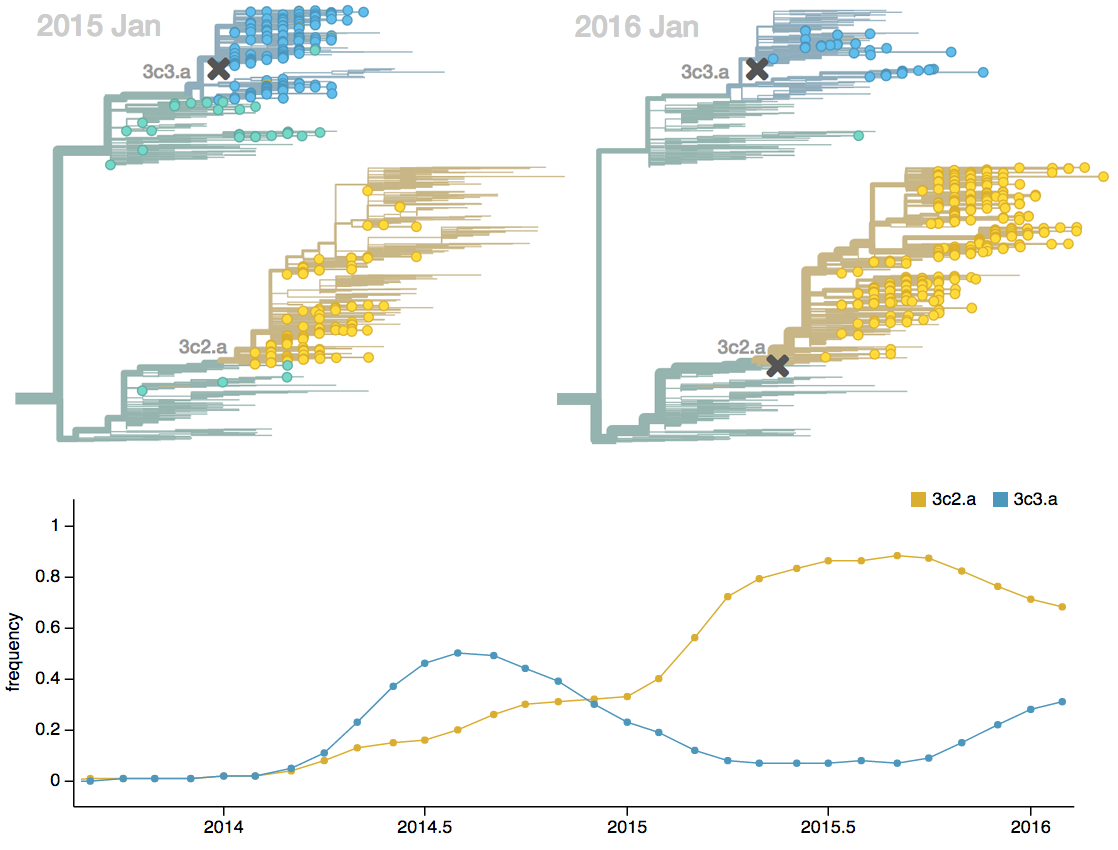
Population turnover (in H3N2) is extremely rapid
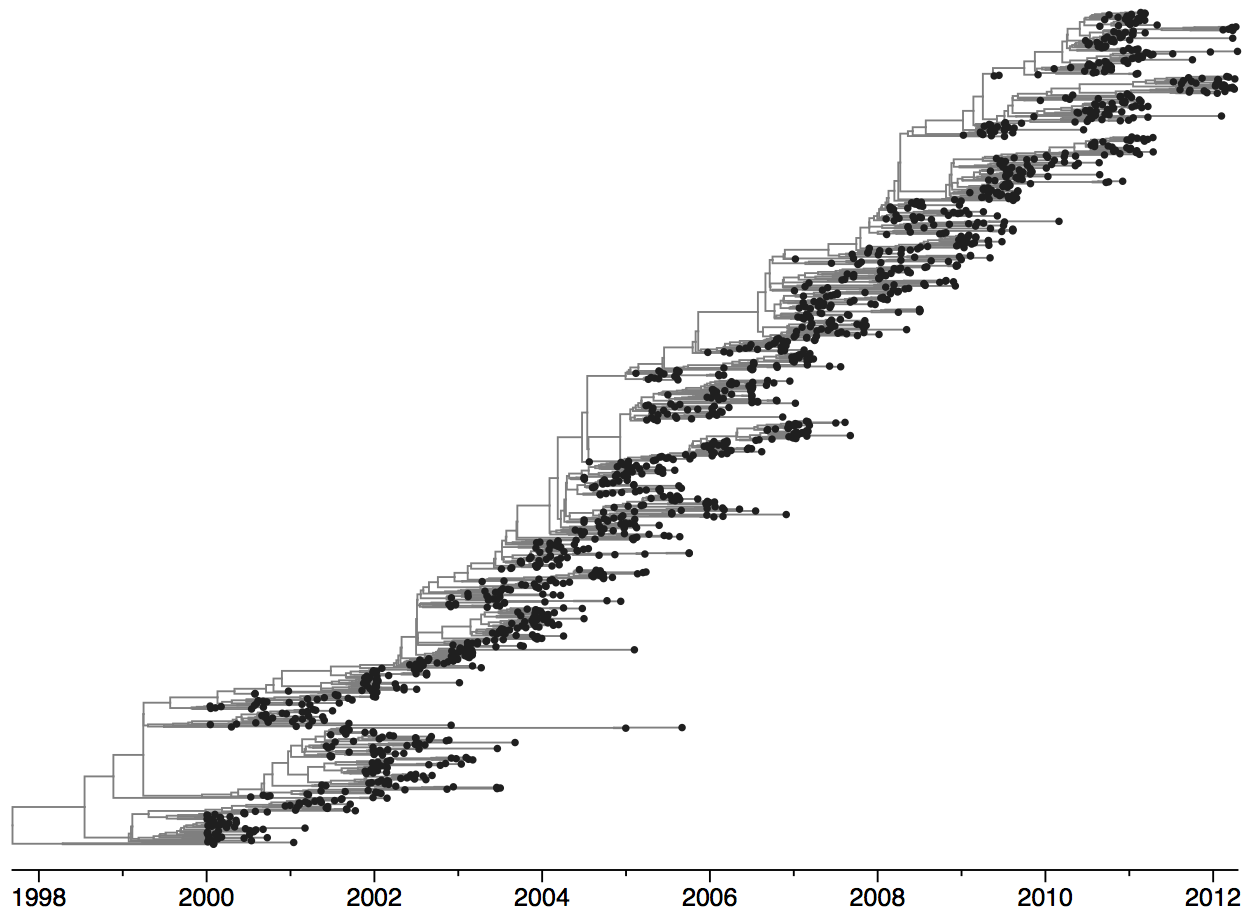
Clades emerge, die out and take over
Clades show rapid turnover

Dynamics driven by antigenic drift
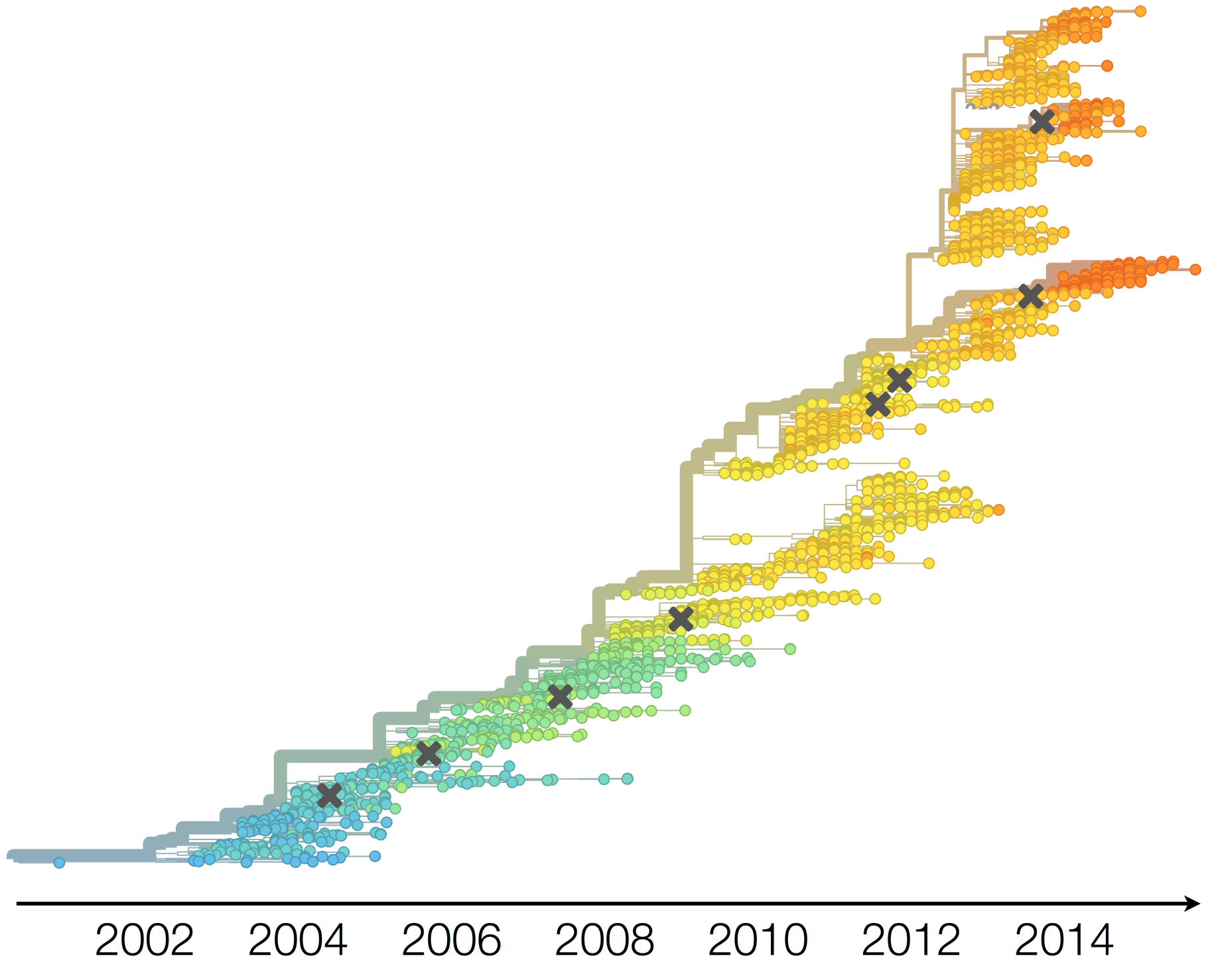
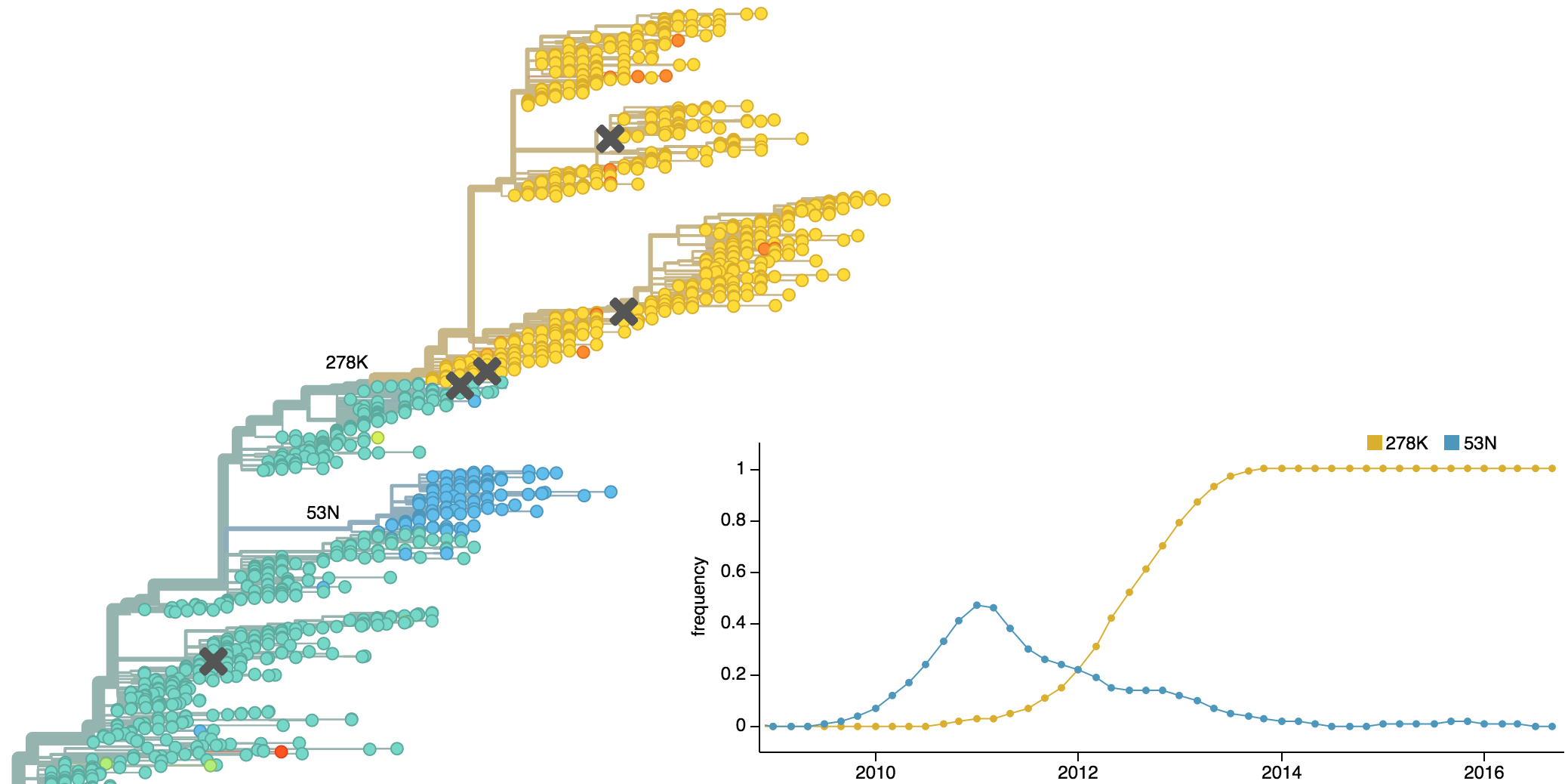
Drift variants emerge and rapidly take over in the virus population
This causes the side effect of evading existing vaccine formulations
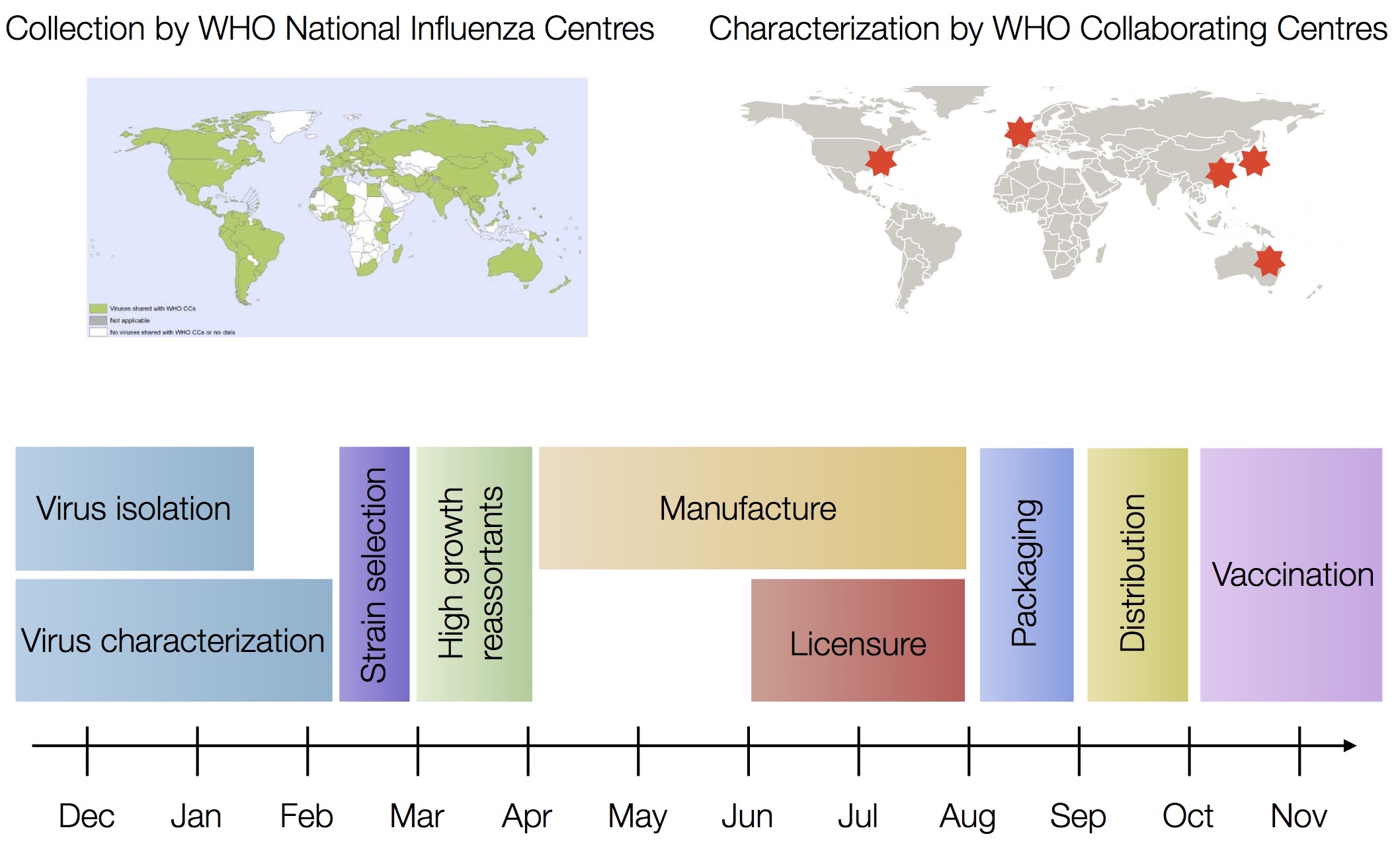
Drift necessitates vaccine updates
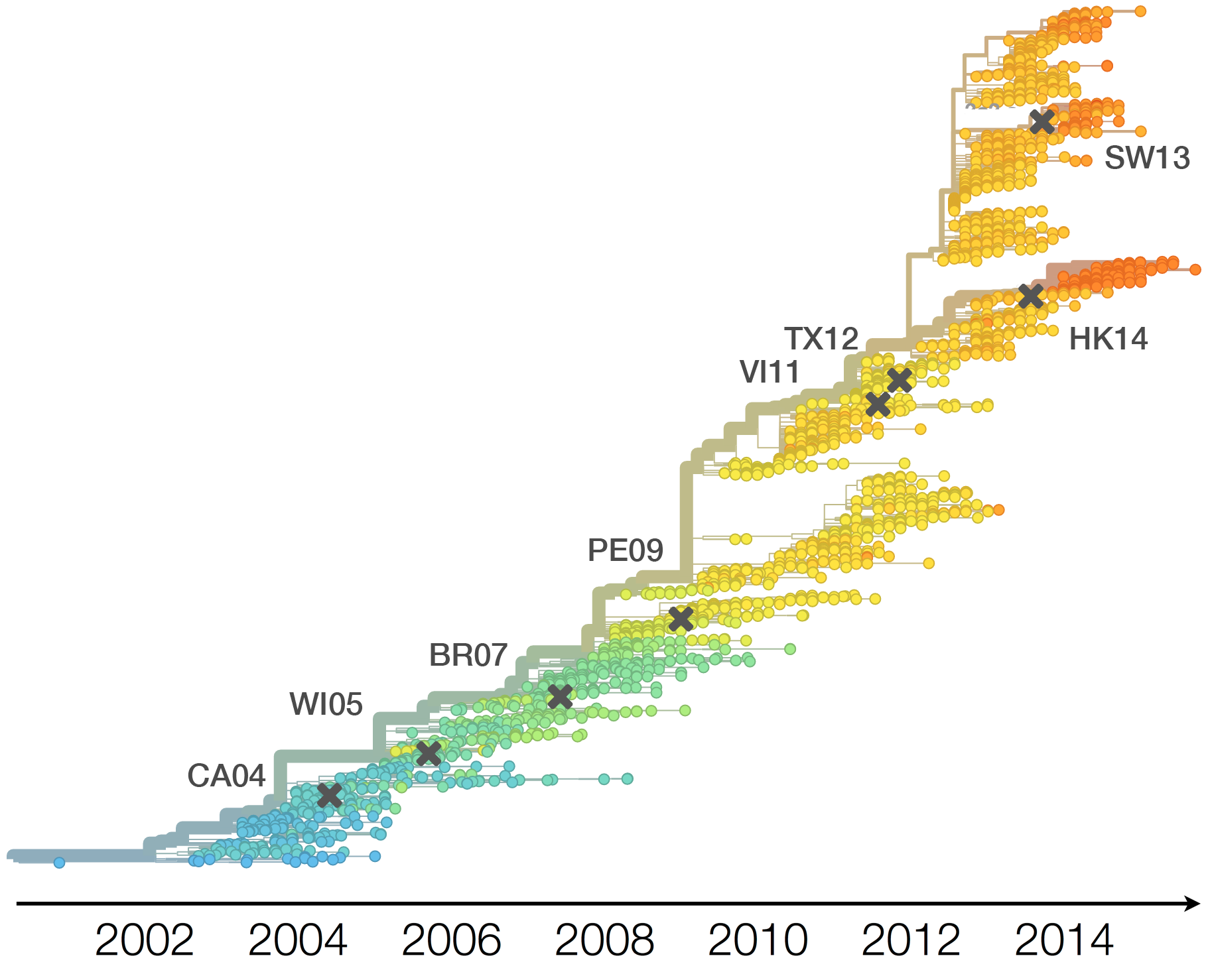
H3N2 vaccine updates occur every ~2 years
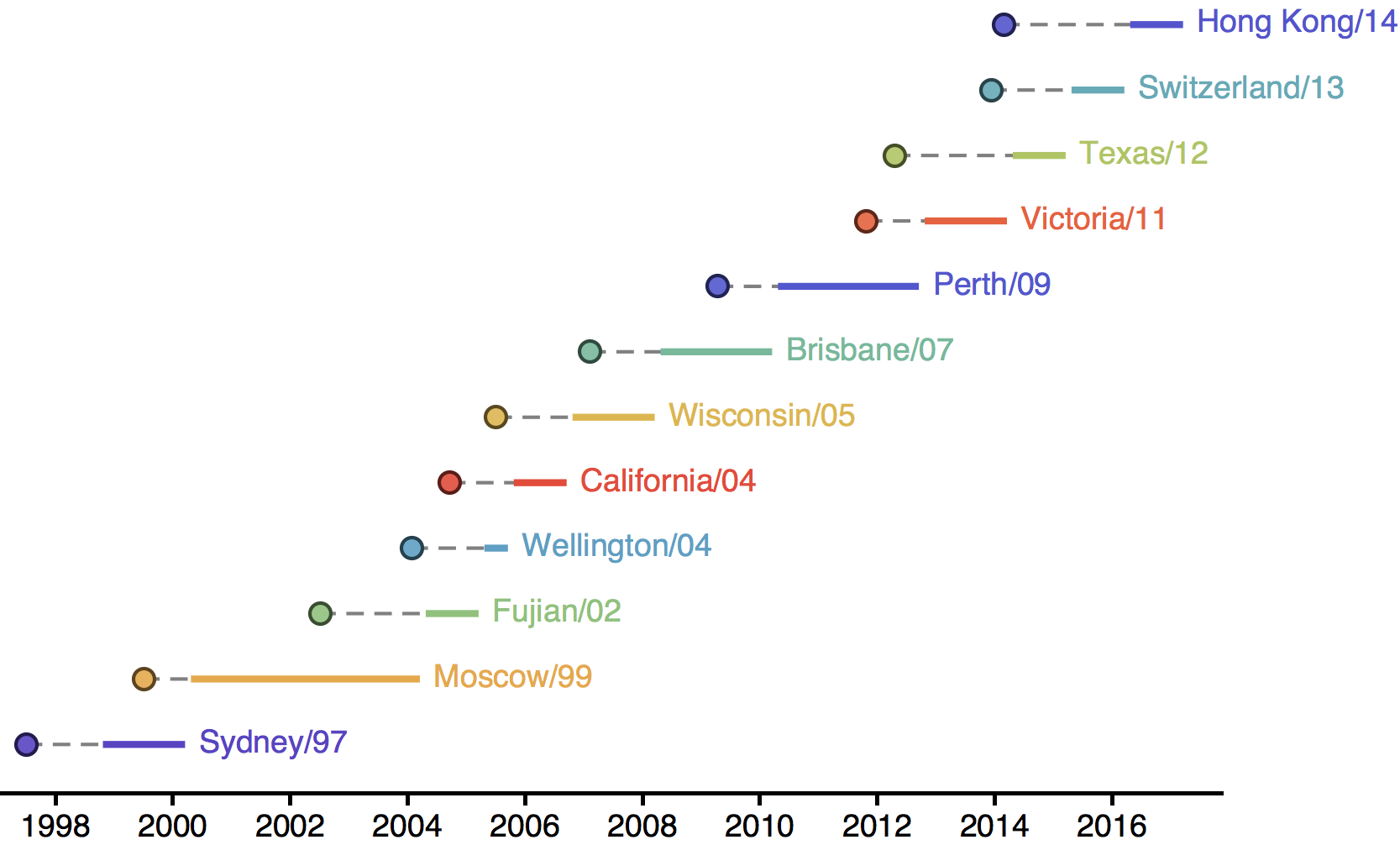
Timely surveillance and rapid analysis essential to vaccine strain selection
nextflu
Project to provide a real-time view of the evolving influenza population
nextflu
Project to provide a real-time view of the evolving influenza population
