Real-time tracking and prediction of influenza virus evolution
Trevor Bedford (@trvrb)
25 May 2017
Immunology and Evolution of Influenza Symposium
Emory University
Want to forecast the make up of the future flu population from the population that exists today
Real-time updates as new information rolls in
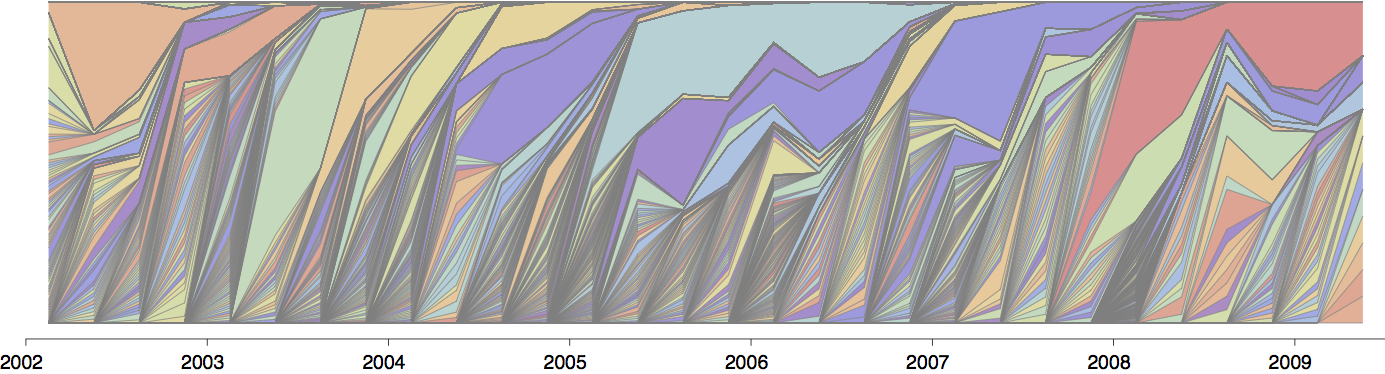
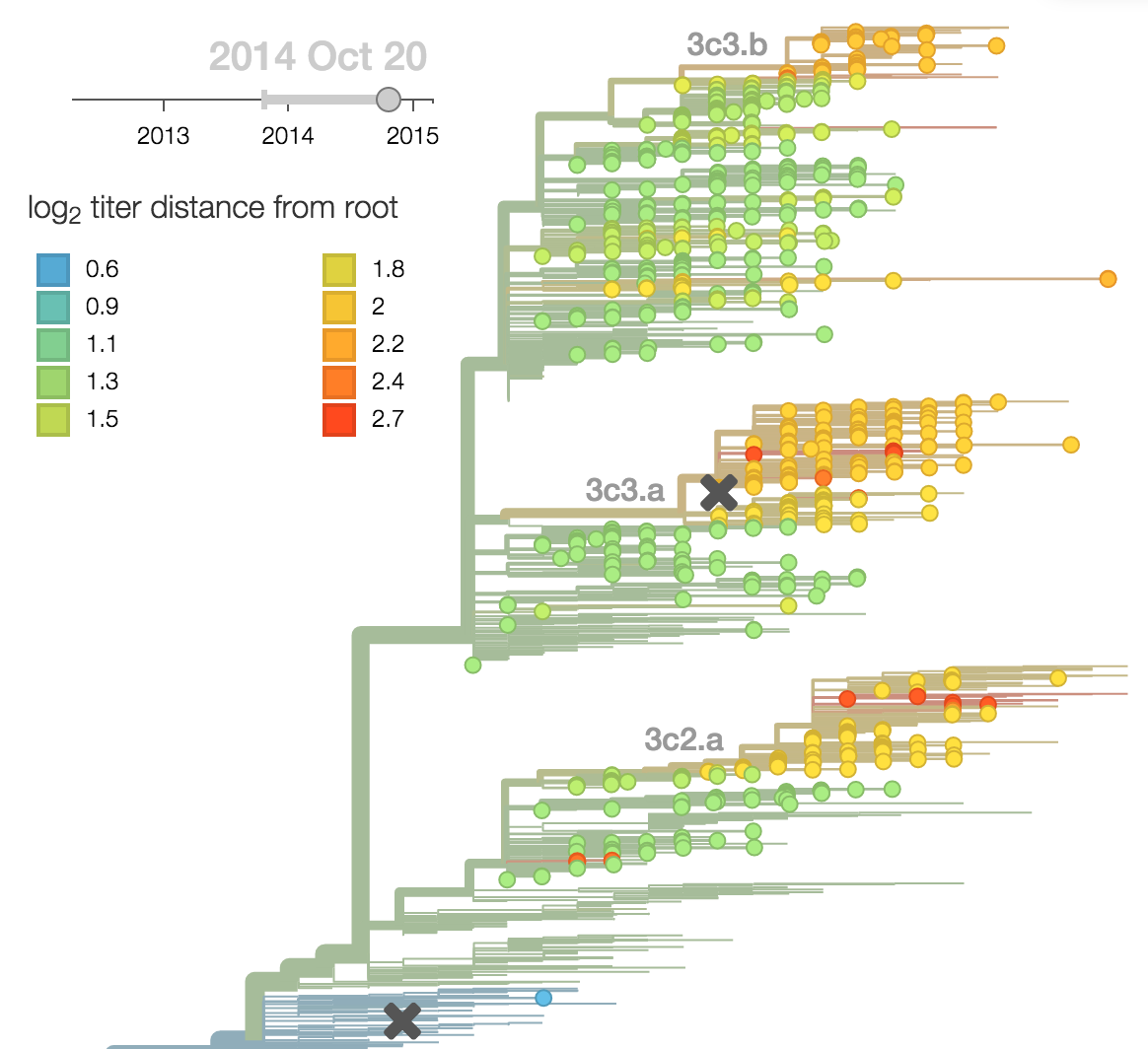
Population turnover (in H3N2) is extremely rapid
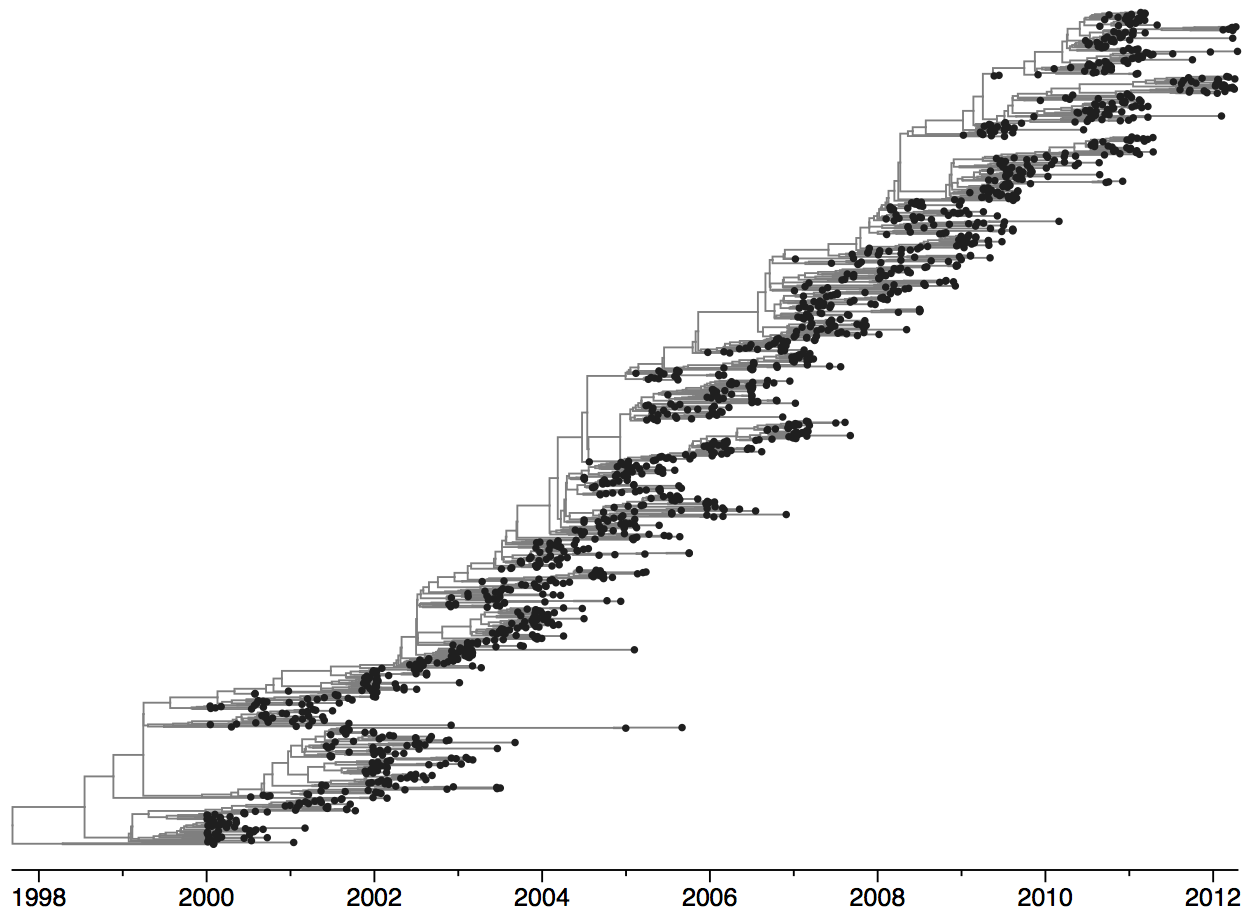
Clades emerge, die out and take over
Clades show rapid turnover

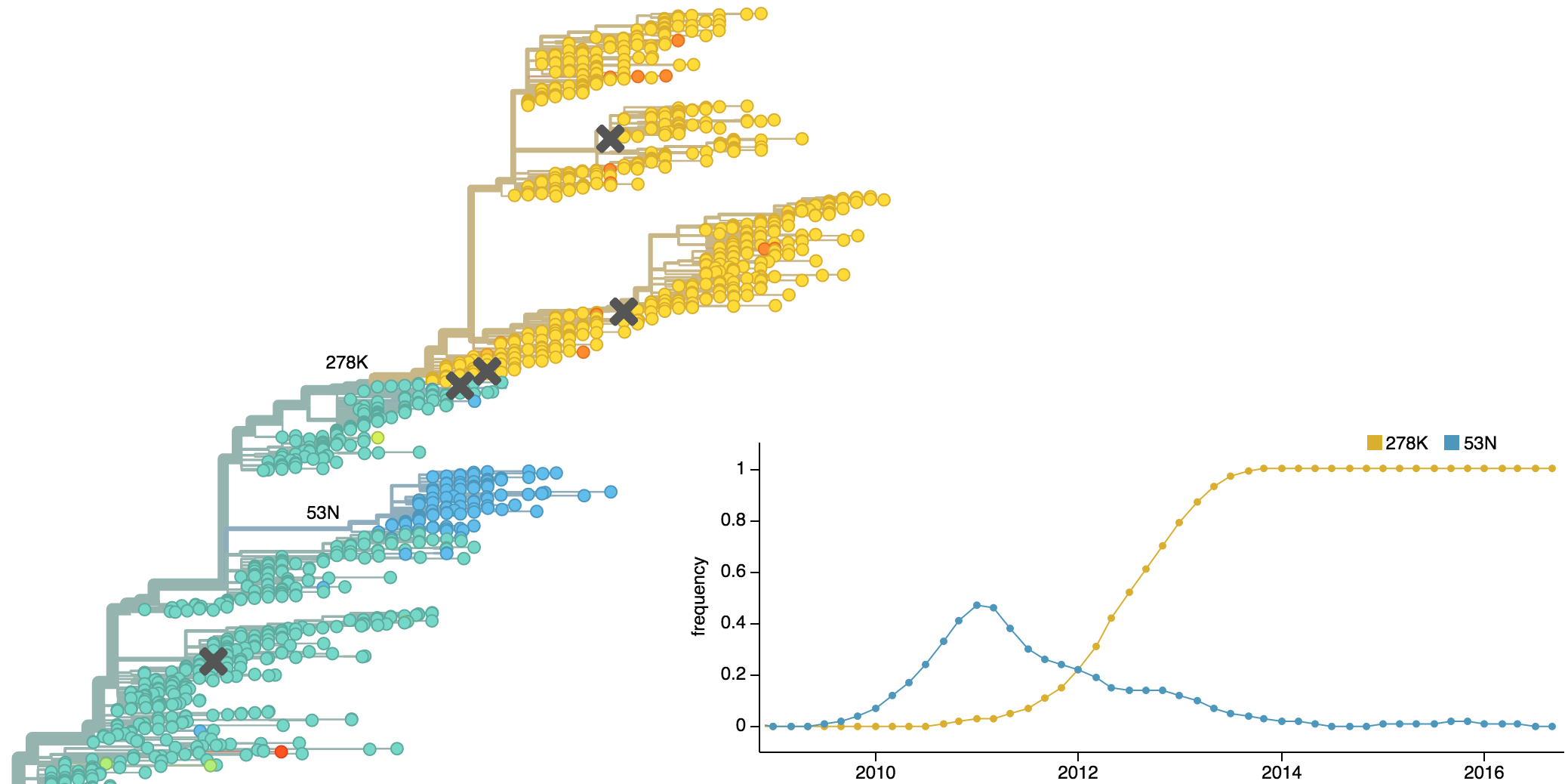
Dynamics driven by antigenic drift
Drift variants emerge and rapidly take over in the virus population
