Tracking and forecasting influenza virus evolution
Trevor Bedford (@trvrb)
29 Aug 2019
Options X
Singapore
Tracking seasonal influenza virus evolution
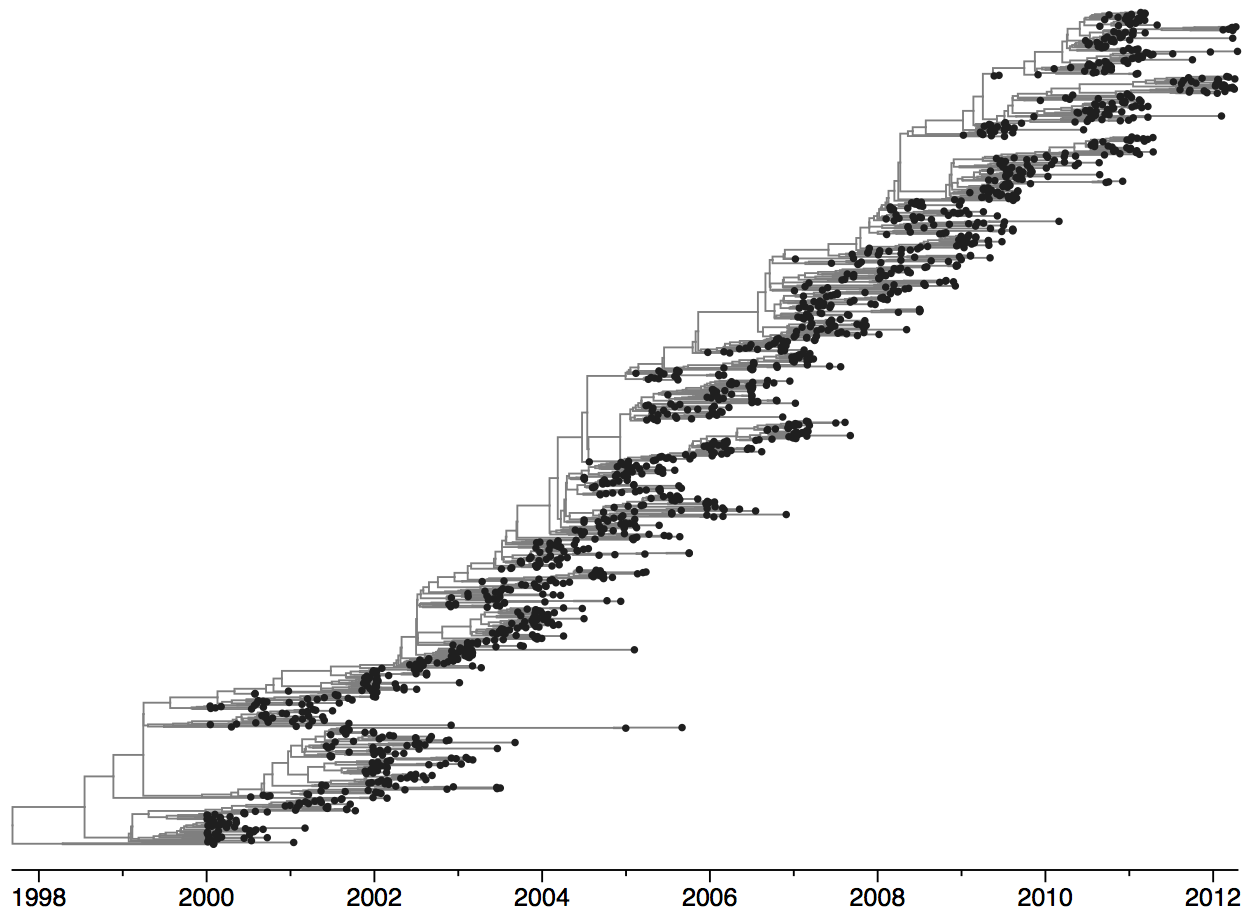
Population turnover of A/H3N2 influenza is extremely rapid
Nextflu
Project to provide a real-time view of the evolving influenza population
All in collaboration with  Richard Neher
Richard Neher
Nextflu → Nextstrain
Real-time tracking of pathogen evolution
with