All in collaboration with Richard Neher

nextflu pipeline
- Download all recent HA sequences from GISAID
- Filter to remove outliers
- Subsample across time and space
- Align sequences
- Build tree
- Estimate clade frequencies
- Infer antigenic phenotypes
- Export for visualization
nextflu.org
Forecasting
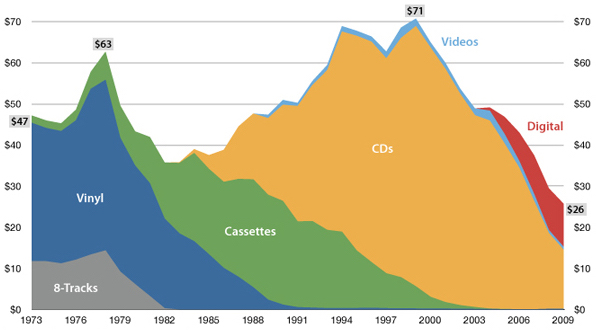
The future is here, it's just not evenly distributed yet
— William Gibson
USA music industry, 2011 dollars per capita
Influenza population turnover
Vaccine strain selection timeline
Seek to explain change in clade frequencies over 1 year
Fitness models can project clade frequencies
Clade frequencies $X$ derive from the fitnesses $f$ and frequencies $x$ of constituent viruses, such that
$$\hat{X}_v(t+\Delta t) = \sum_{i:v} x_i(t) \, \mathrm{exp}(f_i \, \Delta t)$$
This captures clonal interference between competing lineages
Predictive fitness models