This causes the side effect of evading existing vaccine formulations
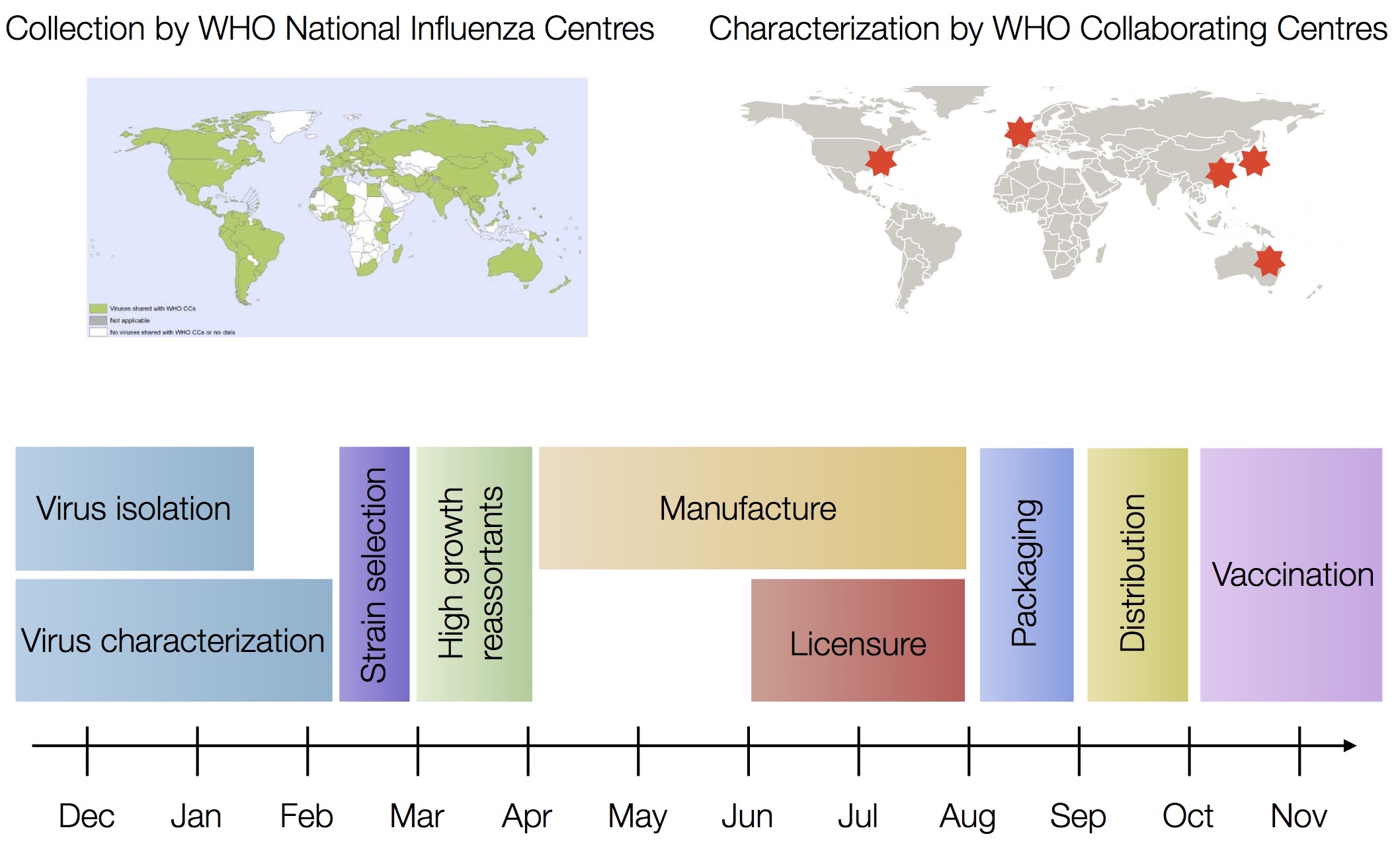
Drift necessitates vaccine updates
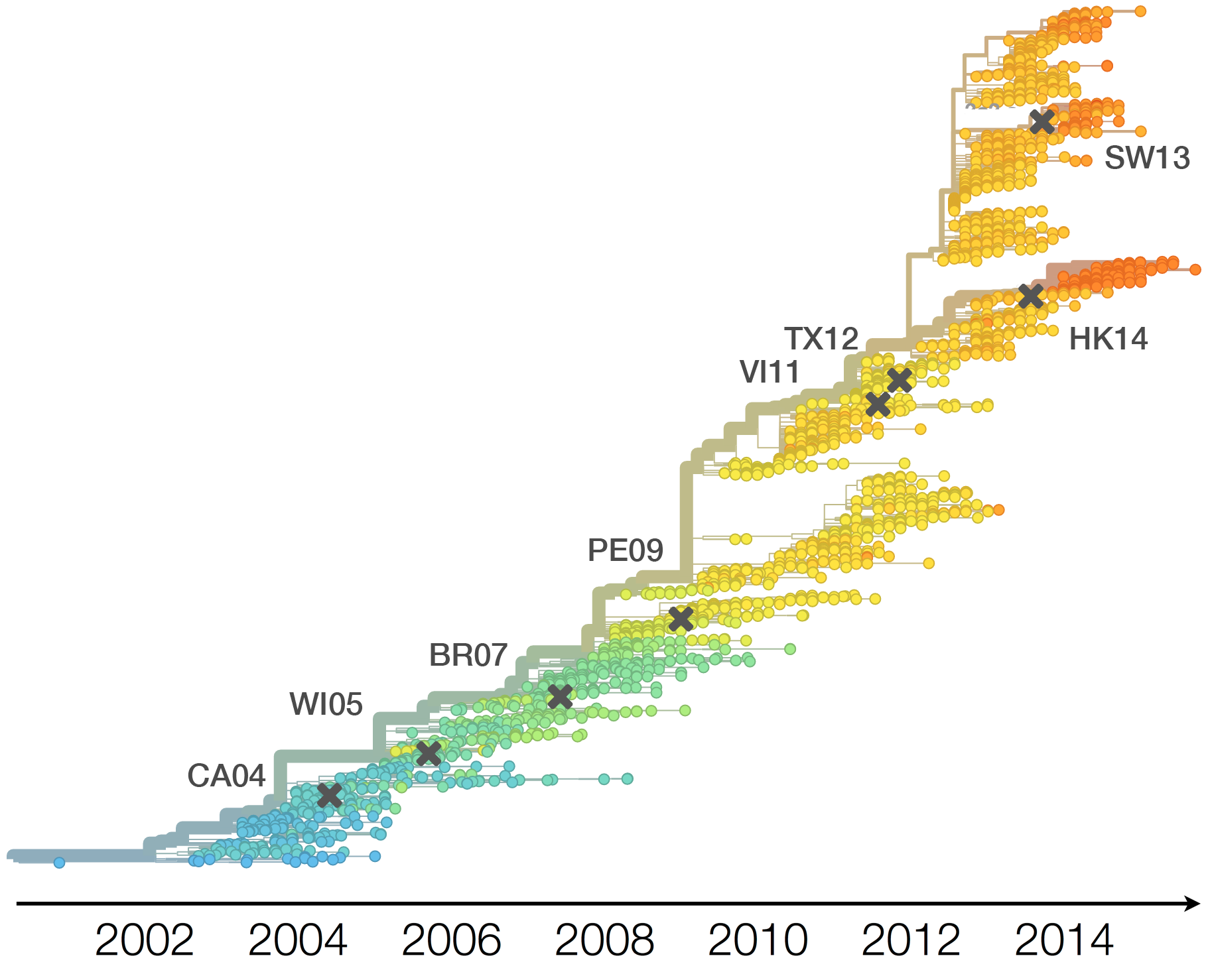
H3N2 vaccine updates occur every ~2 years
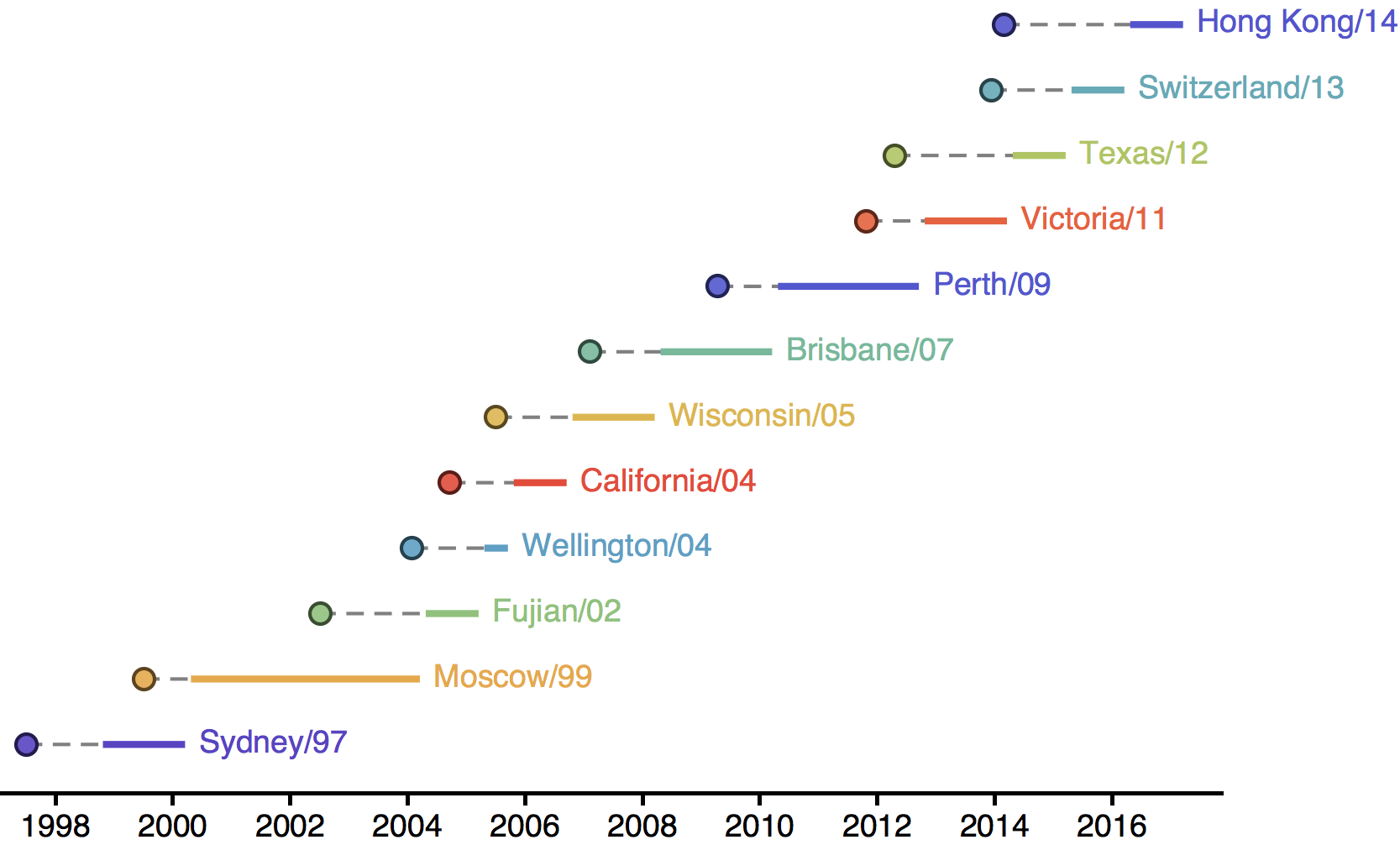
Timely surveillance and rapid analysis essential to vaccine strain selection
nextflu
Project to provide a real-time view of the evolving influenza population
nextflu
Project to provide a real-time view of the evolving influenza population
All in collaboration with Richard Neher

nextflu pipeline
- Download all recent HA sequences from GISAID
- Filter to remove outliers
