Real-time evolutionary forecasting
Trevor Bedford (@trvrb)
13 Jan 2016
Influenza VSDB Meeting
CDC
Slides at bedford.io/talks/
Influenza H3N2 vaccine updates
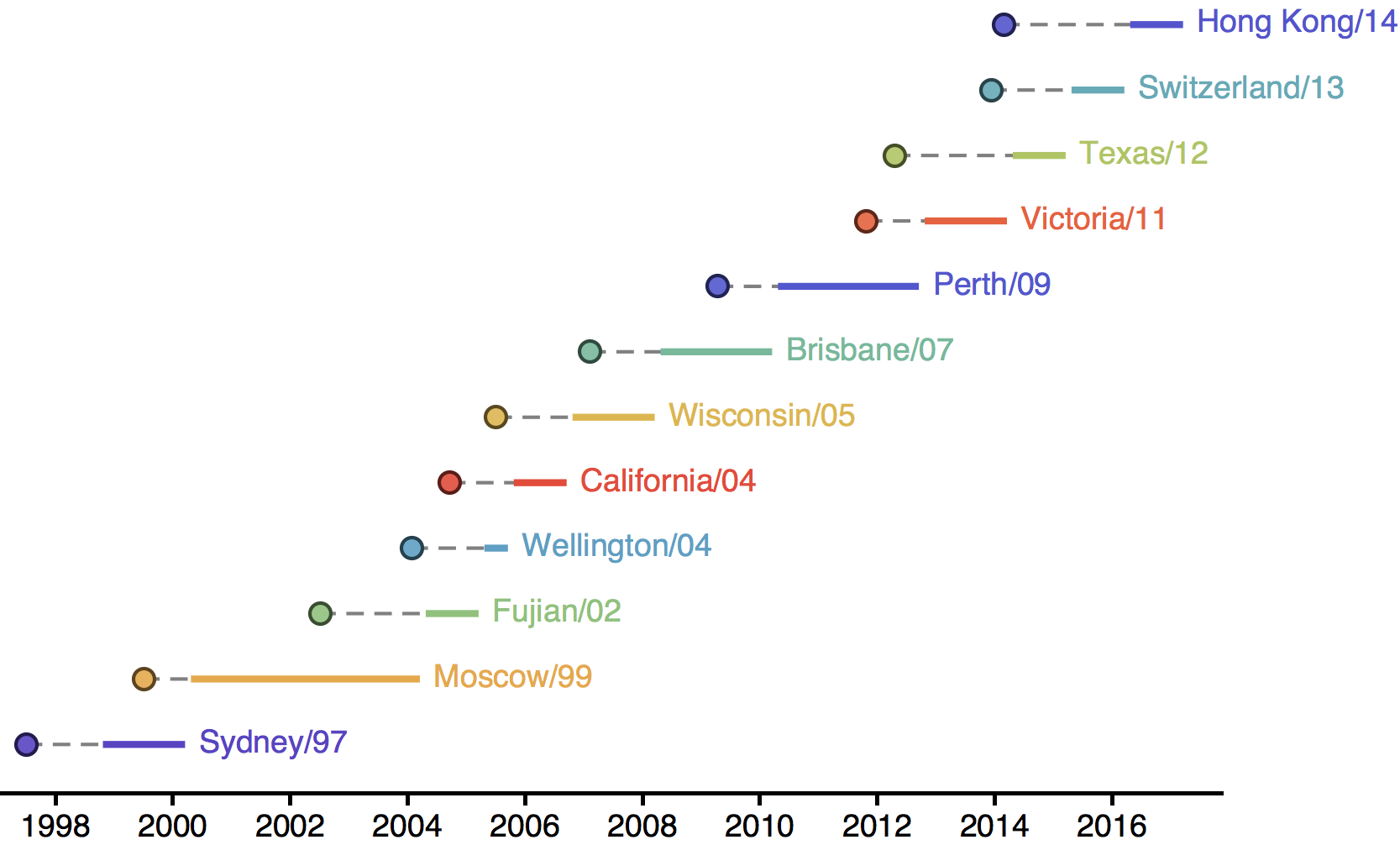
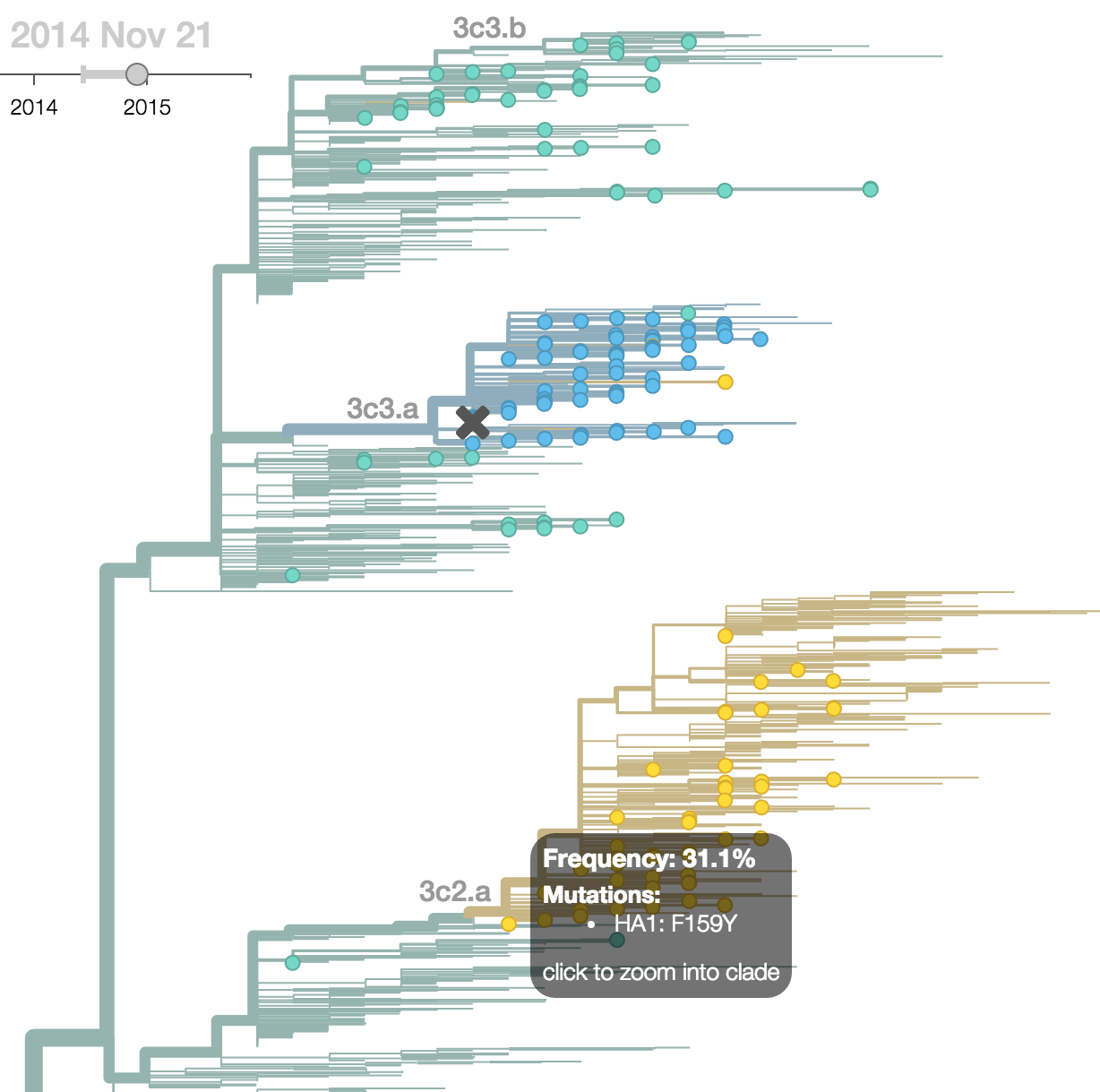
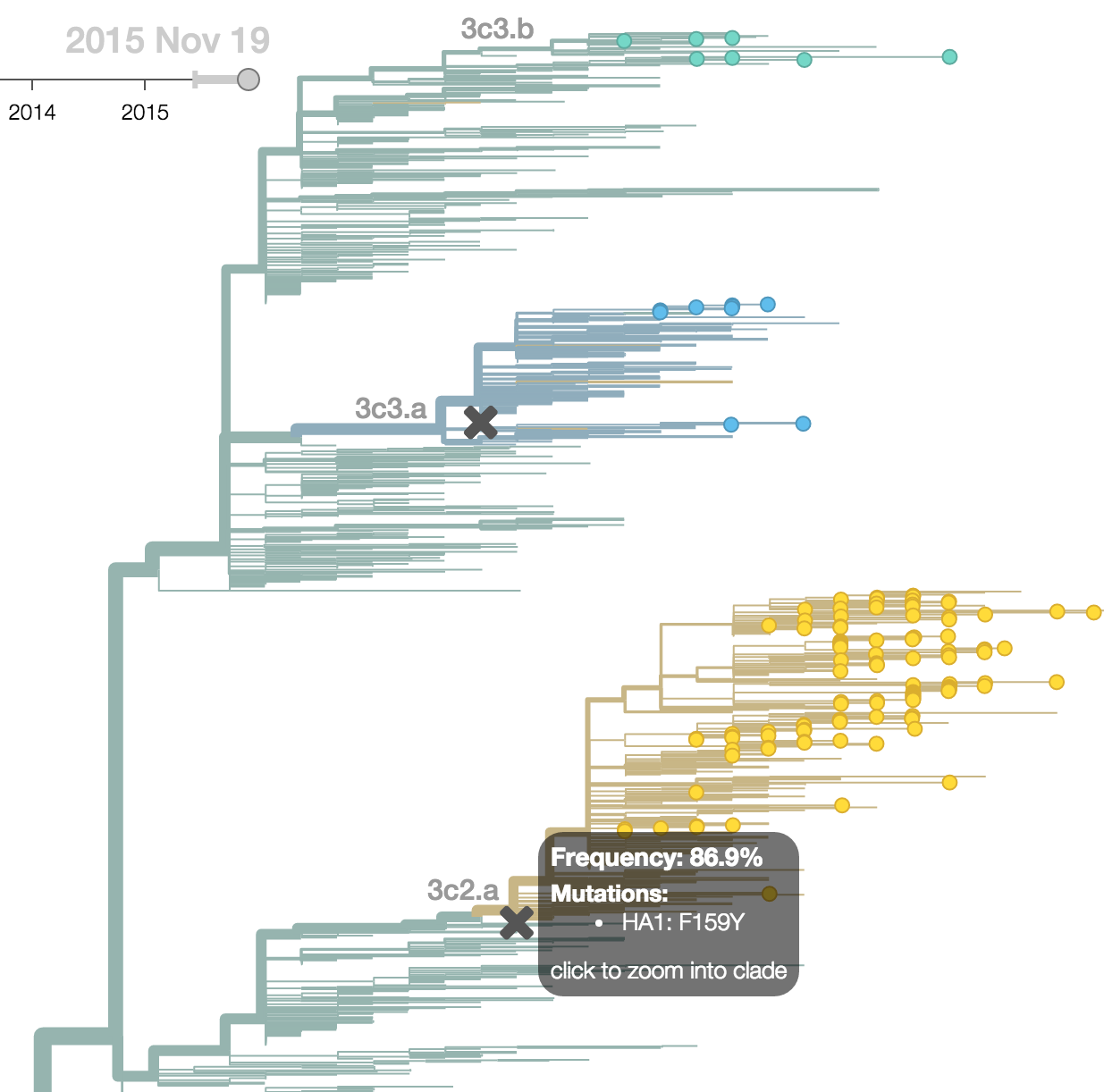
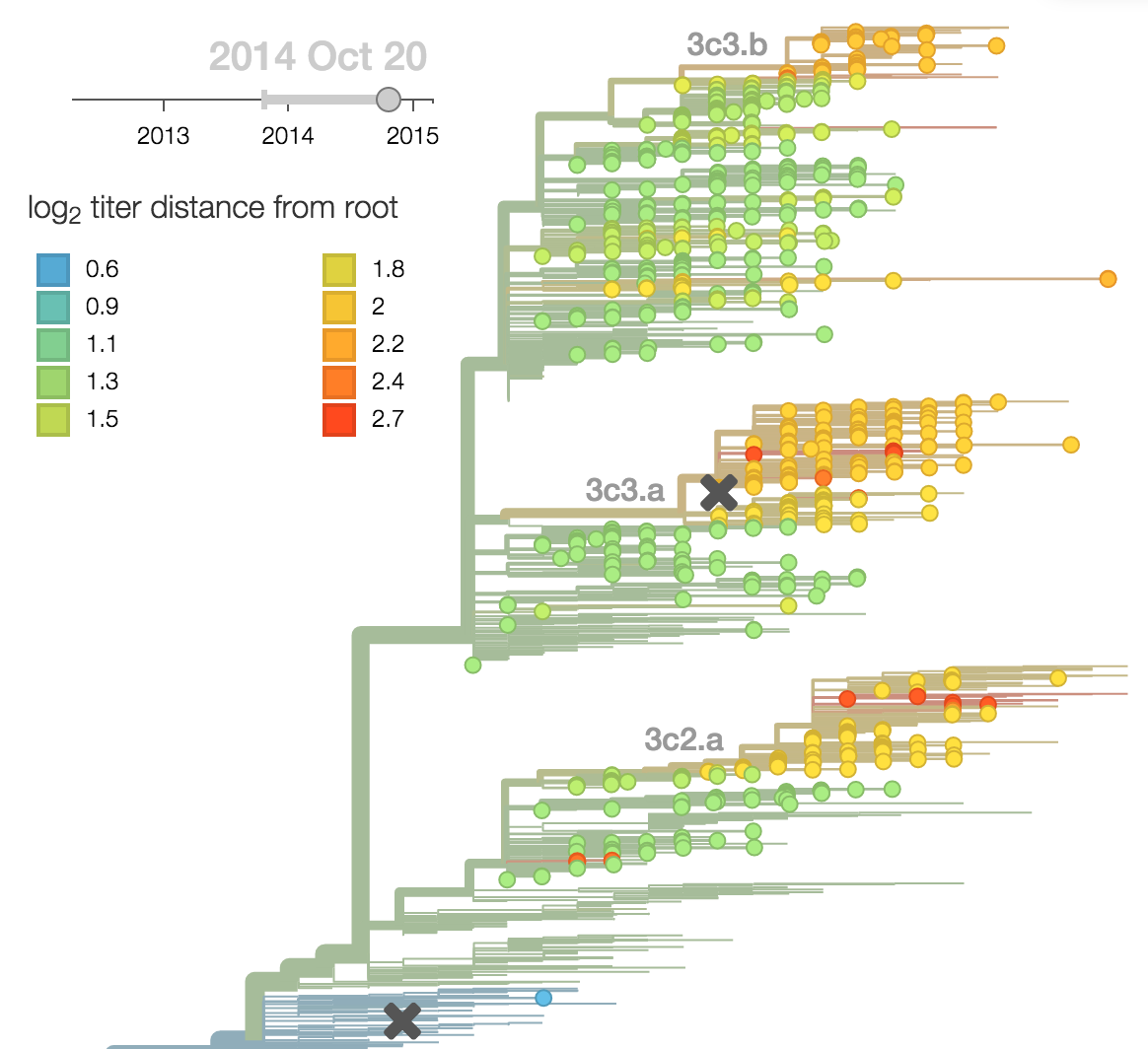
H3N2 phylogeny showing antigenic drift
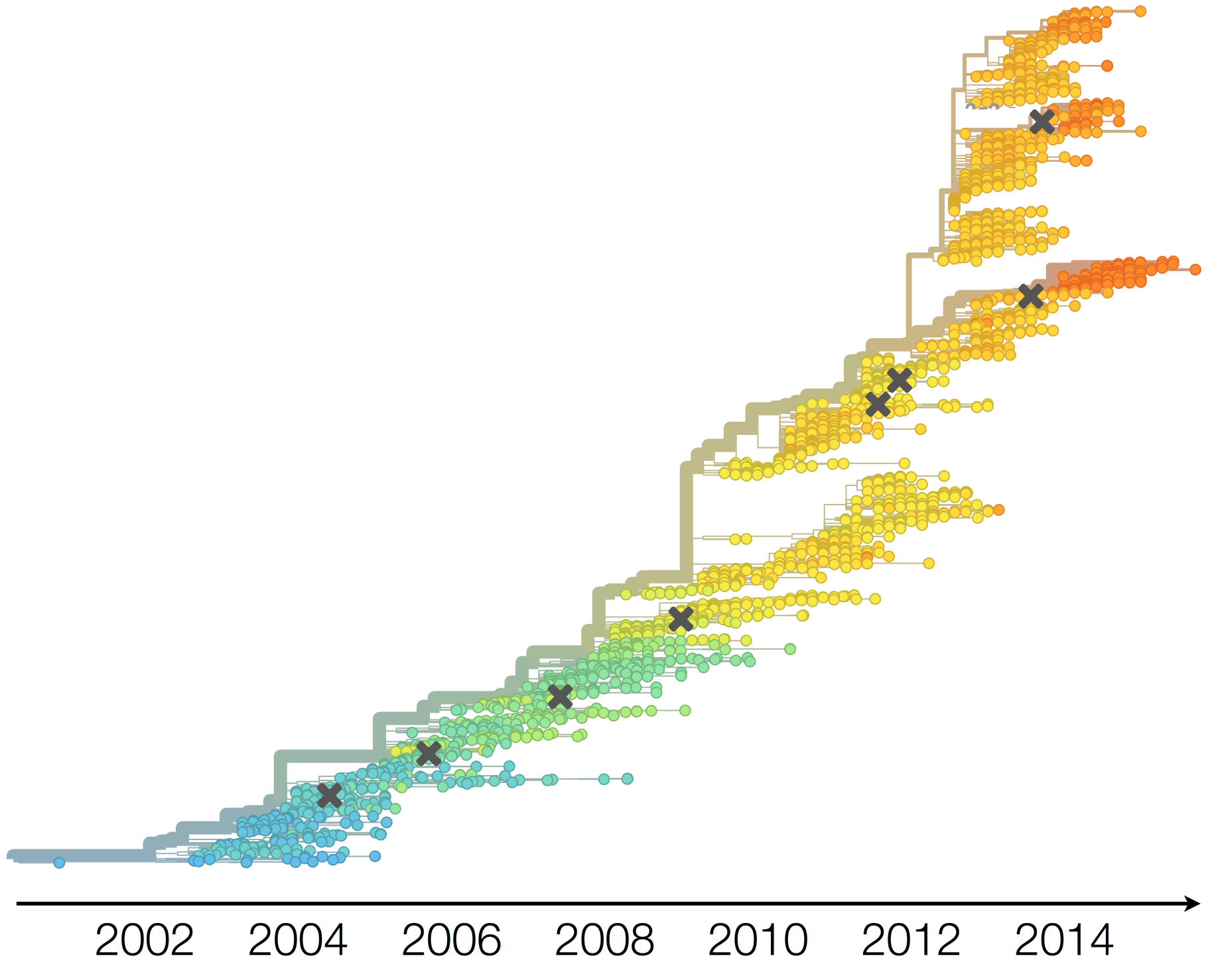
H3N2 phylogeny showing antigenic drift
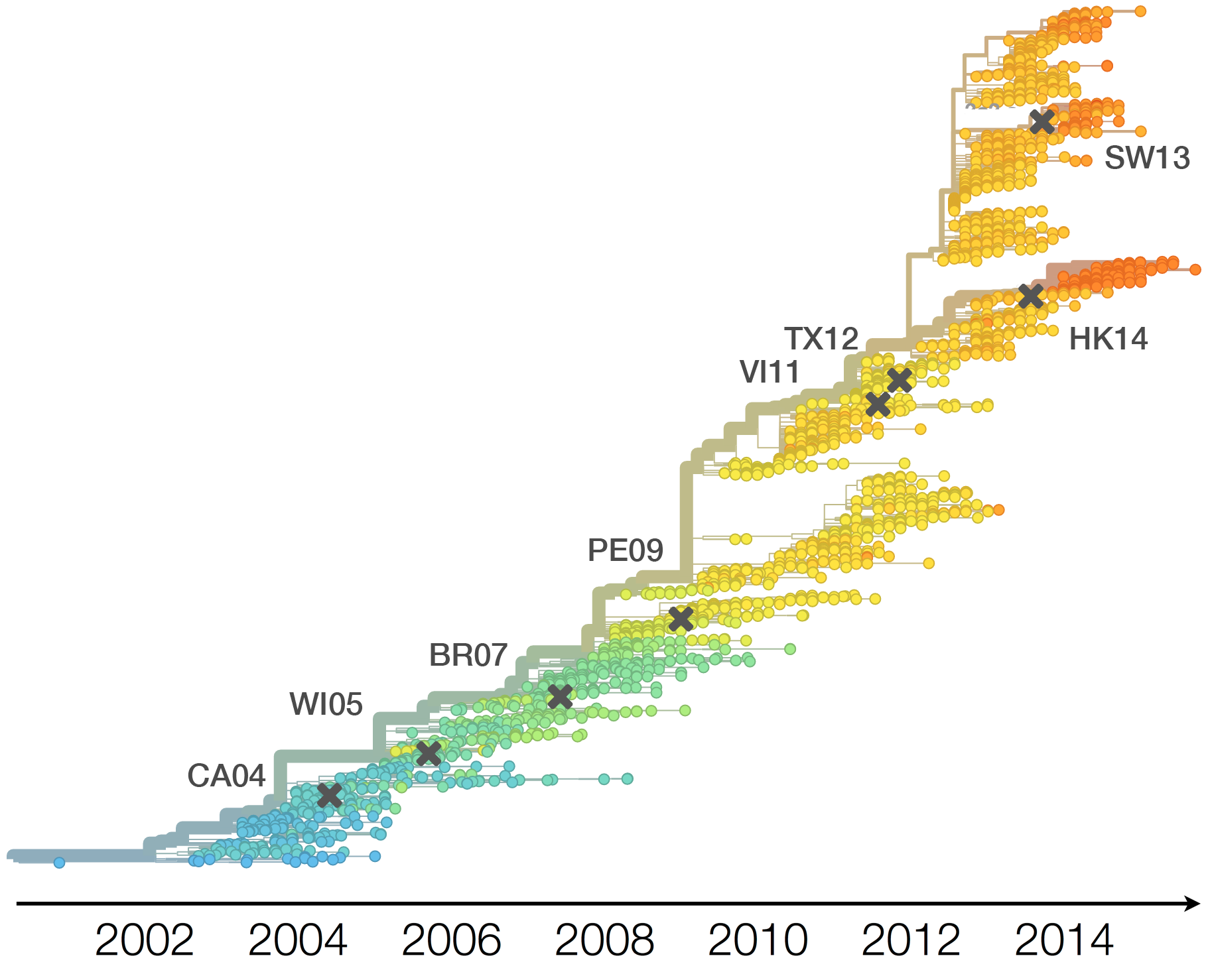
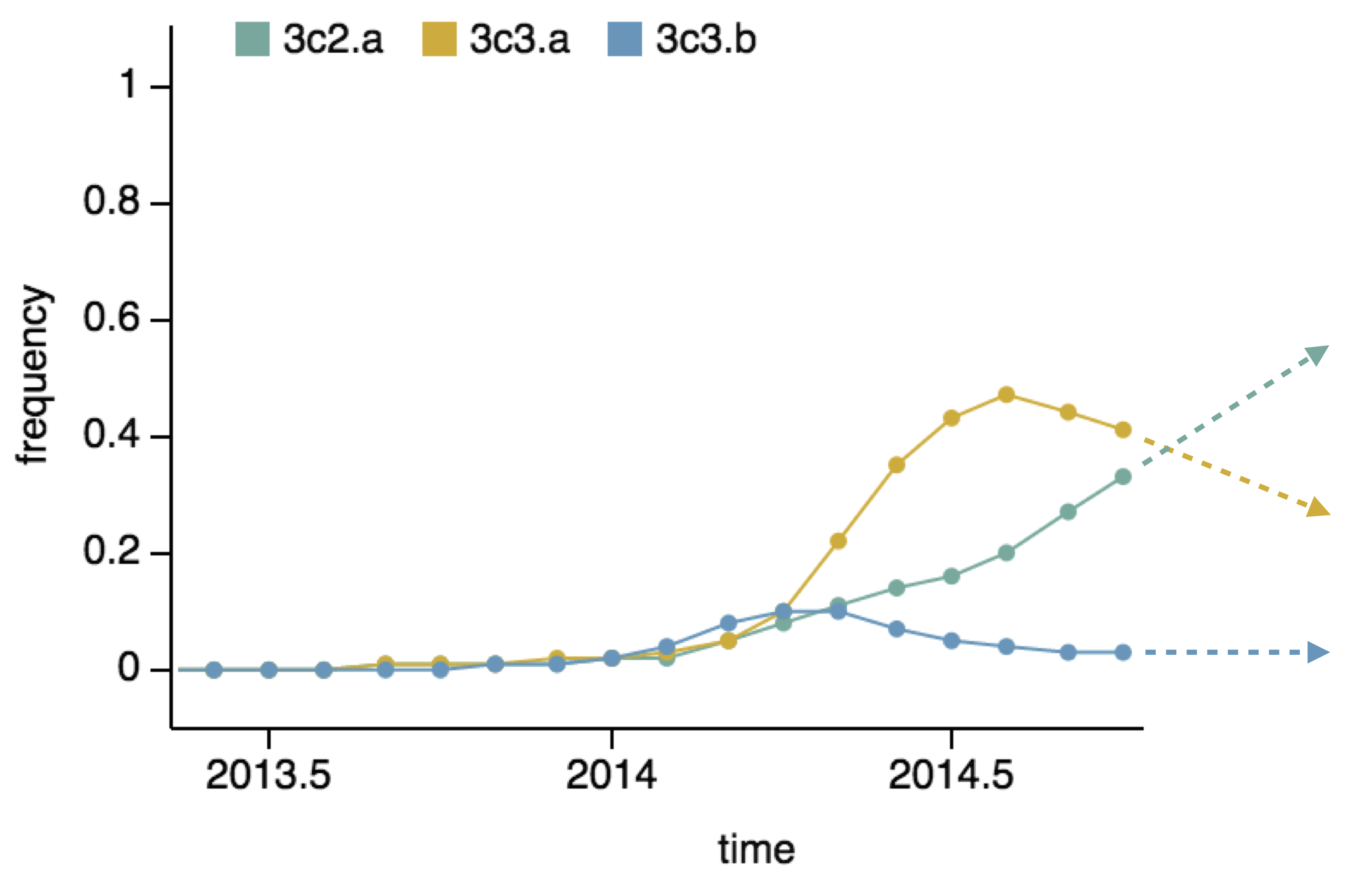
Drift variants rapidly take over the virus population
