Novel coronavirus (nCoV) phylodynamics
Trevor Bedford (@trvrb)
10 Feb 2020
CMDI Seminar Series
Georgia Institute of Technology
Slides at: bedford.io/talks
Sequencing to reconstruct pathogen spread
Epidemic process
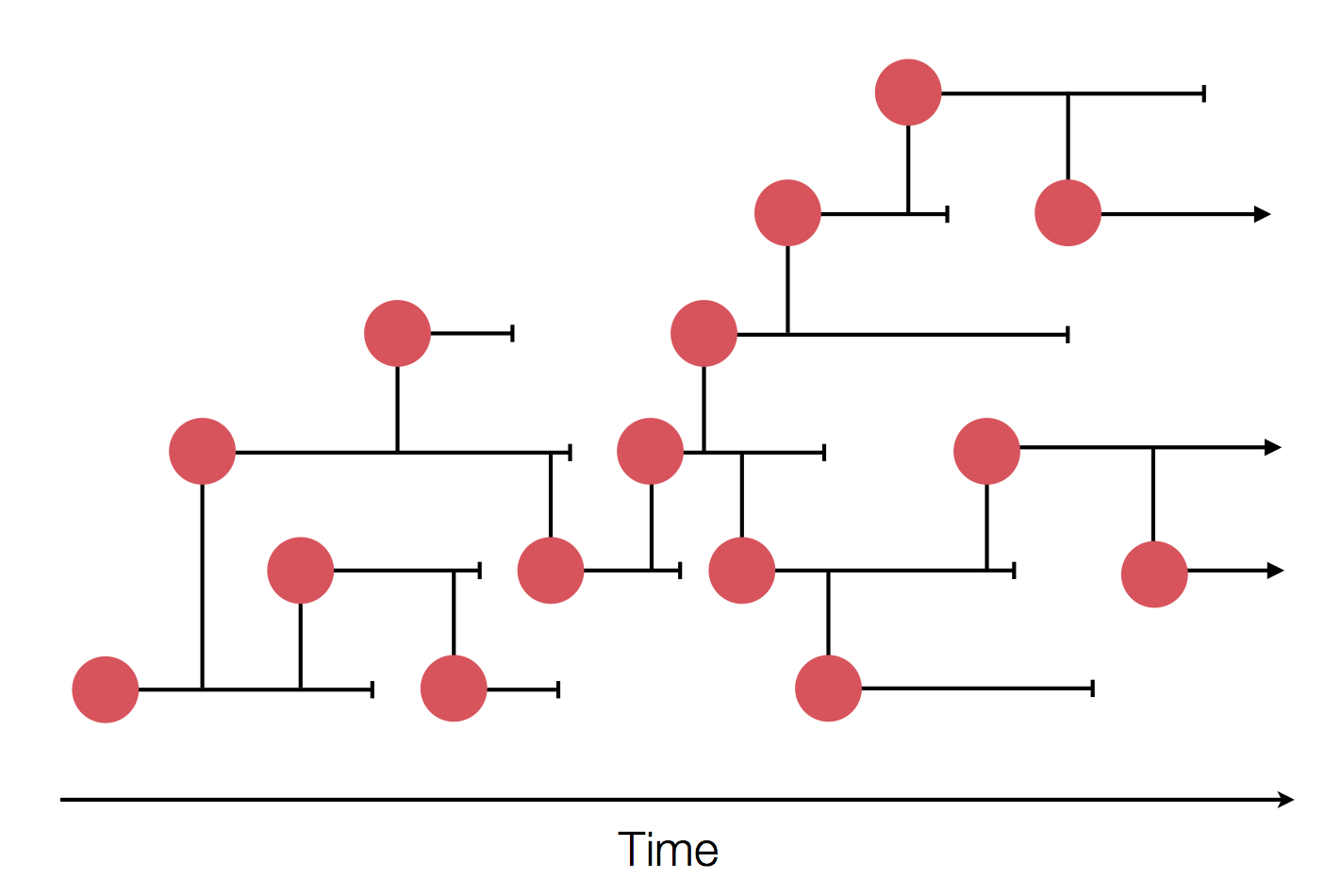
Sample some individuals
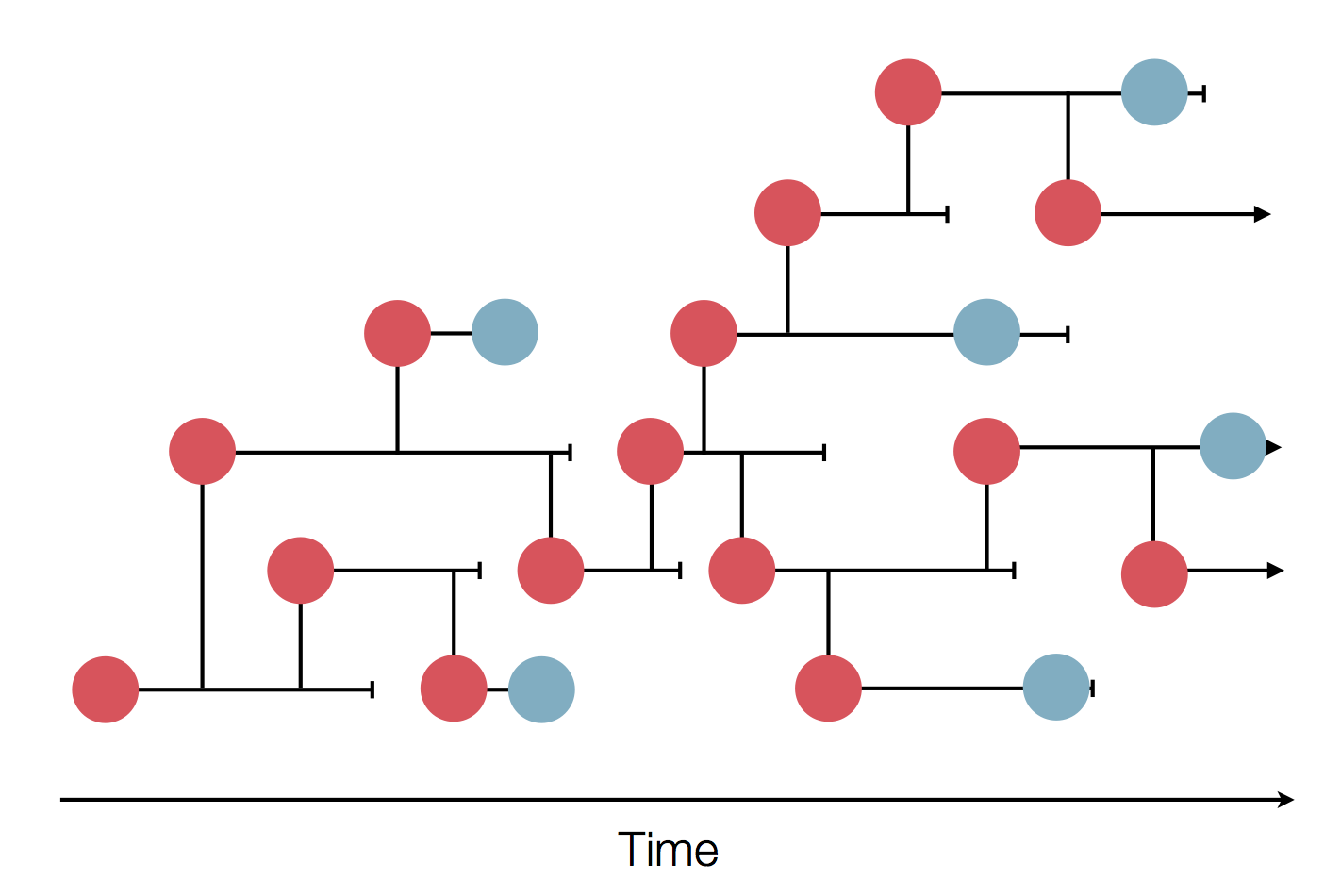
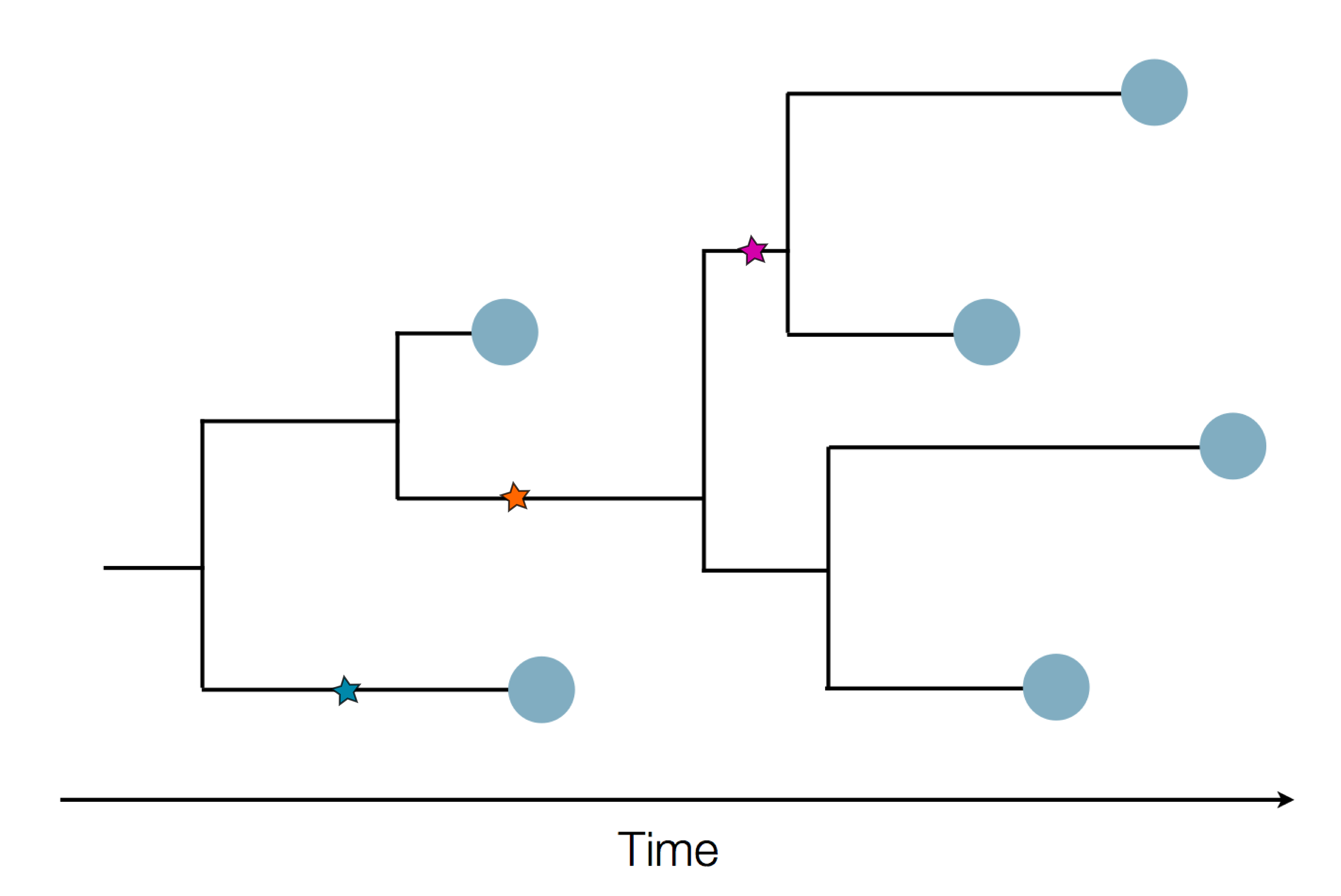
Sequence and determine phylogeny
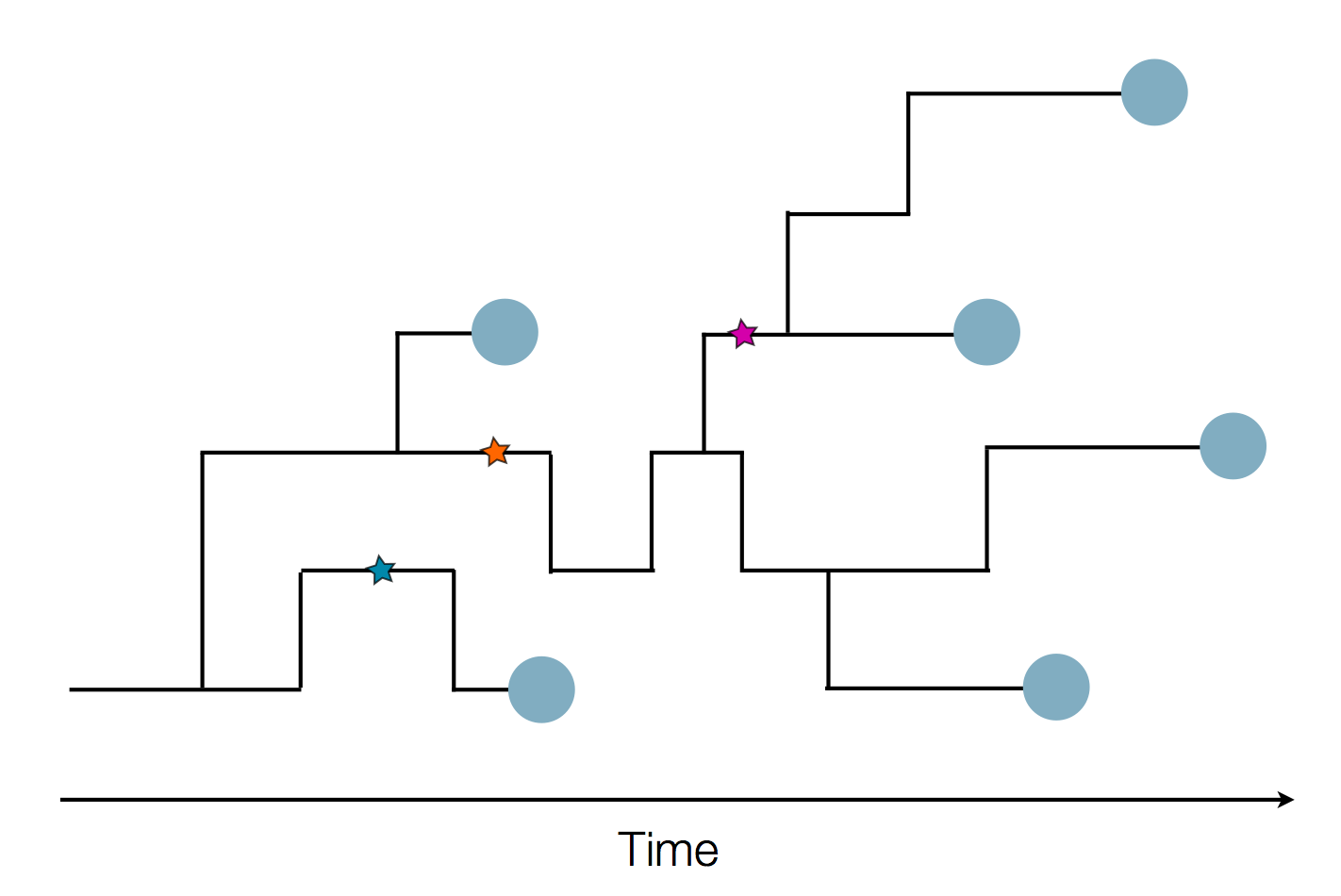
Sequence and determine phylogeny
Nextstrain
Project to conduct real-time molecular epidemiology and evolutionary analysis of emerging epidemics
with
![]() Richard Neher,
Richard Neher,
![]() James Hadfield,
James Hadfield,
![]() Emma Hodcroft,
Emma Hodcroft,