- Subsample across time and space
- Align sequences
- Build tree
- Estimate clade frequencies
- Infer antigenic phenotypes
- Export for visualization
Up-to-date analysis publicly available at:
nextflu.org
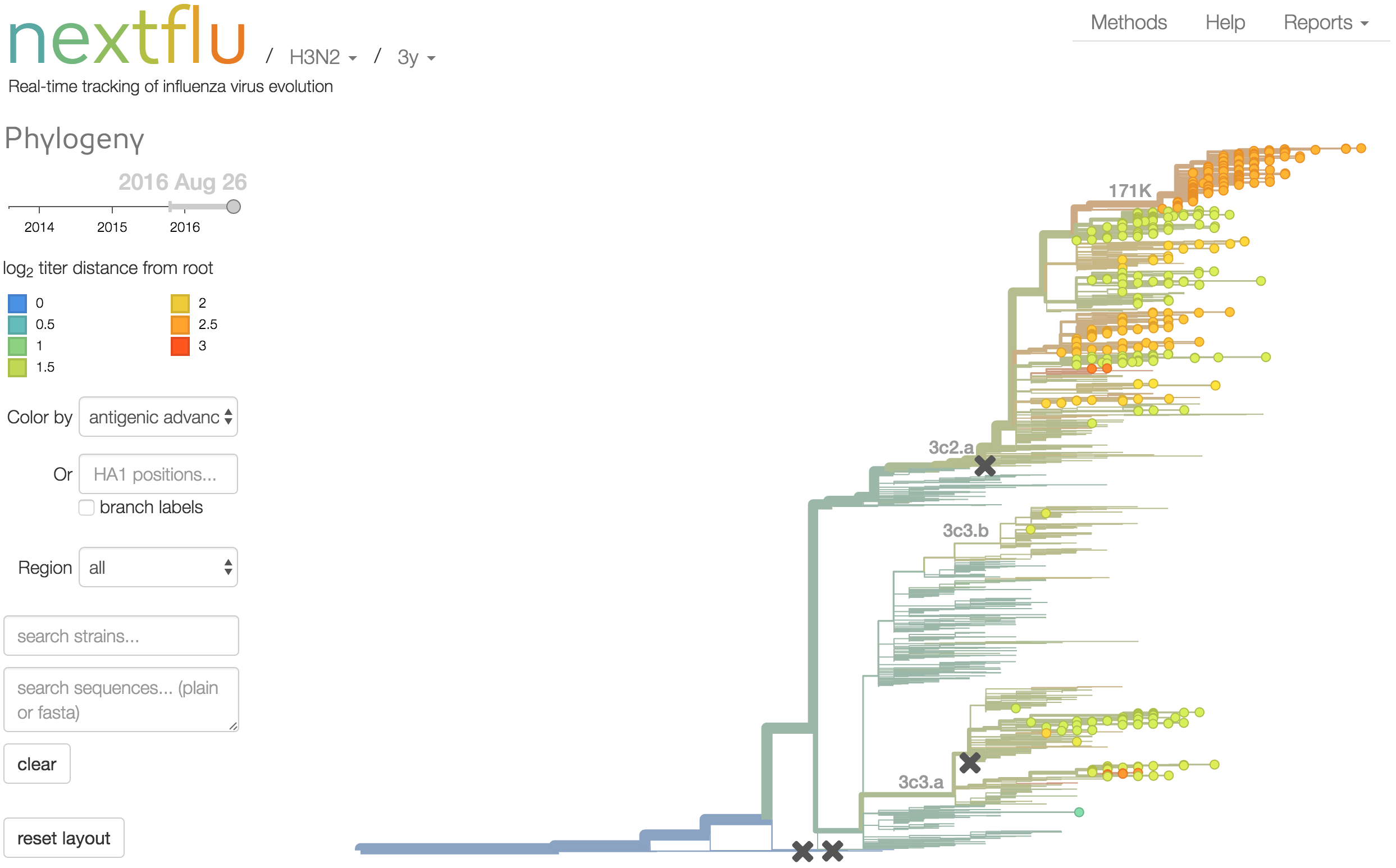
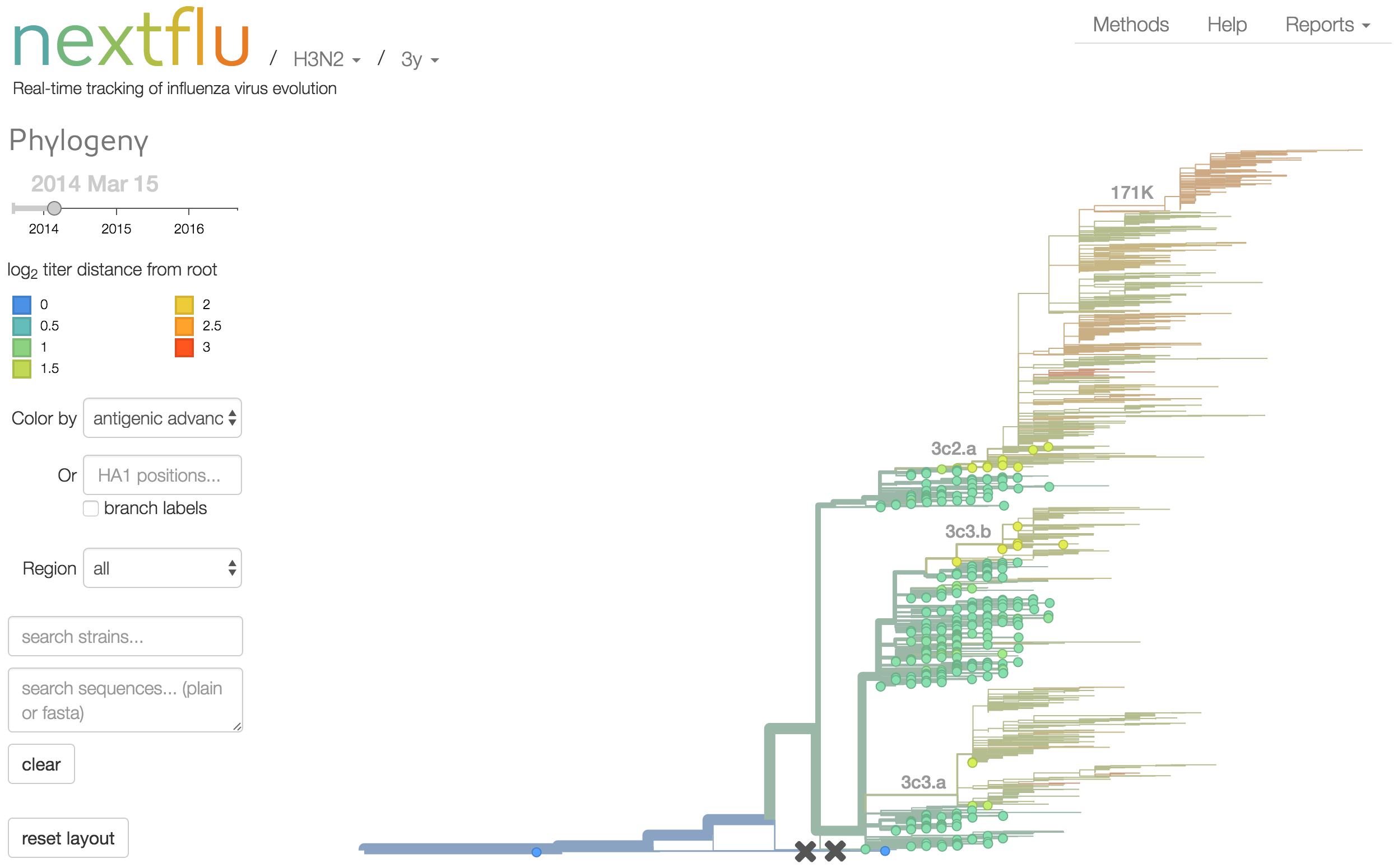
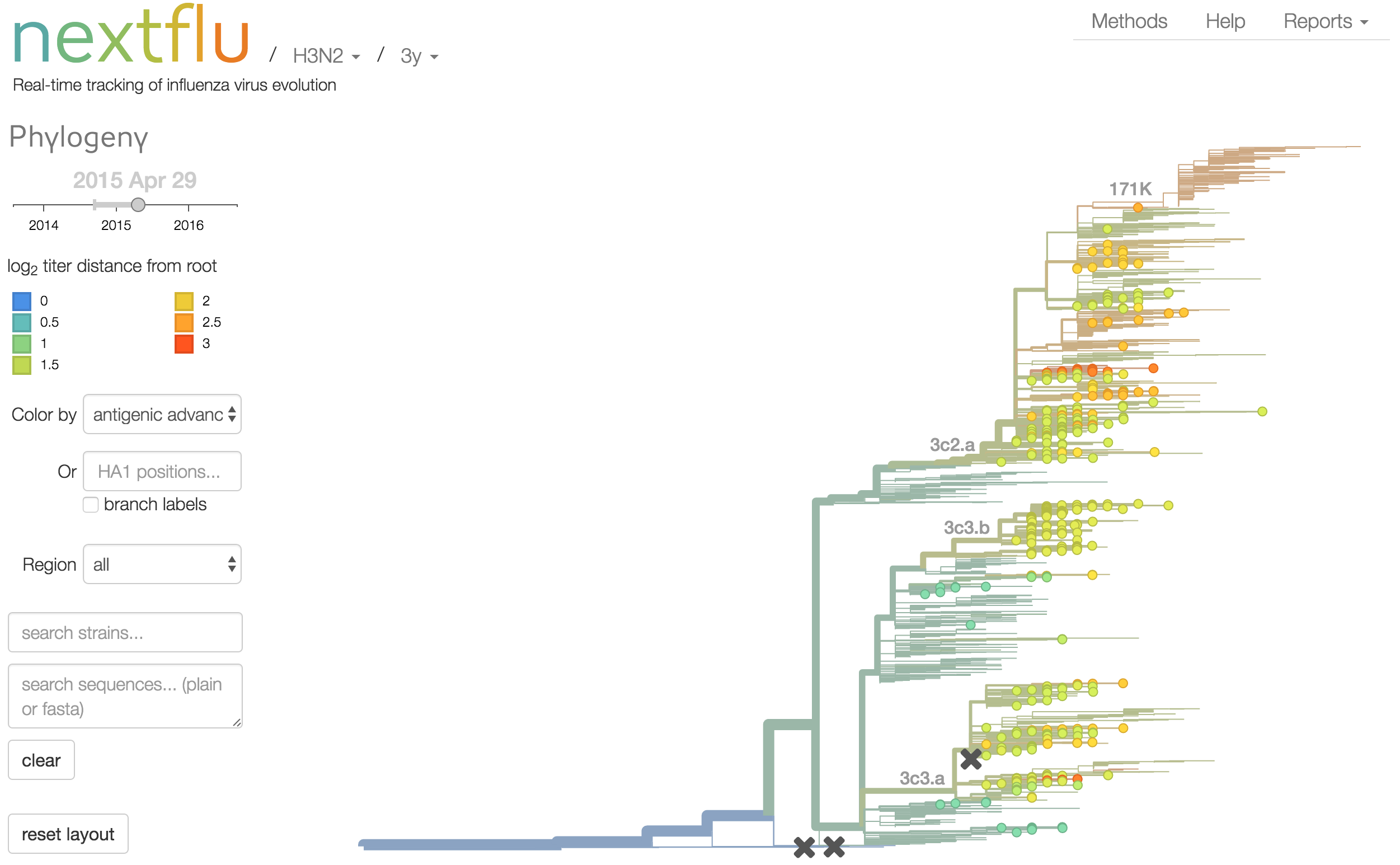
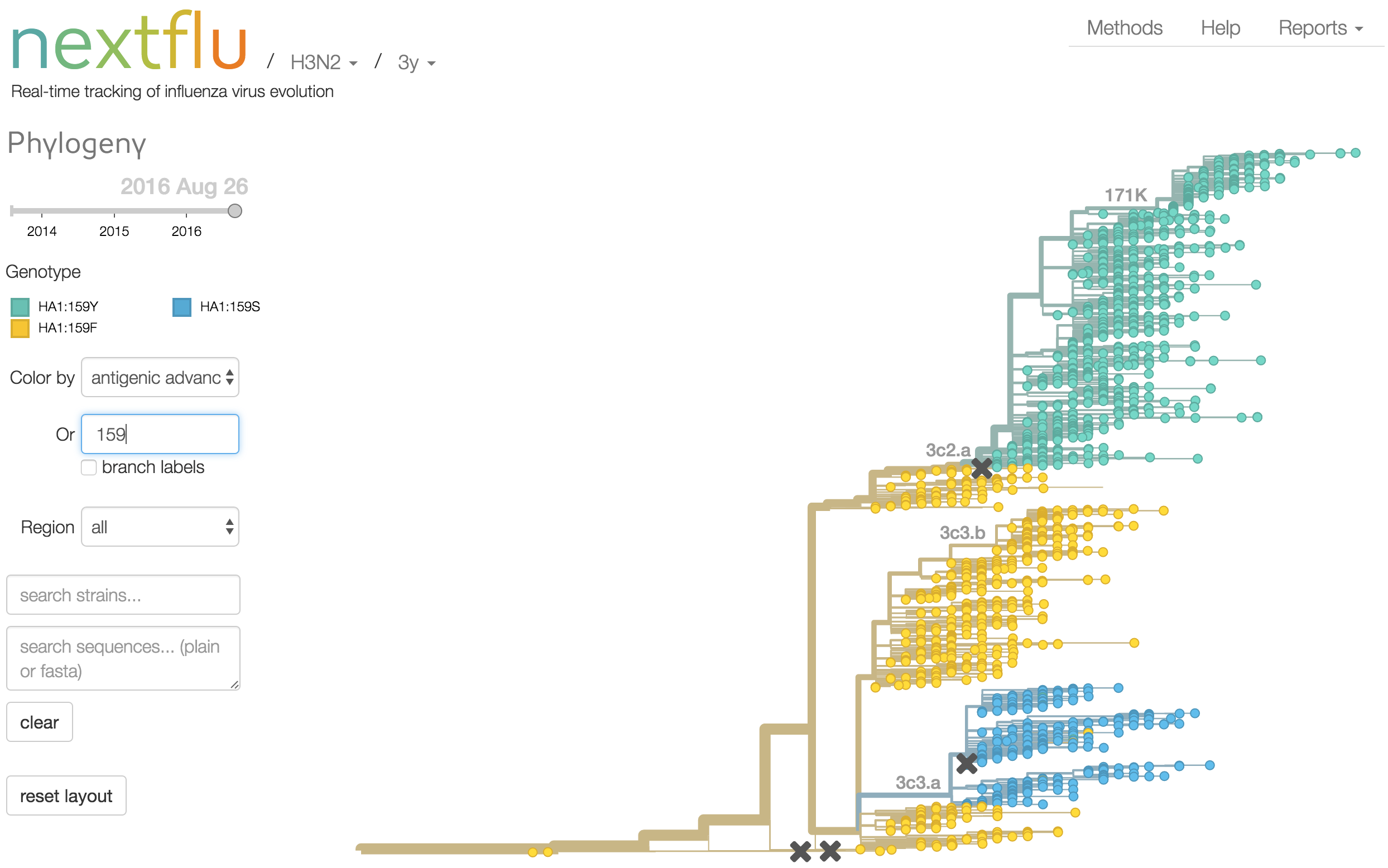
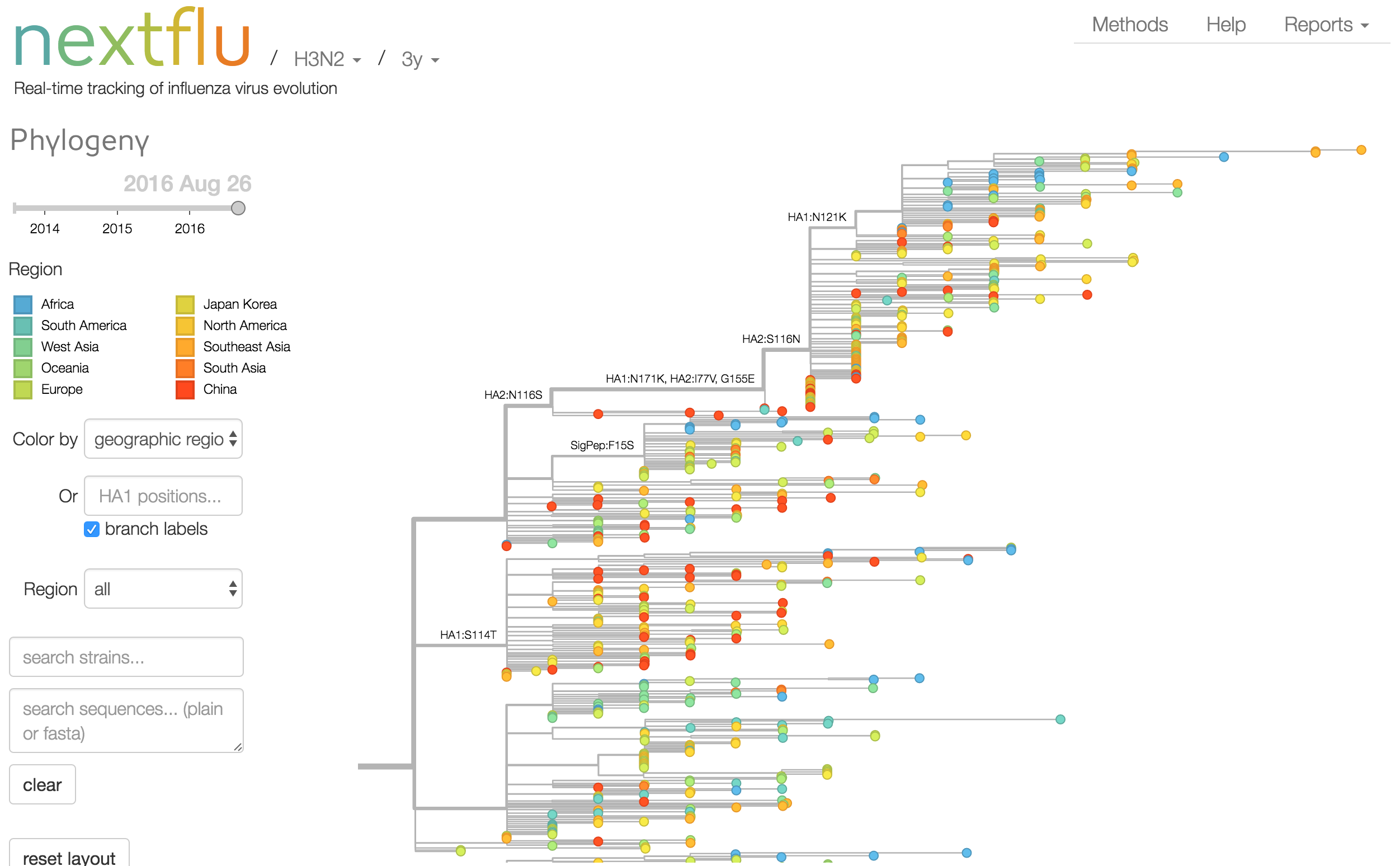
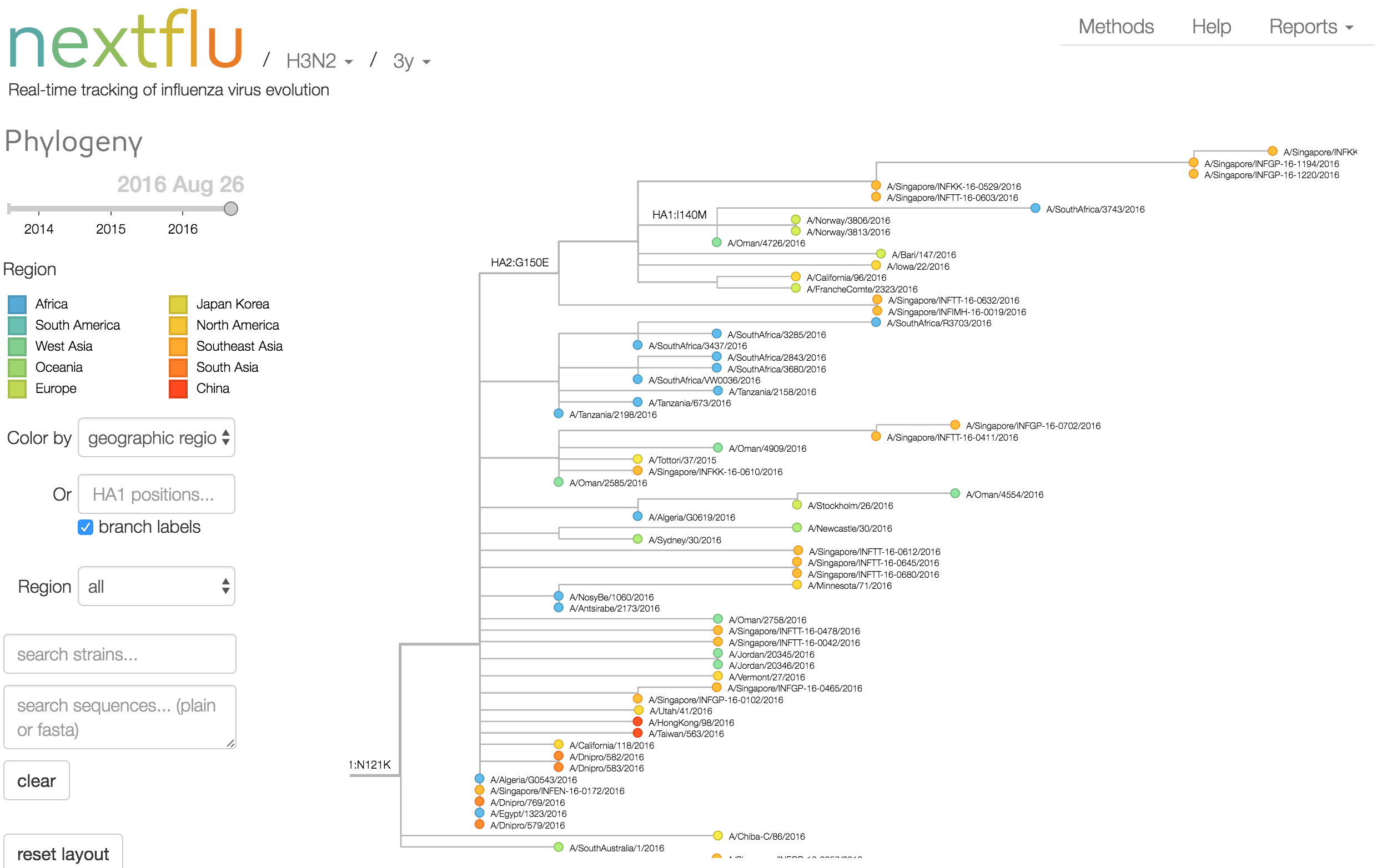
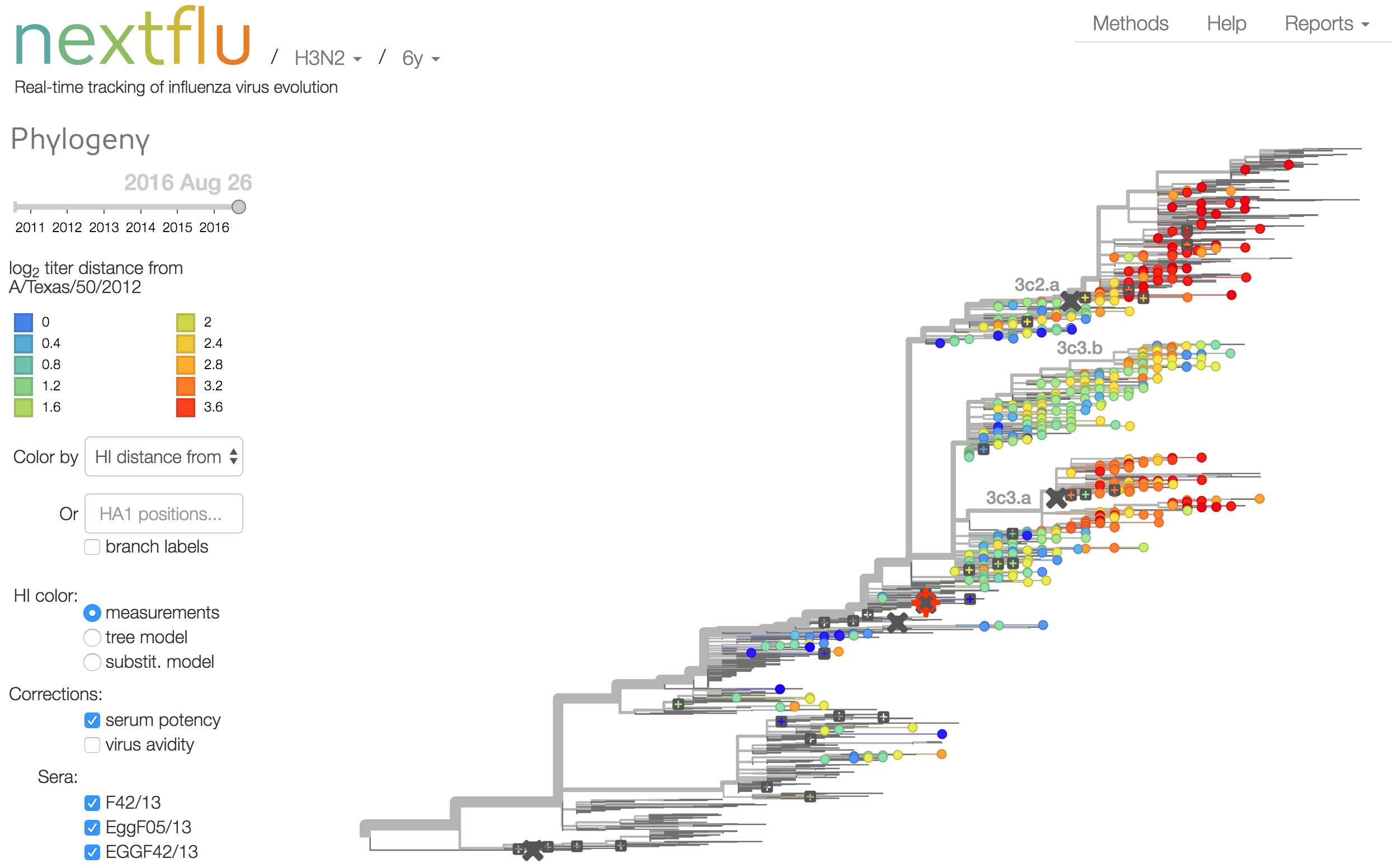
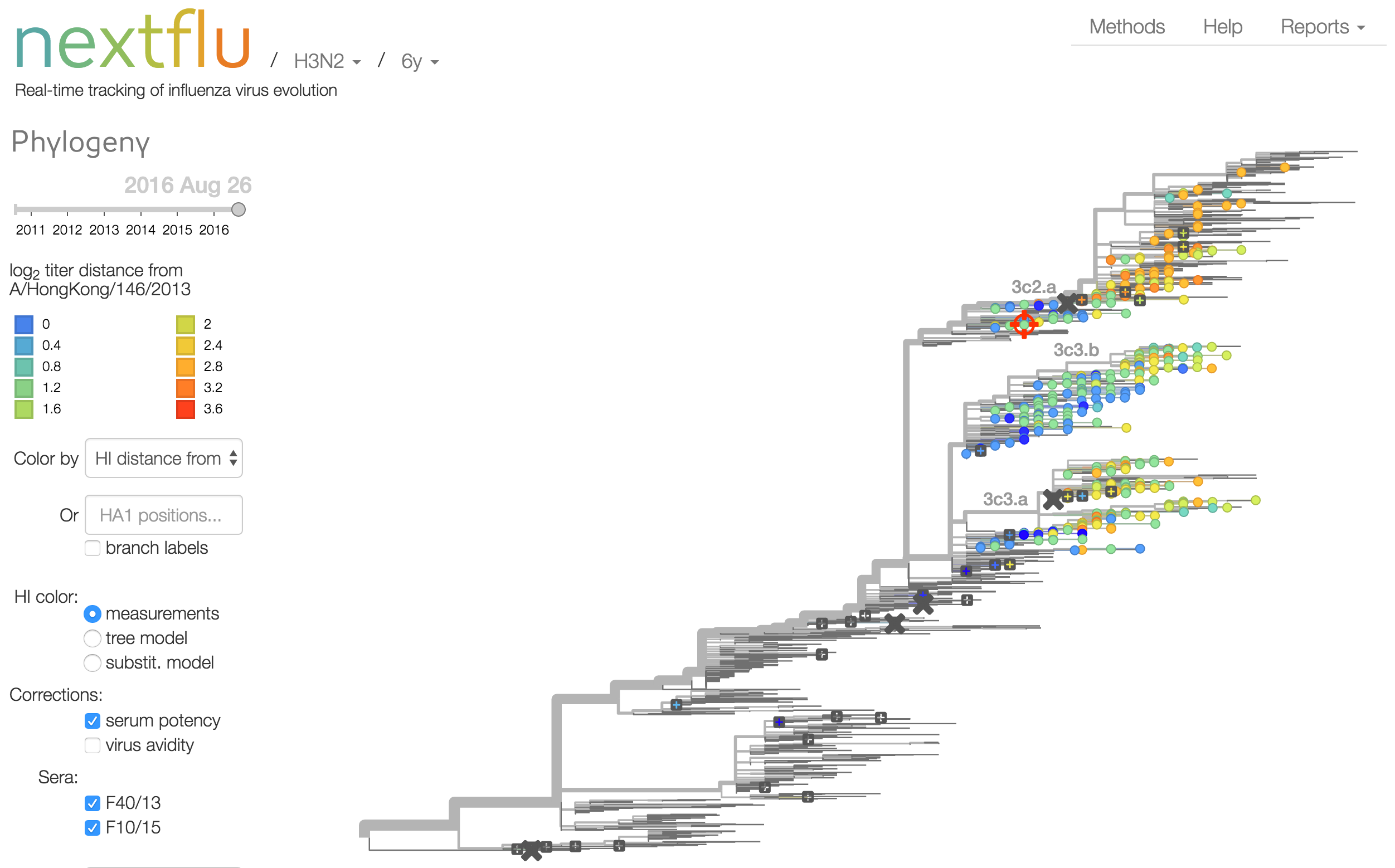
Antigenic analysis
Influenza hemagglutination inhibition (HI) assay
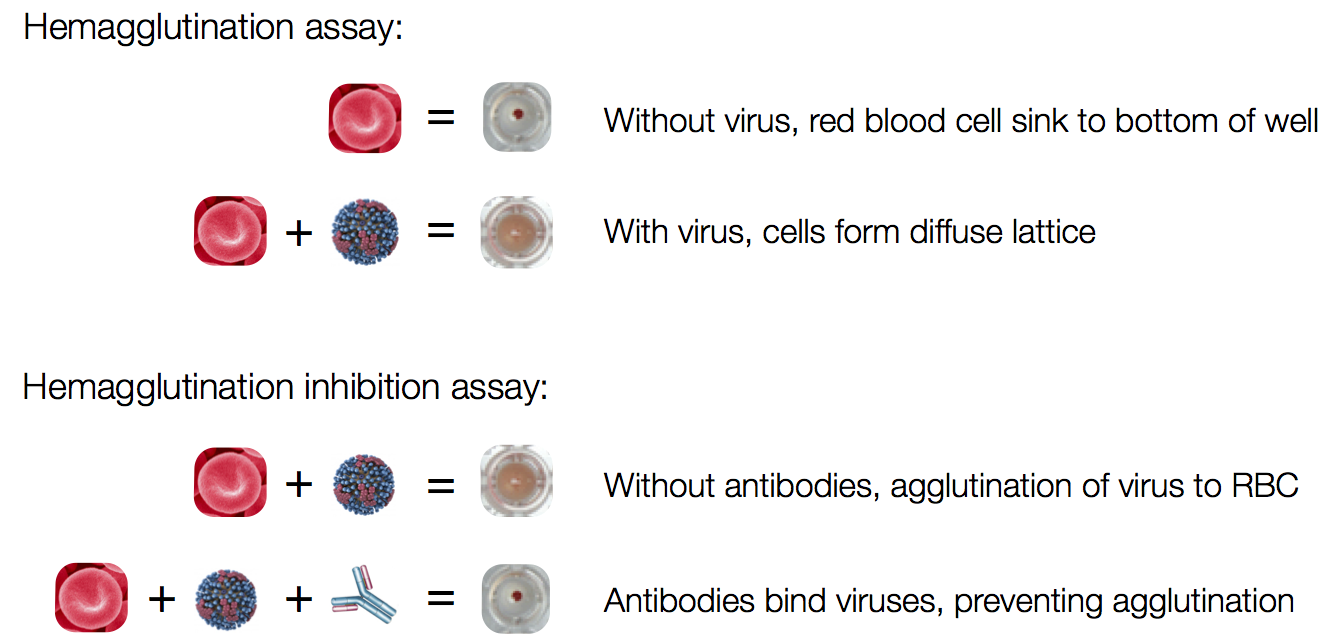
HI measures cross-reactivity across viruses