Timely surveillance and rapid analysis essential to understand ongoing influenza evolution
nextflu
Project to provide a real-time view of the evolving influenza population
All in collaboration with Richard Neher

nextflu pipeline
- Download all recent HA sequences from GISAID
- Filter to remove outliers
- Subsample across time and space
- Align sequences
- Build tree
- Estimate frequencies
- Export for visualization
Up-to-date analysis publicly available at:
nextflu.org
Antigenic evolution
Influenza hemagglutination inhibition (HI) assay
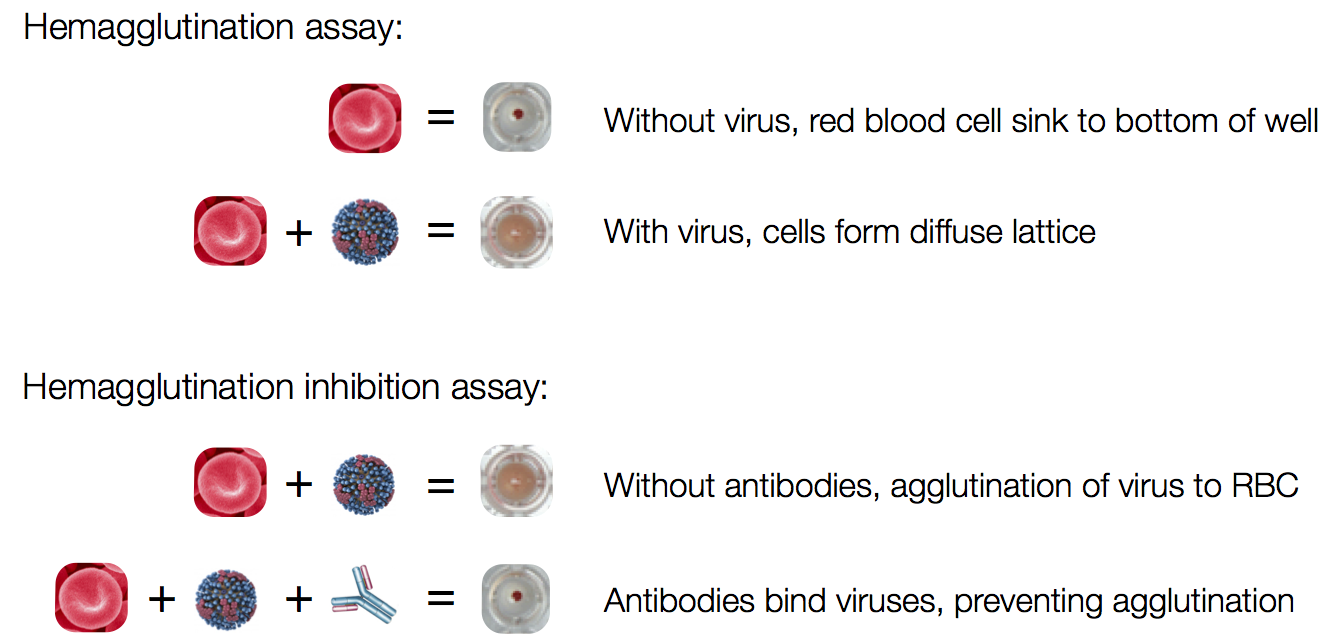
HI measures cross-reactivity across viruses