A simple predictive model estimates the fitness $f$ of virus $i$ as
$$\hat{f}_i = \beta^\mathrm{ep} \, f_i^\mathrm{ep} + \beta^\mathrm{ne} \, f_i^\mathrm{ne}$$
where $f_i^\mathrm{ep}$ measures cross-immunity via substitutions at epitope sites and $f_i^\mathrm{ep}$ measures mutational load via substitutions at non-epitope sites
We implement a similar model based on two predictors
- Clade frequency change
- Antigenic advancement
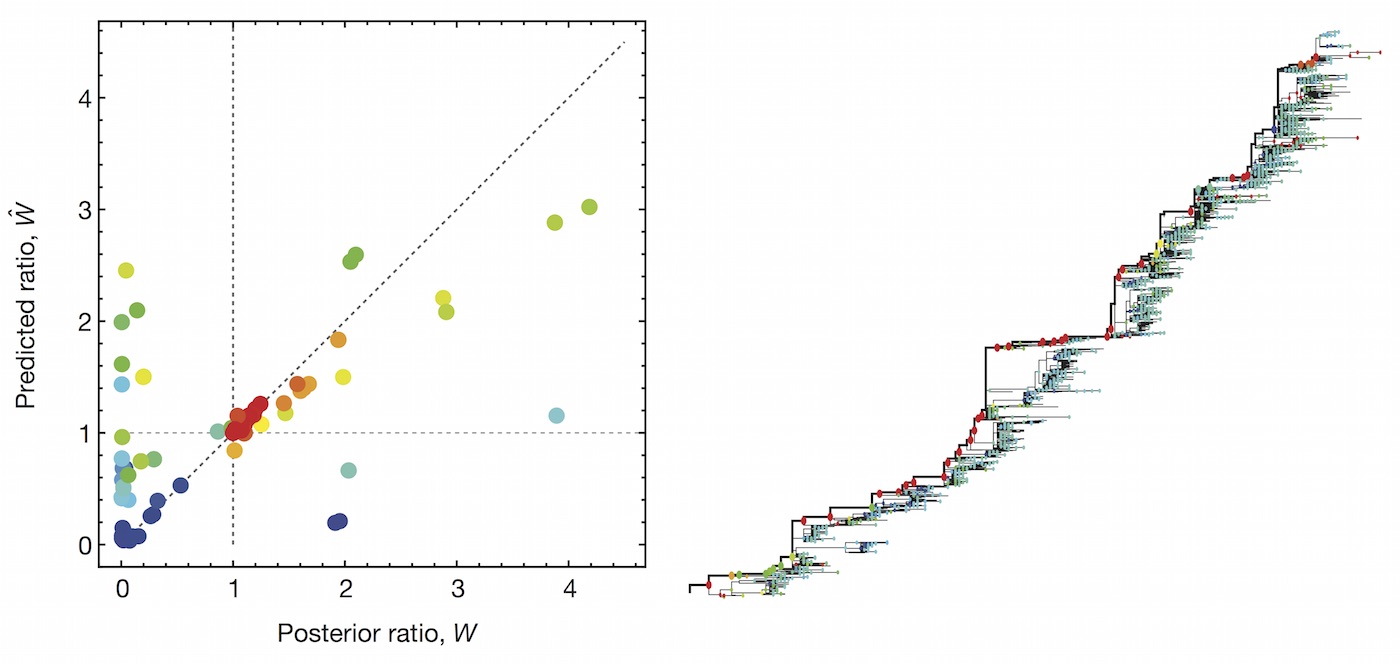
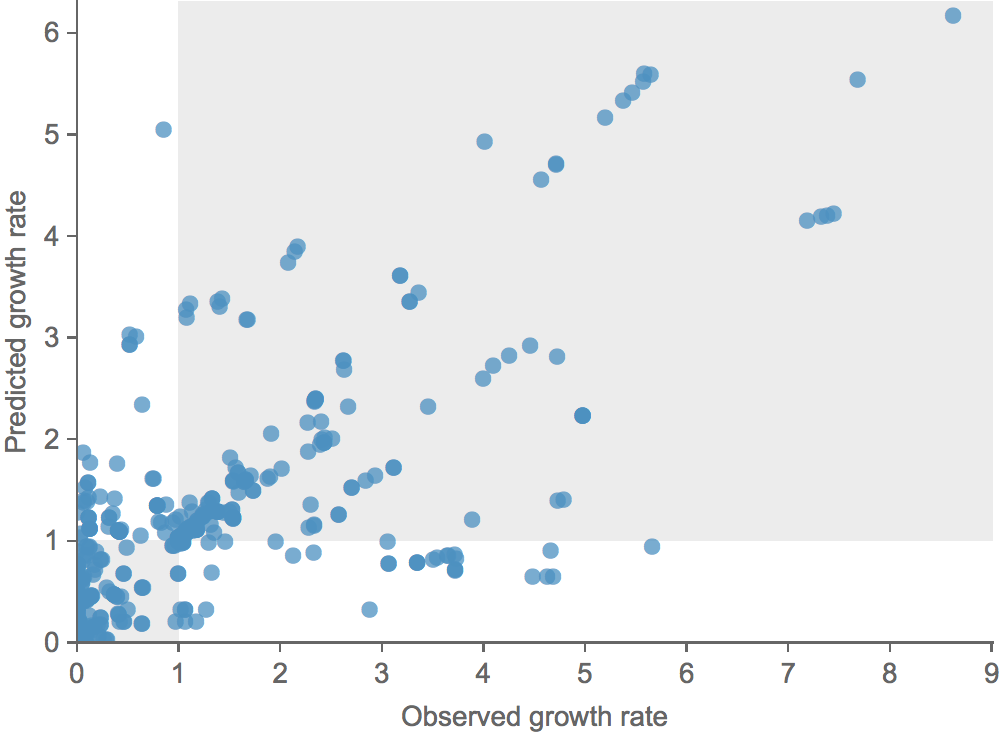
Clade growth rate is well correlated (ρ = 0.66)
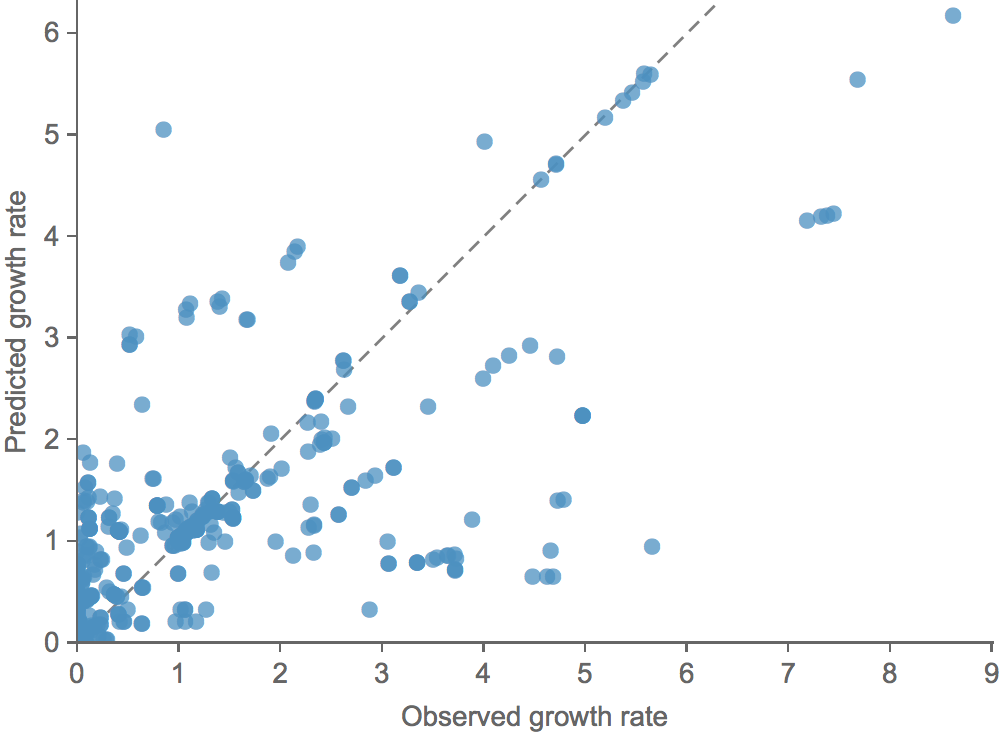
Growth vs decline correct in 84% of cases
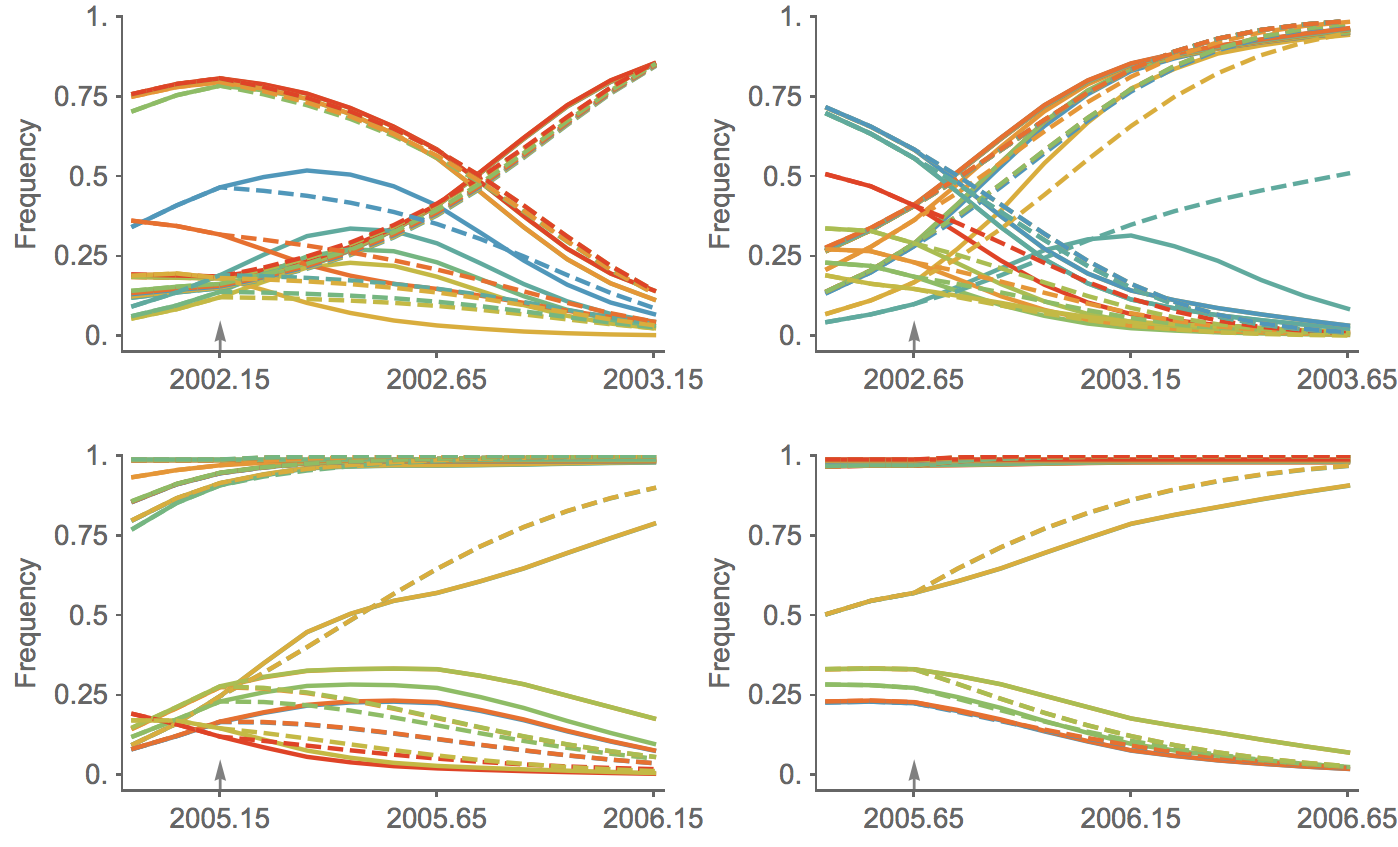
Trajectories show more detailed congruence
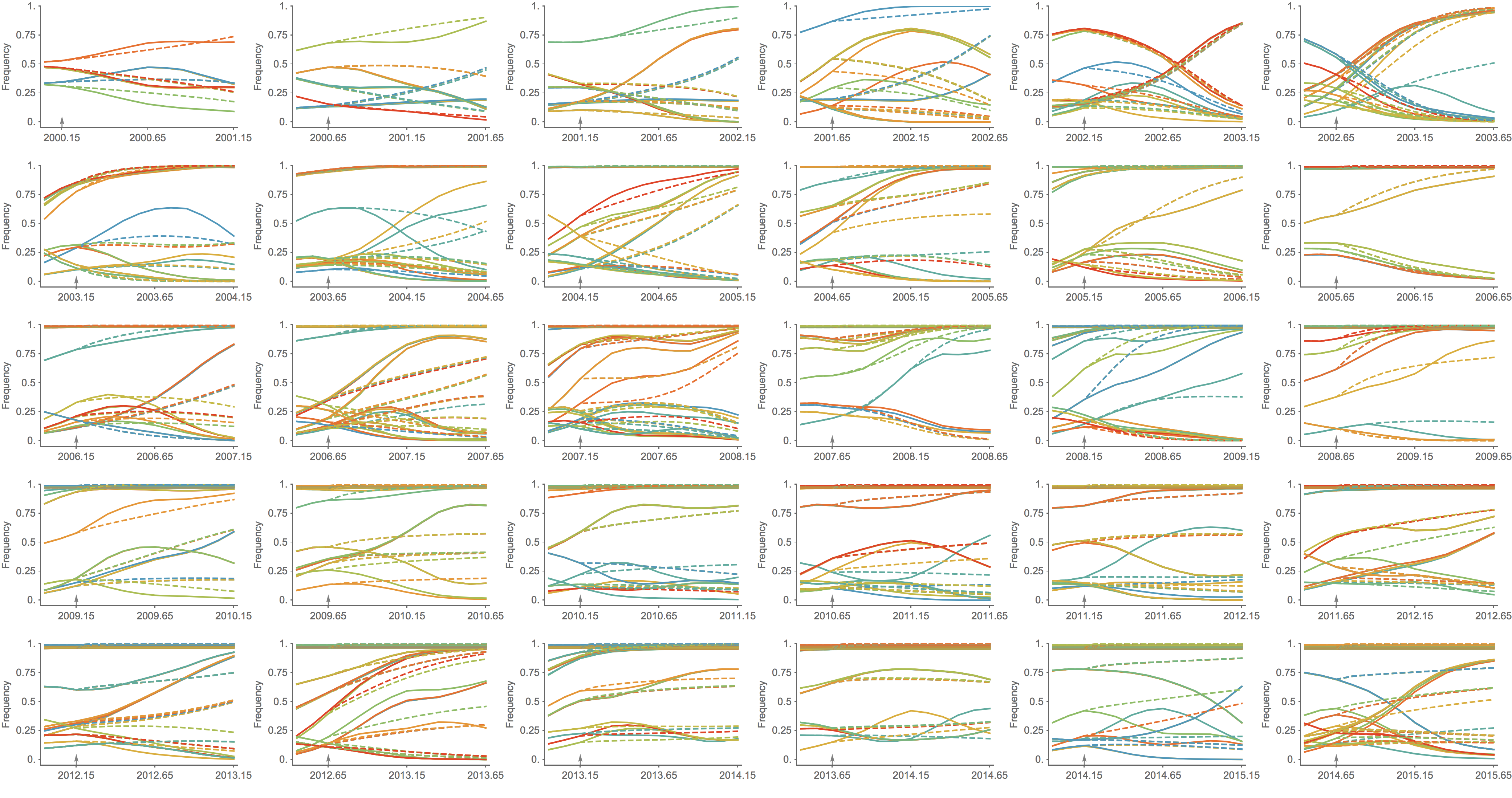
Trajectories show more detailed congruence
Real-time analyses are actionable and may inform influenza vaccine strain selection