![]() Thomas Sibley,
Thomas Sibley,
![]() John Huddleston,
John Huddleston,
![]() Louise Moncla,
Louise Moncla,
![]() Misja Ilcisin,
Misja Ilcisin,
![]() Kairsten Fay,
Kairsten Fay,
![]() Jover Lee,
Jover Lee,
![]() Allison Black,
Allison Black,
![]() Colin Megill,
Colin Megill,
![]() Sidney Bell,
Sidney Bell,
![]() Barney Potter,
Barney Potter,
![]() Charlton Callender
Charlton Callender
Nextstrain architecture
All code open source at github.com/nextstrain
Two central aims: (1) rapid and flexible phylodynamic analysis and
(2) interactive visualization
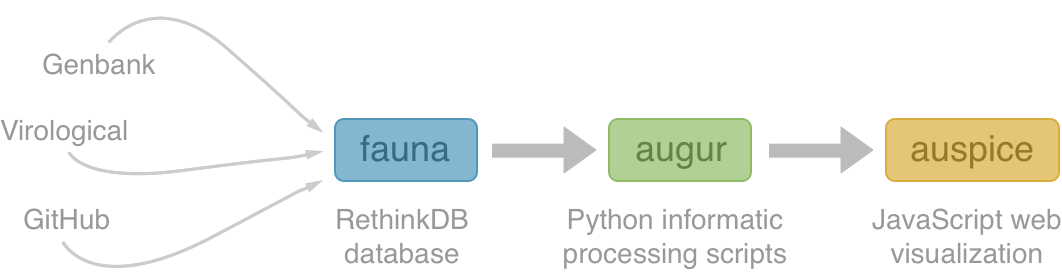
Nextstrain is two things
- a bioinformatics toolkit and visualization app, which can be used for a broad range of datasets