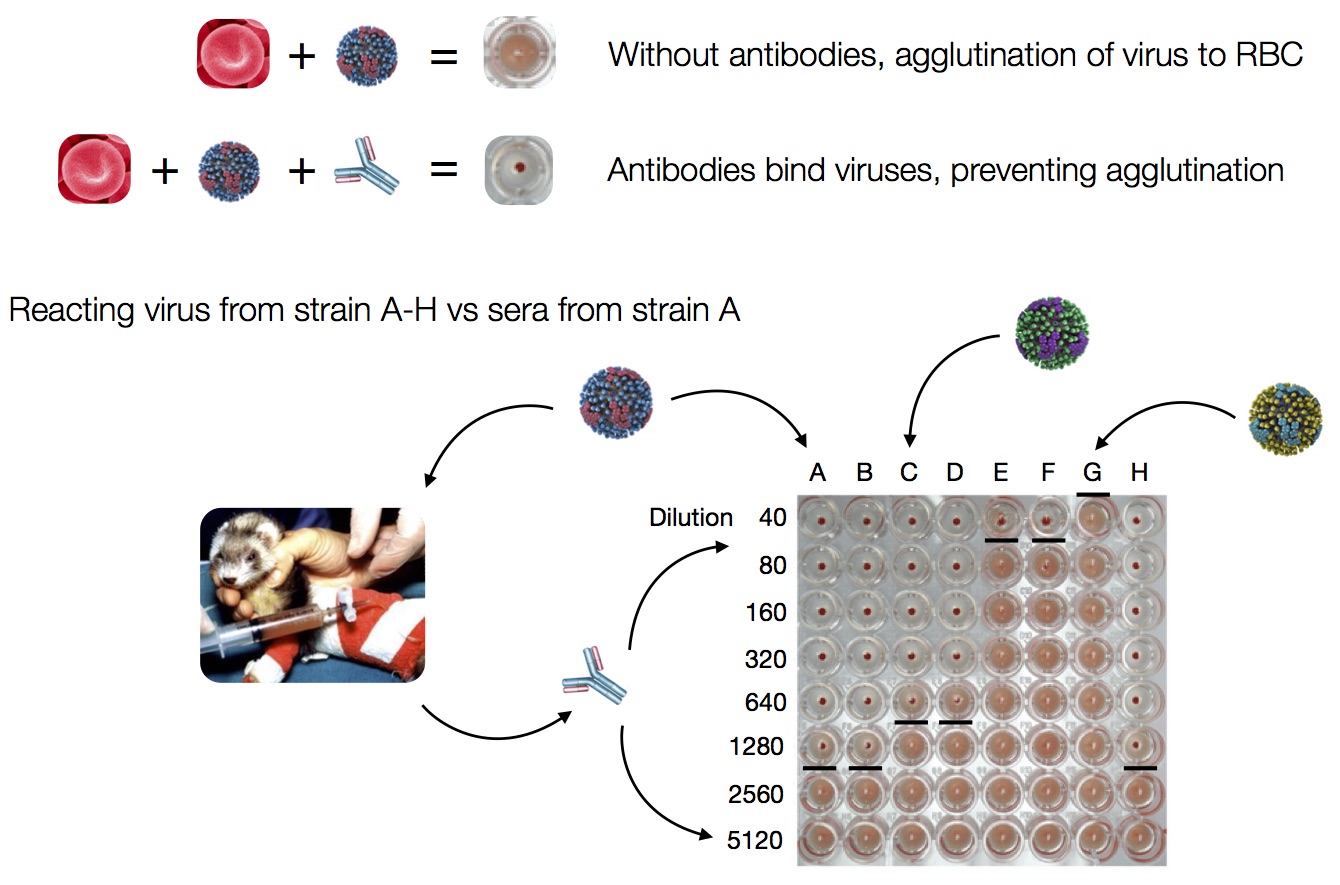
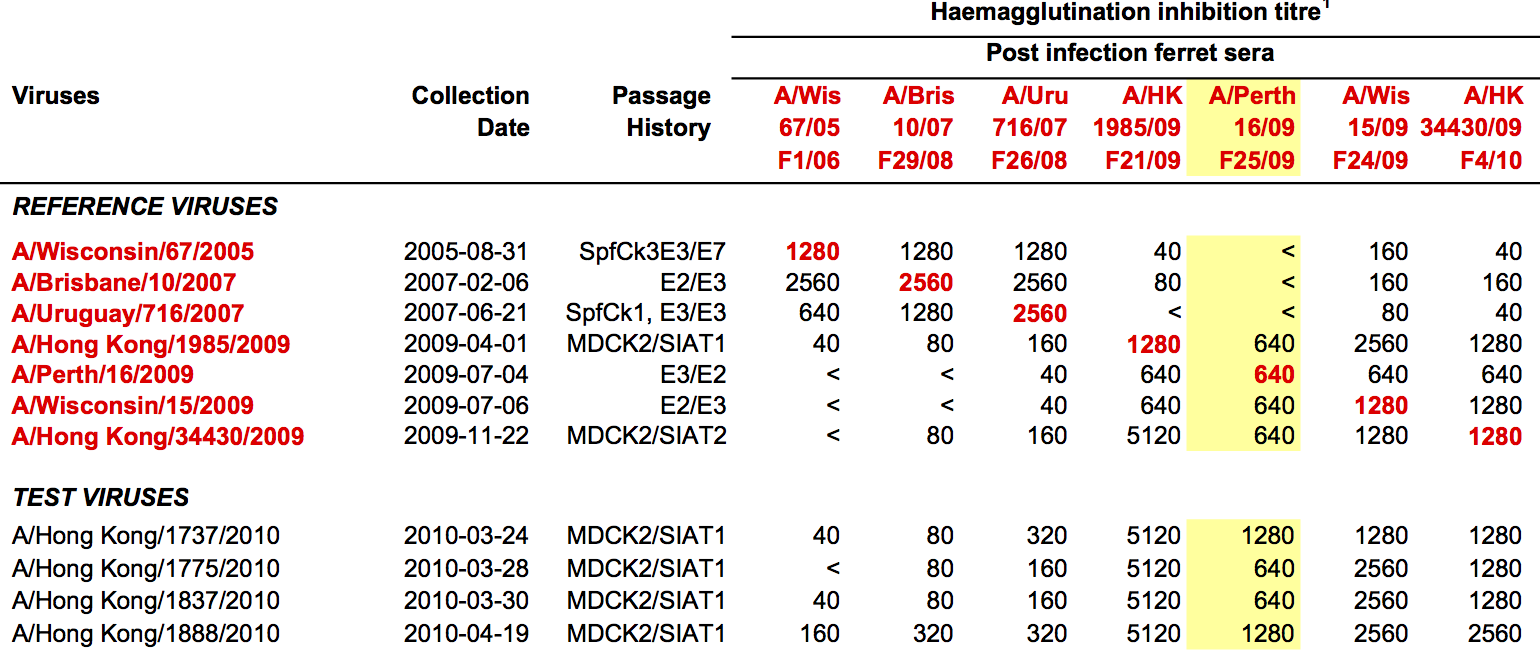
Data in the form of table of maximum inhibitory titers
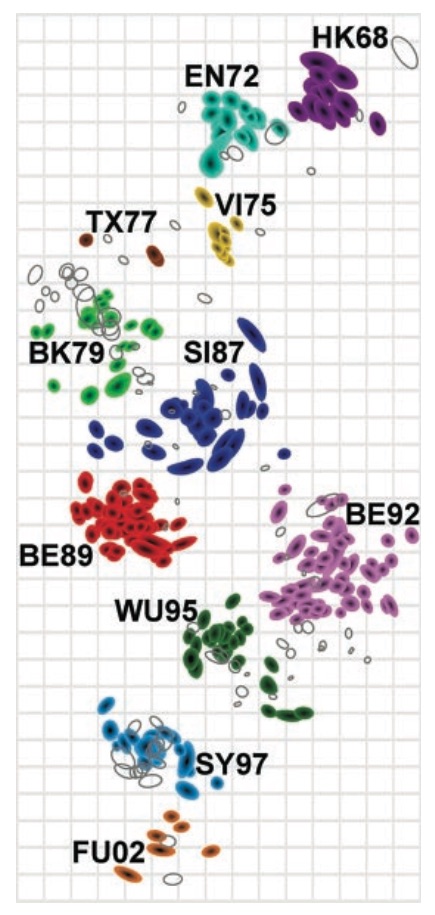
Antigenic cartography compresses HI measurements into an interpretable diagram
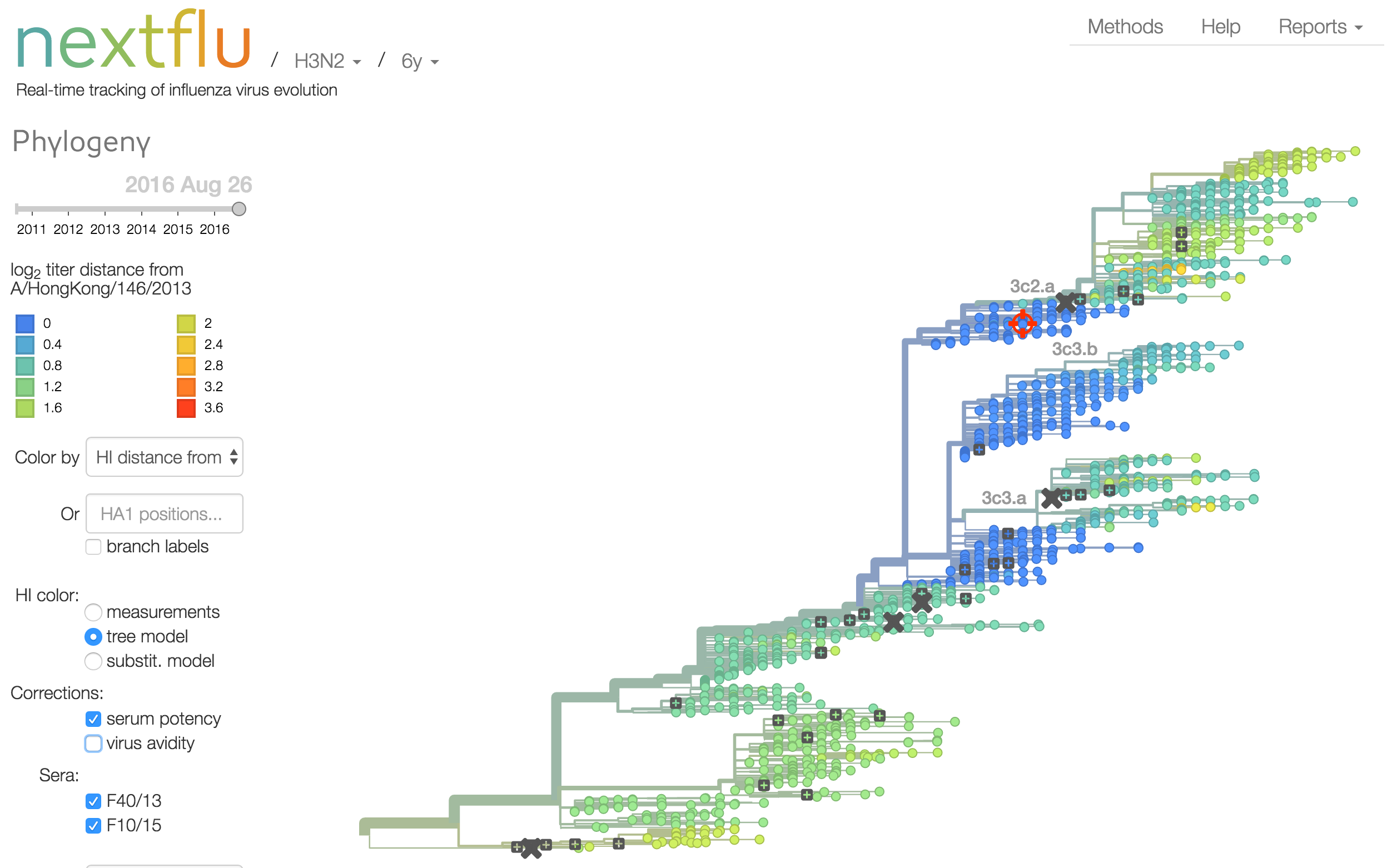
Instead of a geometric model, we sought a phylogenetic model of HI titer data
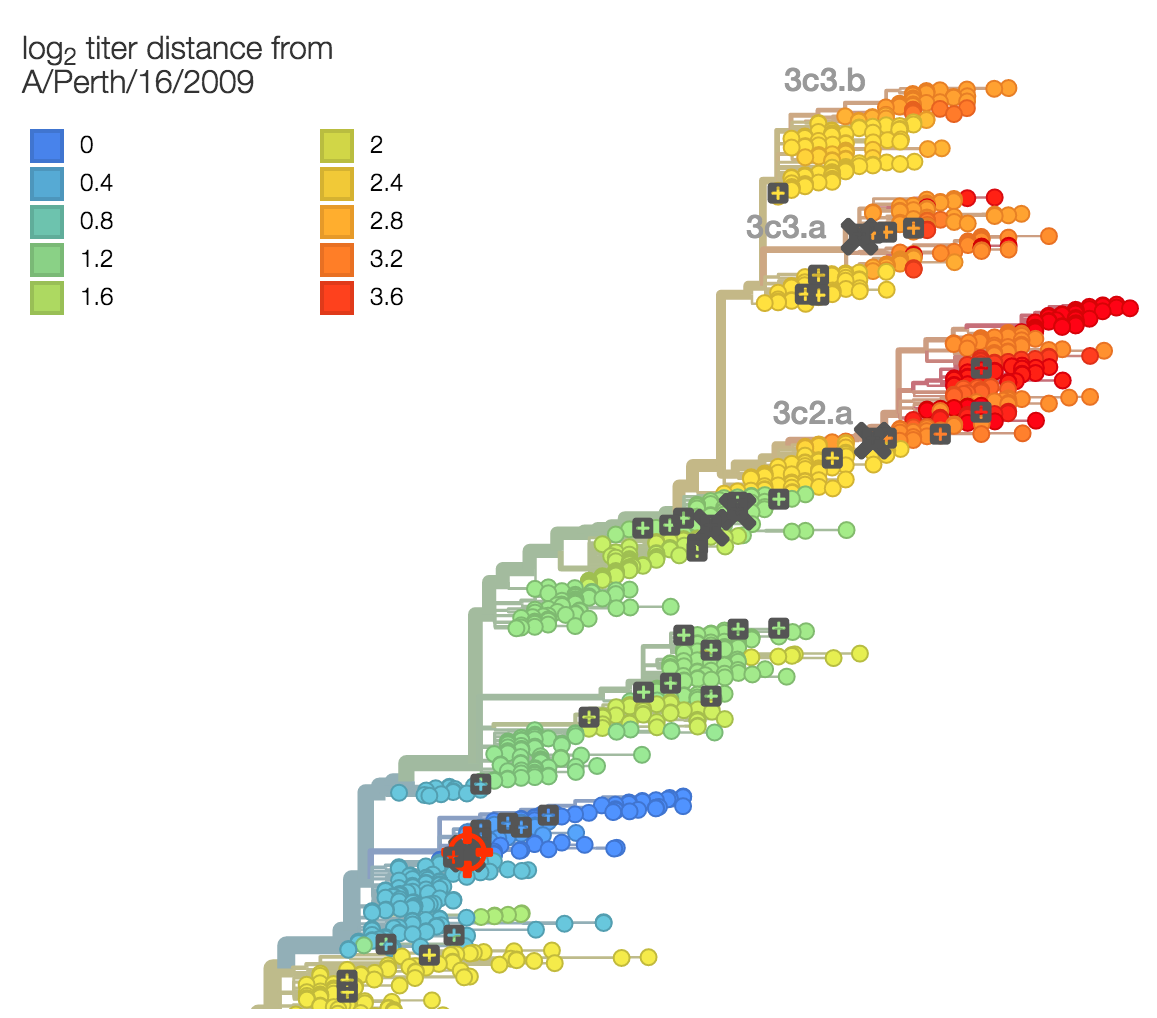
Identify phylogeny branches associated with drops in HI titer
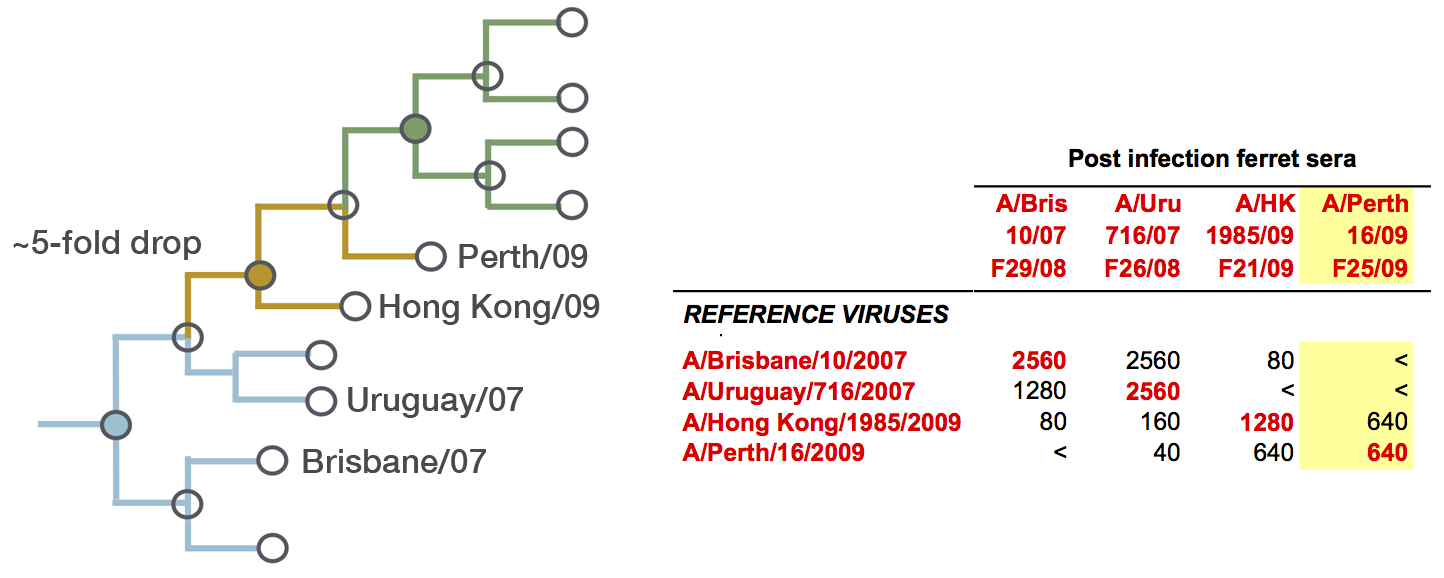
Model can be used to interpolate across tree and predict phenotype of untested viruses
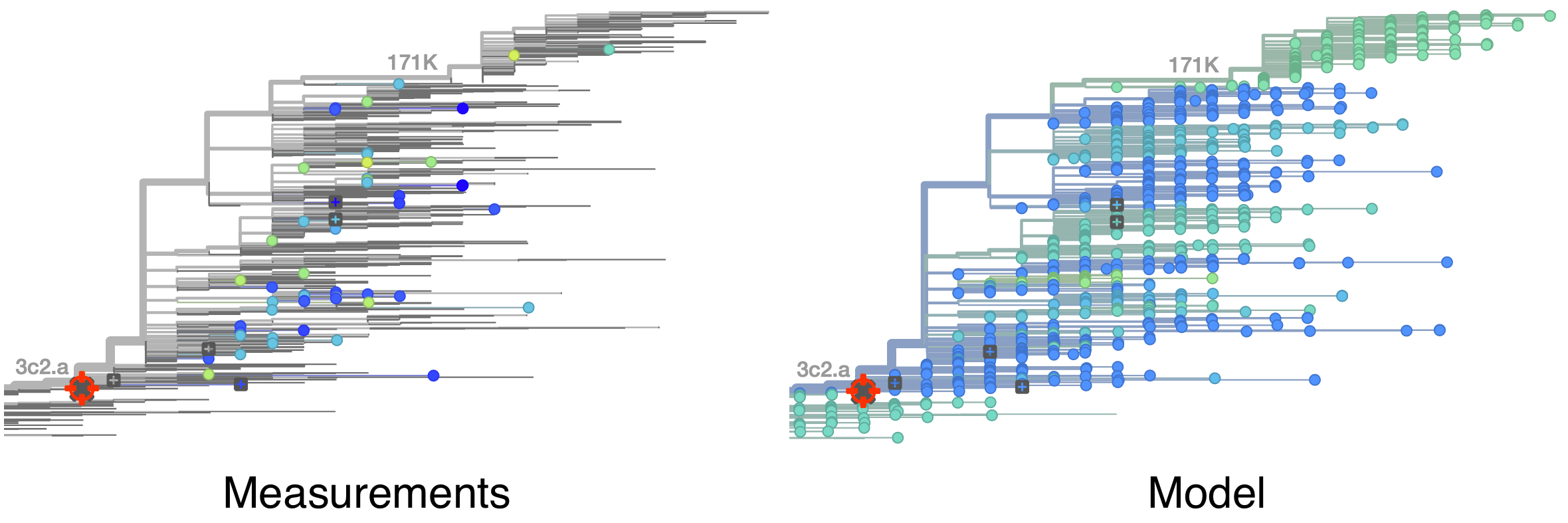