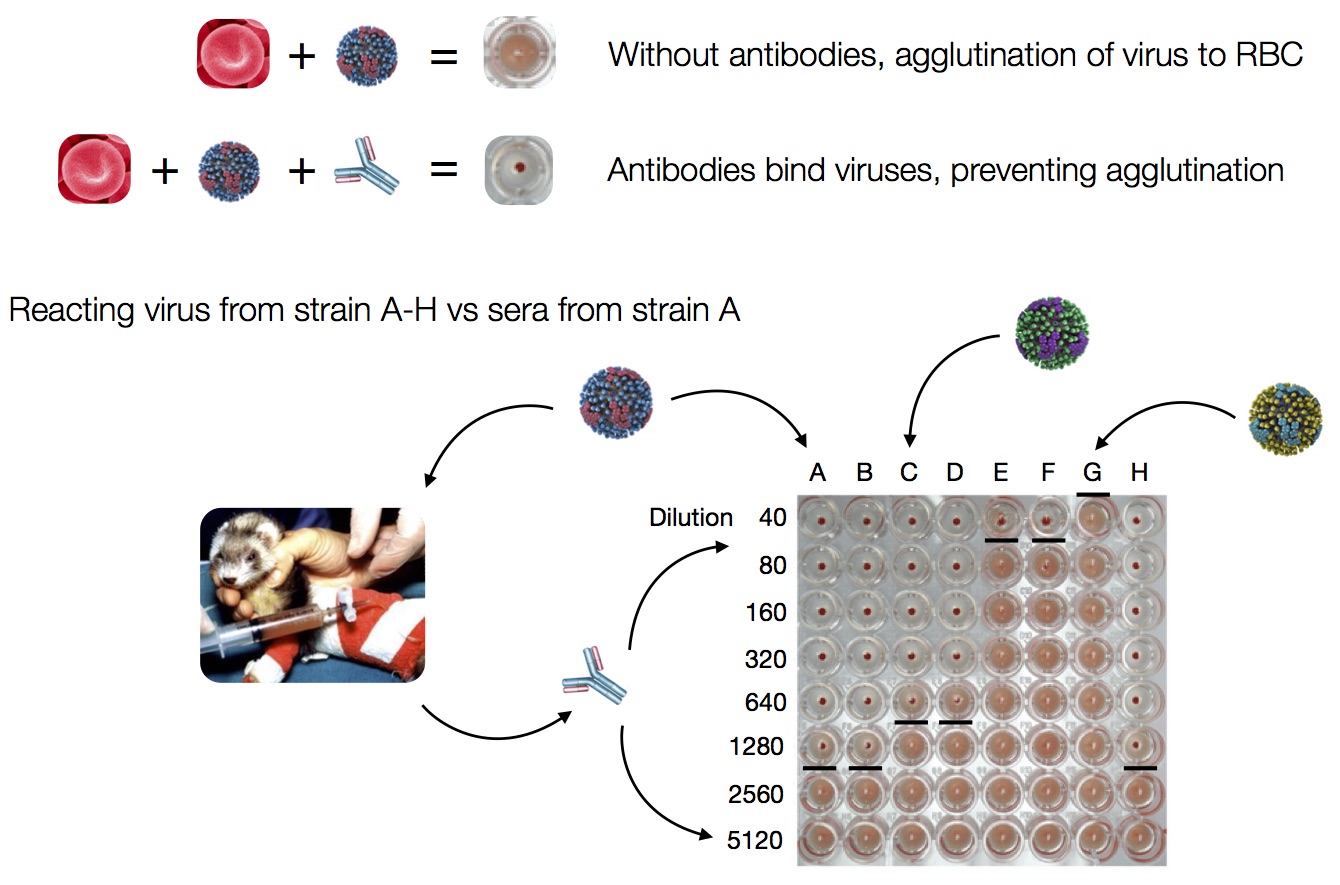
Data in the form of table of maximum inhibitory titers
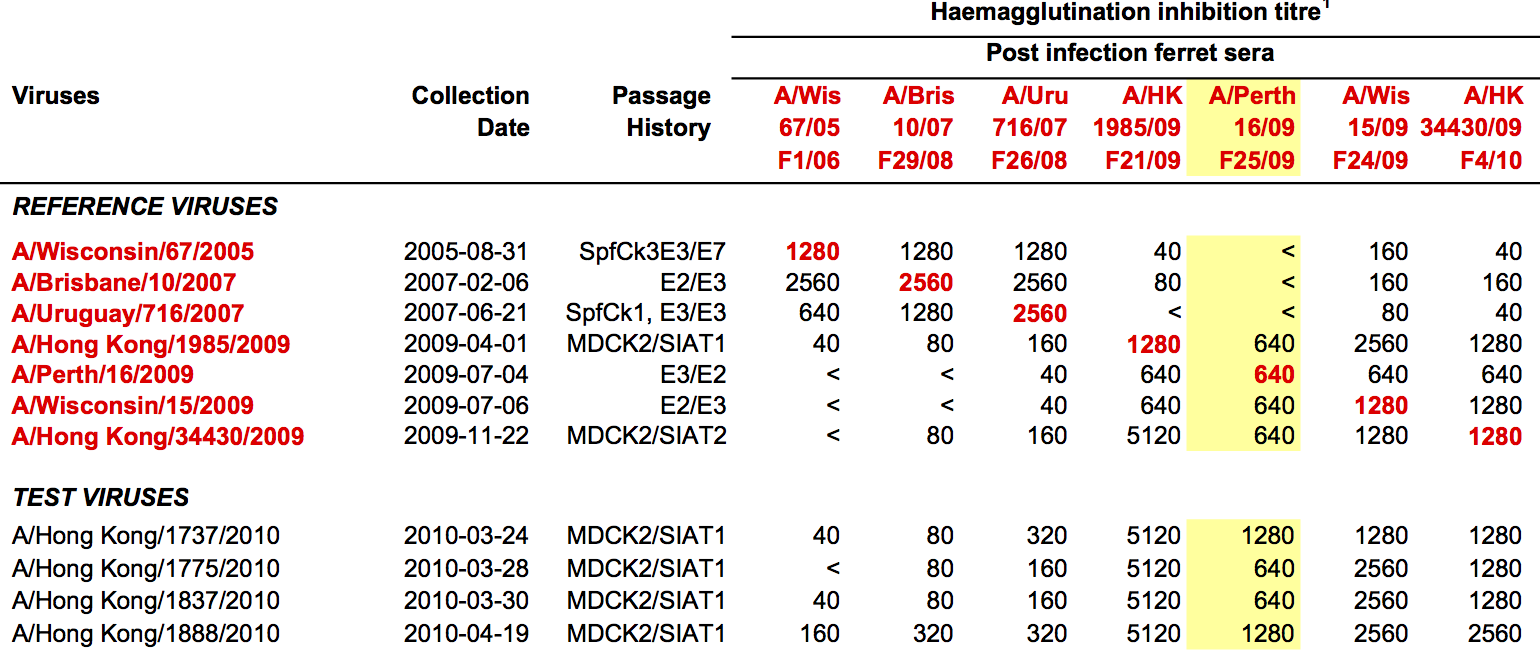
Fit HI titer drops to phylogeny branches
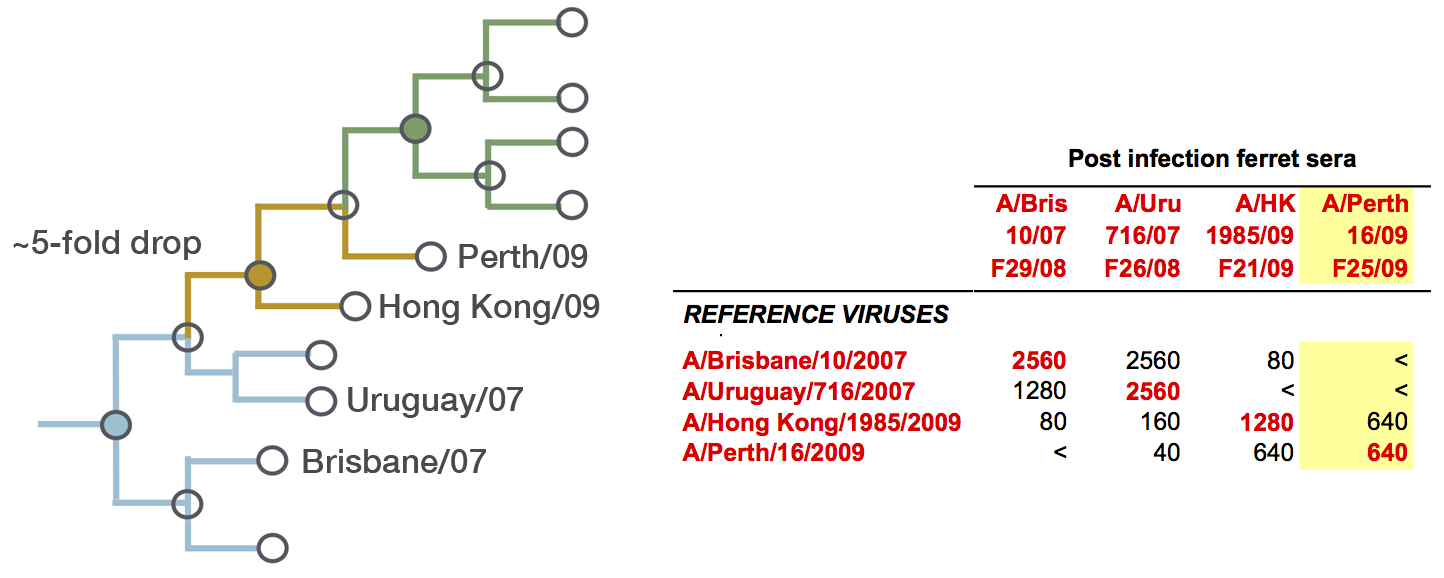
Model is highly predictive of missing titer values
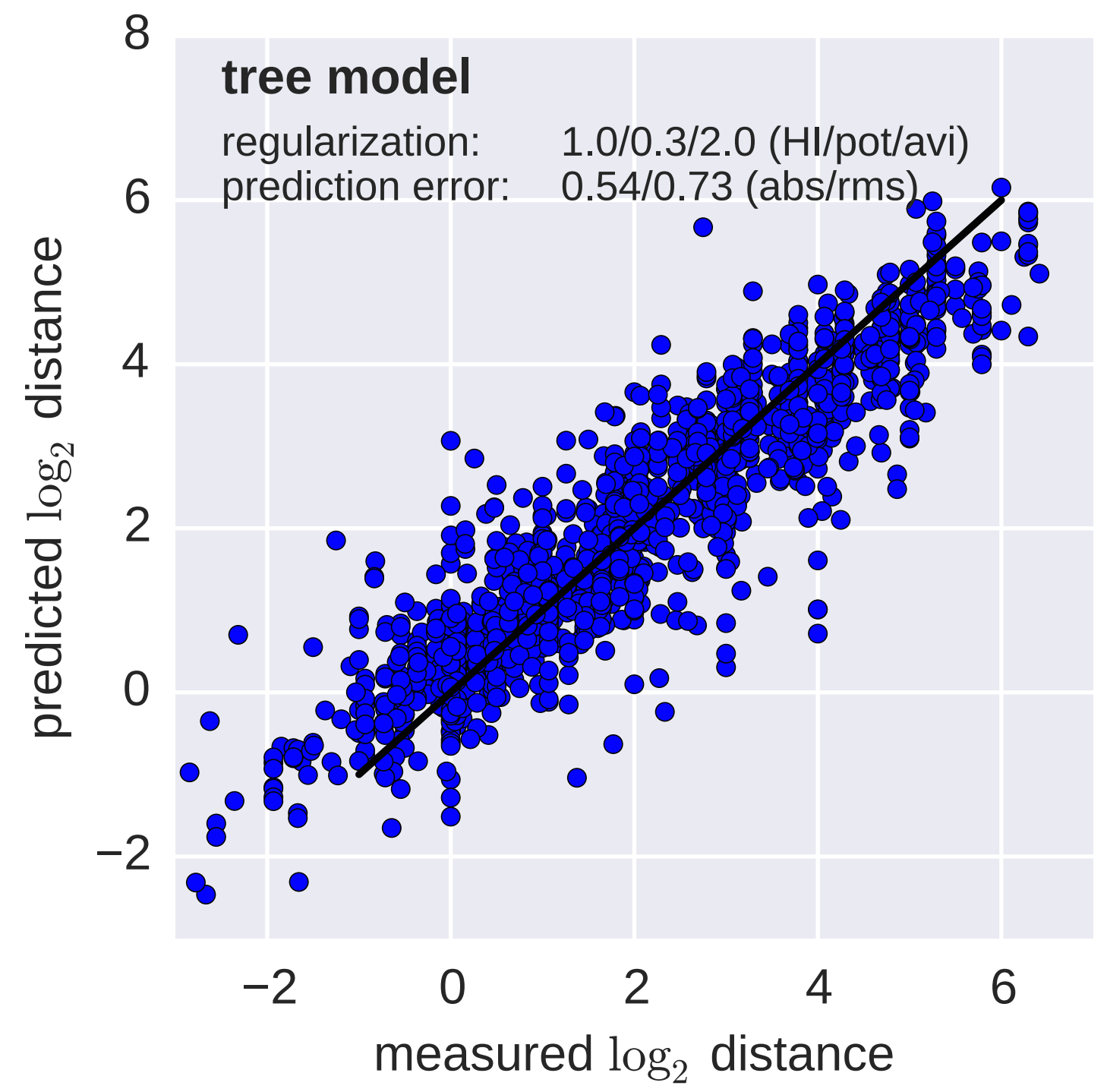
Broad patterns agree with cartographic analyses
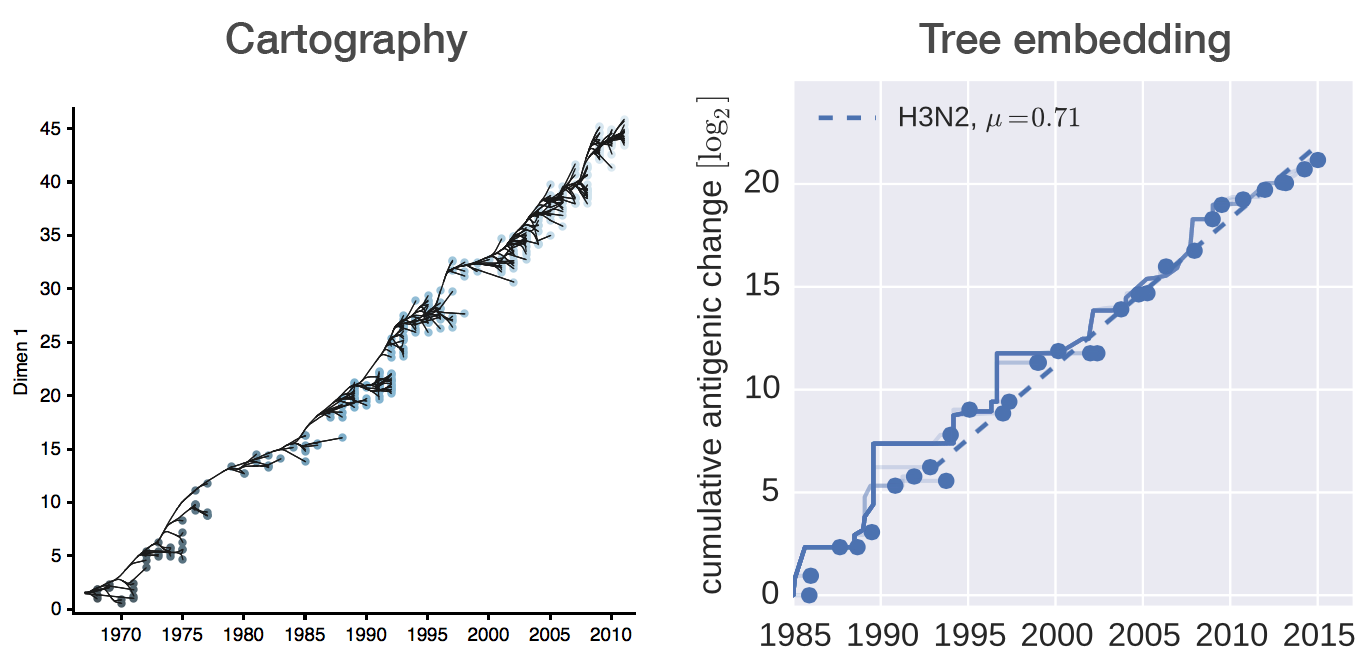
Recent HI data from WHO CC London annual and interim reports
Up-to-date analysis at:
nextflu.org
Forecasting
The future is here, it's just not evenly distributed yet
— William Gibson