Ebola
Tracking geographic spread of the Ebola epidemic
with Gytis Dudas, Andrew Rambaut, Luiz Carvalho, Marc Suchard, Philippe Lemey,
and many others
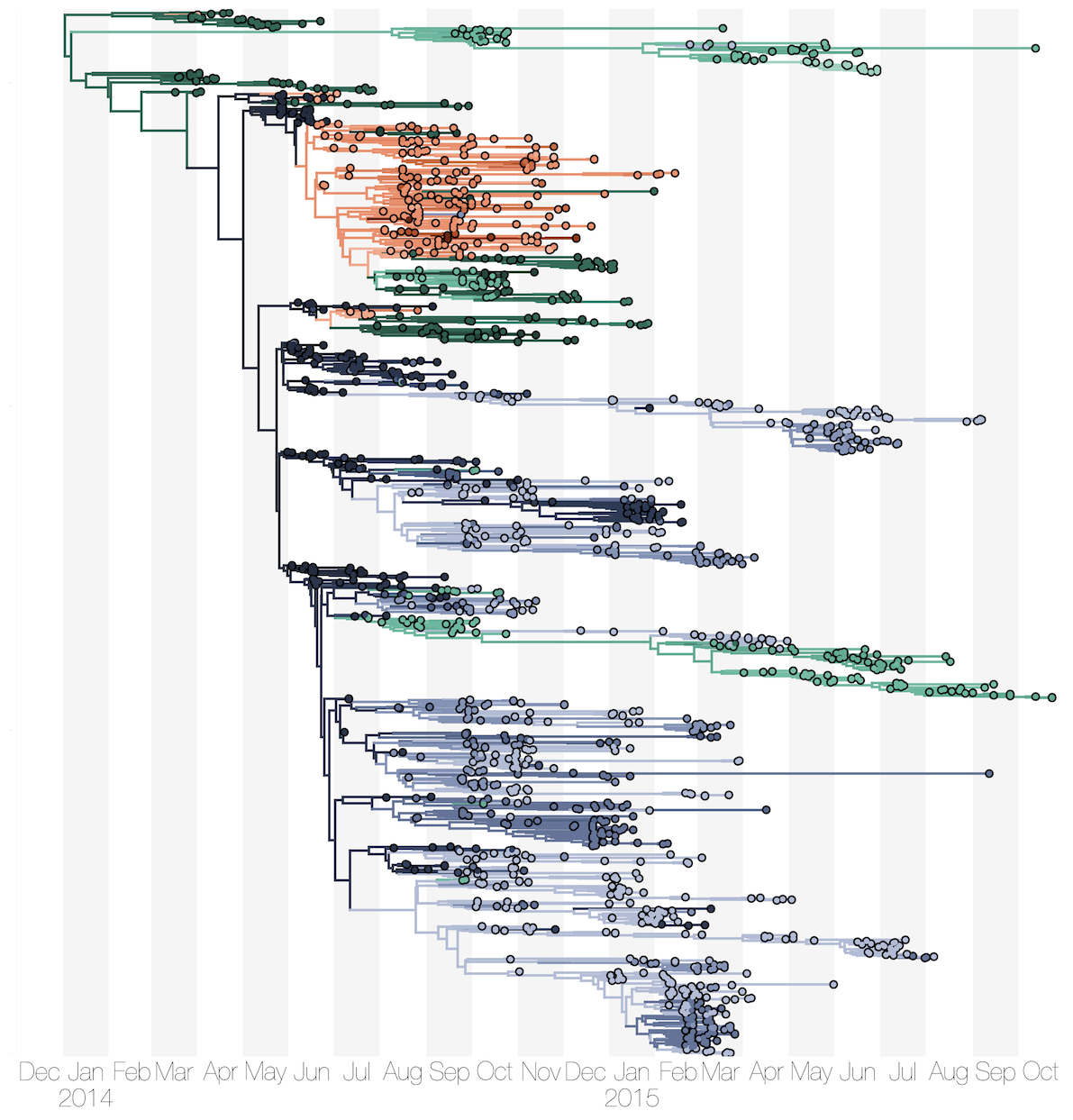
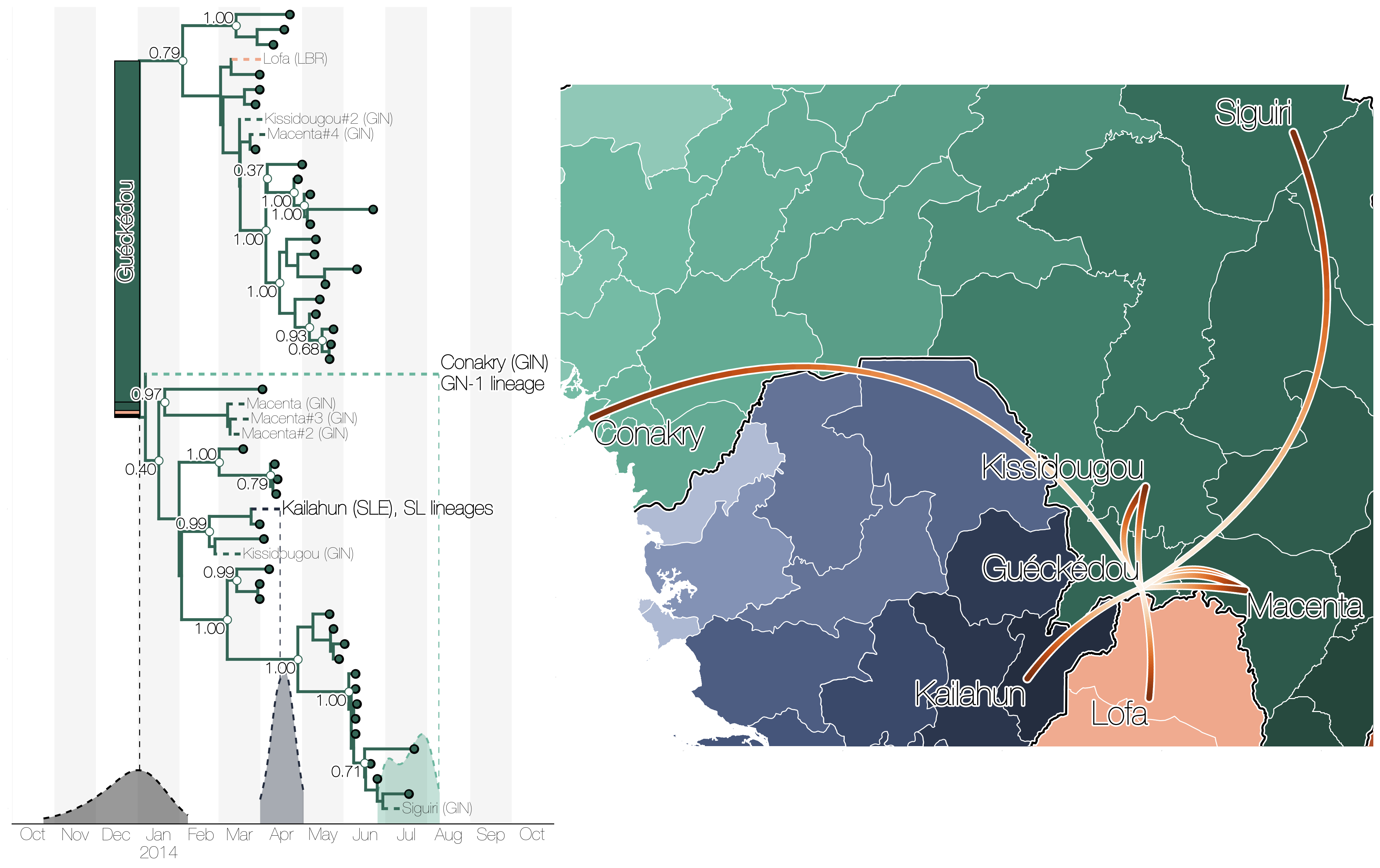
Sequencing of 1610 Ebola virus genomes collected during the 2013-2016 West African epidemic
Phylogenetic reconstruction of evolution and spread
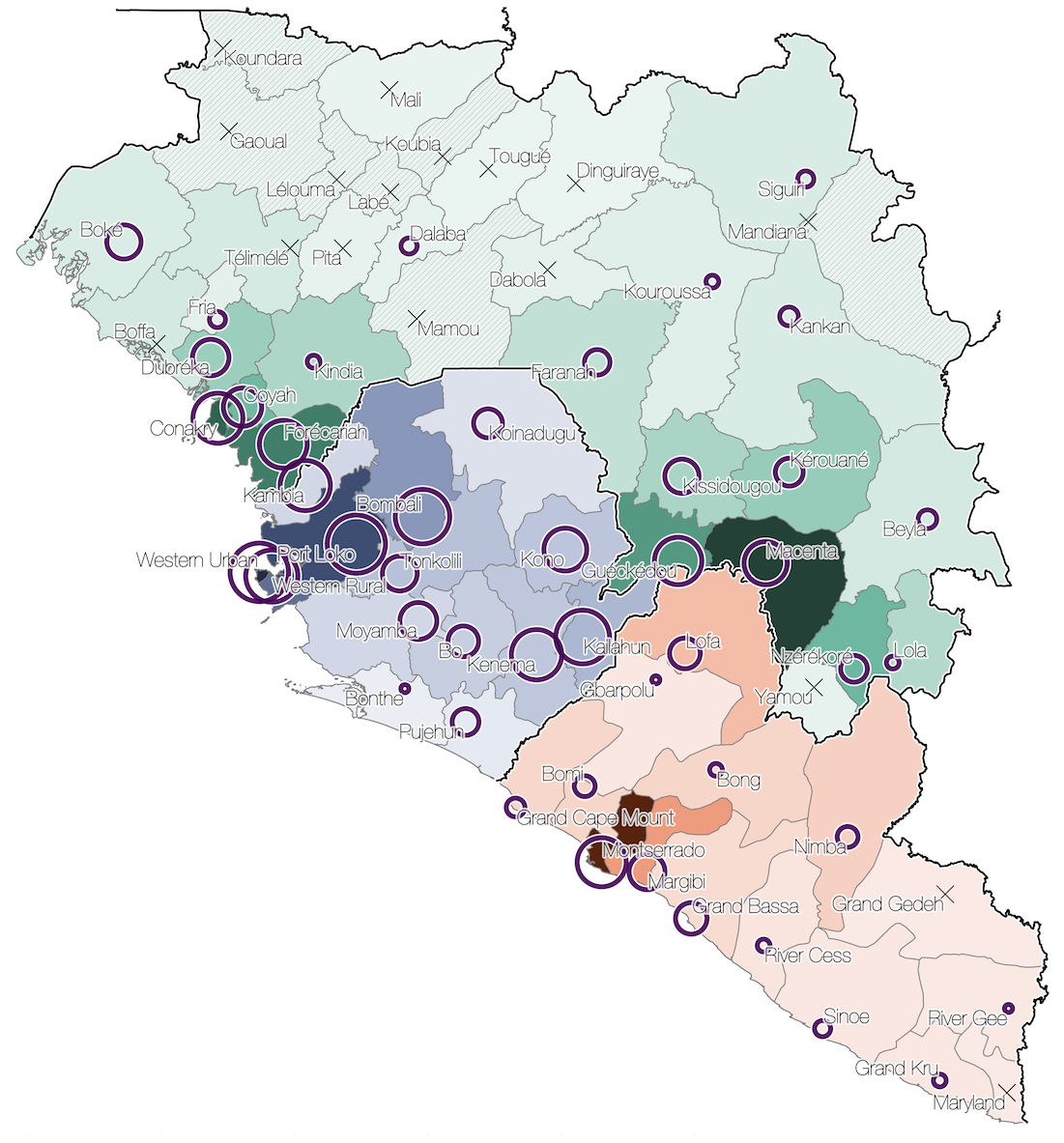
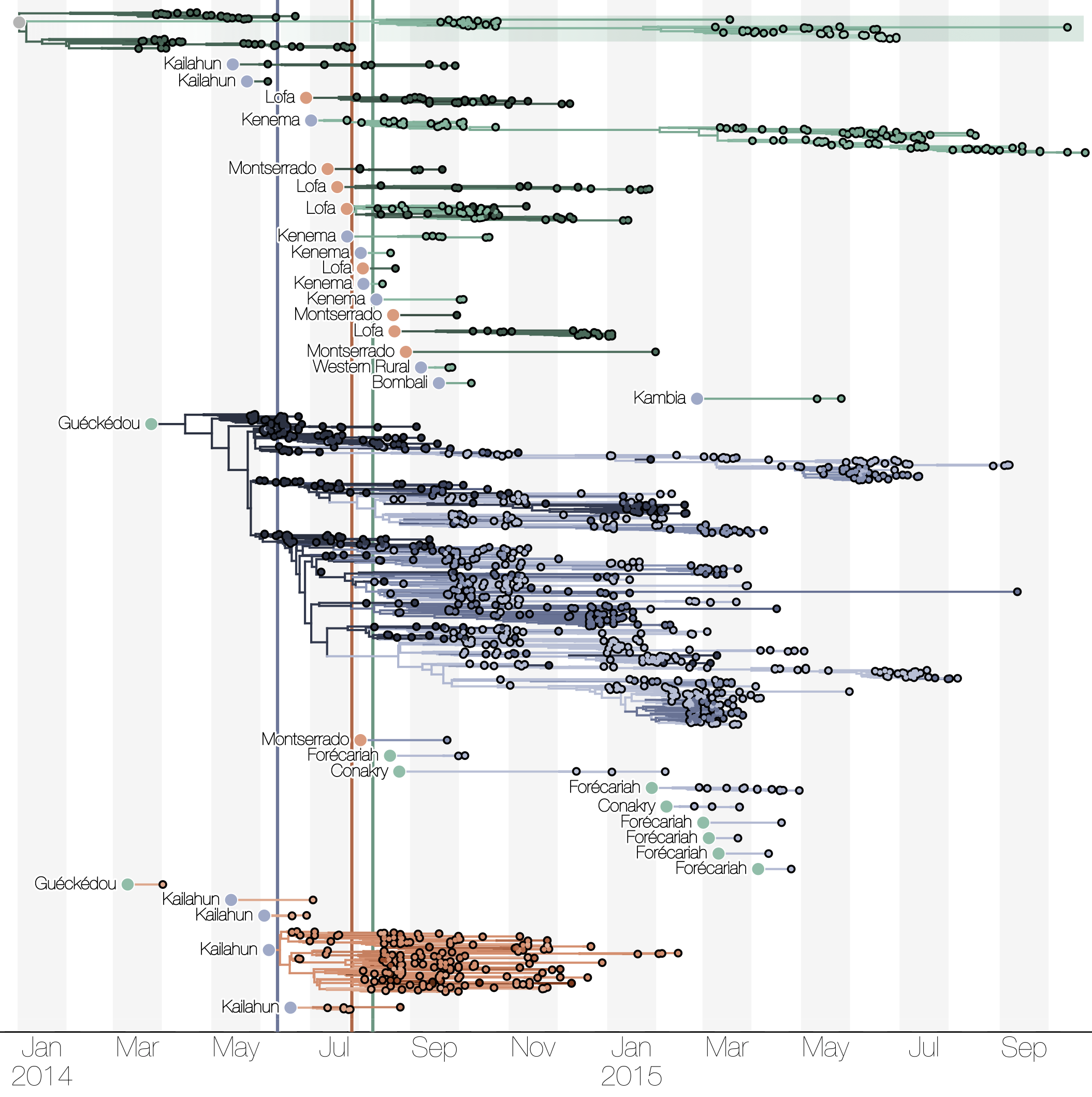
Initial emergence from Guéckédou
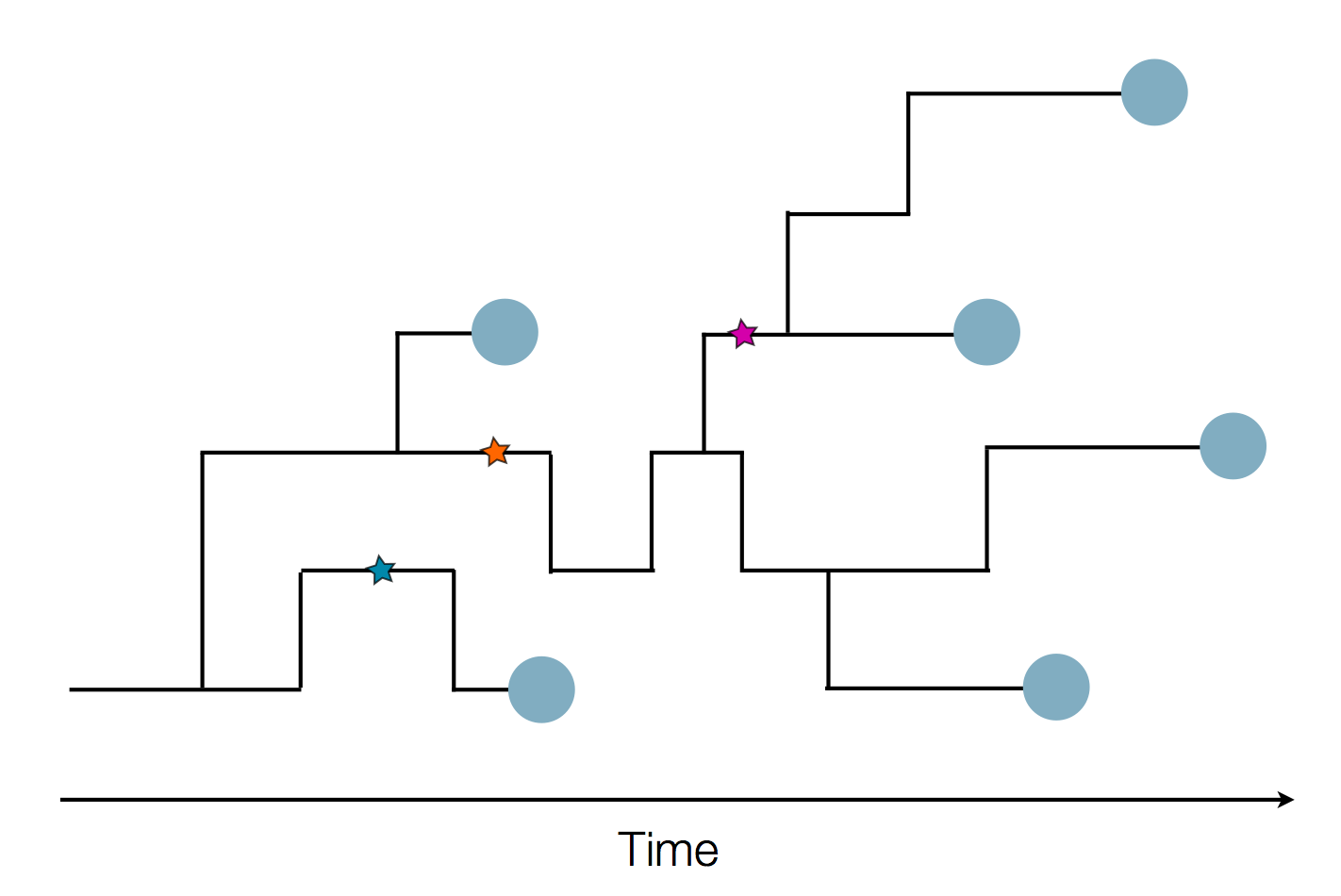
Tracking migration events
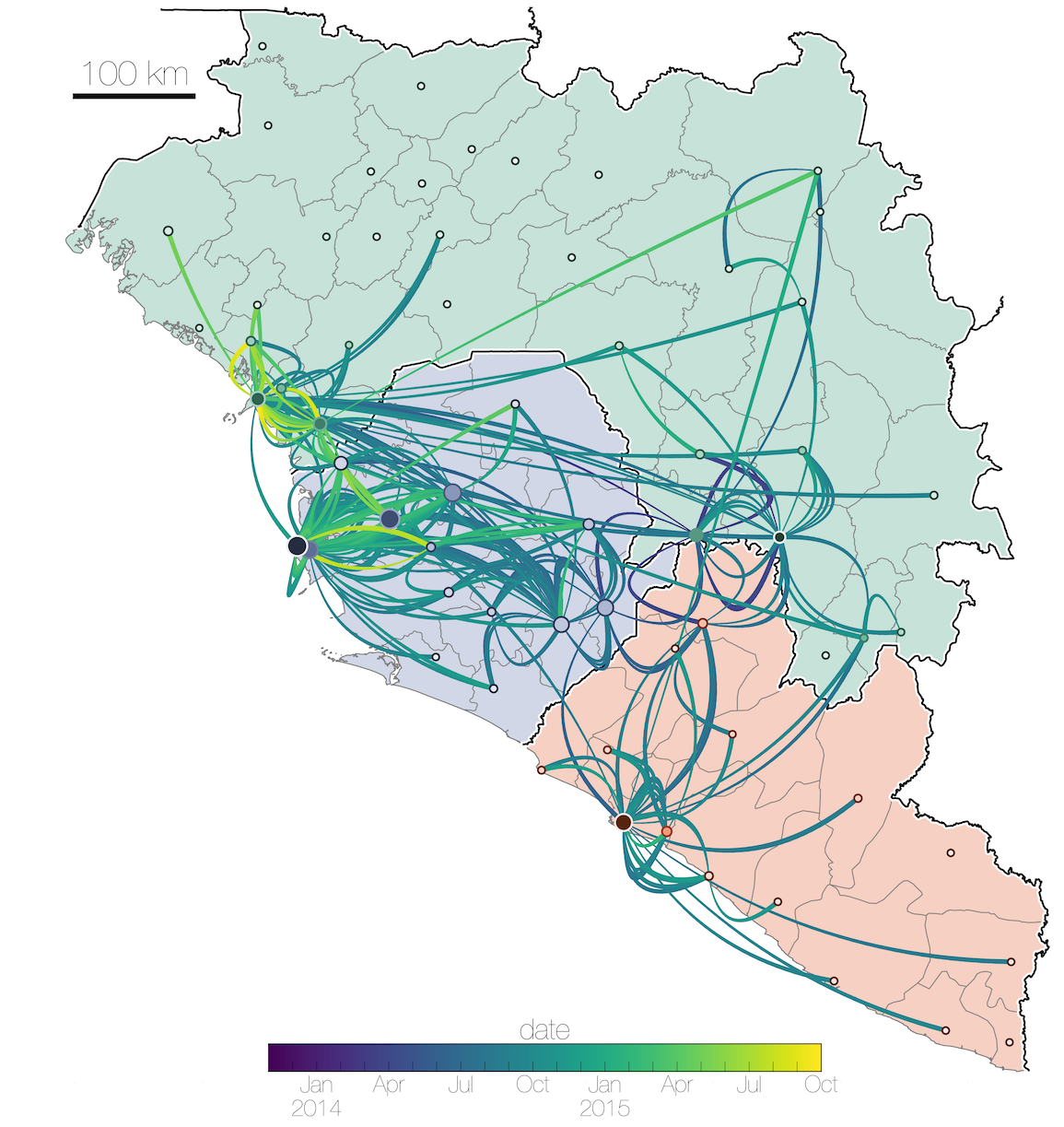
Factors influencing migration rates
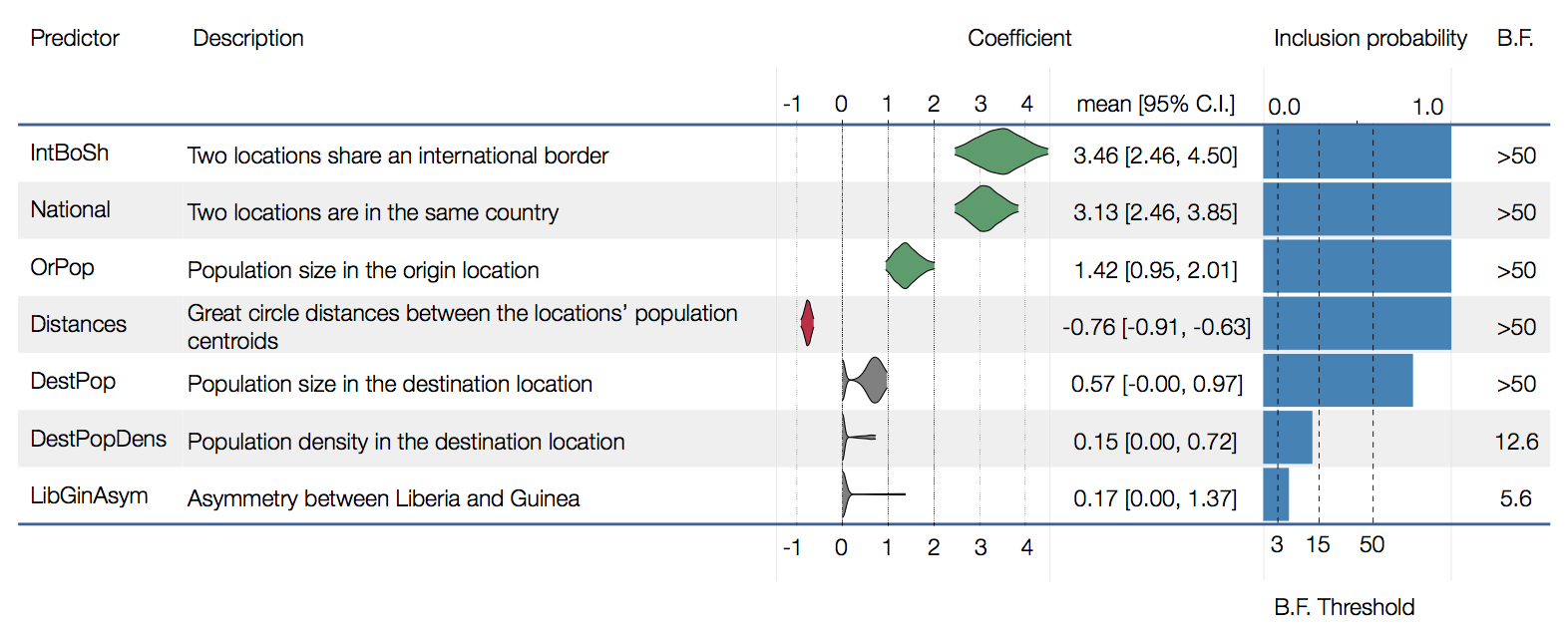
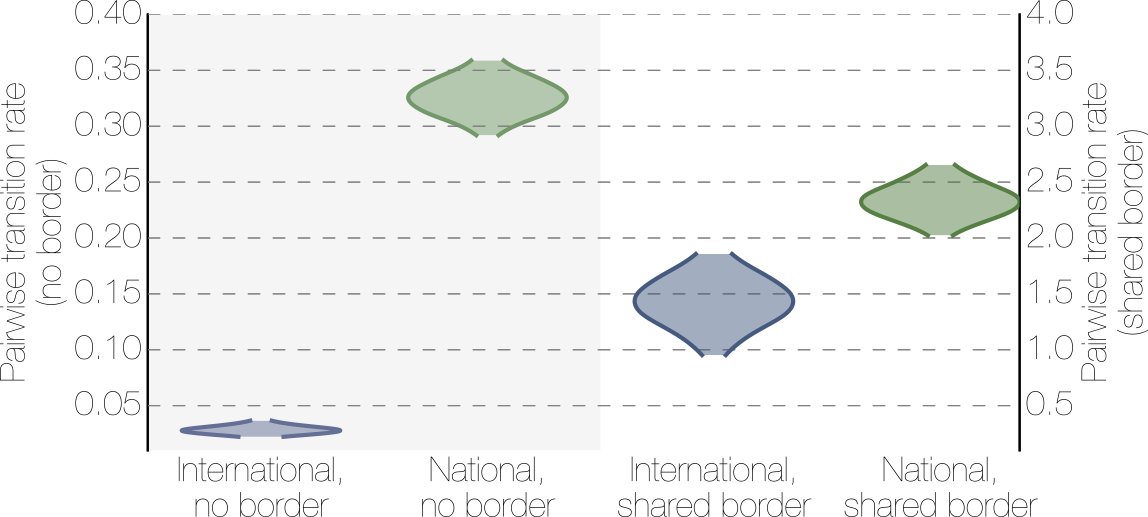
Effect of borders on migration rates
Spatial structure at the country level
Substantial mixing at the regional level
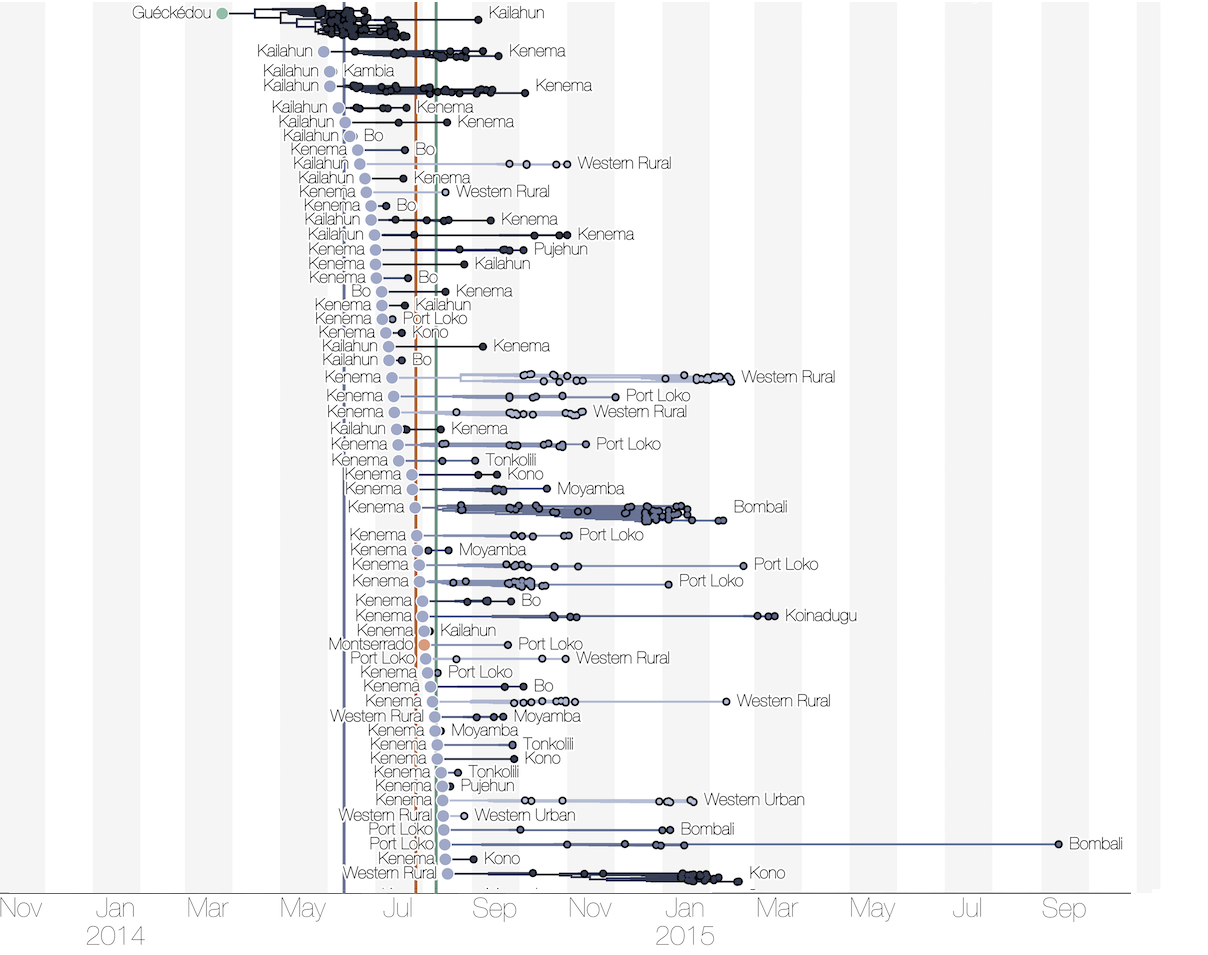