- a collection of real-time pathogen analyses kept up-to-date on the website nextstrain.org
nextstrain.org
Novel coronavirus (nCoV)
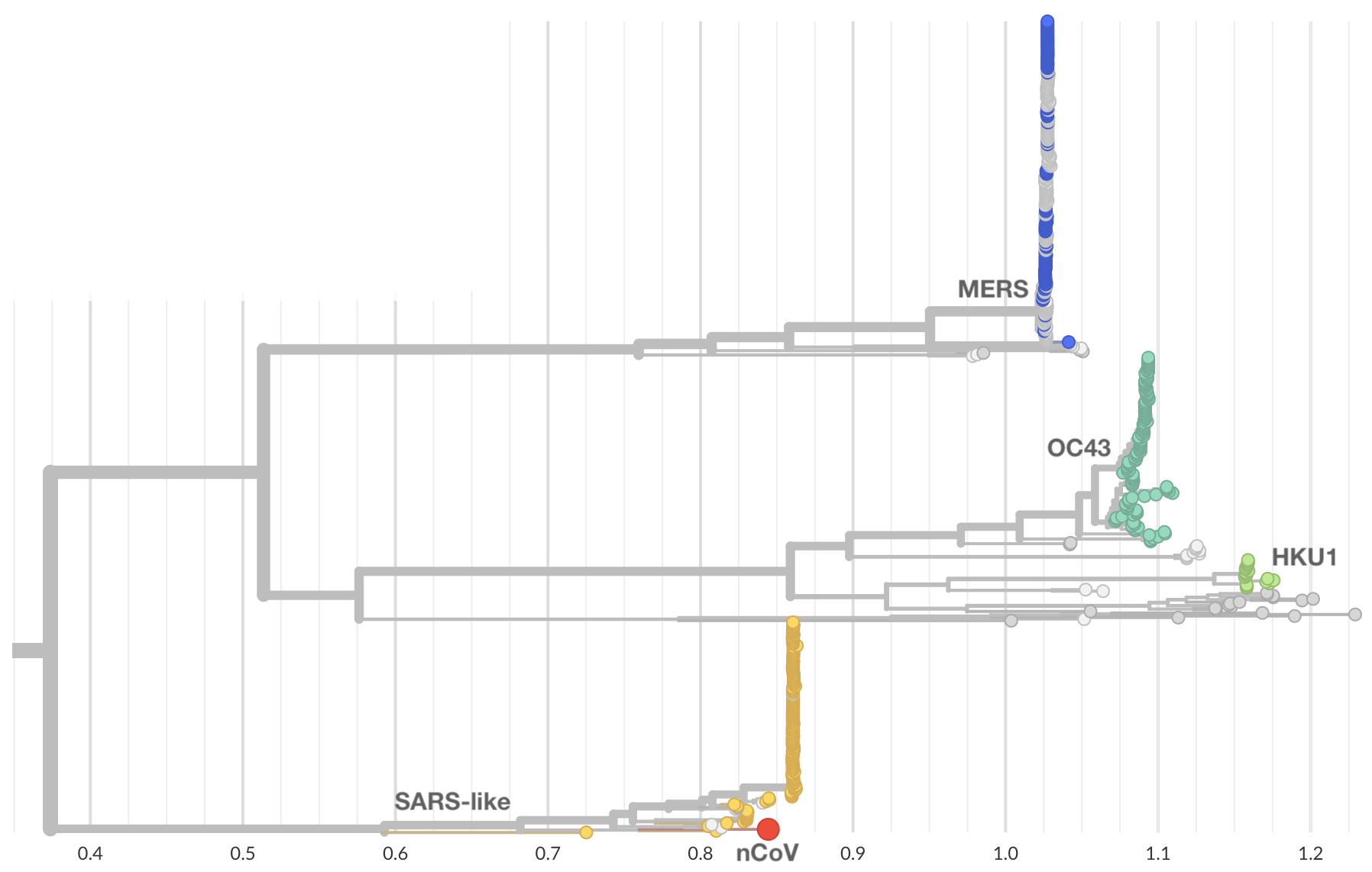
Jan 10: nCoV is a betacoronavirus
Jan 10: nCoV is a betacoronavirus
Jan 11: And belongs to SARS-like coronaviruses
Jan 11: And belongs to SARS-like coronaviruses
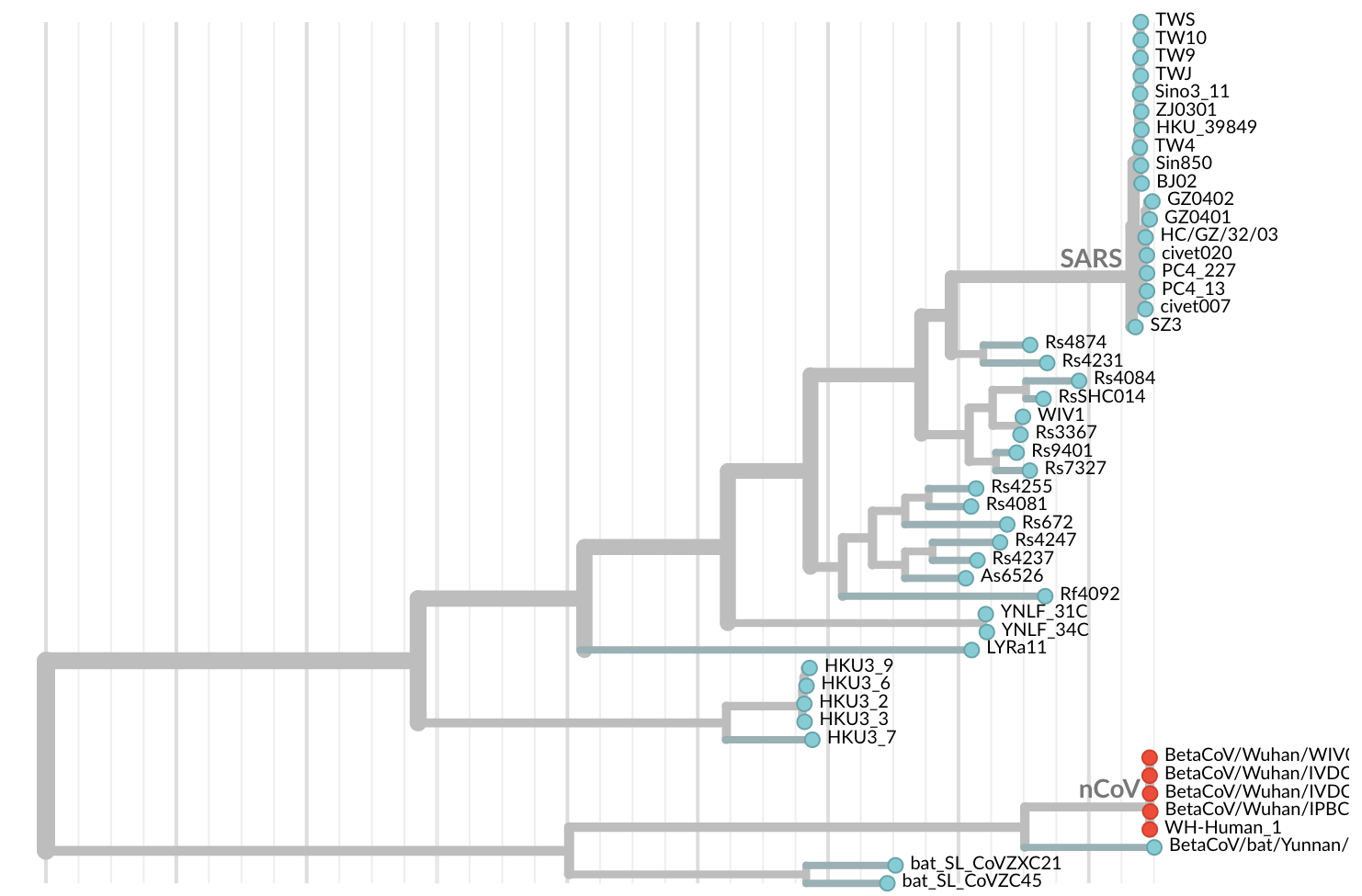
Jan 11: These viruses have a natural reservoir in bats
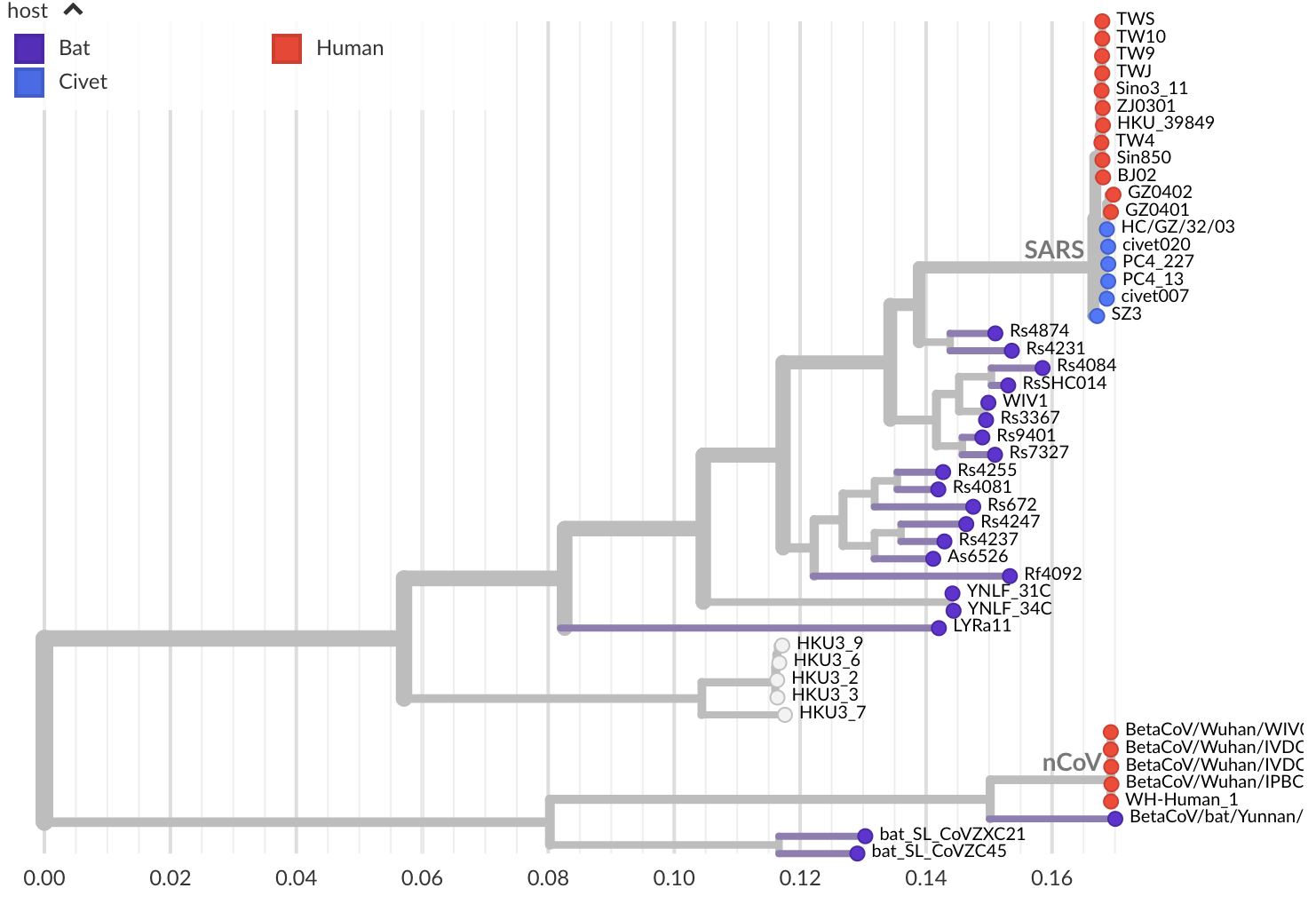
Jan 11: Initial 5 nCoV genomes from Wuhan showed highly restricted genetic diversity
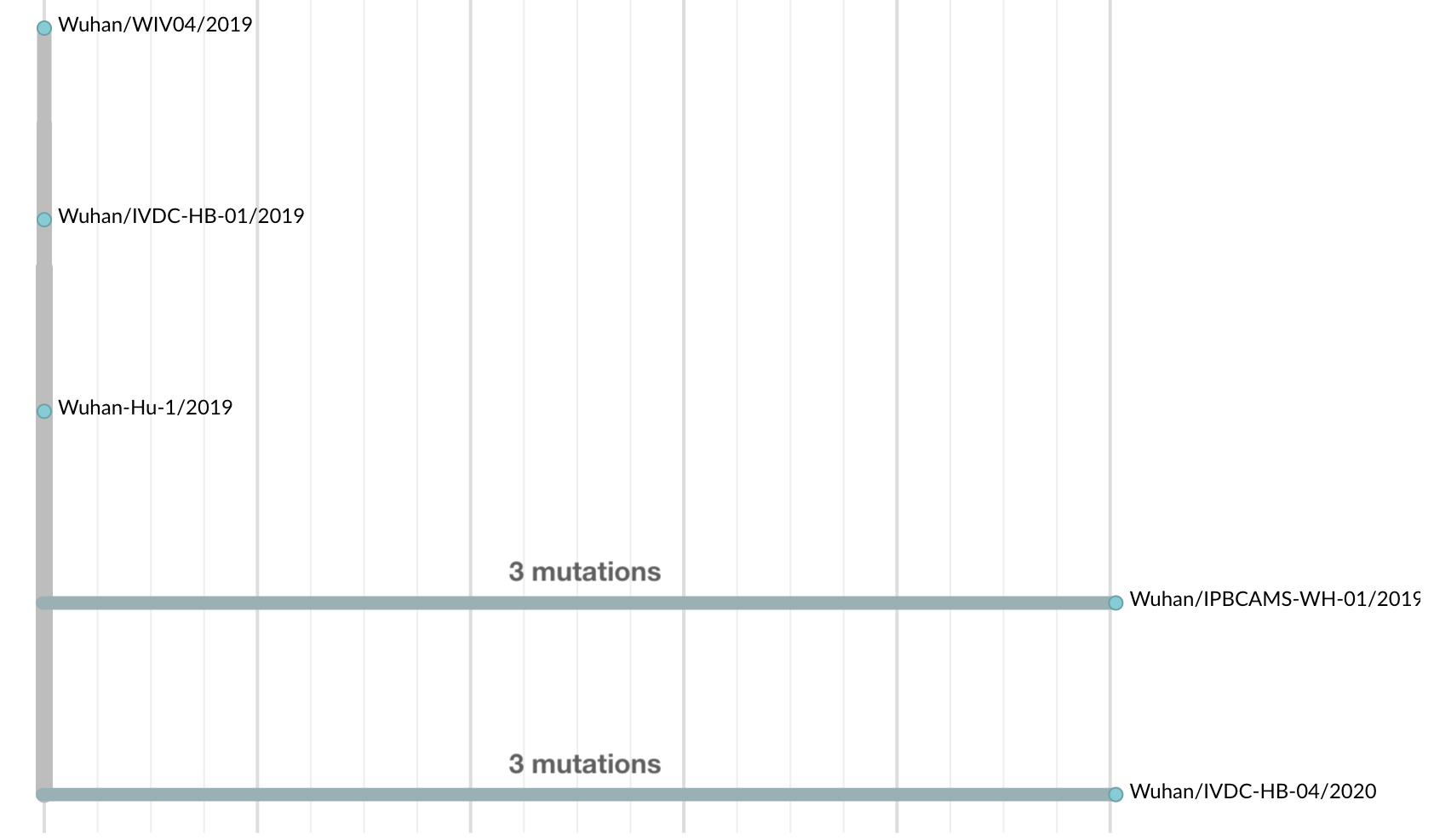
Initially thought clustering due to epi investigation of linked cases at Huanan seafood market