USA music industry, 2011 dollars per capita
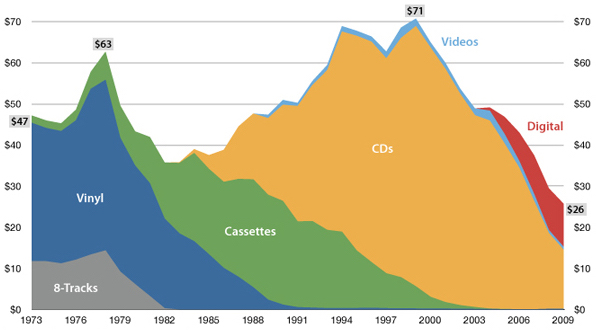
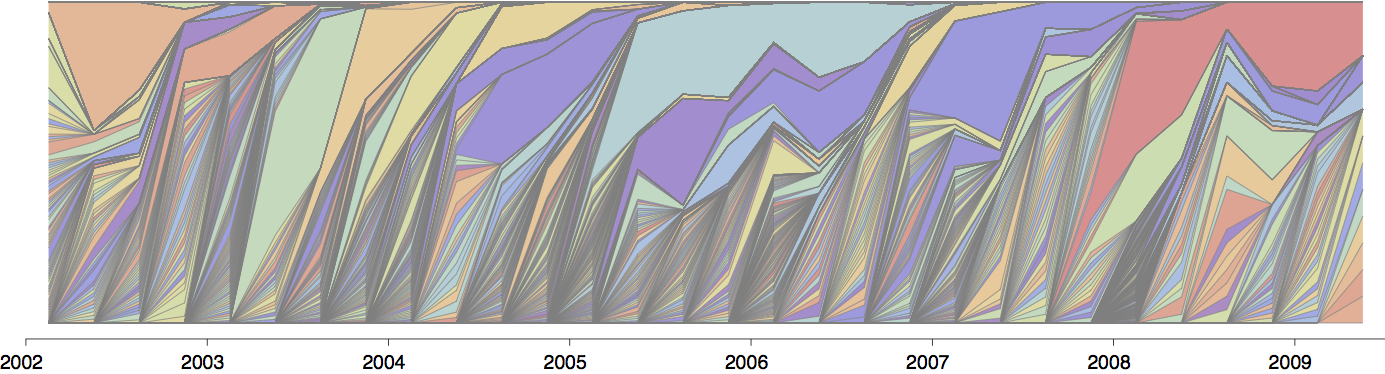
Influenza population turnover
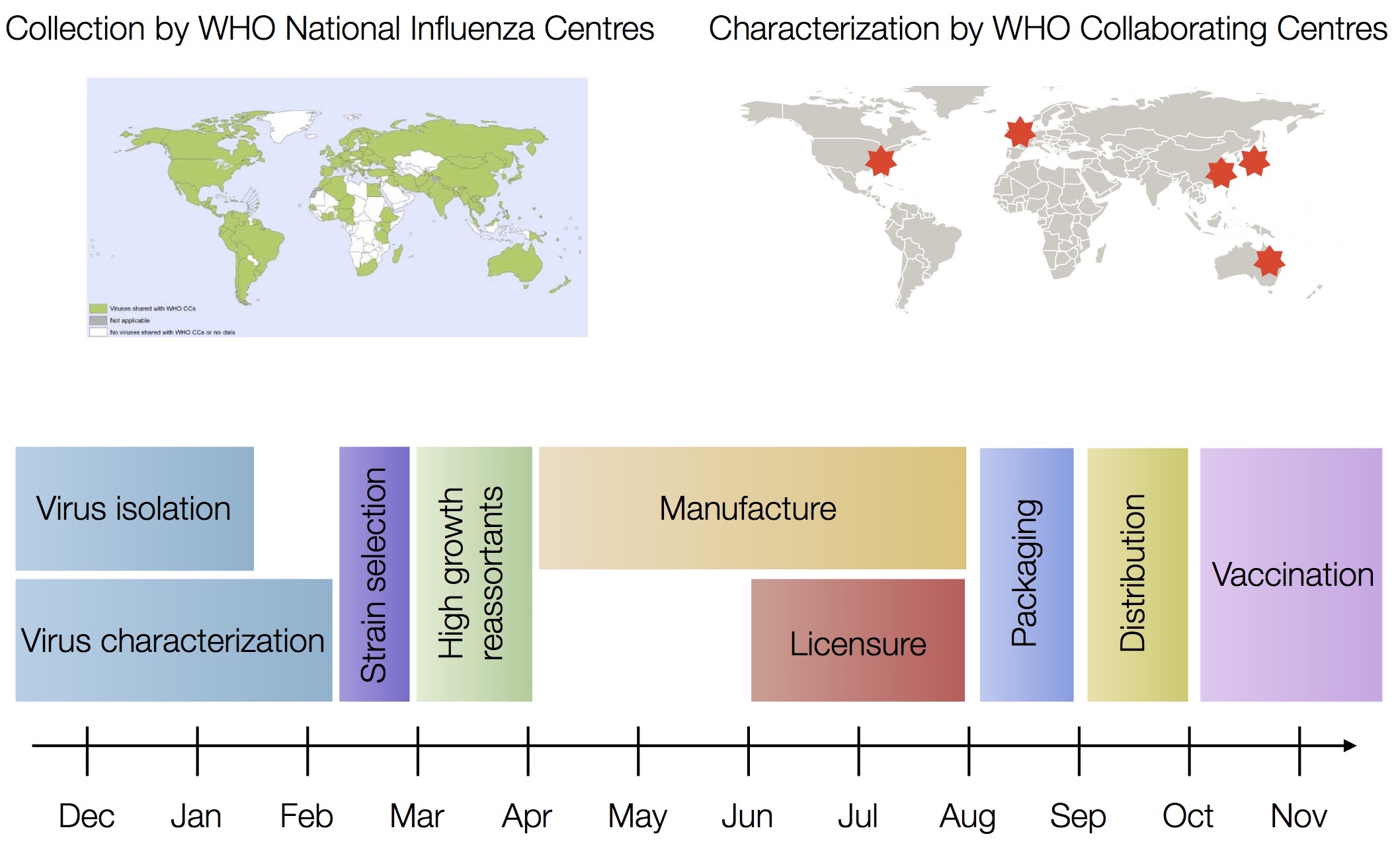
Vaccine strain selection timeline
Seek to explain change in clade frequencies over 1 year
Fitness models can project clade frequencies
Clade frequencies $X$ derive from the fitnesses $f$ and frequencies $x$ of constituent viruses, such that
$$\hat{X}_v(t+\Delta t) = \sum_{i:v} x_i(t) \, \mathrm{exp}(f_i \, \Delta t)$$
This captures clonal interference between competing lineages
Predictive fitness models