Regional outbreaks due to multiple introductions
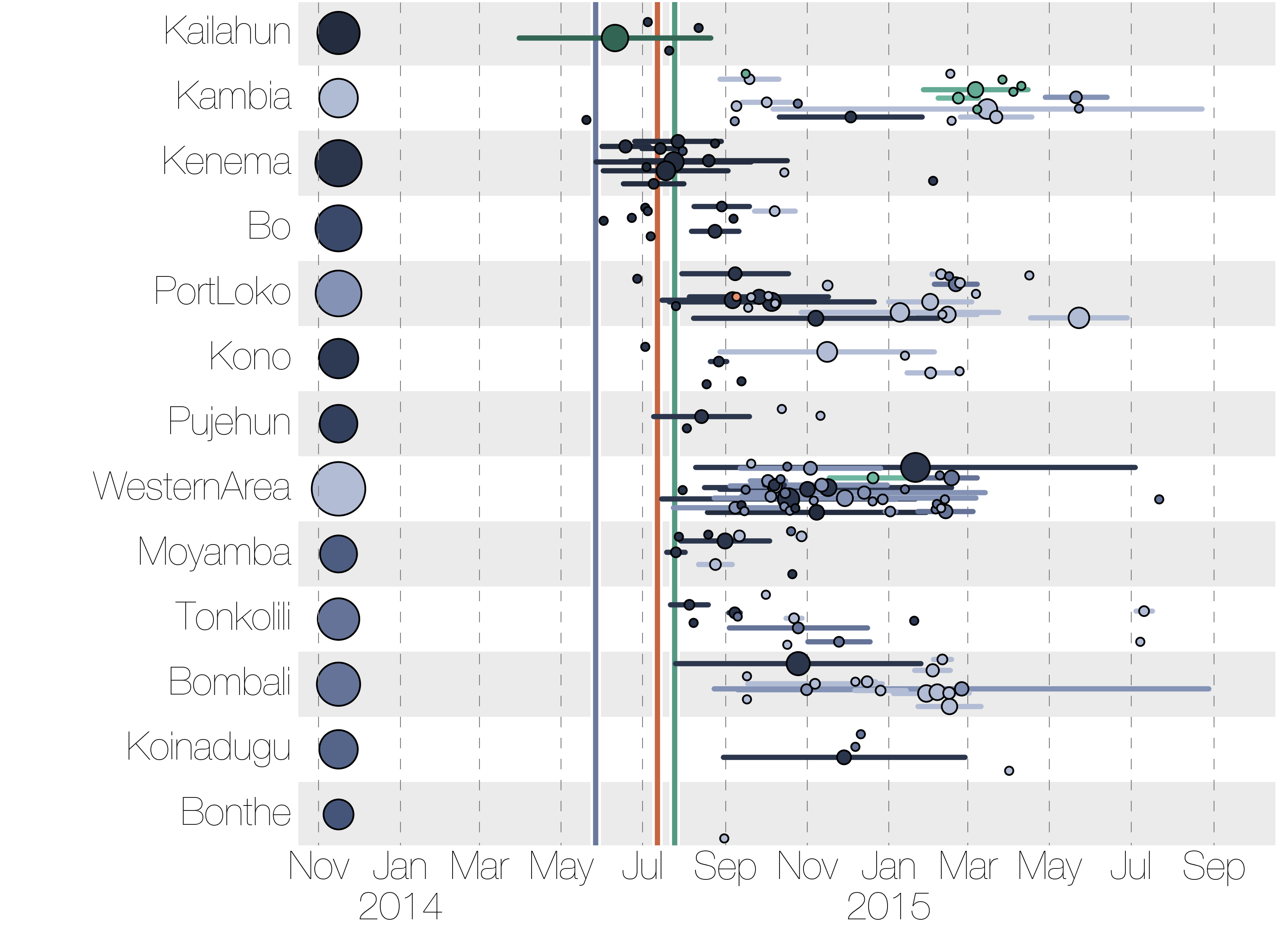
Each introduction results in a minor outbreak
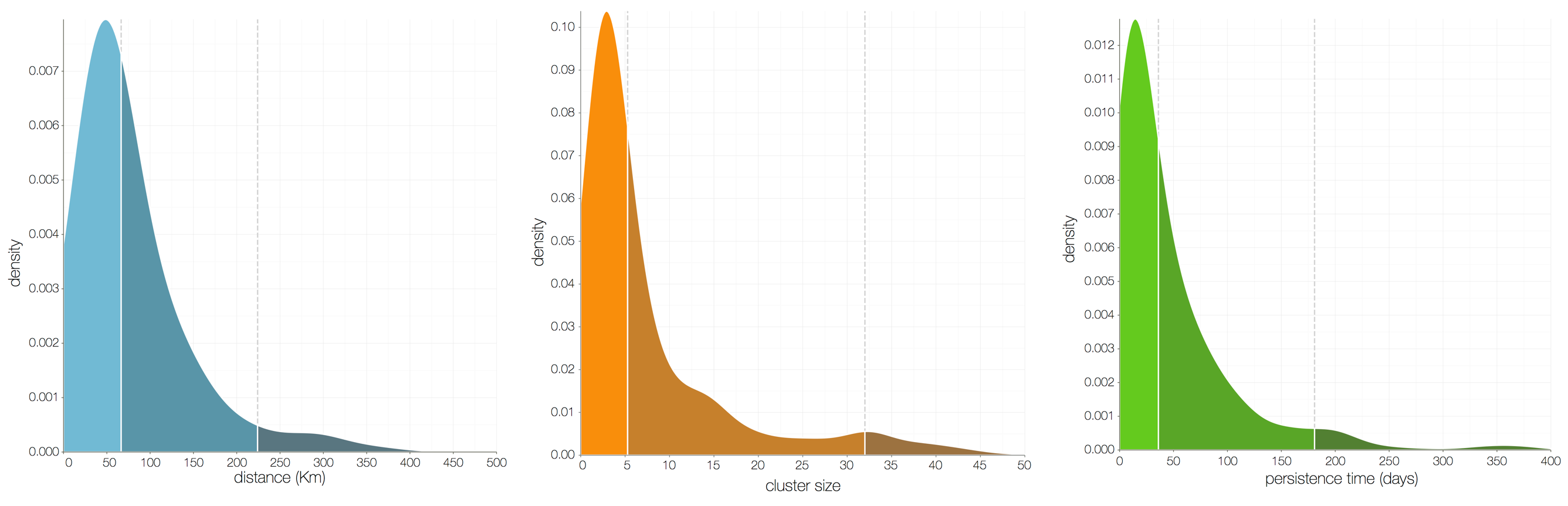
Ebola spread in West Africa followed a gravity model with moderate slowing by international borders, in which spread is driven by short-lived migratory clusters
Zika
Zika's arrival and spread in the Americas
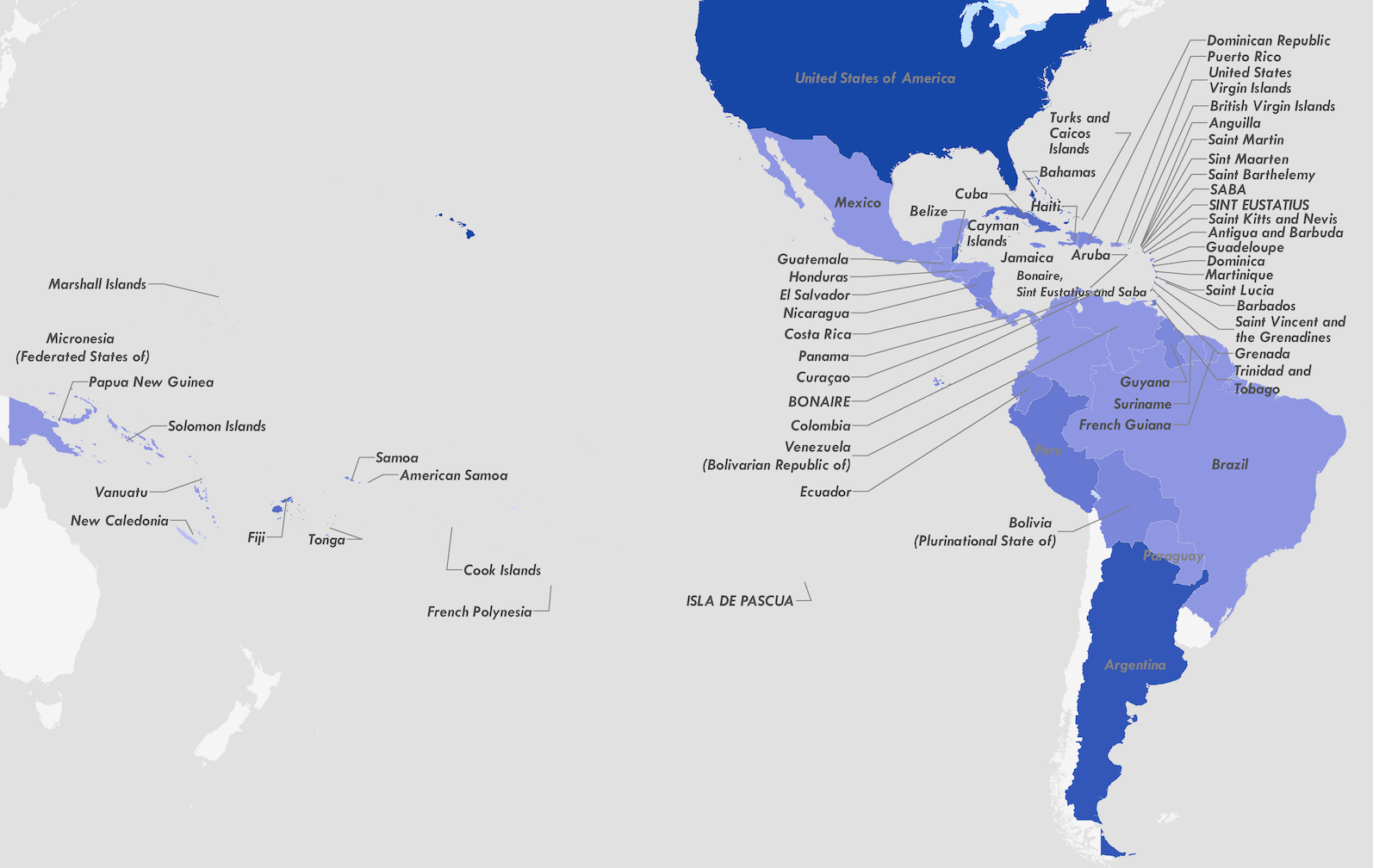
Tracking origins of the Zika epidemic
with Nuno Faria, Nick Loman, Oli Pybus, Luiz Alcantara, Ester Sabino, Josh Quick, Allison Black, Ingra Morales, Julien Thézé, Marcio Nunes, Jacqueline de Jesus, Marta Giovanetti, Moritz Kraemer, Sarah Hill and many others
Road trip through northeast Brazil to collect samples and sequence
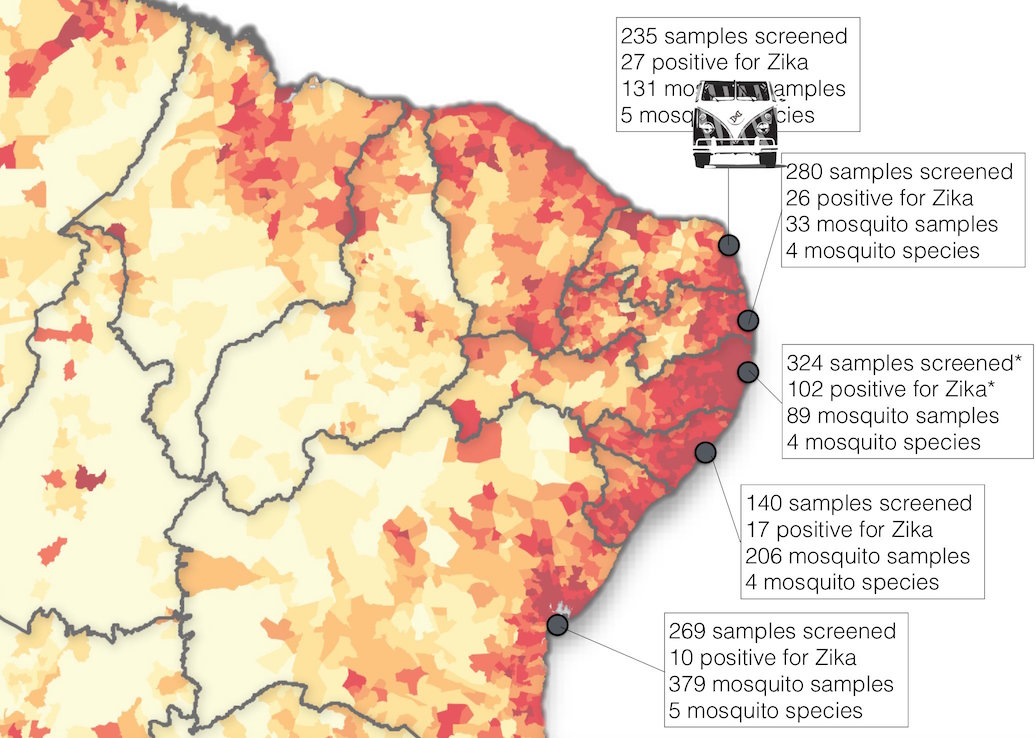
Case reports and diagnostics suggest initiation in northeast Brazil
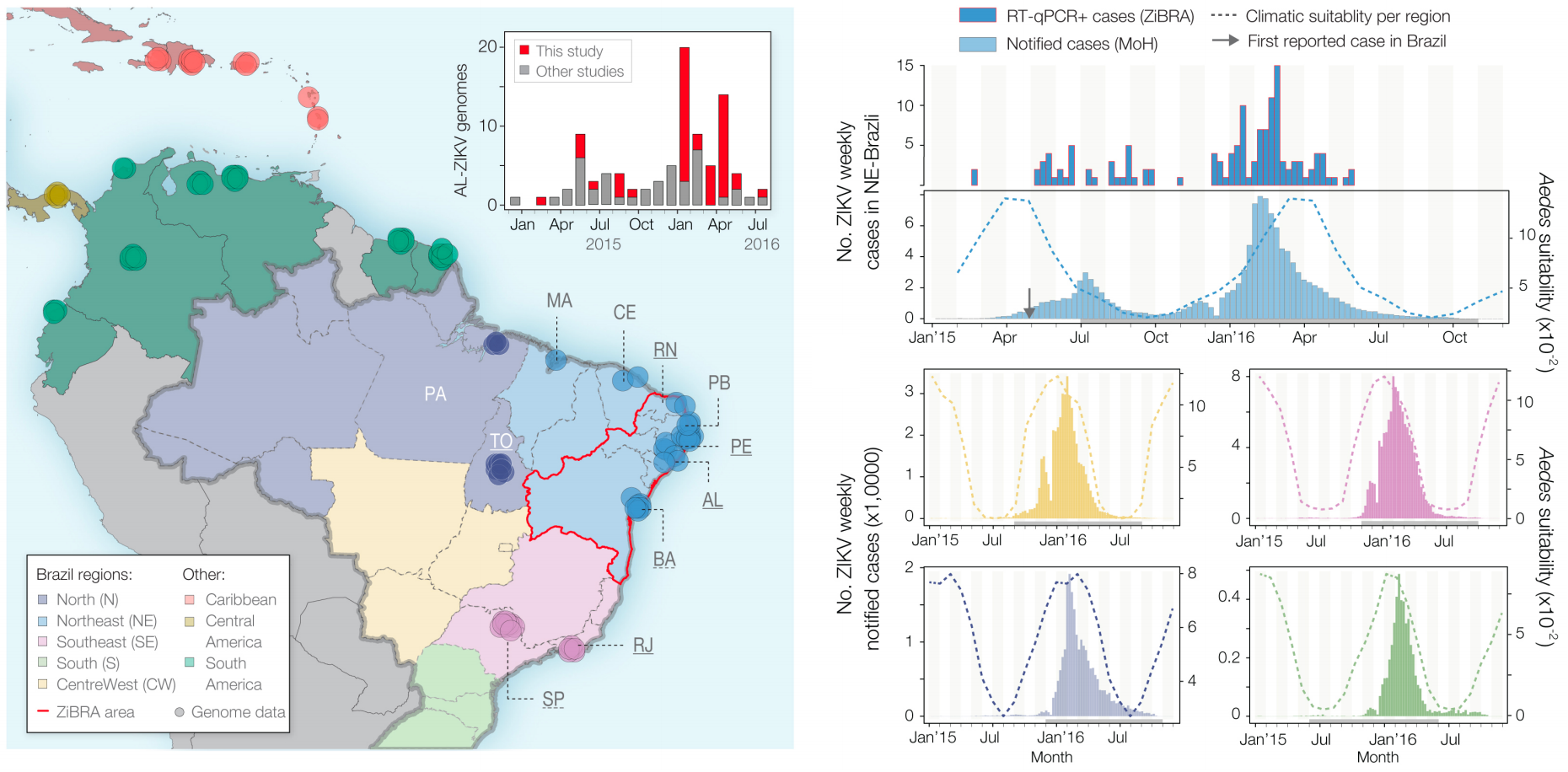
