H5N1 sequence data update
Trevor Bedford
Fred Hutchinson Cancer Center / Howard Hughes Medical Institute
12 Jun 2024
Monthly SPHERES call
CDC
Slides at: bedford.io/talks
Genomic data available as of today
H5N1 genomes from samples collected in 2024 from the USA
- GISAID
- 424 consensus genomes with metadata (274 cattle sequences)
- GenBank
- 383 consensus genomes with metadata available via this NCBI Virus query (239 cattle sequences)
- SRA
- Additional 399 samples lacking collection date and collection state available from BioProject PRJNA1102327 (additional 230 cattle samples)
Curated sequence data
- We fetch data from Genbank via combination of NCBI Datasets and NCBI Virus, including matching across segments and annotating admin division from strain name
- Data from SRA BioProject PRJNA1102327 is assembled into consensus genomes and shared to GitHub by the Andersen Lab at Scripps. We fetch this data from GitHub and merge with GenBank based on SRA accession.
- This ingest routine is run daily and resulting sequence data available at: and metadata available at:
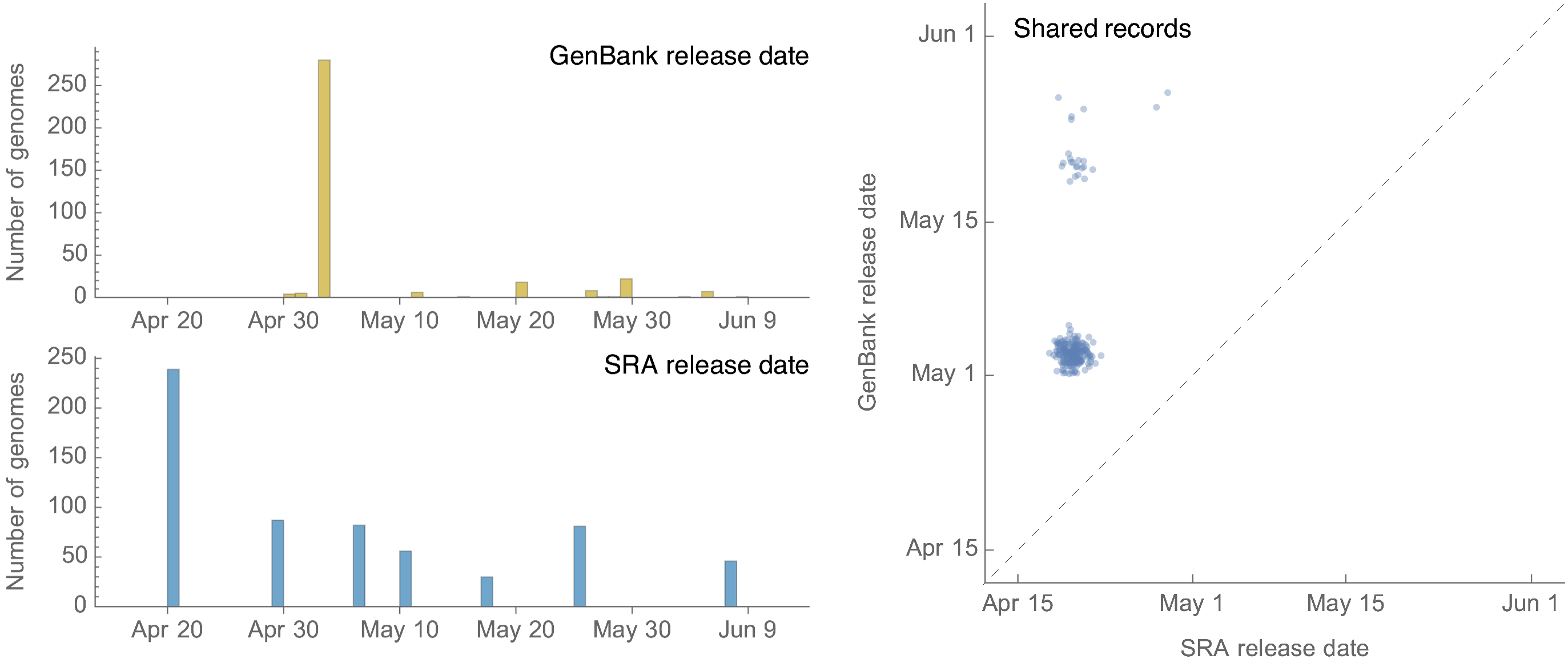
GenBank vs SRA release dates