A simple predictive model estimates the fitness $f$ of virus $i$ as
$$\hat{f}_i = \beta^\mathrm{ep} \, f_i^\mathrm{ep} + \beta^\mathrm{ne} \, f_i^\mathrm{ne}$$
where $f_i^\mathrm{ep}$ measures cross-immunity via substitutions at epitope sites and $f_i^\mathrm{ep}$ measures mutational load via substitutions at non-epitope sites
We implement a similar model based on two predictors
- Clade frequency change
- Antigenic advancement
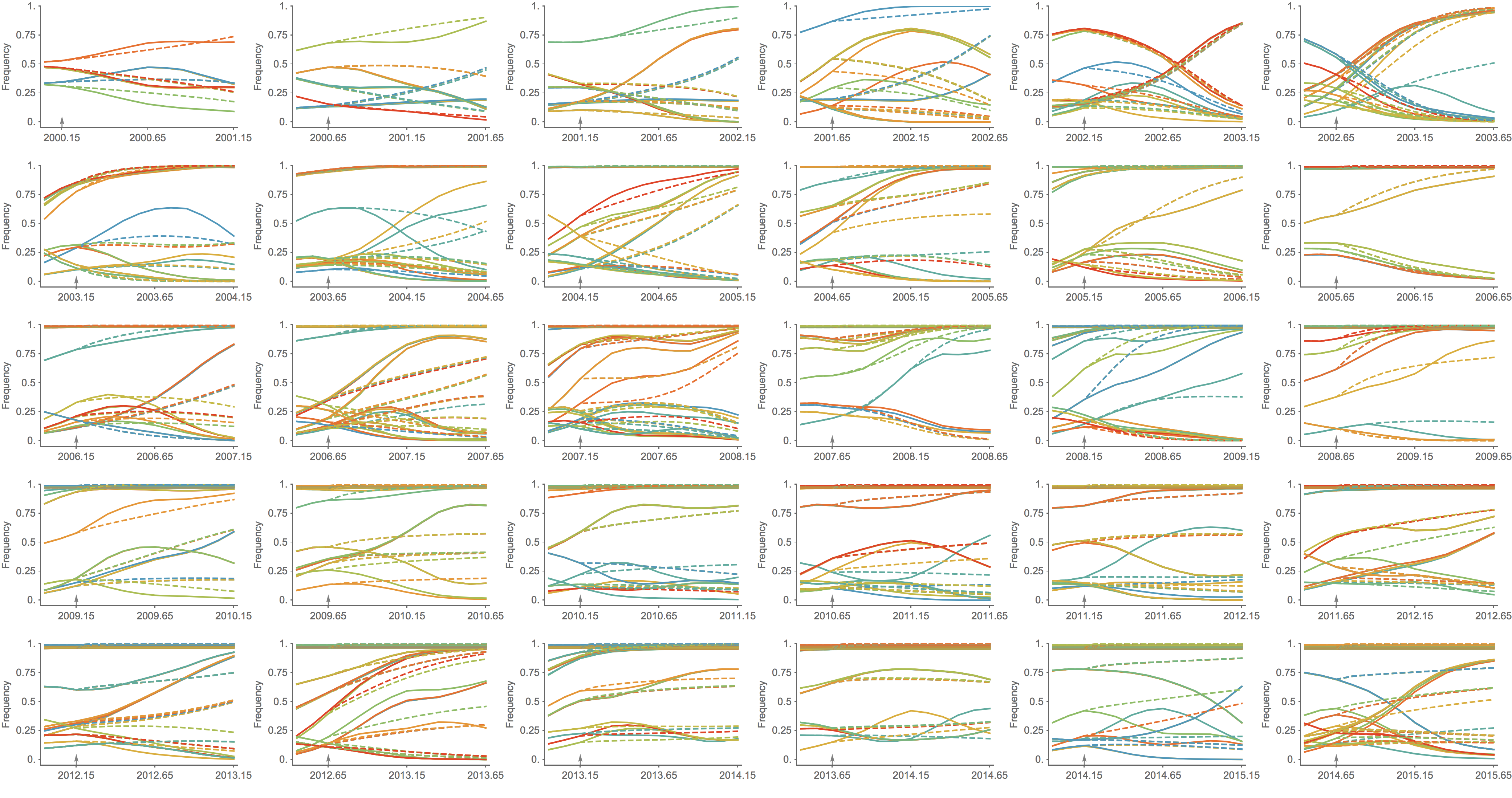
Project frequencies forward,
growing clades have high fitness
Calculate HI drop from ancestor,
drifted clades have high fitness