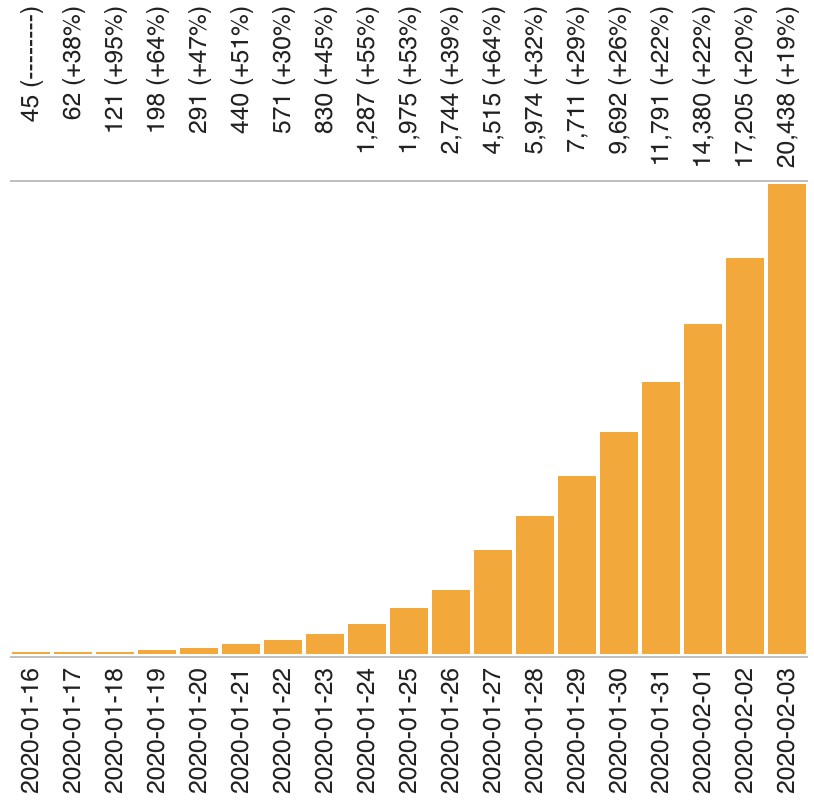
Jan 17: Additional 2 nCoV genomes from Thailand travel cases also lacked diversity
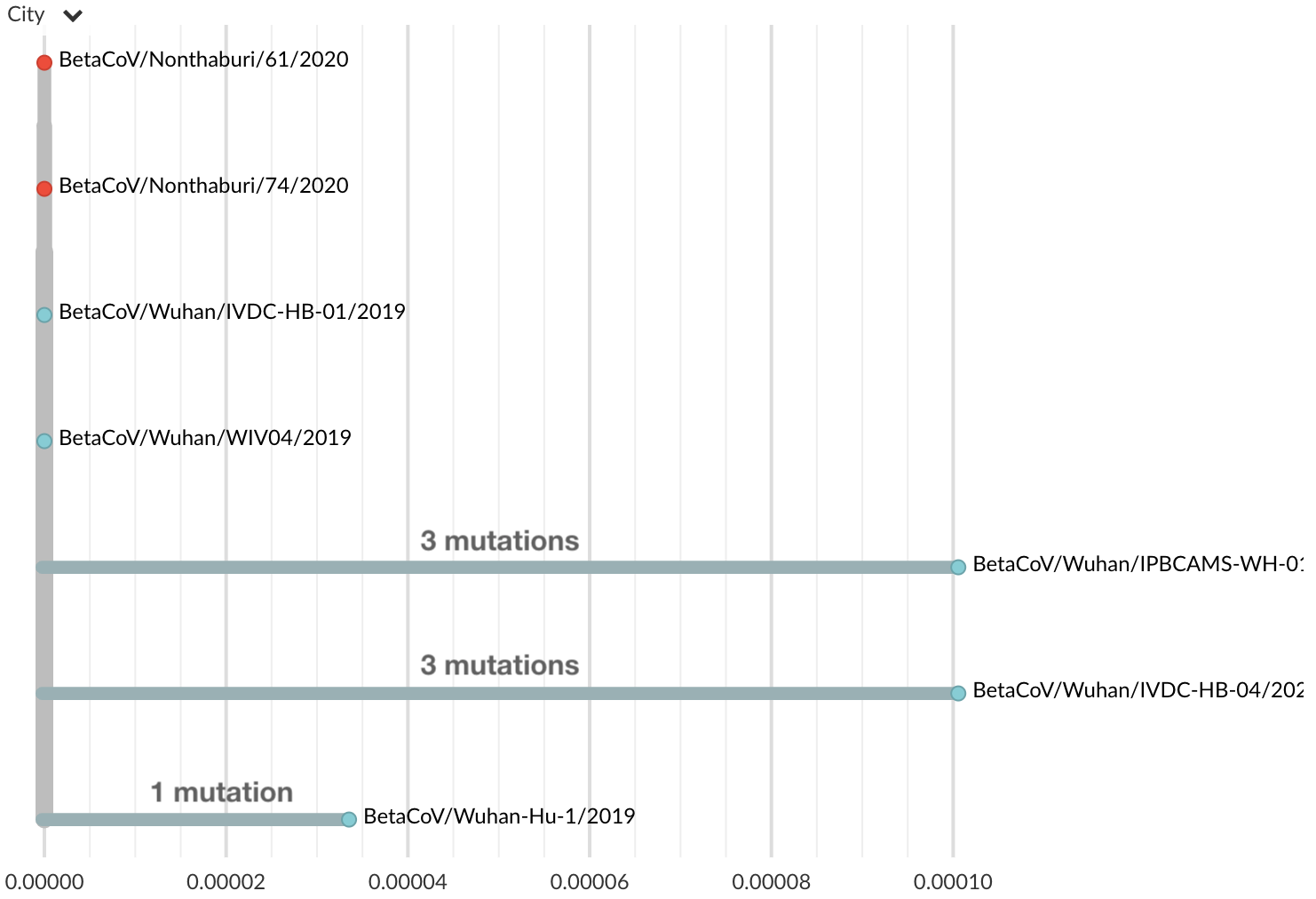
Jan 19: Additional 5 nCoV genomes from Wuhan still lacked diversity
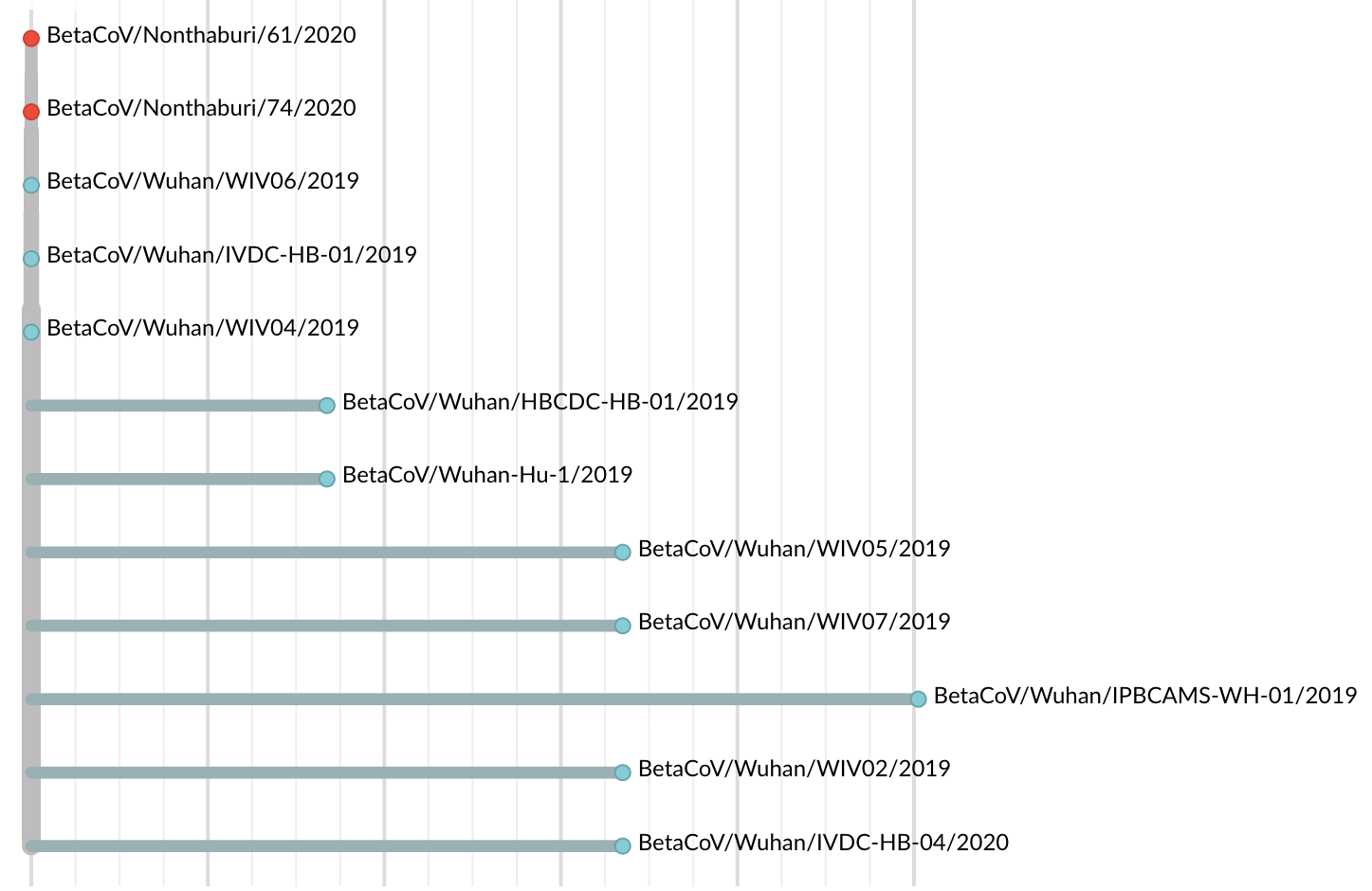
😧
Single introduction into the human population between Nov 15 and Dec 15 and human-to-human epidemic spread from this point forward
Spent the week of Jan 20 alerting public health officials, and since then have aimed to keep nextstrain.org/ncov updated within ~1hr of new sequences being deposited
Data sharing through GISAID
- Shanghai Public Health Clinical Center, Fudan University, Shanghai, China
