Genomic epidemiology of the H5N1 outbreak in US cattle
Trevor Bedford
Fred Hutchinson Cancer Center / Howard Hughes Medical Institute
14 May 2024
Monthly Meeting
Northwest PGCoE
Slides at: bedford.io/talks
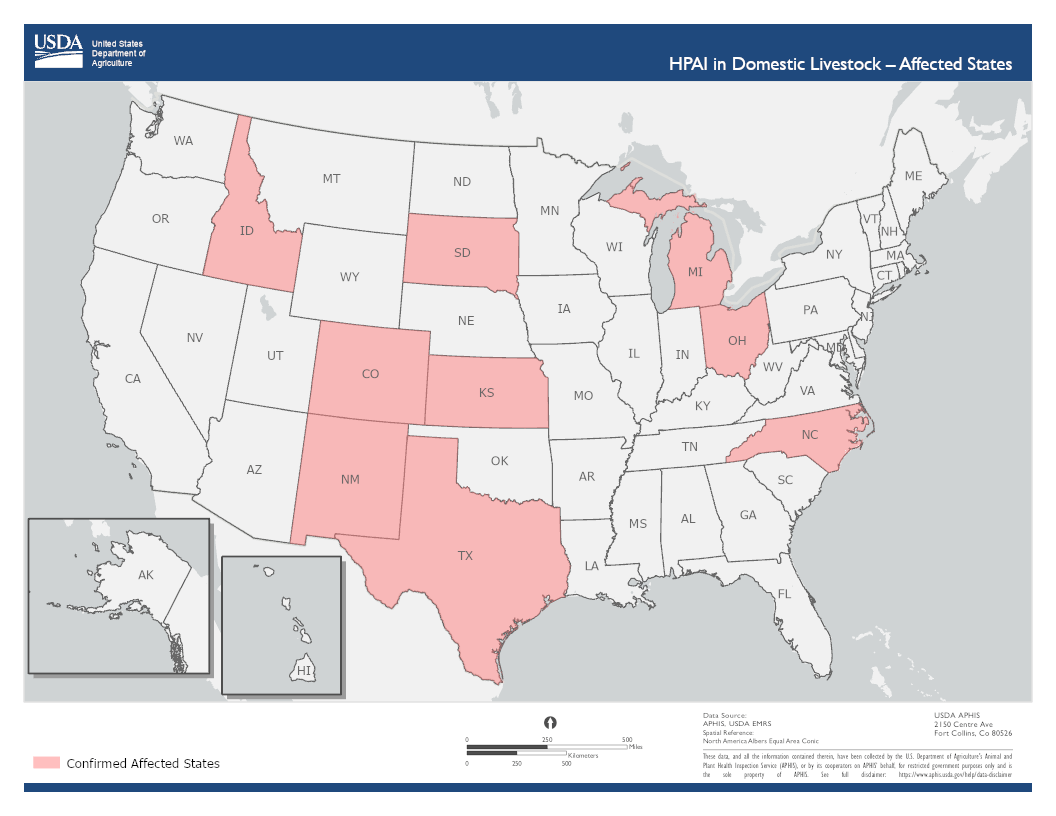
Confirmed infections in cattle in nine states with first detection on March 25
Genomic analysis
with  Louise Moncla, James Hadfield and John Huddleston
Louise Moncla, James Hadfield and John Huddleston
Broader context of H5N1 evolution and spread
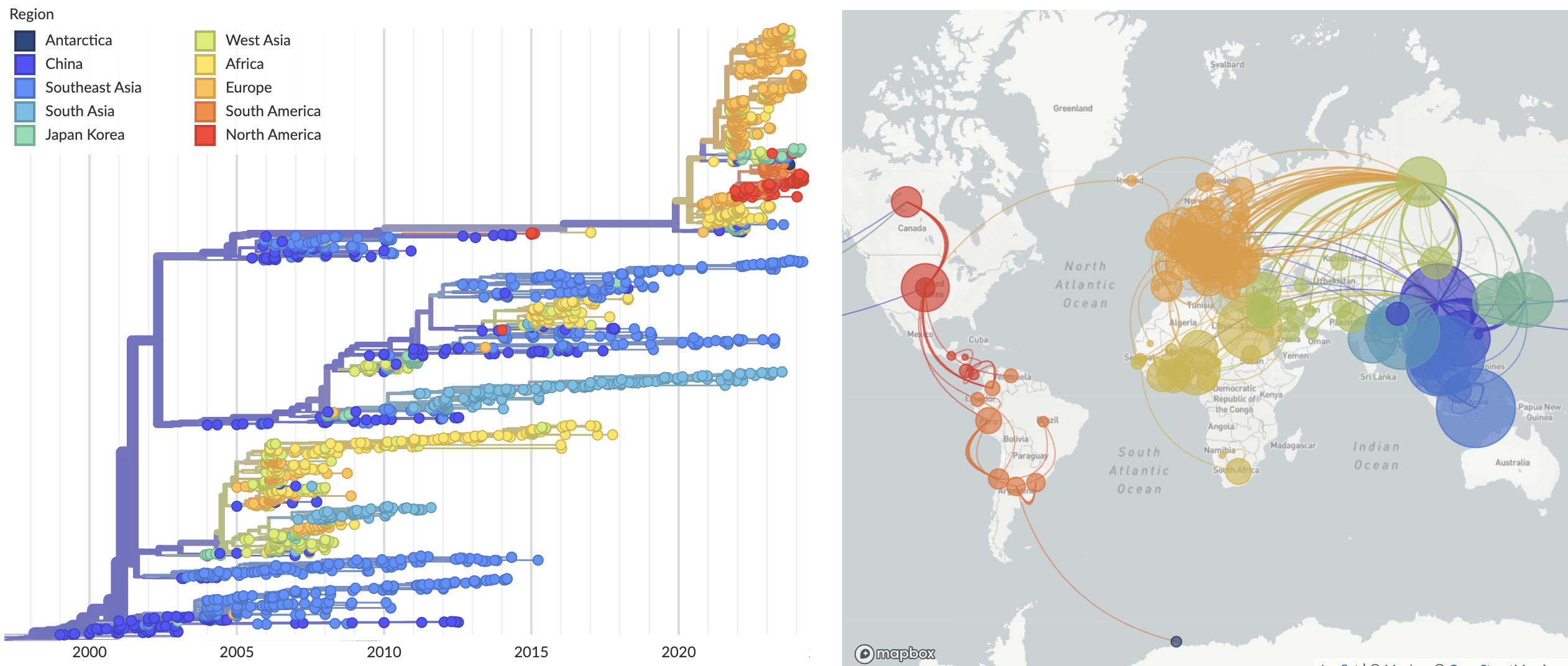