Fitness model parameterization
Our predictive model estimates the fitness $f$ of virus $i$ as
$$\hat{f}_i = \beta^\mathrm{freq} \, f_i^\mathrm{freq} + \beta^\mathrm{HI} \, f_i^\mathrm{HI}$$
We learn coefficients and validate model based on previous 15 H3N2 seasons
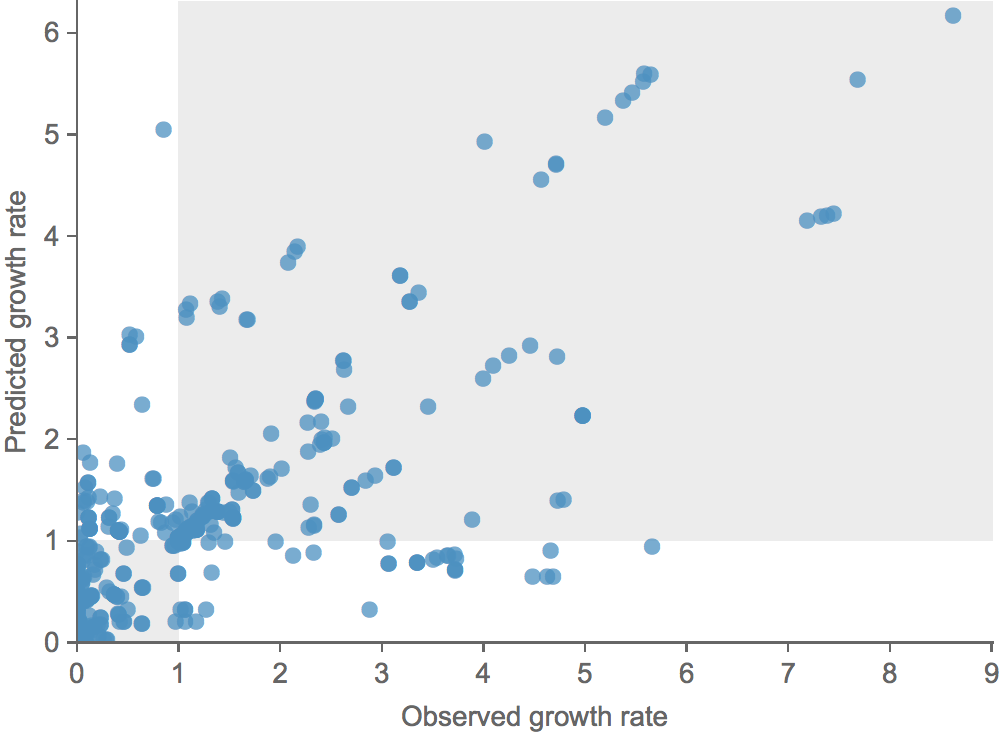
Clade growth rate is well predicted
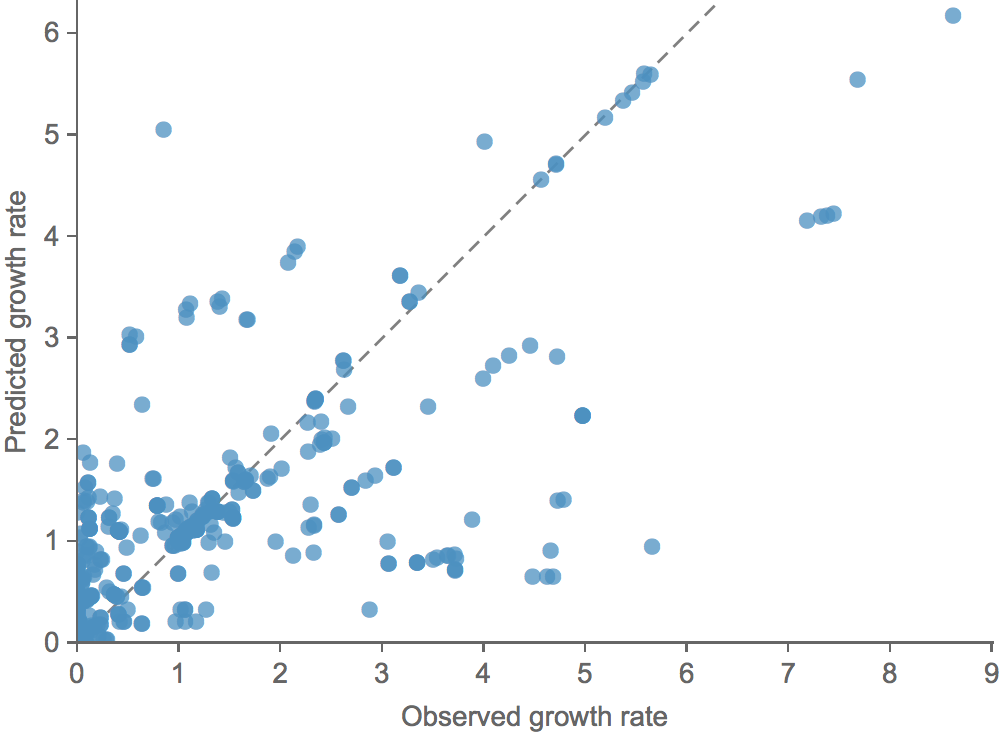
Growth vs decline correct in 83% of cases
Clade error increases steadily over time
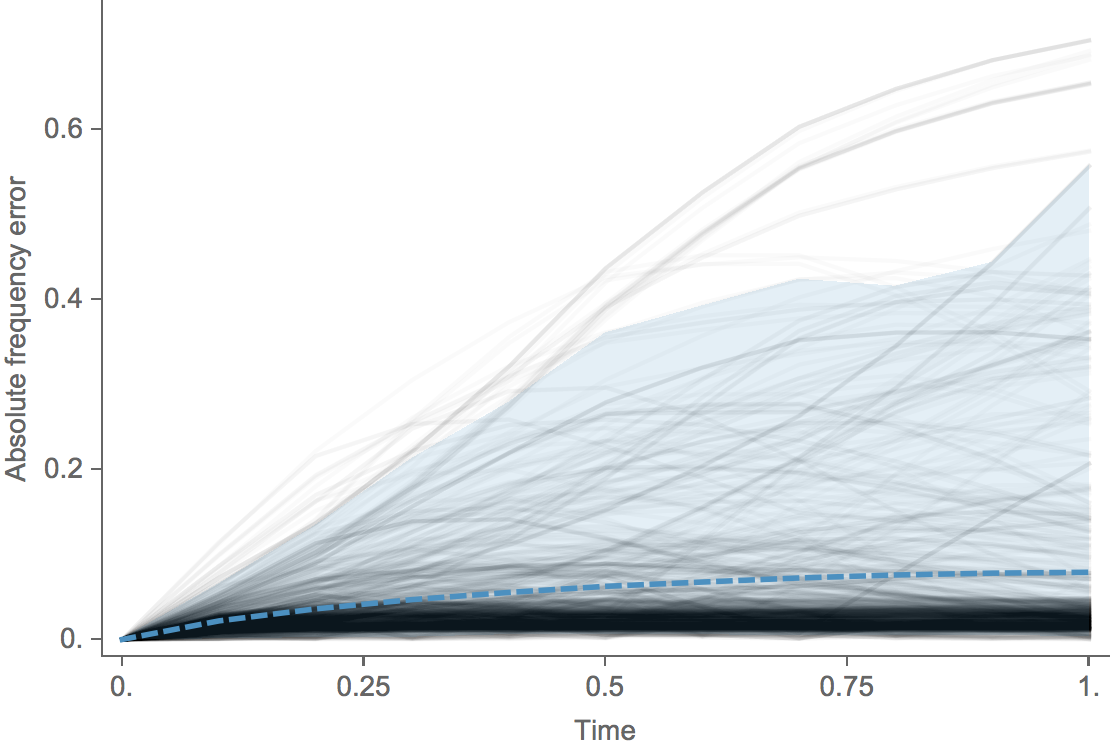
Trajectories show more detailed congruence
Formalizes intuition about drivers of influenza dynamics