- National Institute for Viral Disease Control and Prevention, China CDC, Beijing, China
- Institute of Pathogen Biology, Chinese Academy of Medical Sciences, Beijing, China
- Wuhan Institute of Virology, Chinese Academy of Sciences, Wuhan, China
- Department of Microbiology, Zhejiang Provincial CDC, Hangzhou, China
- Guangdong Provincial CDC, Guangzhou, China
- Shenzhen Key Laboratory of Pathogen and Immunity, Shenzhen, China
- Hangzhou Center for Disease and Control Microbiology Lab, Zhejiang, China
- National Institute of Health, Nonthaburi, Thailand
Data sharing through GISAID (continued)
- National Institute of Infectious Diseases, Tokyo, Japan
- Korea Centers for Disease Control & Prevention, Cheongju, Korea
- National Public Health Laboratory, Singapore
- US Centers for Disease Control and Prevention, Atlanta, USA
- Institut Pasteur, Paris, France
- Respiratory Virus Unit, Microbiology Services Colindale, Public Health England
- Department of Virology, University of Helsinki and Helsinki University Hospital, Helsinki, Finland
- University of Melbourne, Peter Doherty Institute for Infection & Immunity, Melbourne, Australia
- Victorian Infectious Disease Reference Laboratory, Melbourne, Australia
Almost real-time with many genomes shared within 3-6 days of sampling
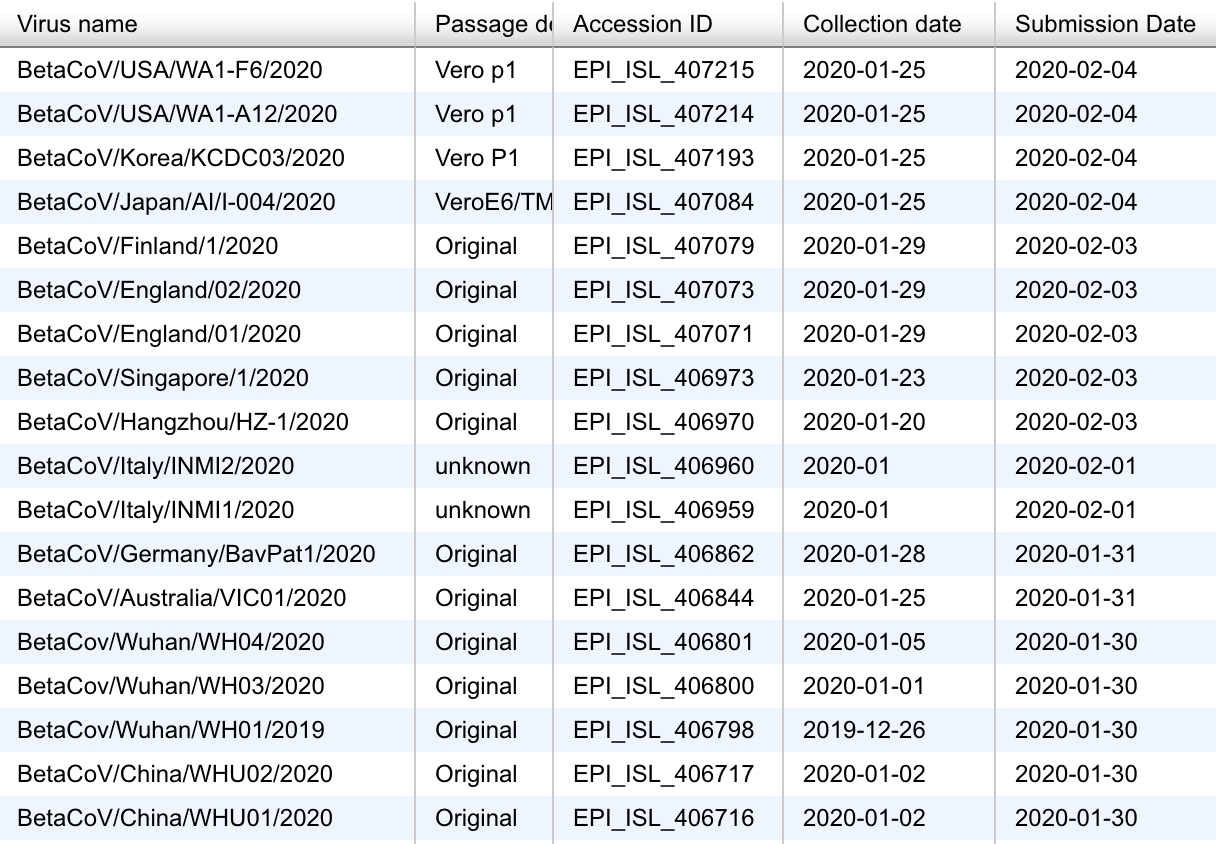
Current state of nextstrain.org/ncov with 80 genomes as of Feb 10