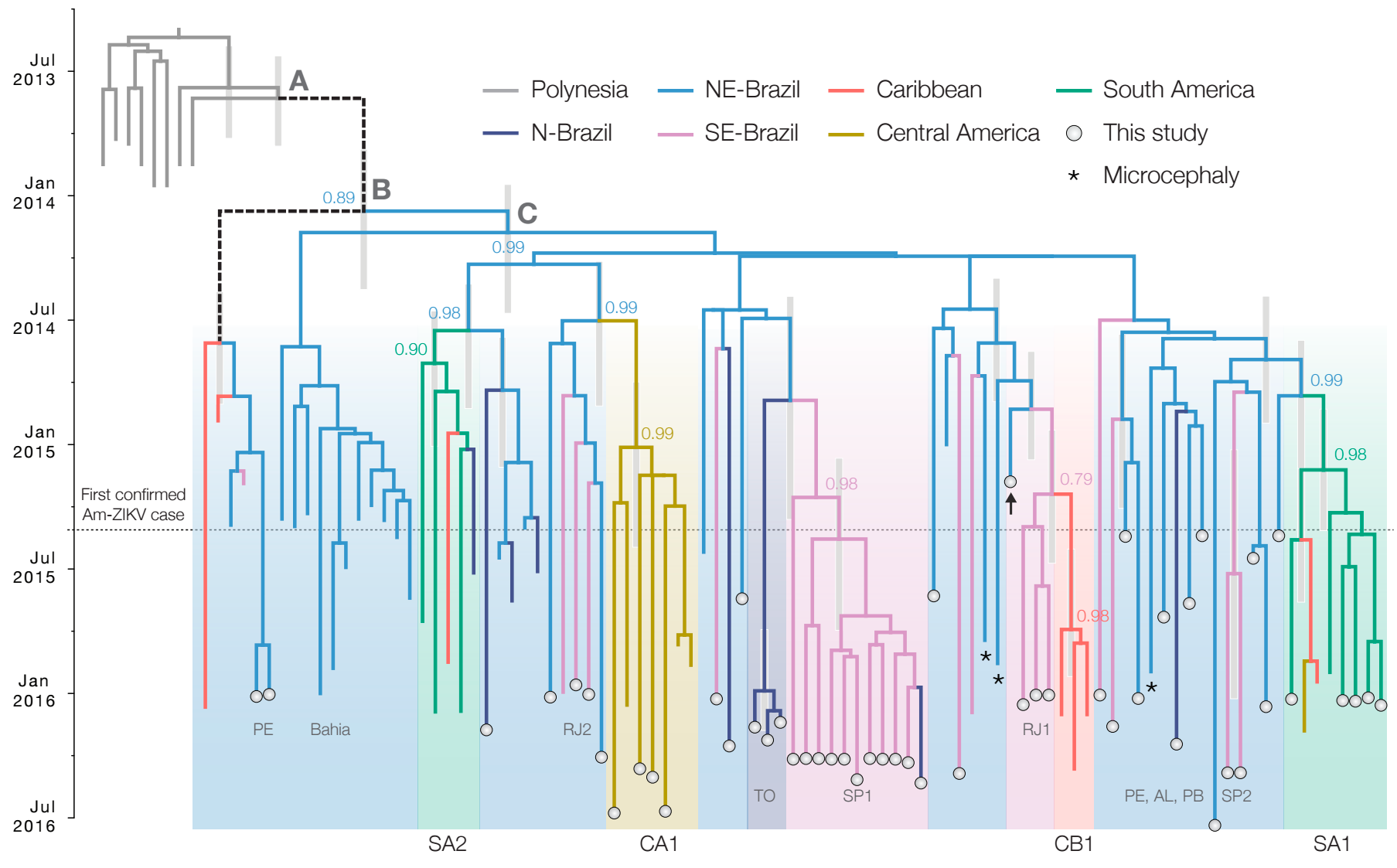
Phylogeny infers an origin in northeast Brazil
Local spread of Zika in Florida
with Nathan Grubaugh, Kristian Andersen, Jason Ladner, Gustavo Palacios, Sharon Isern, Oli Pybus, Moritz Kraemer, Gytis Dudas, Amanda Tan, Karthik Gangavarapu, Michael Wiley, Stephen White, Julien Thézé, Scott Michael, Leah Gillis, Pardis Sabeti, and many others
Outbreak of locally-acquired infections focused in Miami-Dade county
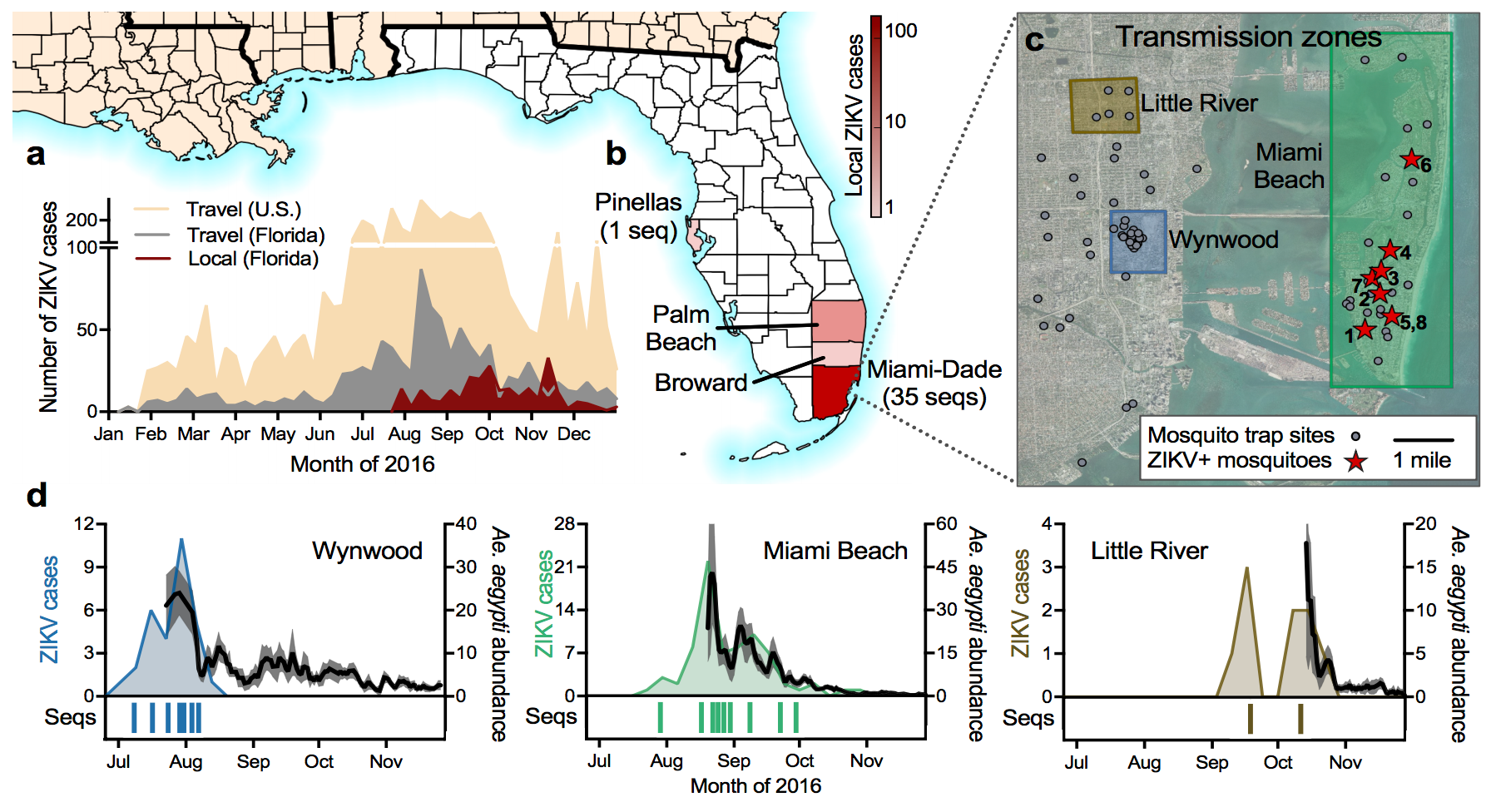
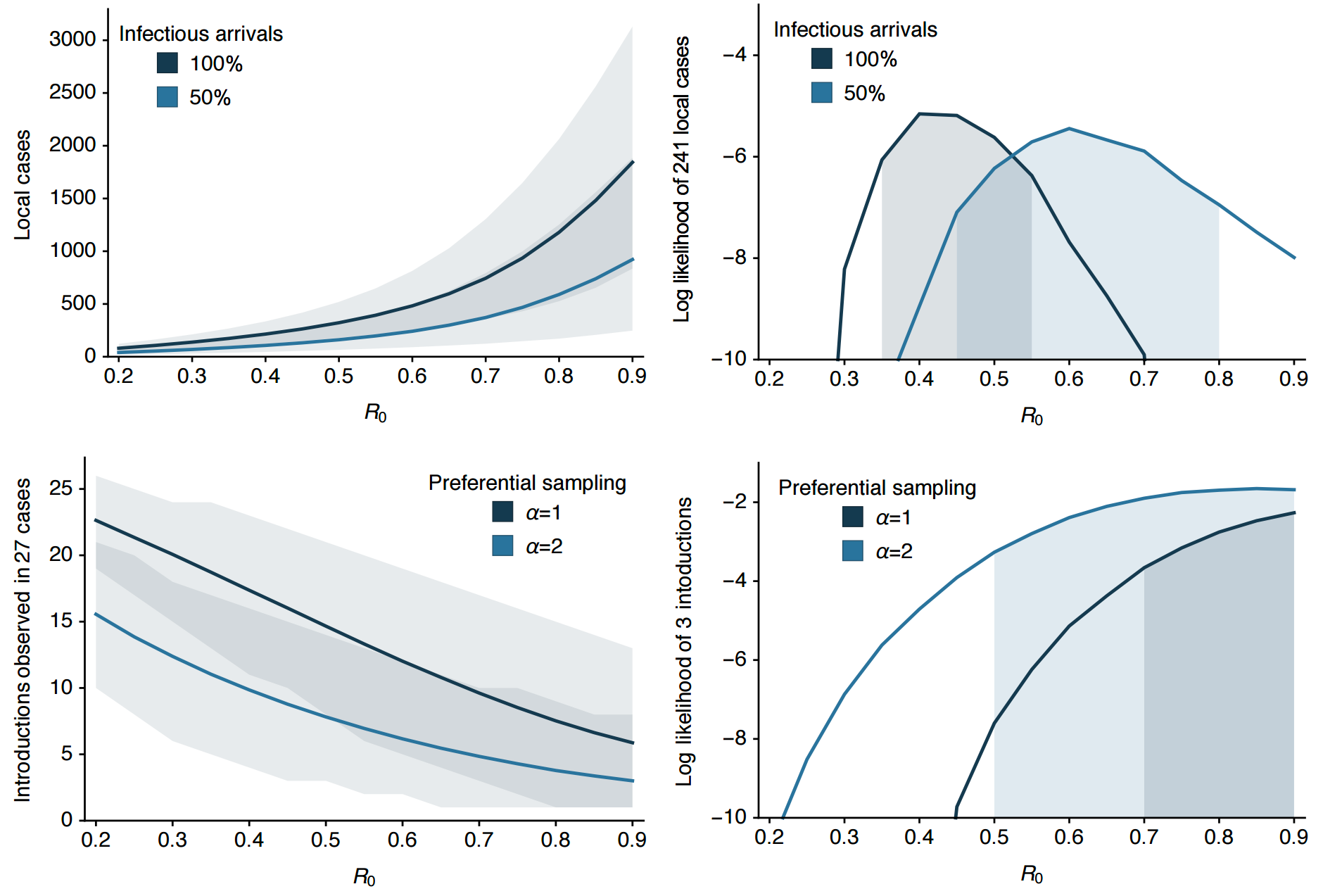
Phylogeny shows a surprising degree of clustering
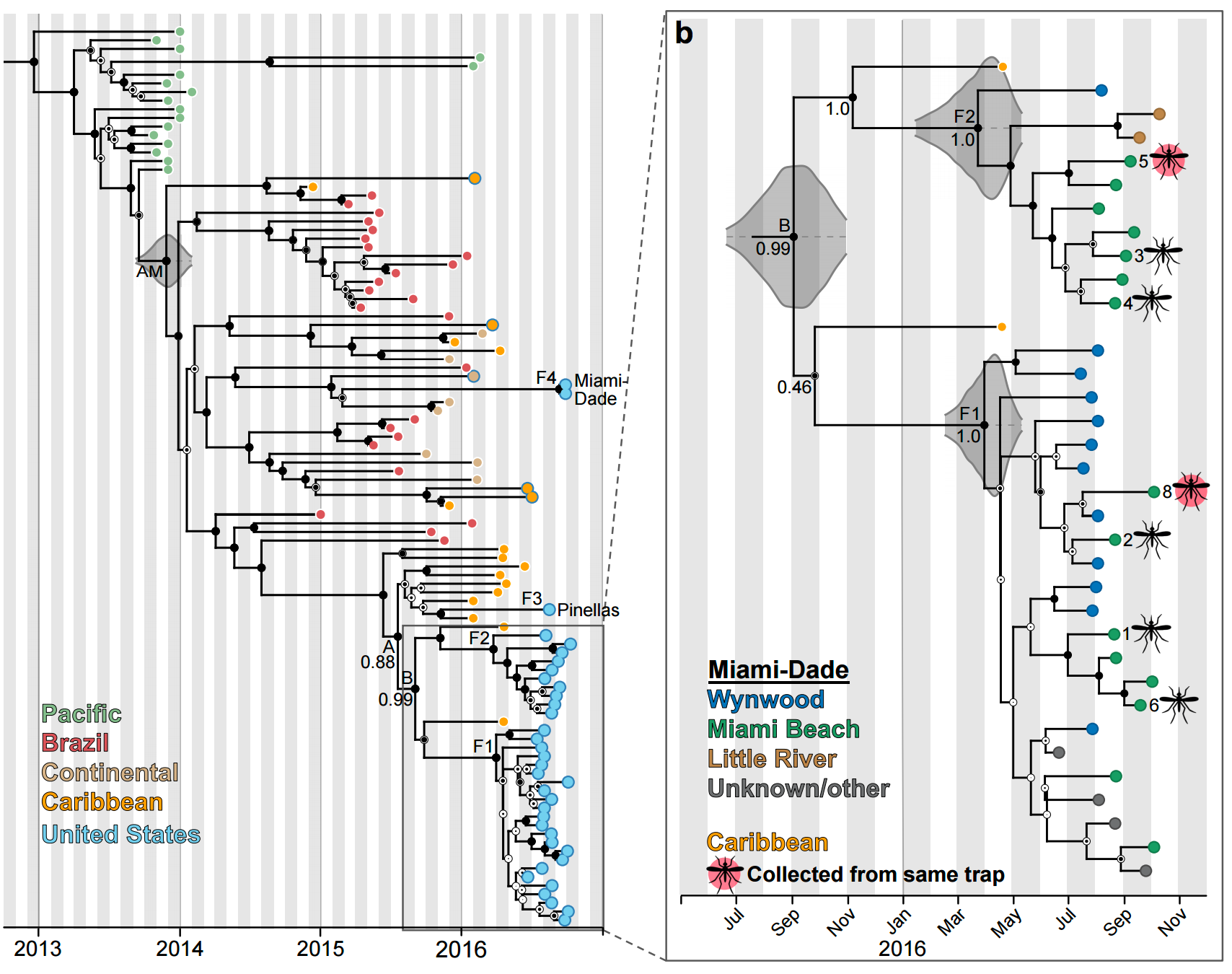
Clustering suggests fewer, longer transmission chains and higher R0
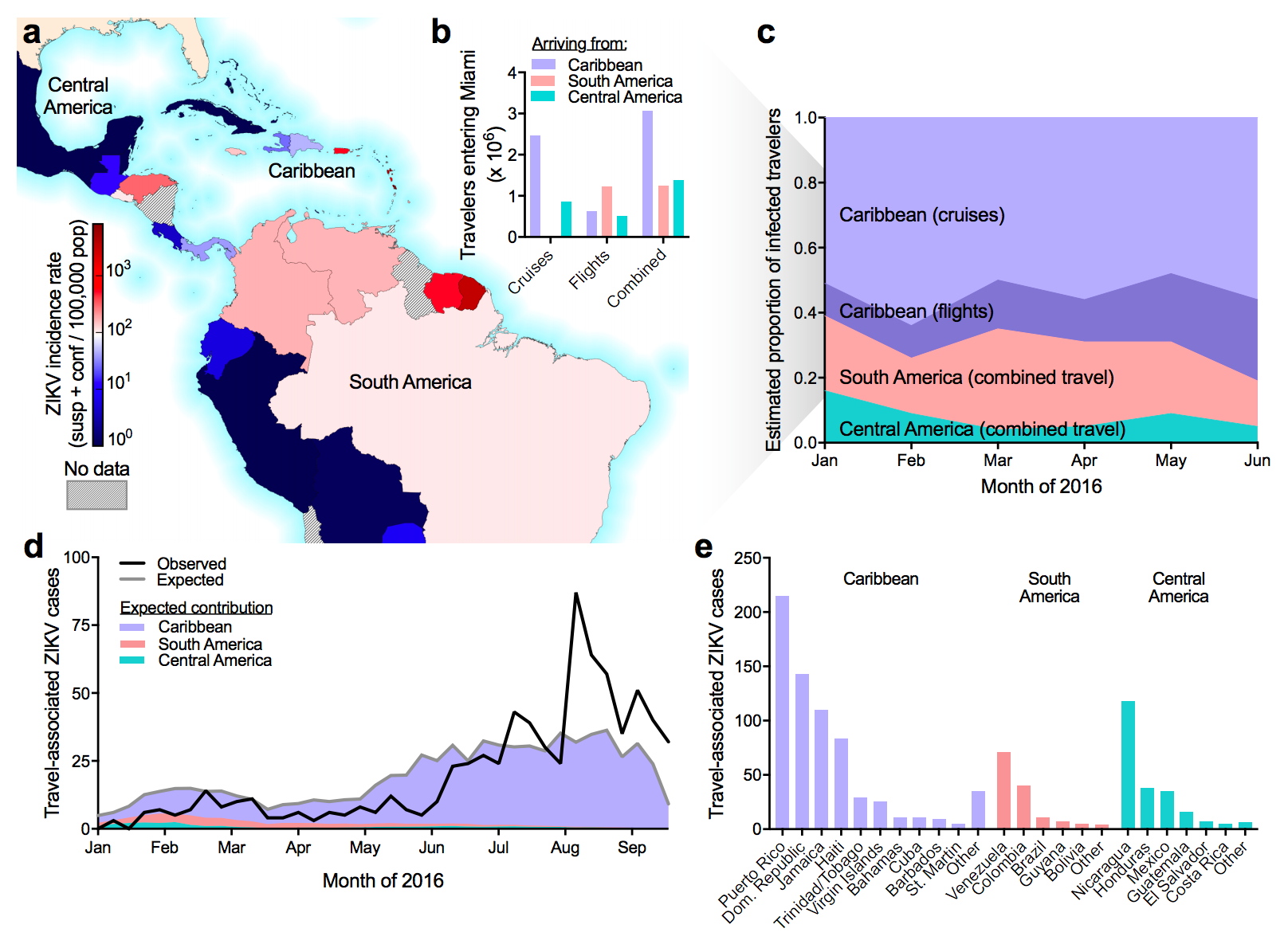
Flow of infected travelers greatest from Caribbean
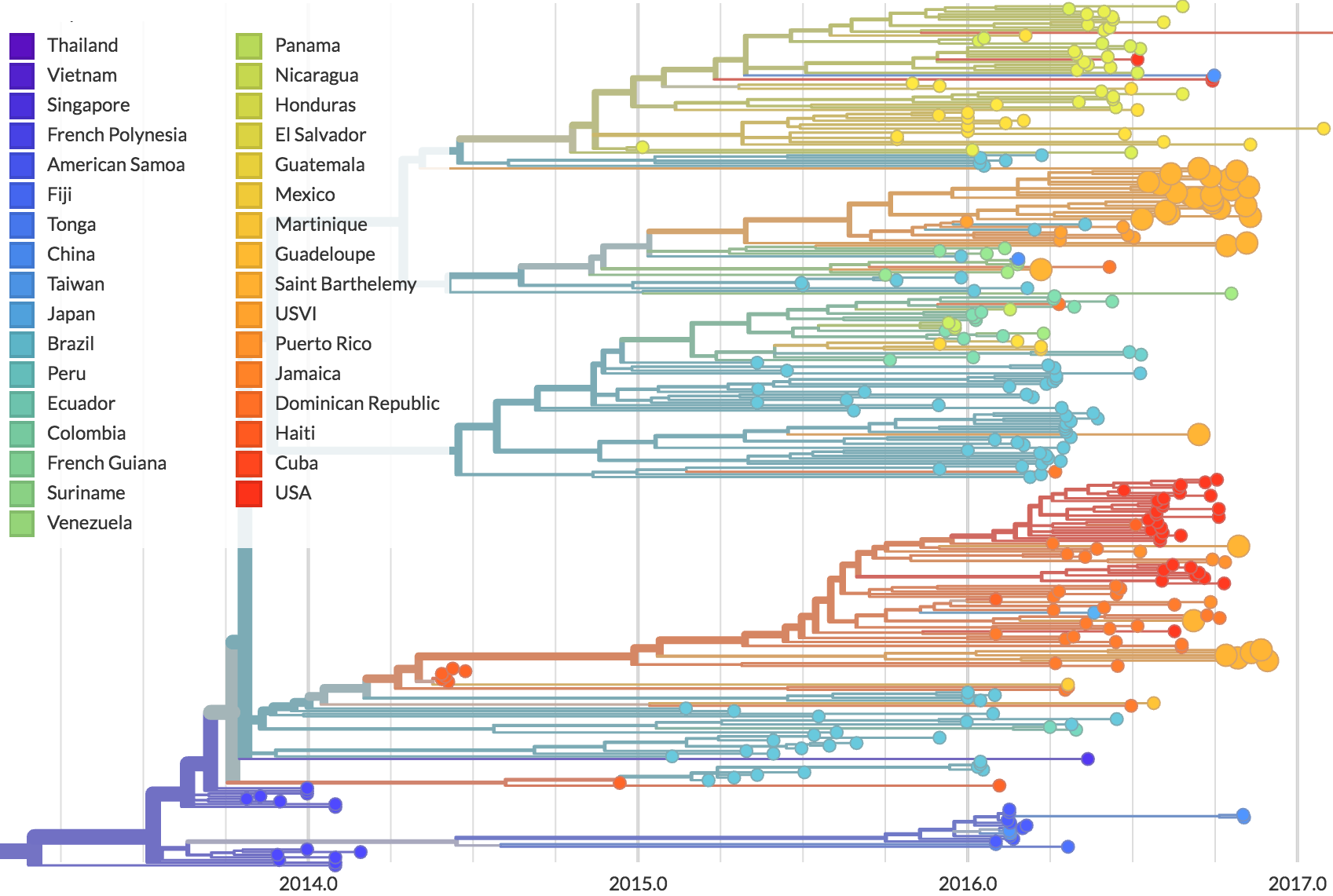
Genomic epidemiology of Zika in the US Virgin Islands