GenBank has seen 66 submissions since May 4 (with the 27 from USDA representing backfill of April SRA releases), while SRA has seen 295 submissions
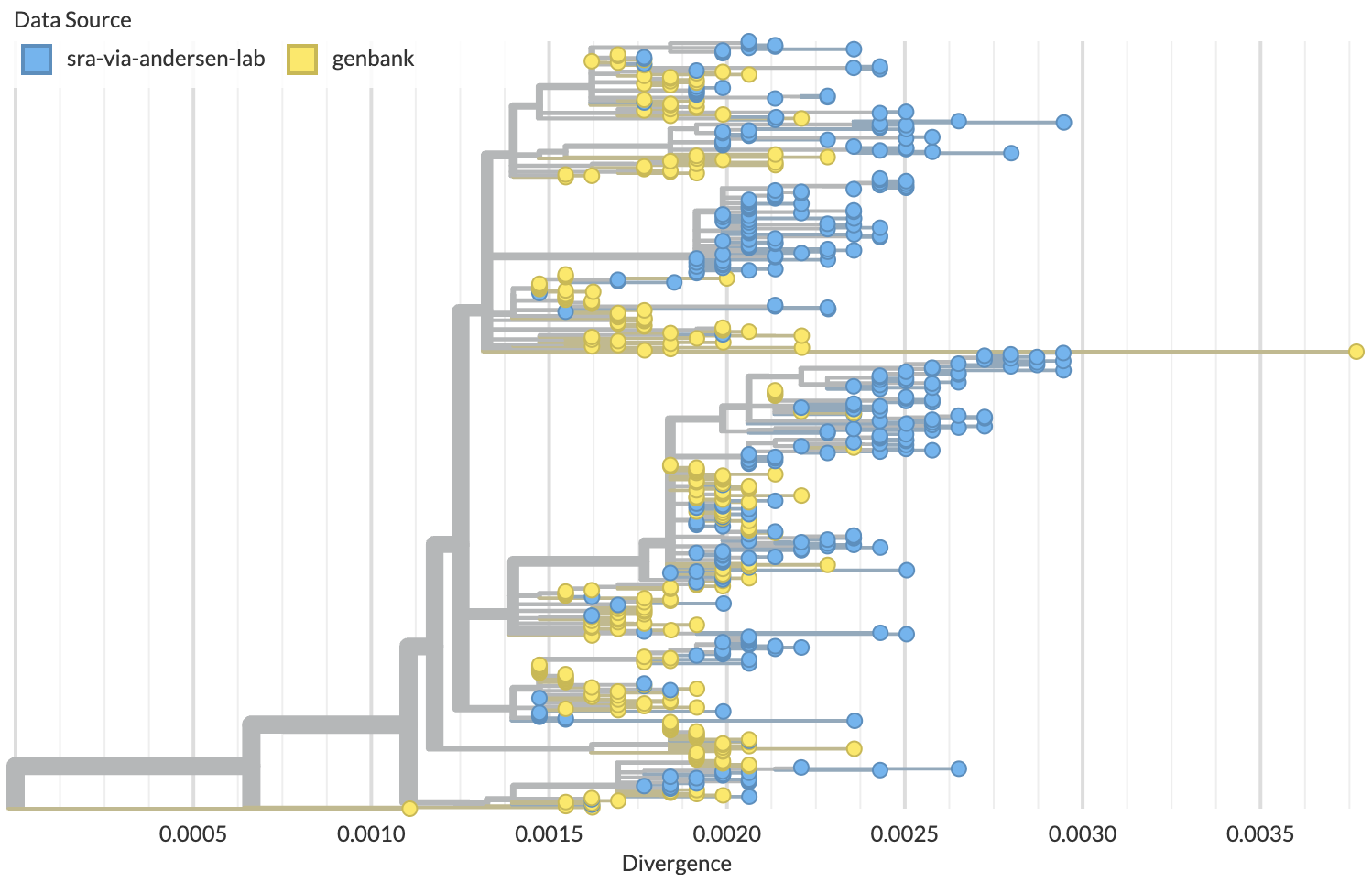
Full genome phylogenetic analysis shows SRA samples are more derived than GenBank samples
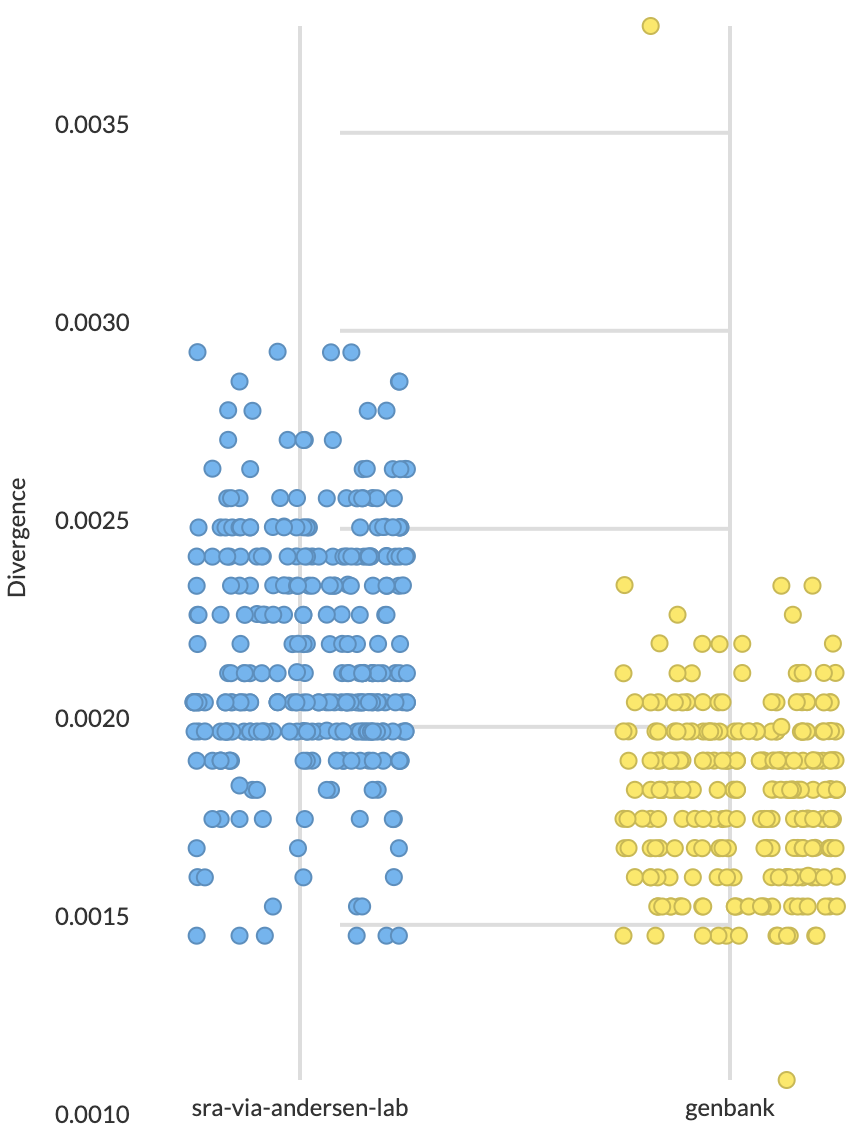
Molecular clock estimates SRA samples as primarily collected in April
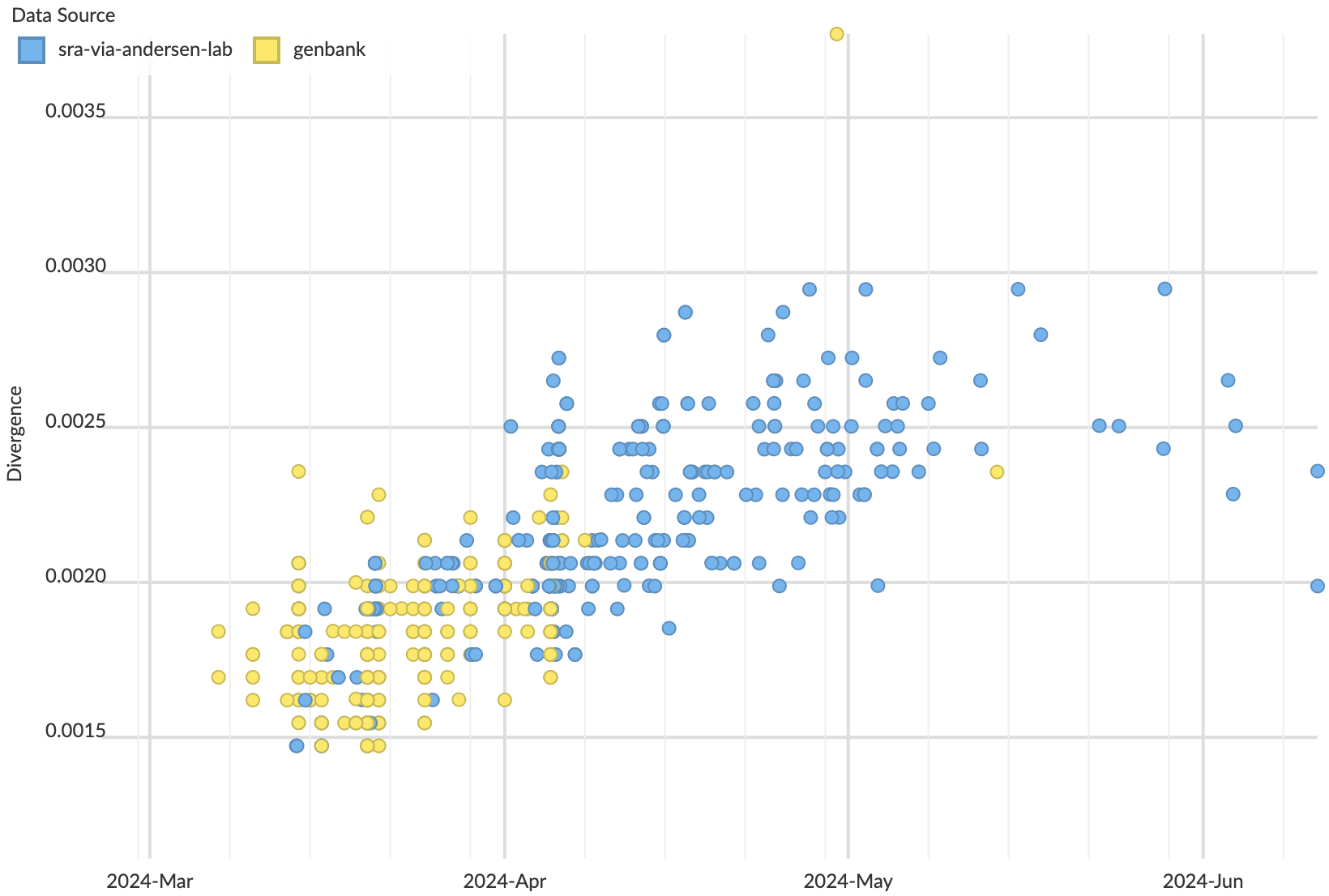
Continuing evolution of H5N1 with new lineages revealed in the SRA data
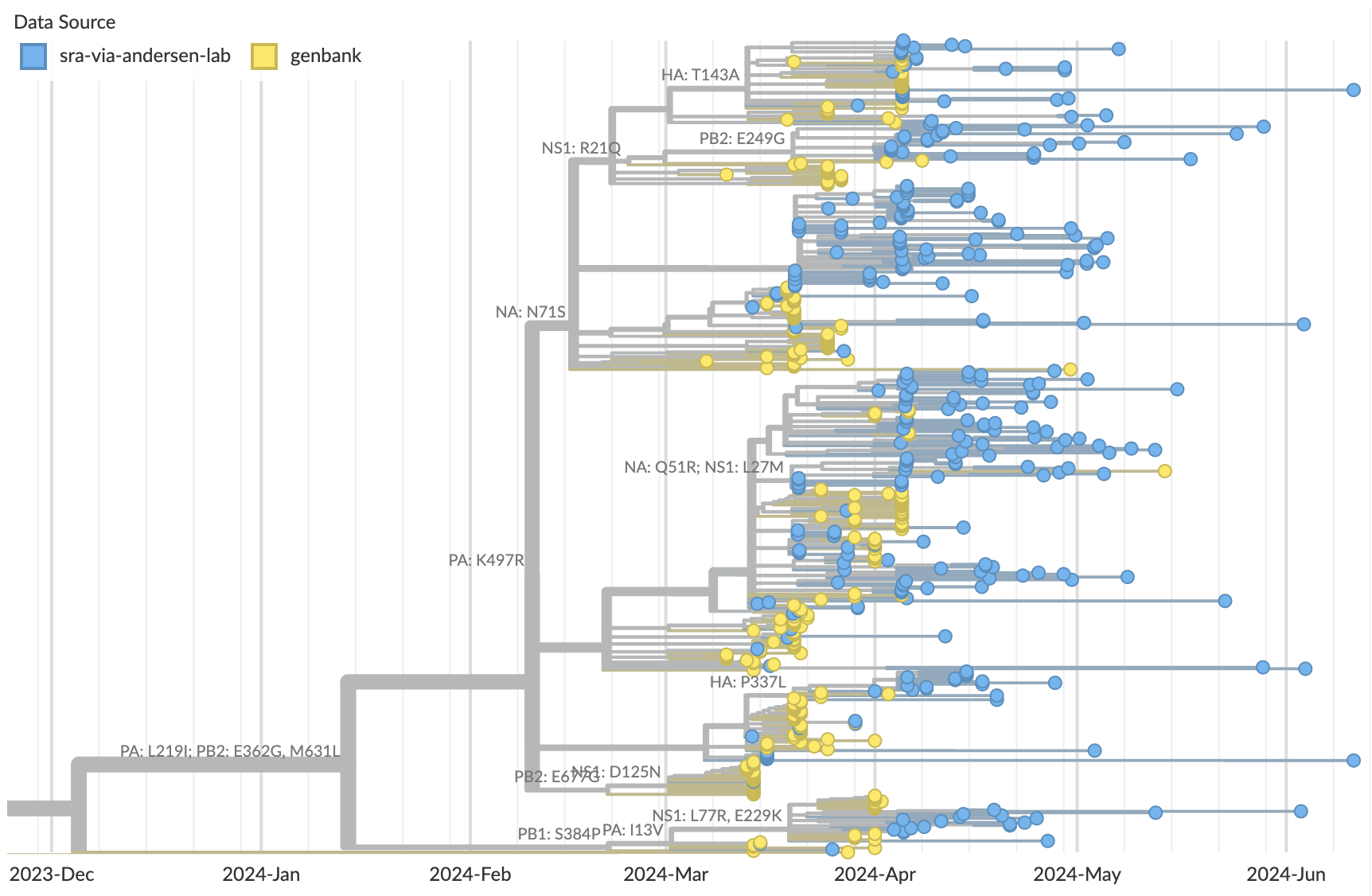
Timely analyses of H5N1 require SRA data
Acknowledgements