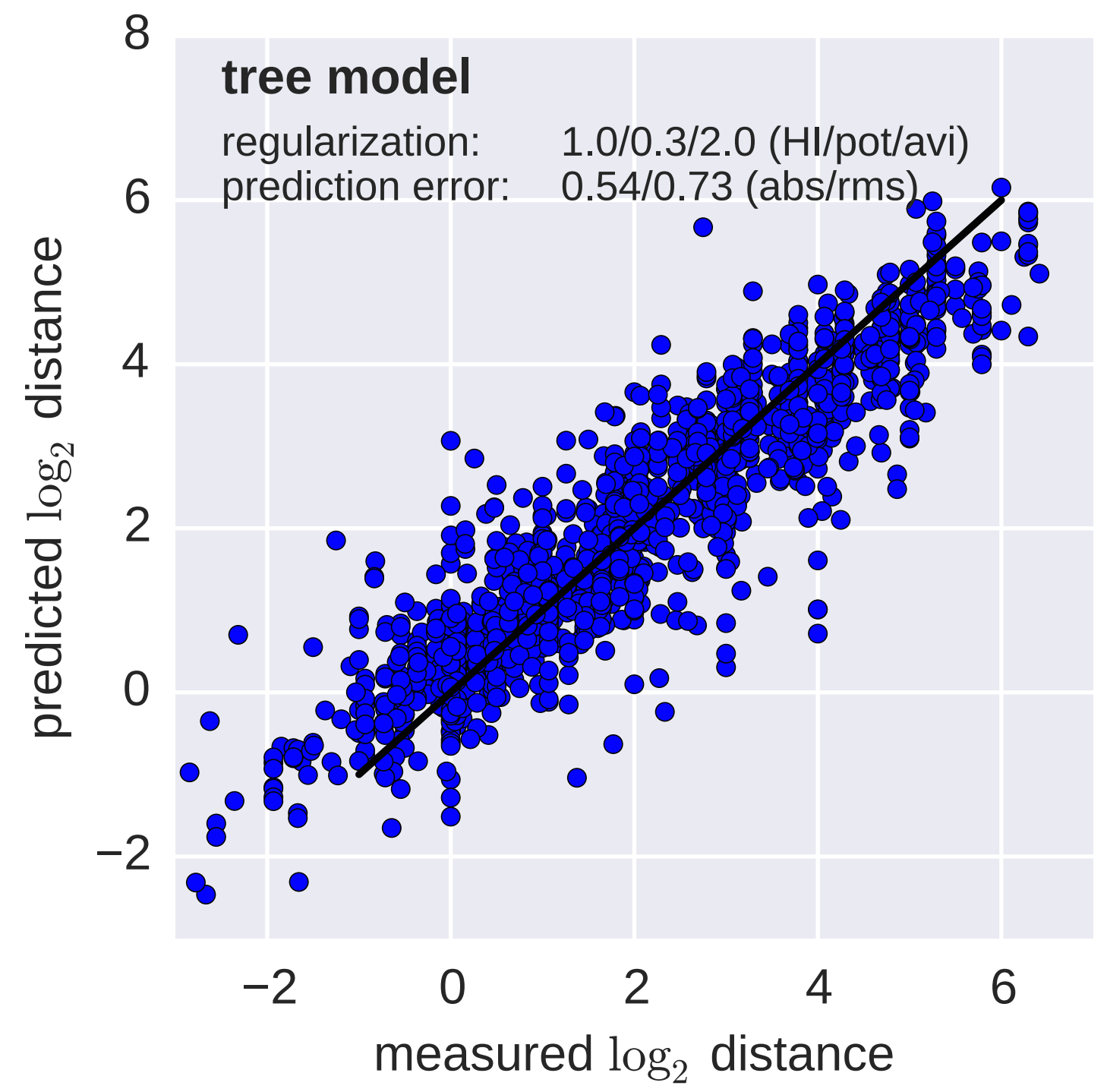
Model is highly predictive of missing titer values
Recent HI data from London WHO Collaborating Center
Up-to-date analysis at:
nextflu.org
Mapping genotype to phenotype changes
Proliferation of "epitope masks" in the literature
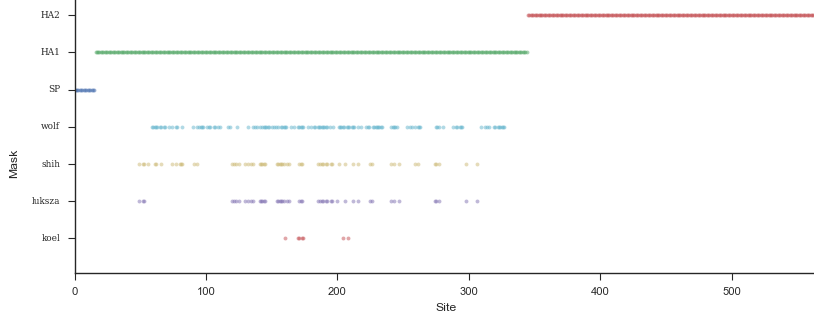
Inferences of branch-specific amino acid changes and branch-specific phenotype changes
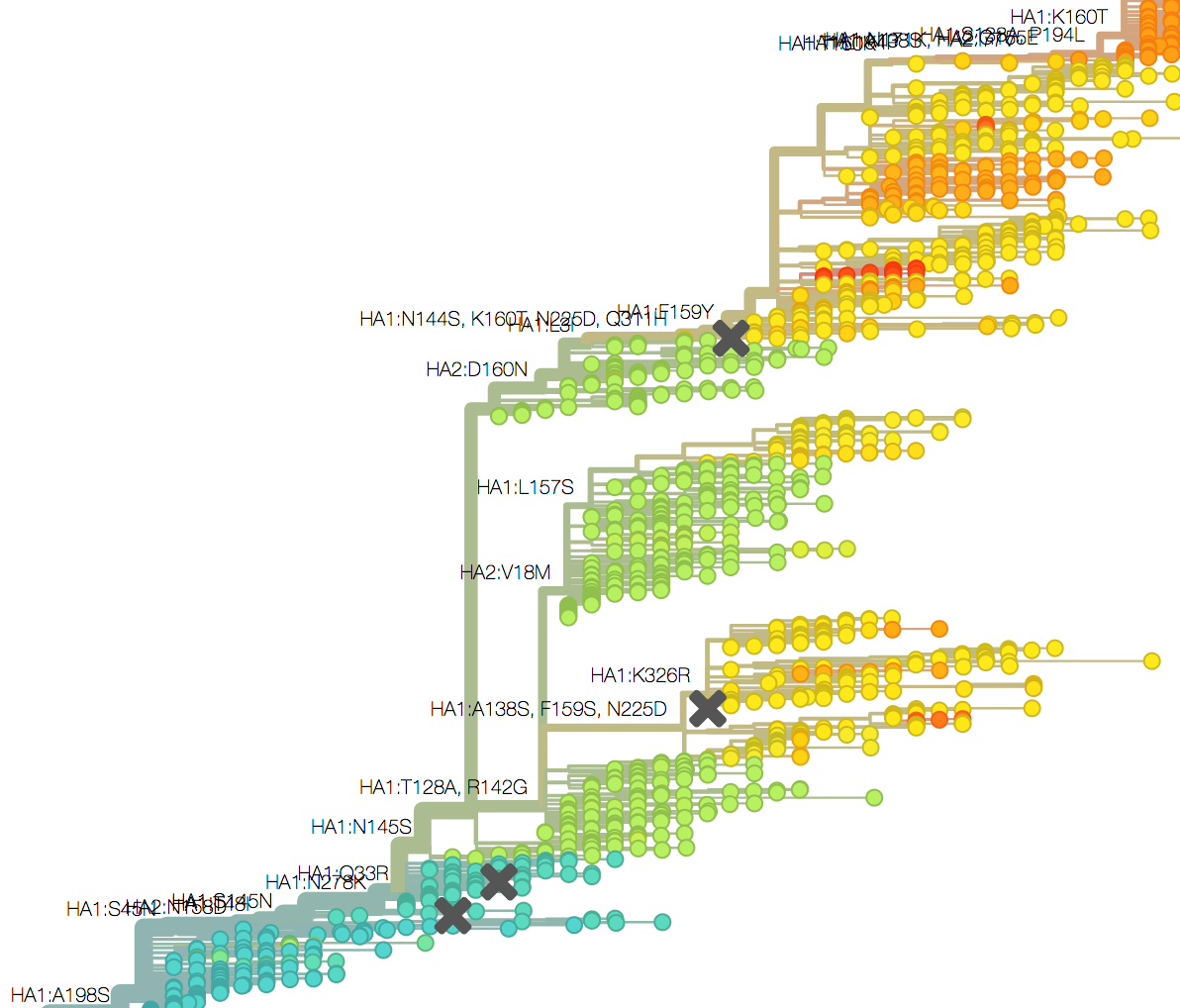
Can start to validate these masks
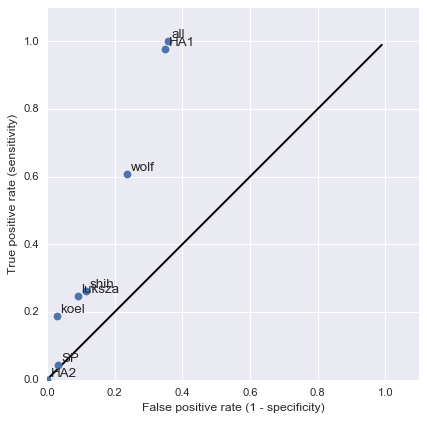
And infer masks that better map to HI data
Forecasting

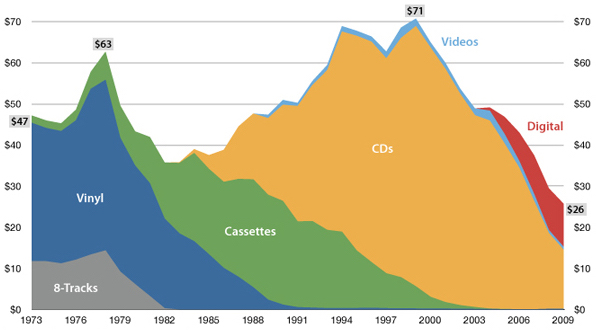
"The future is here, it's just not evenly