Introduction into Europe and the Americas of clade 2.3.4.4b
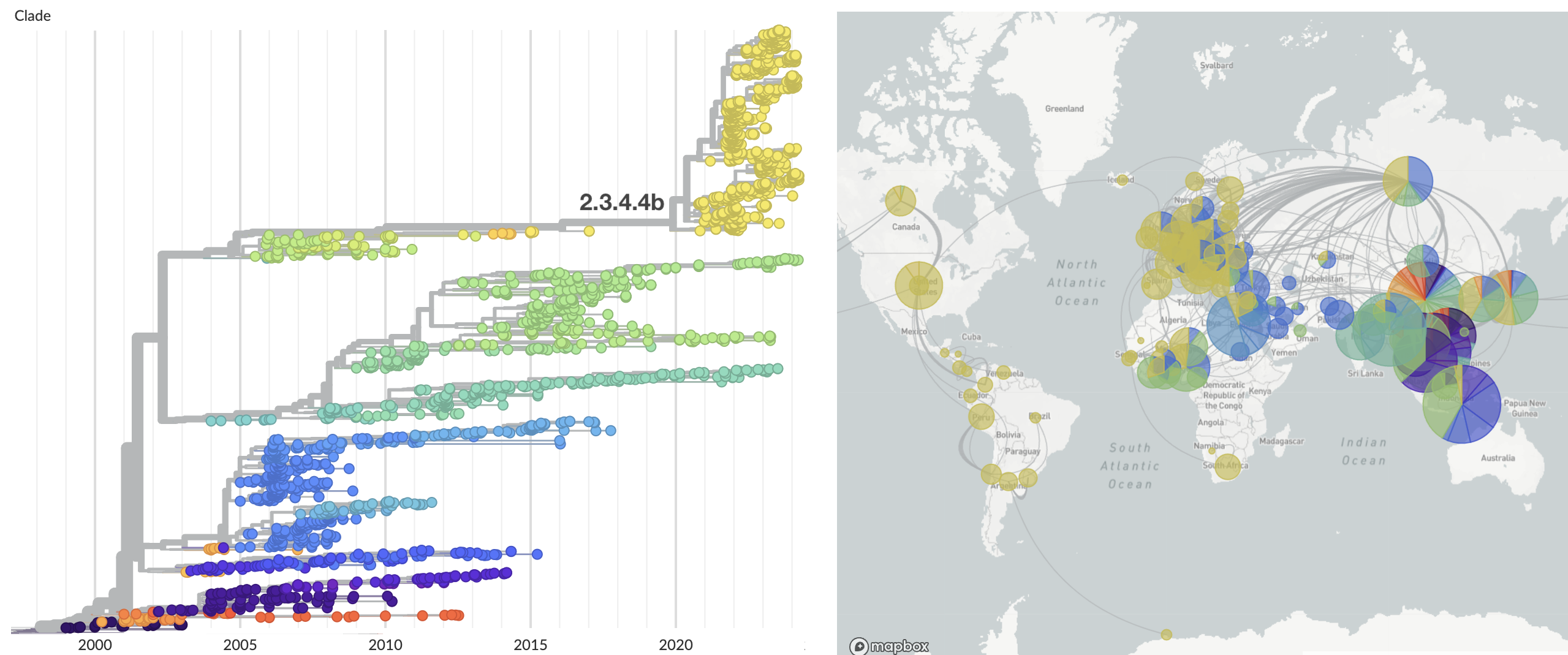
Spread in the Americas characterized by spillover to multiple mammalian hosts
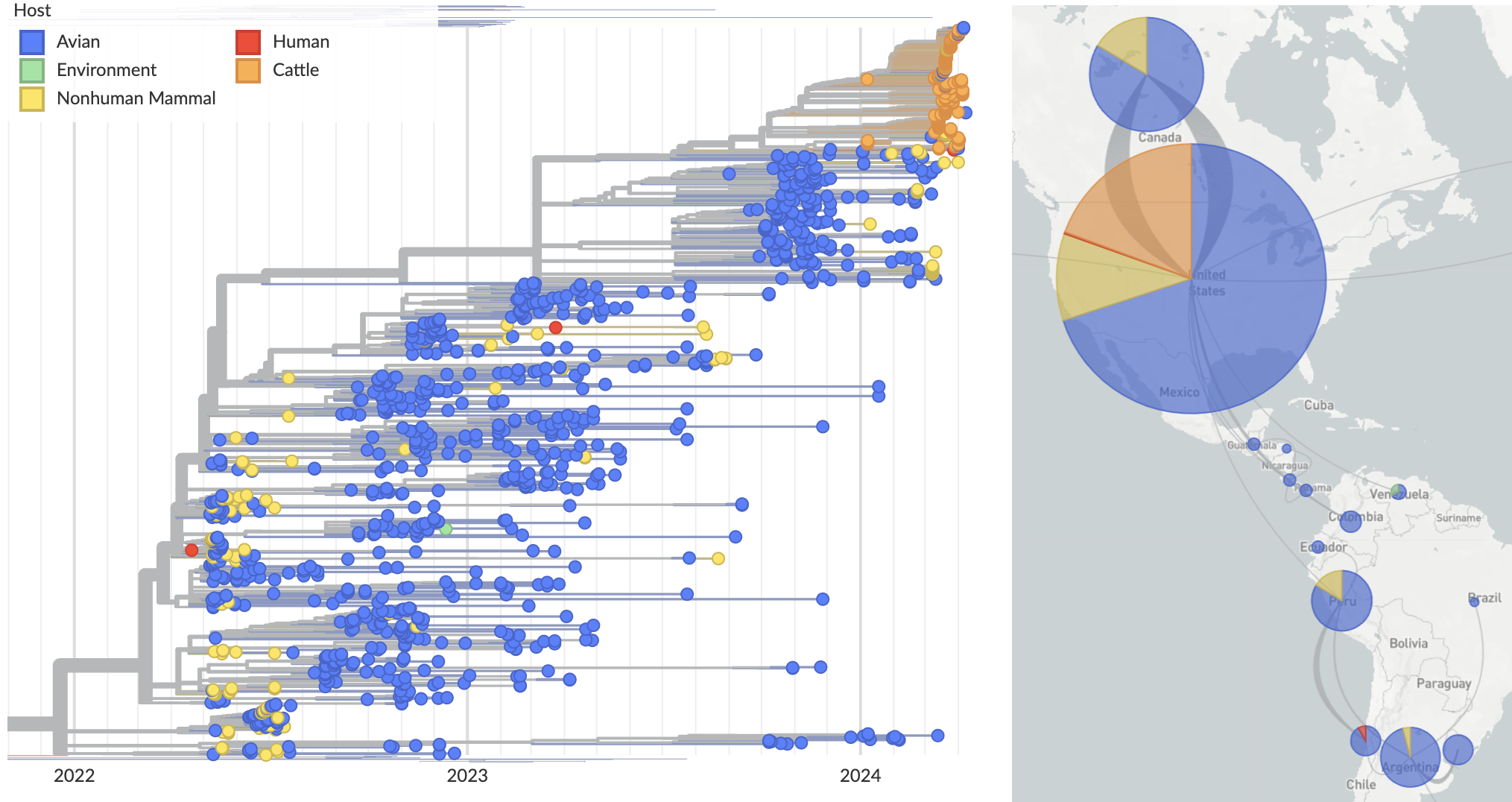
Cattle infections form distinct clade across segments indicative of single spillover and cow-to-cow spread
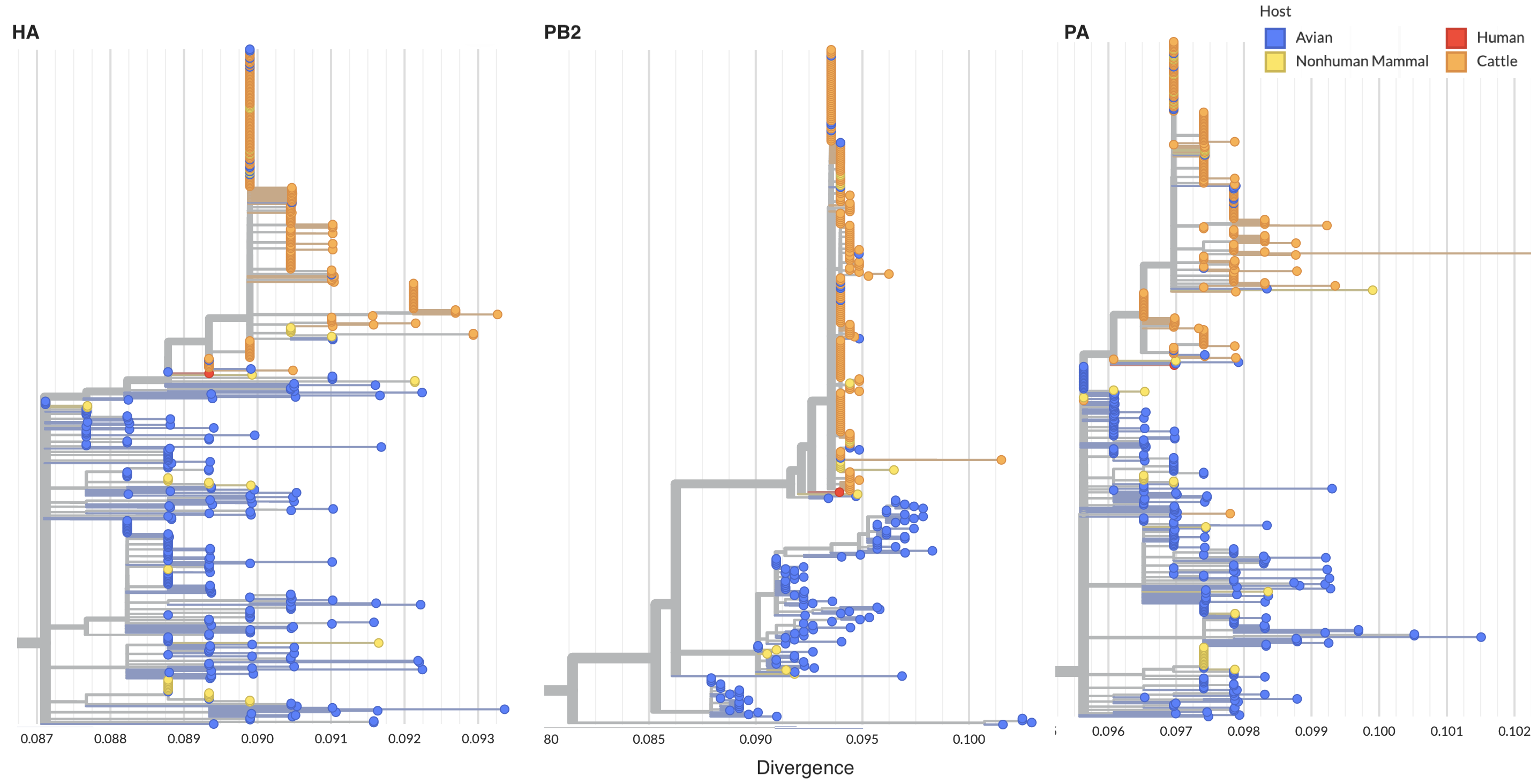
Reassortment in birds prior to cattle spillover, but cattle clade appears non-reassorting
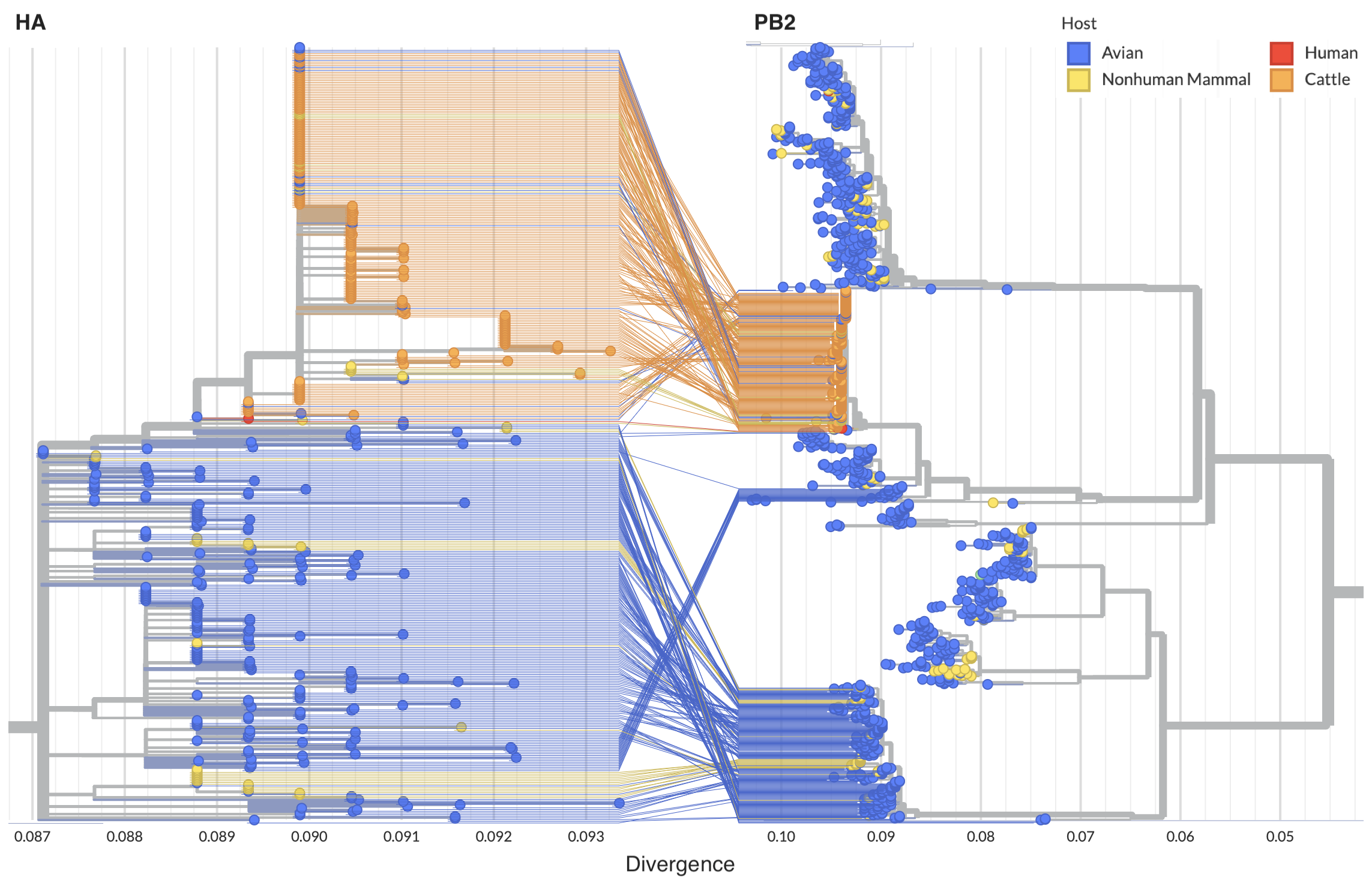
Full genome analysis of concatenated segments provides substantially finer resolution