Providing updated genomic situation reports at nextstrain.org/narratives/ncov/sit-rep/
Scientific communication surrounding outbreak has completely flipped with everything posted to bioRxiv, modeling groups posting live analyses and crowd-sourced line lists 🙌🏻
This communication between academics and public health officials has spilled over with huge interest from general public
This is having knock-on effects on science communication and spread of misinformation
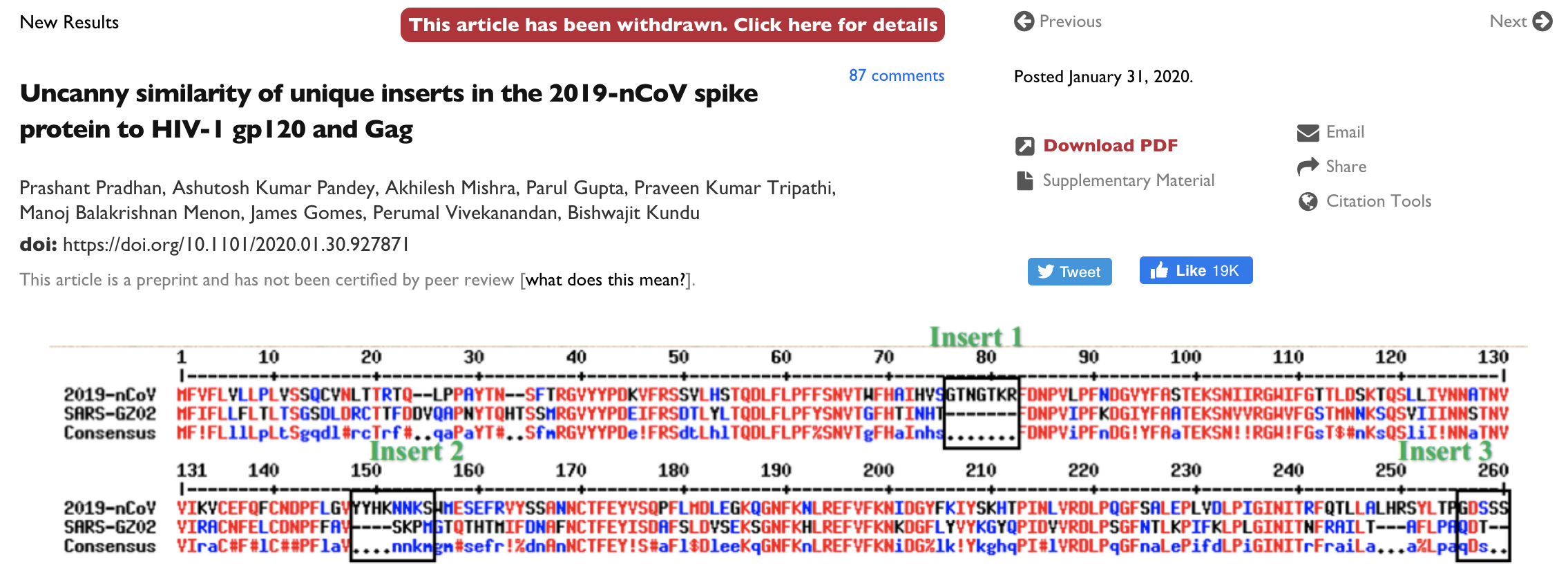
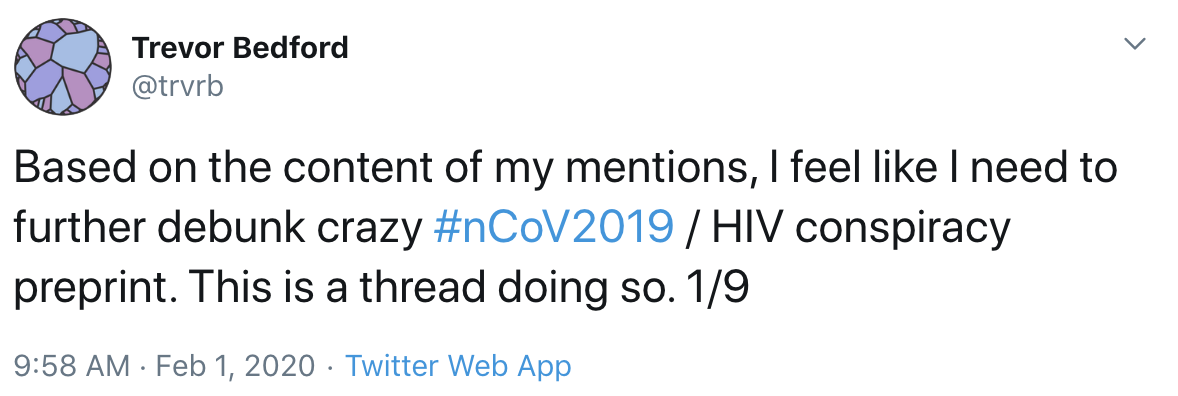
Moving forward
- Epidemic spread still appears to be sustained with estimates of R0 between 1.5-3.5 and epidemic doubling time of ~6 days
- We have not yet had time to ascertain effects of intervention measures, but my hope for containment is slim
- Biggest question for me now surrounds infection-to-fatality ratio
- I expect genomic data to be most immediately useful to help pin down emerging community transmission
Acknowledgements
Bedford Lab:
![]() Alli Black,
Alli Black,
![]() John Huddleston,
John Huddleston,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Louise Moncla,
Louise Moncla,
![]() Maya Lewinsohn,
Maya Lewinsohn,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Kairsten Fay,
Kairsten Fay,
![]() Misja Ilcisin,
Misja Ilcisin,
![]() Nicola Müller,
Nicola Müller,
![]() Marlin Figgins
Marlin Figgins
Data producers from all over the world, GISAID, Virological.org