— William Gibson
distributed yet"
USA music industry, 2011 dollars per capita
Influenza population turnover
Vaccine strain selection timeline
Seek to explain change in clade frequencies over 1 year
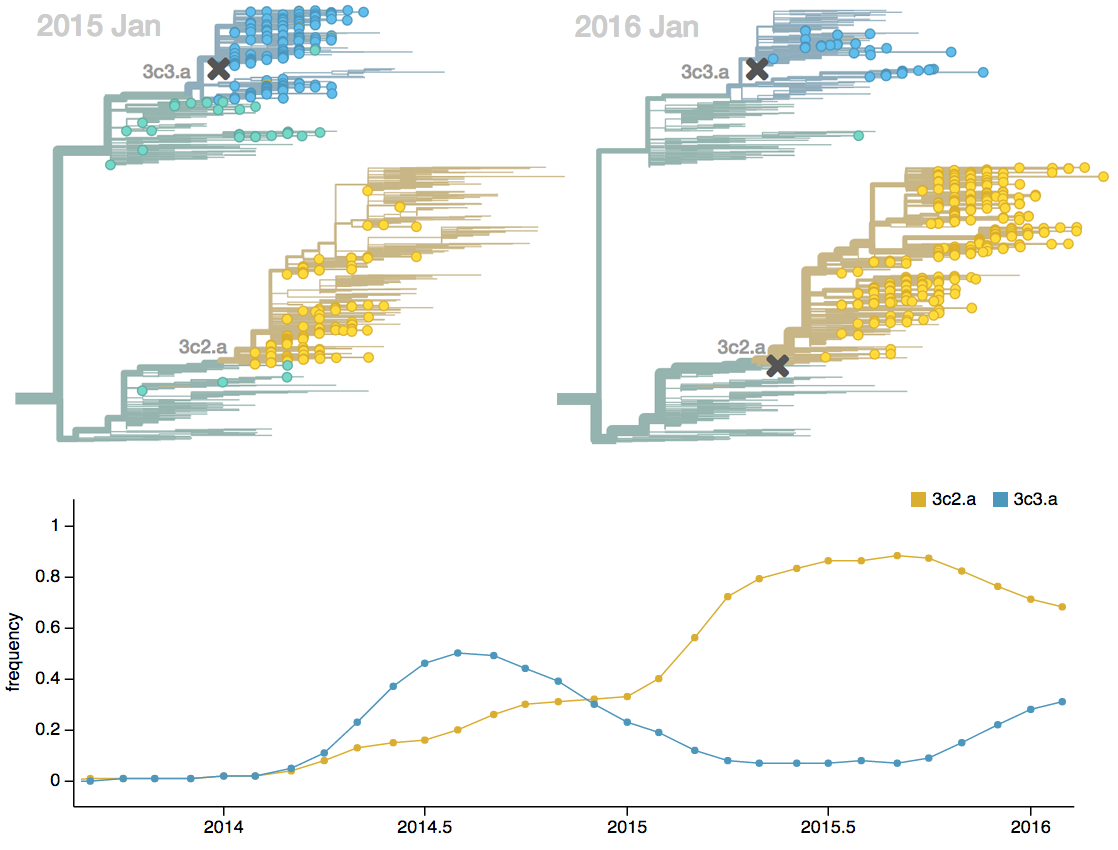
Fitness models can project clade frequencies
Clade frequencies $X$ derive from the fitnesses $f$ and frequencies $x$ of constituent viruses, such that
$$\hat{X}_v(t+\Delta t) = \sum_{i:v} x_i(t) \, \mathrm{exp}(f_i \, \Delta t)$$
This captures clonal interference between competing lineages
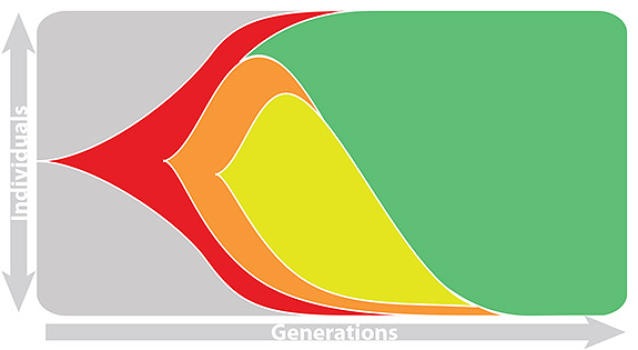
The question of forecasting becomes: how do we accurately estimate fitnesses of circulating viruses?