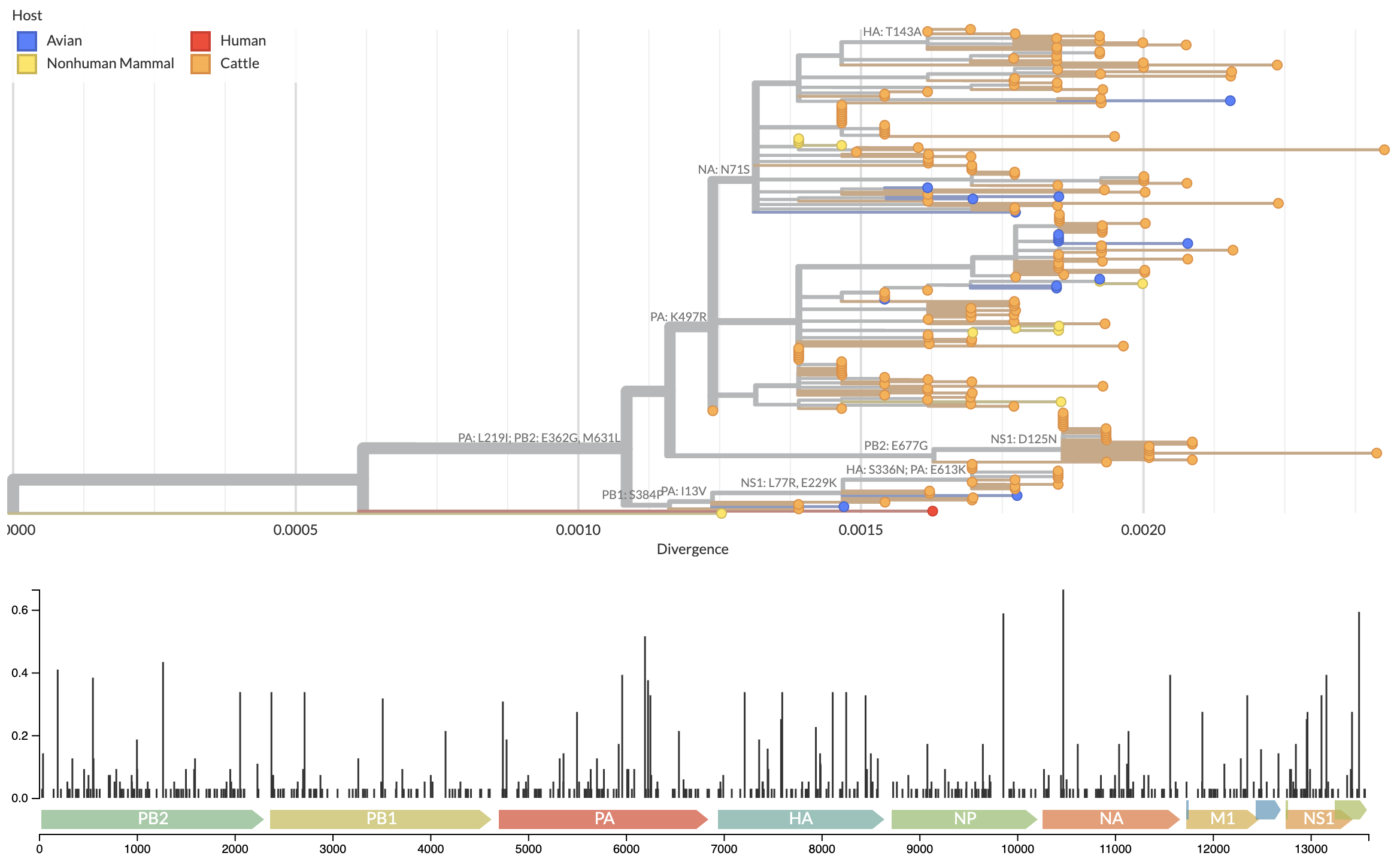
Growing epizootic in cattle after introduction in ~Dec with subsequent spillback into birds
Molecular clock dating introduction into cattle
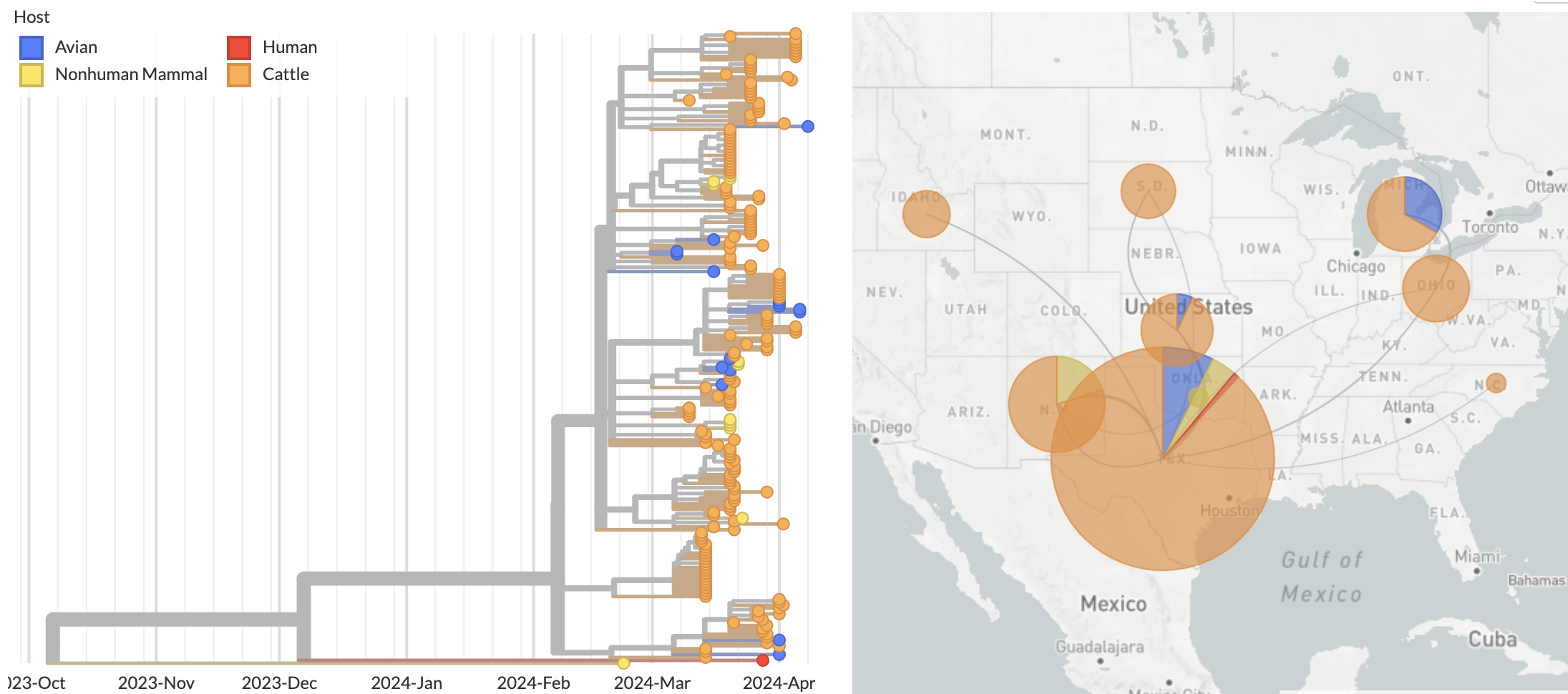
Human case outgroups to cattle infections, suggesting unsampled diversity and potentially a second cattle introduction
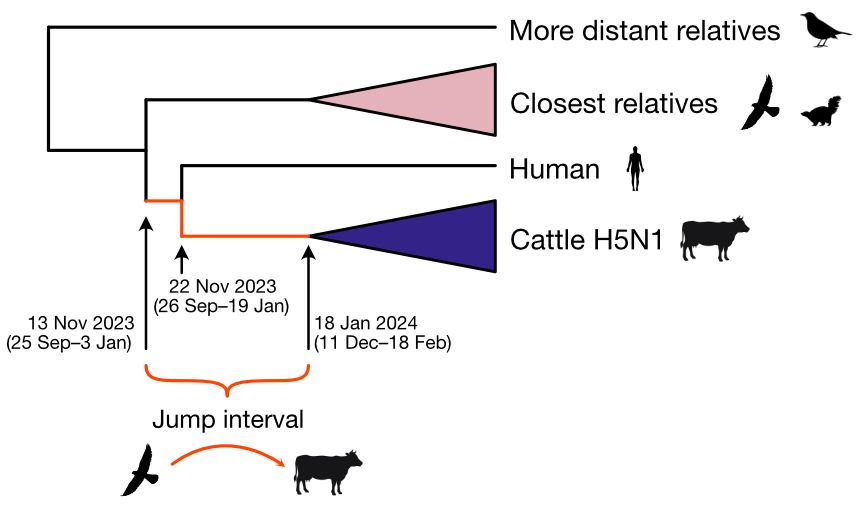
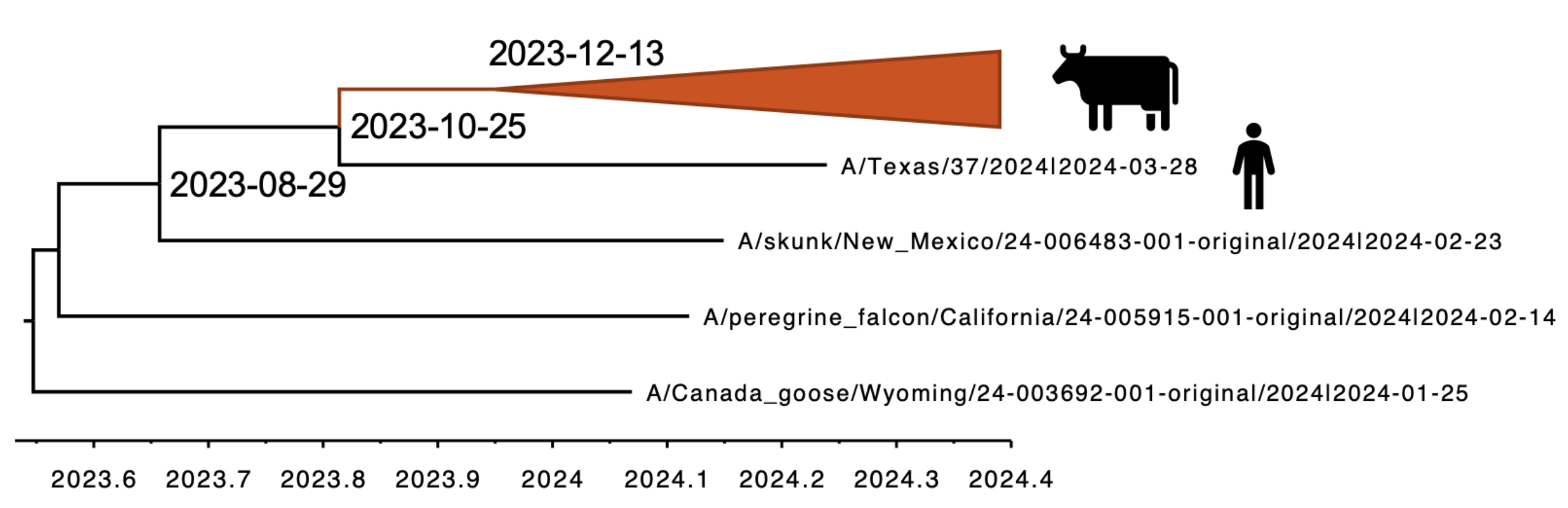
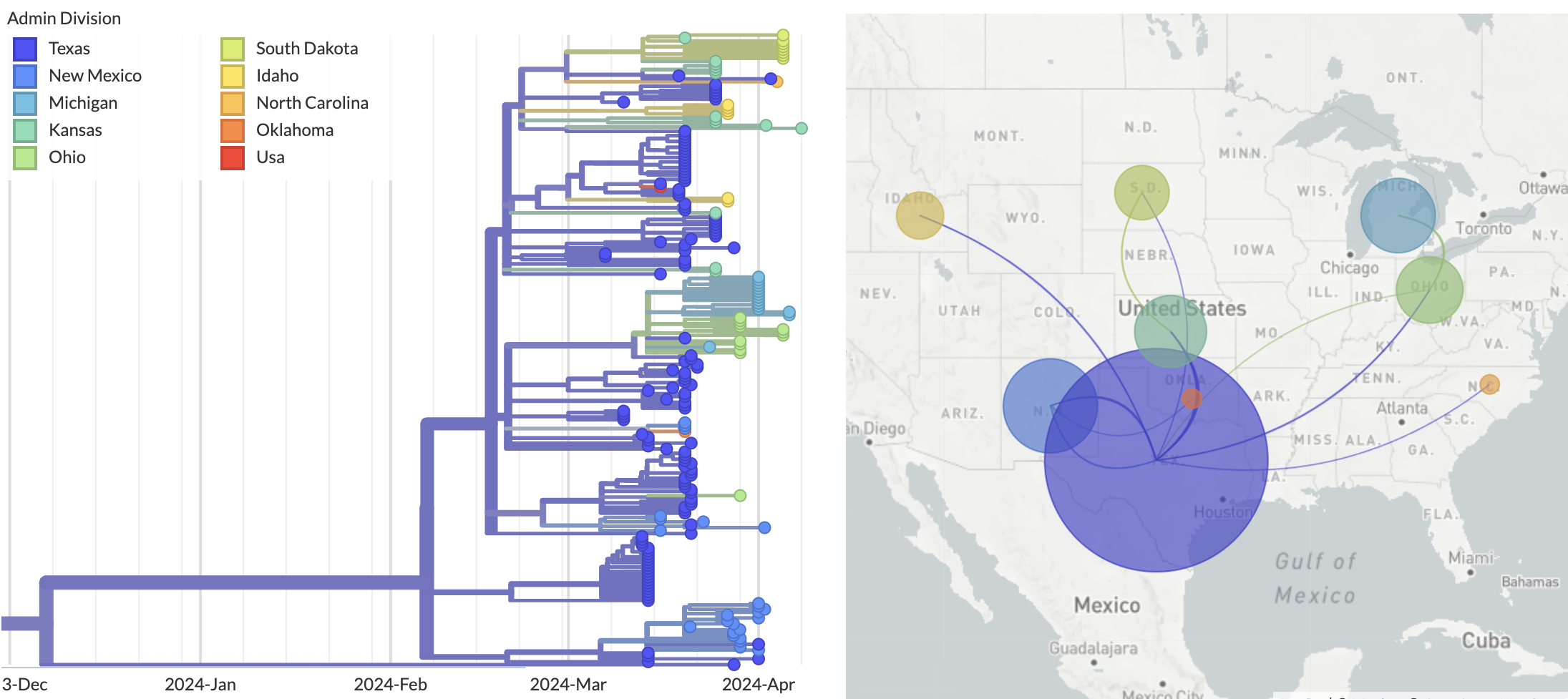
Epizootic origin appears to be Texas congruent with initial cases