| Model | Ep coefficient | HI coefficient | Freq error | Growth corr |
|---|---|---|---|---|
| Epitope only | 2.36 | -- | 0.10 | 0.57 |
| HI only | -- | 2.05 | 0.08 | 0.63 |
| Epitope + HI | -0.11 | 2.15 | 0.08 | 0.67 |
Prototype up at
dev.nextflu.org
Further work on predictive modeling
- Integrate data predictors and data sources, e.g. plan to investigate a geographic predictor
- Possible to build predictive models for H1N1 and B and to forecast NA evolution
Evolutionary analyses can inform influenza vaccine strain selection
Analyses must be rapid and widely available
Predictive models can flag clades for experimental follow-up and creation of vaccine candidates
Acknowledgements
Richard Neher (Max Planck Tübingen), Colin Russell (Cambridge University)
WHO Global Influenza Surveillance Network / GISAID: Ian Barr, Shobha Broor, Mandeep Chadha, Nancy Cox, Rod Daniels, Palani
Gunasekaran, Aeron Hurt, Jacqueline Katz, Anne Kelso, Alexander Klimov, Nicola Lewis, Xiyan Li, John McCauley, Takato Odagiri, Varsha Potdar, Yuelong Shu, Eugene Skepner, Masato Tashiro, Dayan Wang, Xiyan Xu



Contact
- Website: bedford.io
- Twitter: @trvrb
- Slides: bedford.io/talks/real-time-forecasting-cdc/