Fortunately, there's lots of training data and previously successful strains have had:
- Amino acid changes at epitope sites
- Antigenic novelty based on HI
- Rapid phylogenetic growth
Predictor: calculate HI drop from ancestor,
drifted clades have high fitness
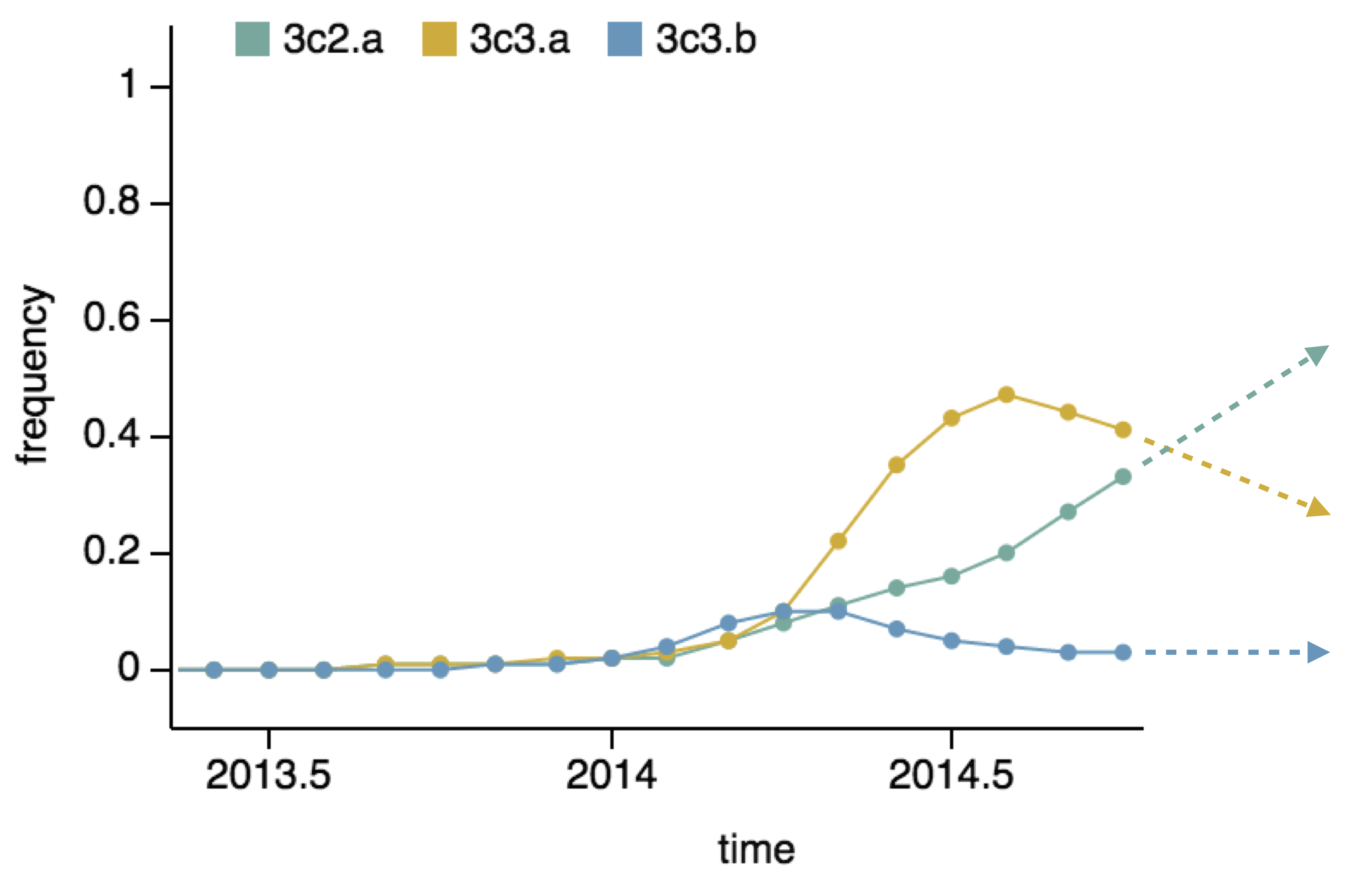
Predictor: project frequencies forward,
growing clades have high fitness
We predict fitness based on a simple formula
where the fitness $f$ of virus $i$ is estimated as
$$\hat{f}_i = \beta^\mathrm{HI} \, f_i^\mathrm{HI} + \beta^\mathrm{freq} \, f_i^\mathrm{freq}$$
where $f_i^\mathrm{HI}$ measures antigenic drift via HI and $f_i^\mathrm{freq}$ measures clade growth/decline
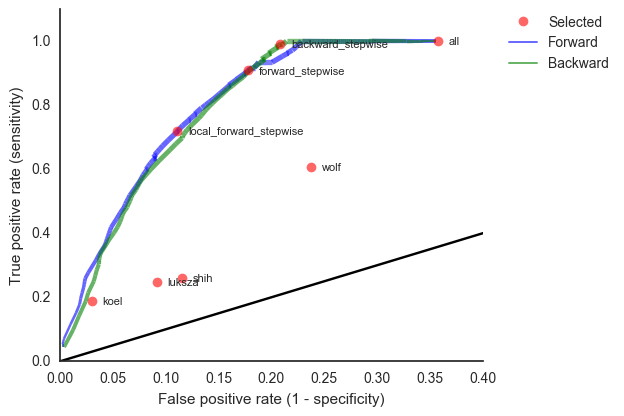
We learn coefficients and validate model based on previous 15 H3N2 seasons
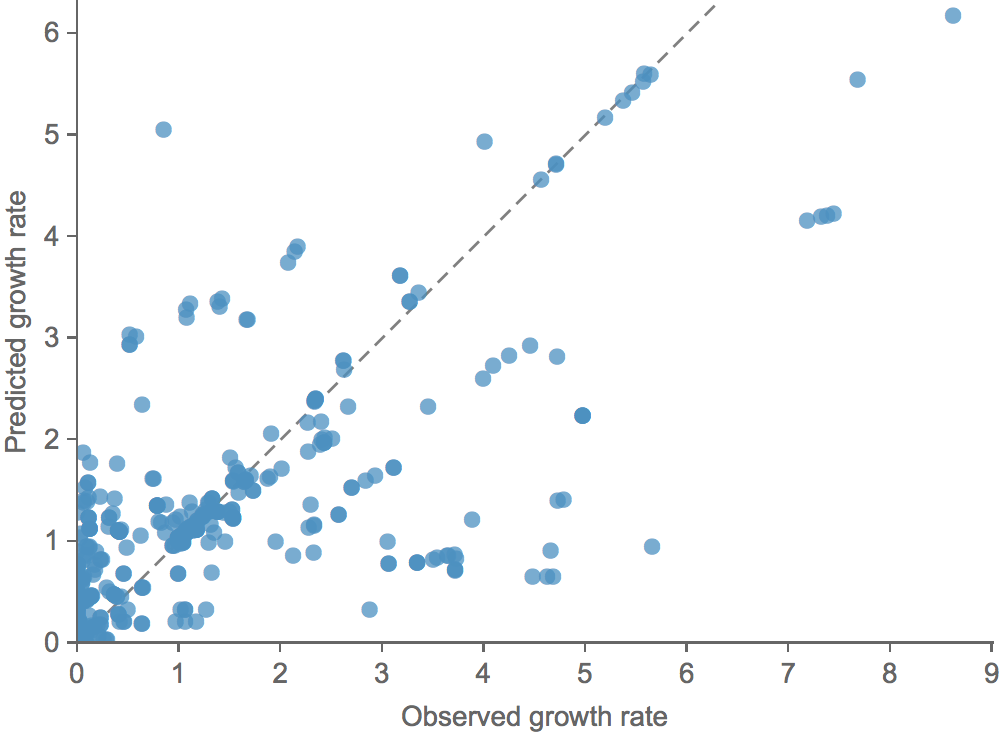
Clade growth rate is well predicted (ρ = 0.66)
Growth vs decline correct in 84% of cases
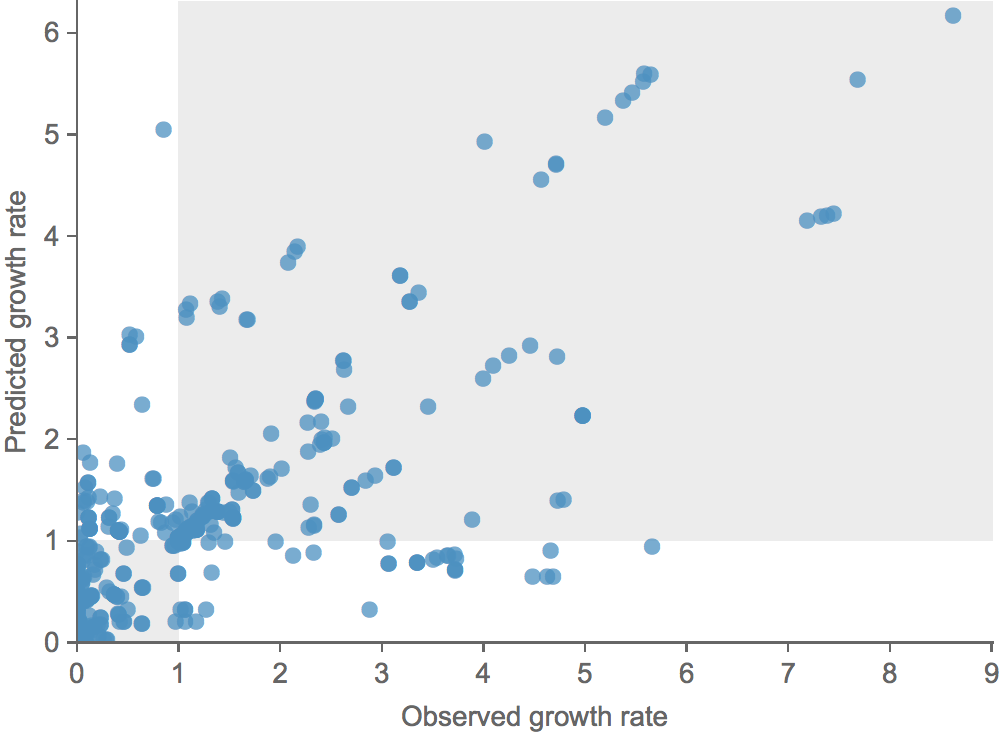
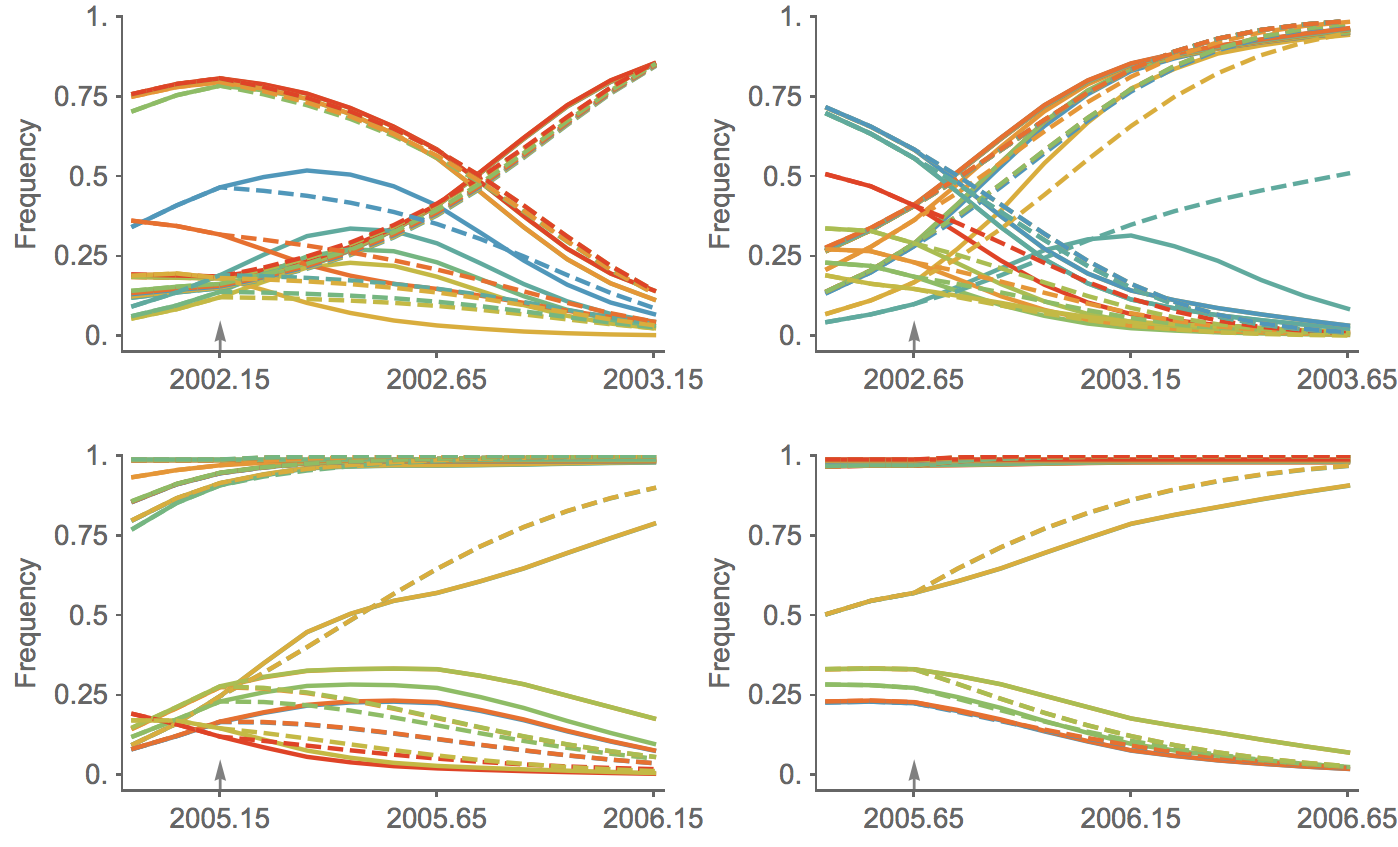
Trajectories show more detailed congruence
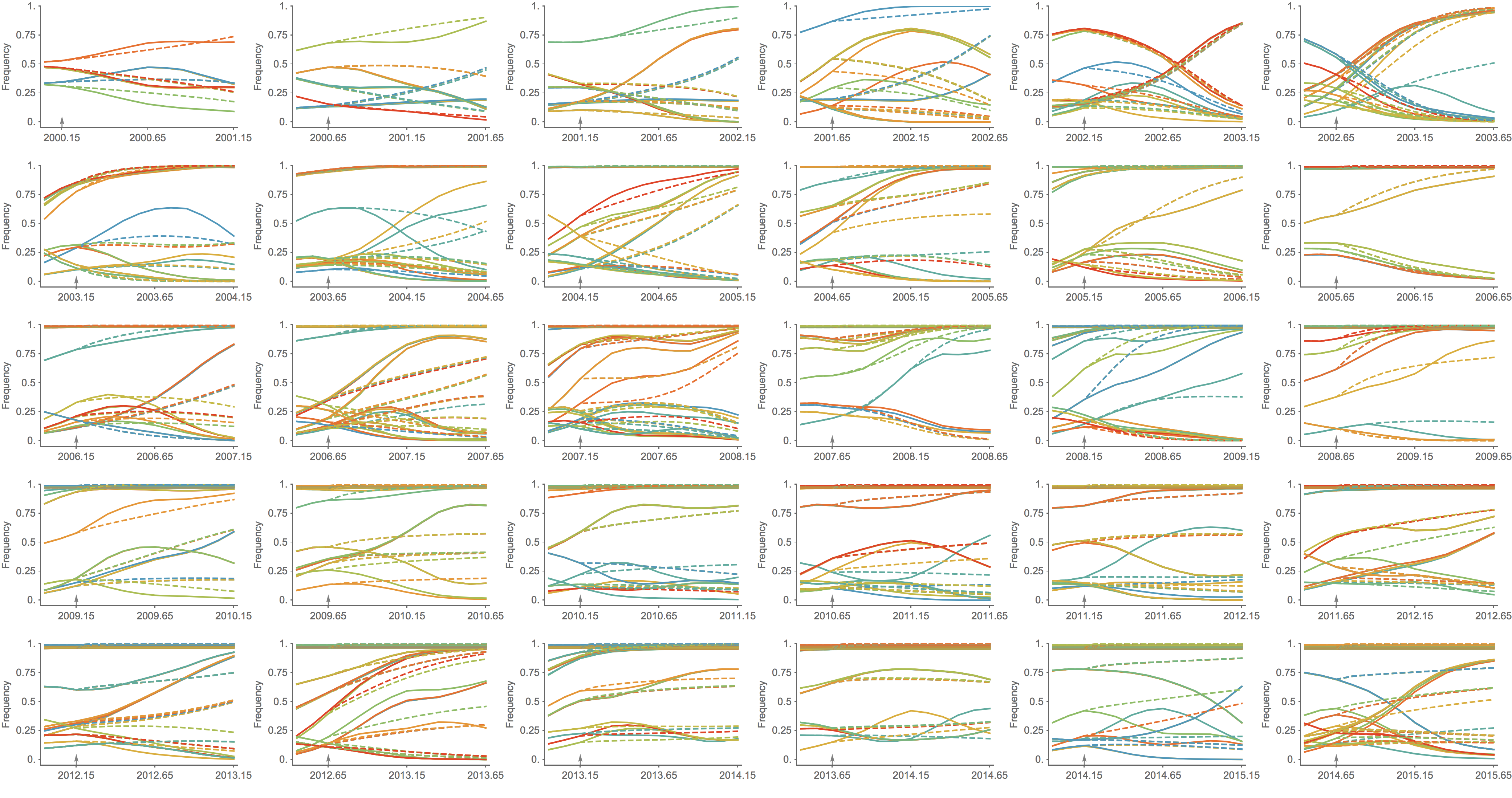
Trajectories show more detailed congruence
This model is similar in formulation and performance to Łuksza and Lässig
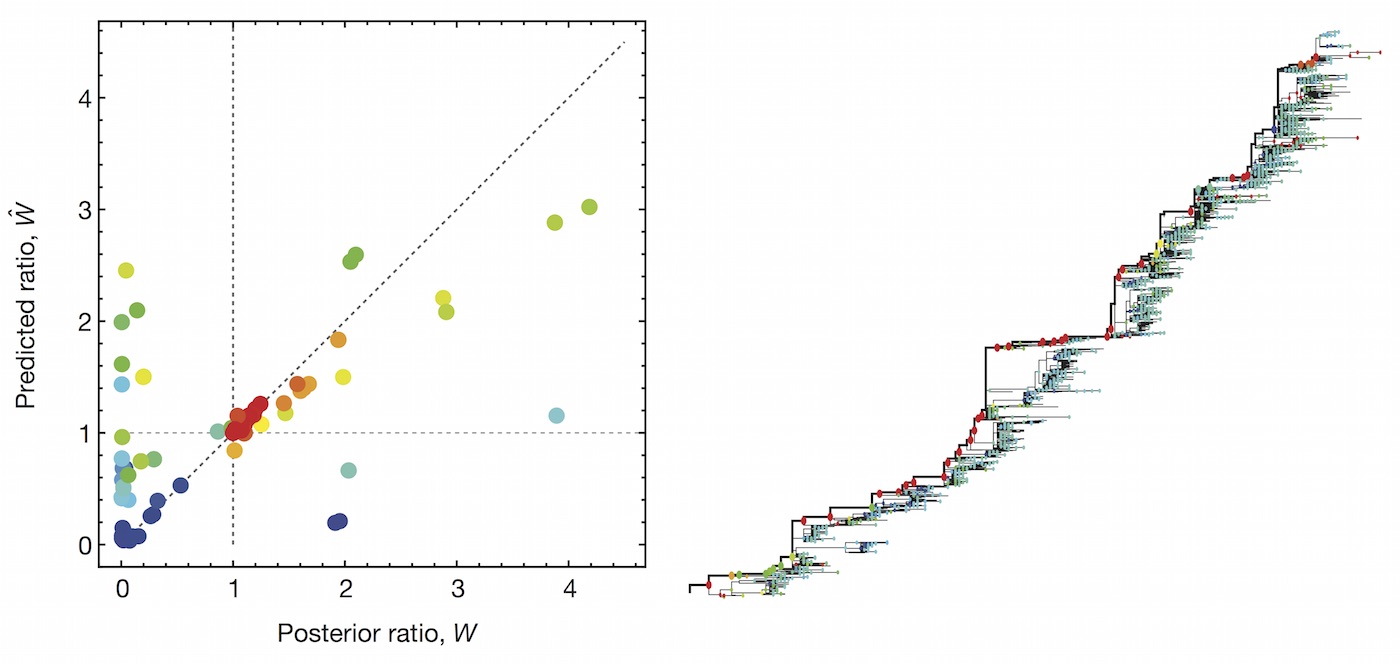
We find that HI is preferred over epitope mutations
| Model | Ep coefficient | HI coefficient | Freq error | Growth corr |
|---|---|---|---|---|
| Epitope only | 2.36 | -- | 0.10 | 0.57 |
| HI only | -- | 2.05 | 0.08 | 0.63 |
| Epitope + HI | -0.11 | 2.15 | 0.08 | 0.67 |
When does the forecast fail?