But note sampling bias, where 119 of 189 cattle-derived sequences were sampled from Texas
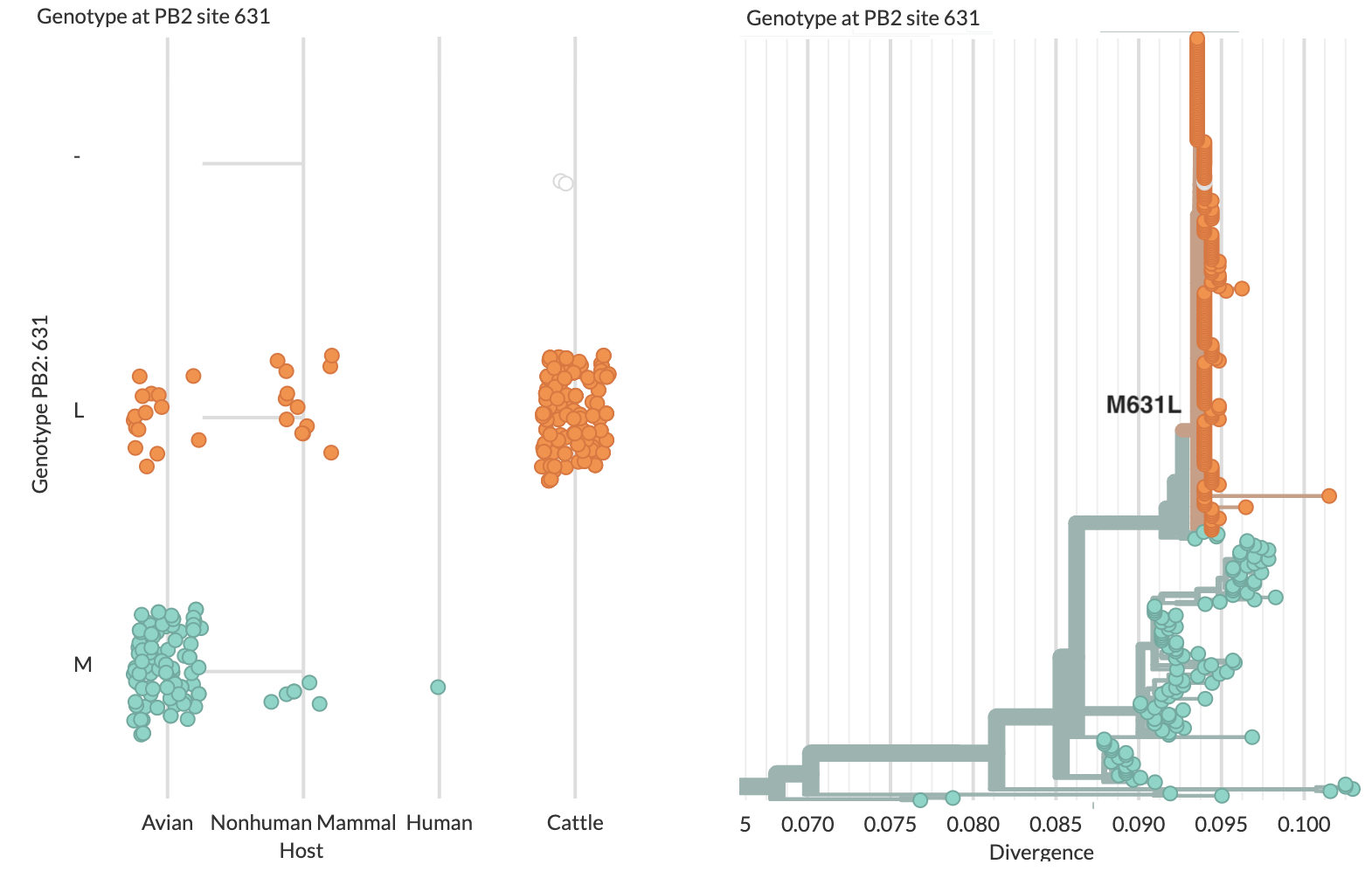
Specific amino acid changes associated with cattle adaptation
Data systems
Timeline of initial genomic data
- Mar 25
- First confirmed detection in Texas, with 10 farms in three states on Mar 30
- Mar 27
- Six viral genomes from Texas shared to GISAID by NVSL and a seventh on Apr 5
- Apr 21
- Raw reads for 239 samples shared to SRA (but lack metadata)
- Apr 22
- Karthik Gangavarapu assembles these reads and shares consensus genomes to GitHub