H5N1 data curation and analysis: ![]() Jover Lee, Kartik Gangavarapu, Praneeth Gangavarapu, Louise Moncla, James Hadfield, John Huddleston,
Kristian Andersen
Jover Lee, Kartik Gangavarapu, Praneeth Gangavarapu, Louise Moncla, James Hadfield, John Huddleston,
Kristian Andersen
H5N1 genomic data: APHIS National Veterinary Services lab at the USDA for sharing cattle sequences, other data producers from all over the world for sharing contextual sequences, GISAID and NCBI for providing data systems
Nextstrain: Richard Neher, Ivan Aksamentov, John Anderson, Kim Andrews, Jennifer Chang, James Hadfield, Emma Hodcroft, John Huddleston, Jover Lee, Victor Lin, Cornelius Roemer, Thomas Sibley
Bedford Lab:
![]() John Huddleston,
John Huddleston,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Miguel Paredes,
Miguel Paredes,
![]() Marlin Figgins,
Marlin Figgins,
![]() Victor Lin,
Victor Lin,
![]() Jennifer Chang,
Jennifer Chang,
![]() Eslam Abousamra,
Eslam Abousamra,
![]() Nashwa Ahmed,
Nashwa Ahmed,
![]() Cécile Tran Kiem,
Cécile Tran Kiem,
![]() Kim Andrews,
Kim Andrews,
![]() Cristian Ovaduic,
Cristian Ovaduic,
![]() Philippa Steinberg,
Philippa Steinberg,
![]() Jacob Dodds,
Jacob Dodds,
![]() John SJ Anderson
John SJ Anderson