Emerging clades are difficult to forecast: little antigenic data and little evidence of "past performance"
Models work well for clades at >10%, but less well for clades <5%
New mutations difficult
Models can project forward from circulating strains, but cannot foresee the appearance of new mutations
Intrinsically limits the timescale of forecasting to ~1 year
Model is only as good as the data
Requires rapid shipping of samples, rapid sequencing and rapid antigenic characterization
Current situation
Looking forward
Further improvements to predictive modeling
- Extend to other seasonal viruses
- Forecast NA evolution
- Integrate neutralization (FRA) assay data
- Model effects of egg adaptation
- Incorporate an explicit geographic model
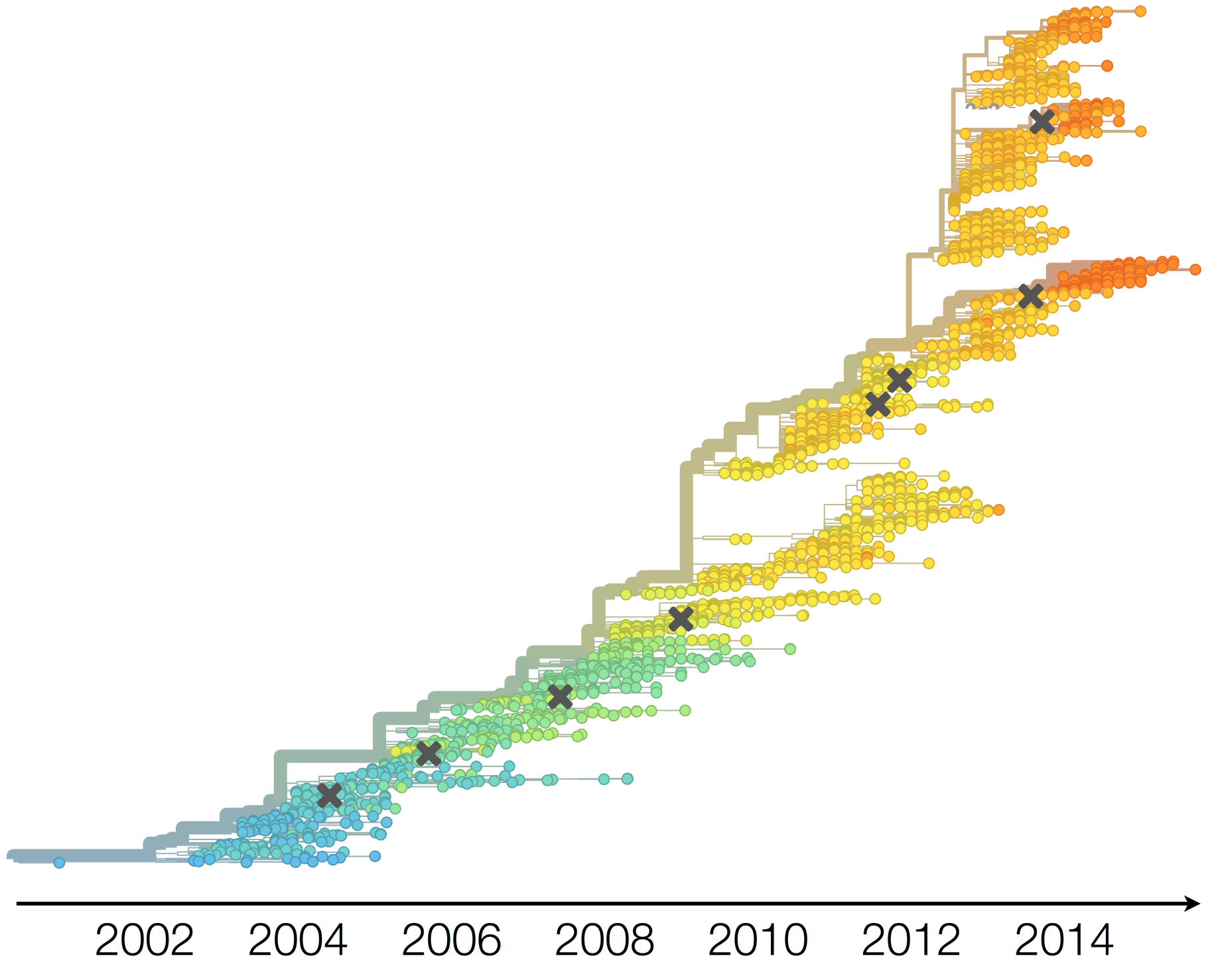
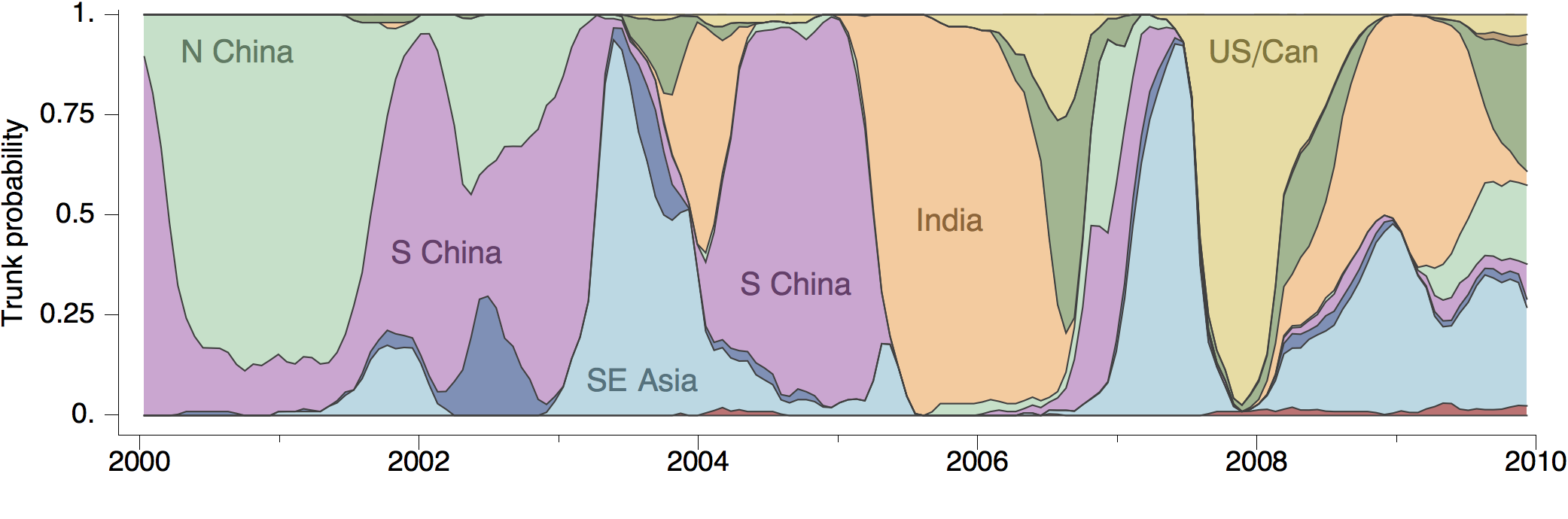
Phylogeny of H3 with geographic history
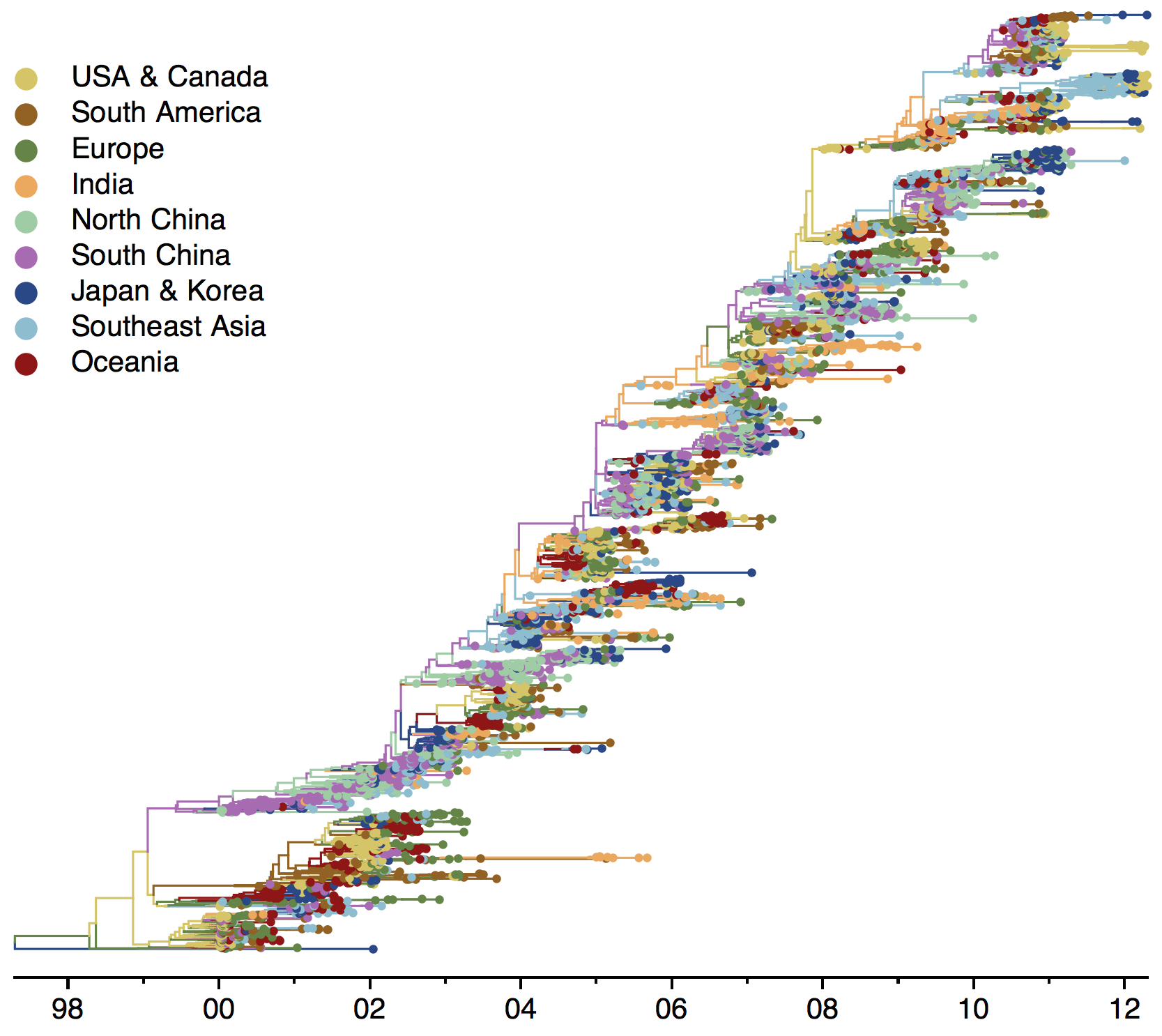
Geographic location of phylogeny trunk
Acknowledgements
Analysis: Richard Neher, Colin Russell, Charlton Callender, John Huddleston, Gytis Dudas, Colin Megill, Andrew Rambaut, Charles Cheung, Marc Suchard, Steven Riley, Philippe Lemey, Boris Shraiman, Marta Łuksza, Michael Lässig
GISRS/GISAID: Ian Barr, Shobha Broor, Mandeep Chadha, Nancy Cox, Rod Daniels, Becky Garten, Palani Gunasekaran, Aeron Hurt, Anne Kelso, Jackie Katz, Nicola Lewis, Xiyan Li, John McCauley, Takato Odagiri, Varsha Potdar, Yuelong Shu, Eugene Skepner, Masato Tashiro, Dayan Wang, Dave Wentworth, Xiyan Xu