- Apr 26
- Florence Débarre shares metadata scraped from USDA YouTube presentation
- May 1
- Nguyen et al preprint shared to bioRxiv
- May 2
- Consensus genomes and metadata for 170 samples from Nguyen et al shared to GISAID by NVSL
- May 3
- Consensus genomes and metadata for Nguyen et al samples shared to GenBank
Current genomic data
Filtered to just cattle sequences
- GISAID
- 177 consensus genomes with metadata (from Nguyen et al)
- GenBank
- 176 consensus genomes with metadata available via this NCBI Virus query (from Nguyen et al)
- SRA
- 211 samples without metadata available via this SRA Run Selector query
Sequences from cattle and other hosts
- SRA
- Currently 460 samples in BioProject PRJNA1102327 with 322 propagated to SRA Run Selector
Current Nextstrain dataflow

Prospective Nextstrain dataflow
Scientific questions addressable by genomic epi
- Is the epizootic rate of growth slowing?
- Are movement restrictions working to keep geographic spread contained?
- Is adaptive evolution to novel cattle host continuing?
Acknowledgements
H5N1 genomic data: APHIS National Veterinary Services lab at the USDA for sharing cattle sequences, other data producers from all over the world for sharing contextual sequences, GISAID for collating data and Karthik Gangavarapu and colleagues for sharing assembled consensus genomes
H5N1 genomic analysis: ![]() Louise Moncla, James Hadfield, John Huddleston
Louise Moncla, James Hadfield, John Huddleston
Nextstrain: Richard Neher, Ivan Aksamentov, John Anderson, Kim Andrews, Jennifer Chang, James Hadfield, Emma Hodcroft, John Huddleston, Jover Lee, Victor Lin, Cornelius Roemer, Thomas Sibley
Bedford Lab:
![]() John Huddleston,
John Huddleston,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Miguel Paredes,
Miguel Paredes,
![]() Marlin Figgins,
Marlin Figgins,
![]() Victor Lin,
Victor Lin,
![]() Jennifer Chang,
Jennifer Chang,
![]() Eslam Abousamra,
Eslam Abousamra,
![]() Nashwa Ahmed,
Nashwa Ahmed,
![]() Cécile Tran Kiem,
Cécile Tran Kiem,
![]() Kim Andrews,
Kim Andrews,
![]() Cristian Ovaduic,
Cristian Ovaduic,
![]() Philippa Steinberg,
Philippa Steinberg,
![]() Jacob Dodds,
Jacob Dodds,
![]() John SJ Anderson
John SJ Anderson