with Allison Black, Barney Potter, Esther Ellis, Brett Ellis, Kristian Andersen,
Nathan Grubaugh, Leora Feldstein, and others
Preliminary analysis of 20 genomes shows multiple introductions to USVI
Important analyses, let's make them more rapid and more automated
Key challenges
- Timely analysis and sharing of results critical
- Dissemination must be scalable
- Integrate many data sources
- Results must be easily interpretable and queryable
nextstrain
Project to conduct real-time molecular epidemiology and evolutionary analysis of emerging epidemics
 Richard Neher,
Richard Neher,
 Trevor Bedford,
Trevor Bedford,
 James Hadfield,
James Hadfield,
 Colin Megill,
Colin Megill,
 Sidney Bell,
Sidney Bell,
 Charlton Callender,
Charlton Callender,
 Barney Potter,
Barney Potter,
 Sarah Murata,
Sarah Murata,
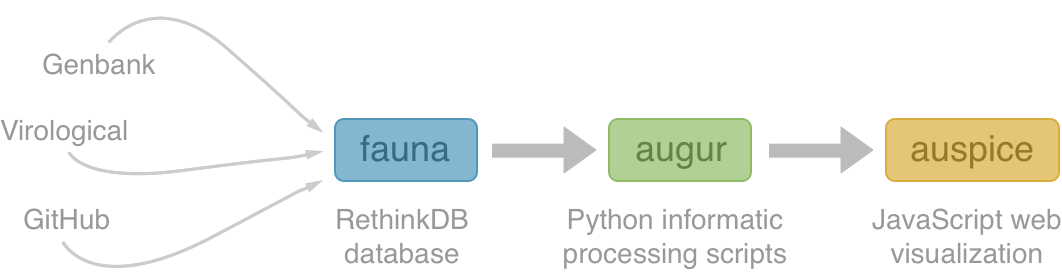
Nextstrain architecture
All code open source at github.com/nextstrain
nextstrain.org
Rapid on-the-ground sequencing by Ian Goodfellow, Matt Cotten and colleagues
Desired analytics are pathogen specific and tied to response measures
Acknowledgements
Influenza: WHO Global Influenza Surveillance Network, GISAID, Worldwide Influenza Centre at the Francis Crick Institute, Richard Neher, Colin Russell, Boris Shraiman
Ebola: Gytis Dudas, Andrew Rambaut, Luiz Carvalho, Philippe Lemey, Marc Suchard, Andrew Tatem, Nick Loman, Ian Goodfellow, Matt Cotten, Paul Kellam, Kristian Andersen, Pardis Sabeti, many others
Zika: Nick Loman, Nuno Faria, Oliver Pybus, Josh Quick, Kristian Andersen, Nathan Grubaugh, Jason Ladner, Gustavo Palacios, Sharon Isern, Gytis Dudas, Allison Black, Barney Potter, Esther Ellis, many others
Nextstrain: Richard Neher, James Hadfield, Colin Megill, Sidney Bell, Charlton Callender, Barney Potter, Sarah Murata