![]() Richard Neher,
Richard Neher,
![]() James Hadfield,
James Hadfield,
![]() Emma Hodcroft,
Emma Hodcroft,
![]() Thomas Sibley,
Thomas Sibley,
![]() Colin Megill,
Colin Megill,
![]() John Huddleston,
John Huddleston,
![]() Barney Potter,
Barney Potter,
![]() Sidney Bell,
Sidney Bell,
![]() Louise Moncla,
Louise Moncla,
![]() Charlton Callender,
Charlton Callender,
![]() Misja Ilcisin,
Misja Ilcisin,
![]() Kairsten Fay,
Kairsten Fay,
![]() Jover Lee
Jover Lee
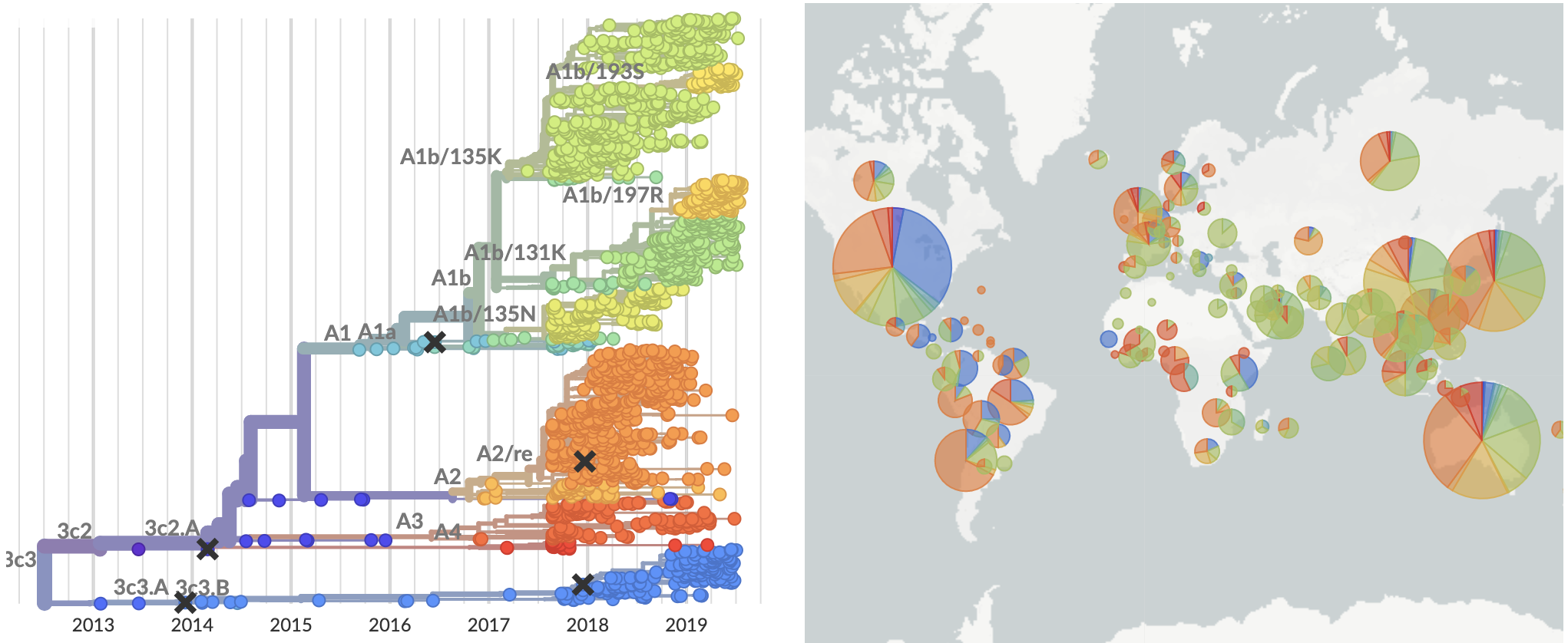
Current view of H3N2 from nextstrain.org/flu
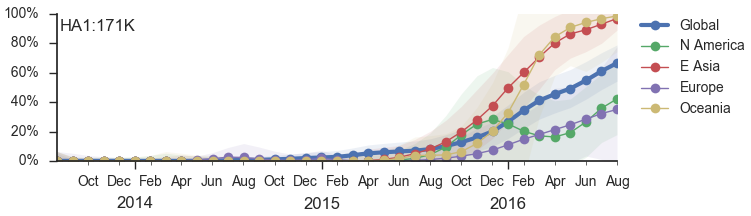
Forecasting has been a goal for a while
At Options IX in 2016, I stated:
We predict the 171K clade will continue to be successful (unless supplanted by a novel mutant)
![]()
Instead, momentum behind 171K (clade 3c2.A1) waned and co-circulation has increased
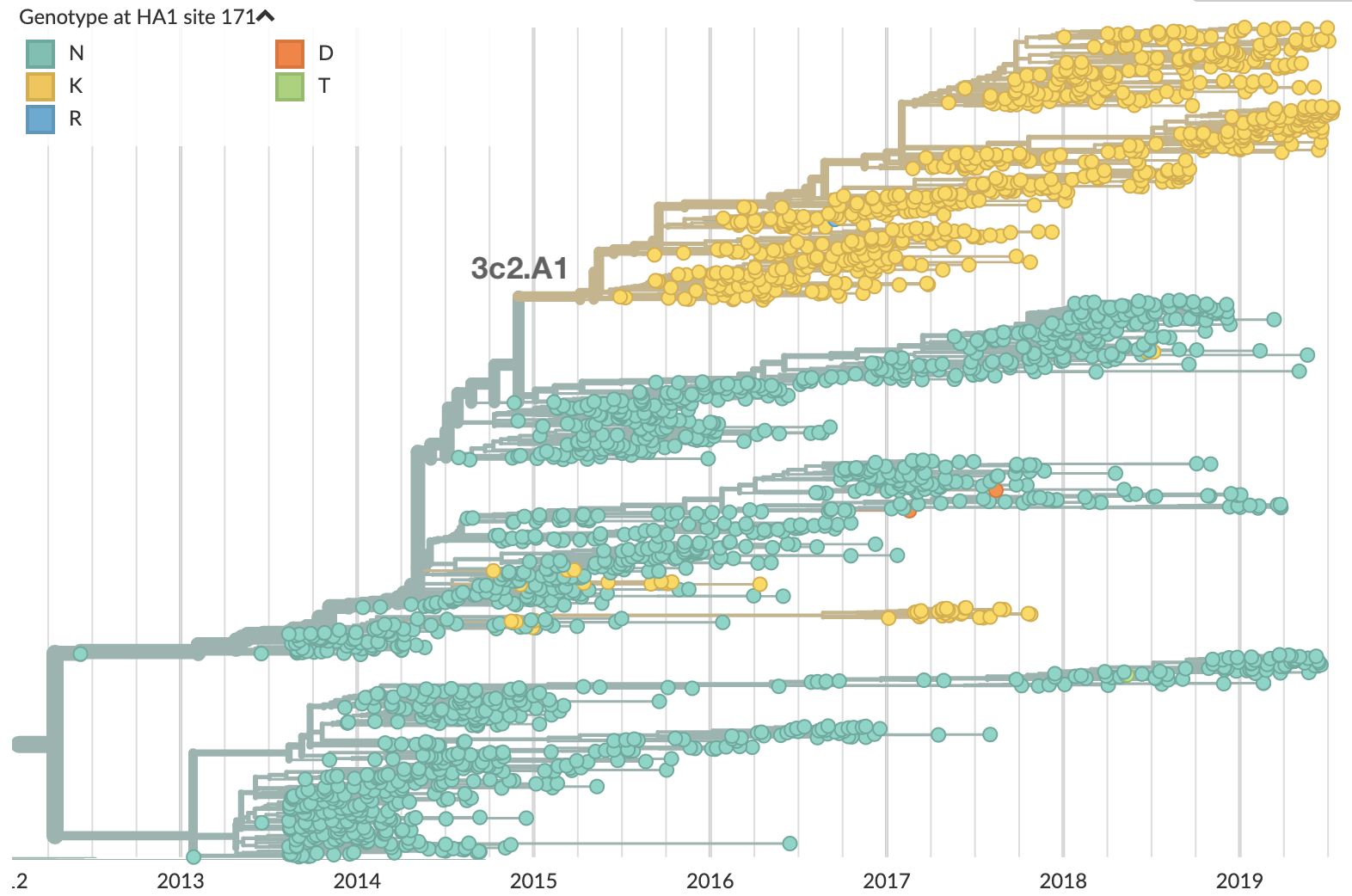
What's going on with this co-circulation?
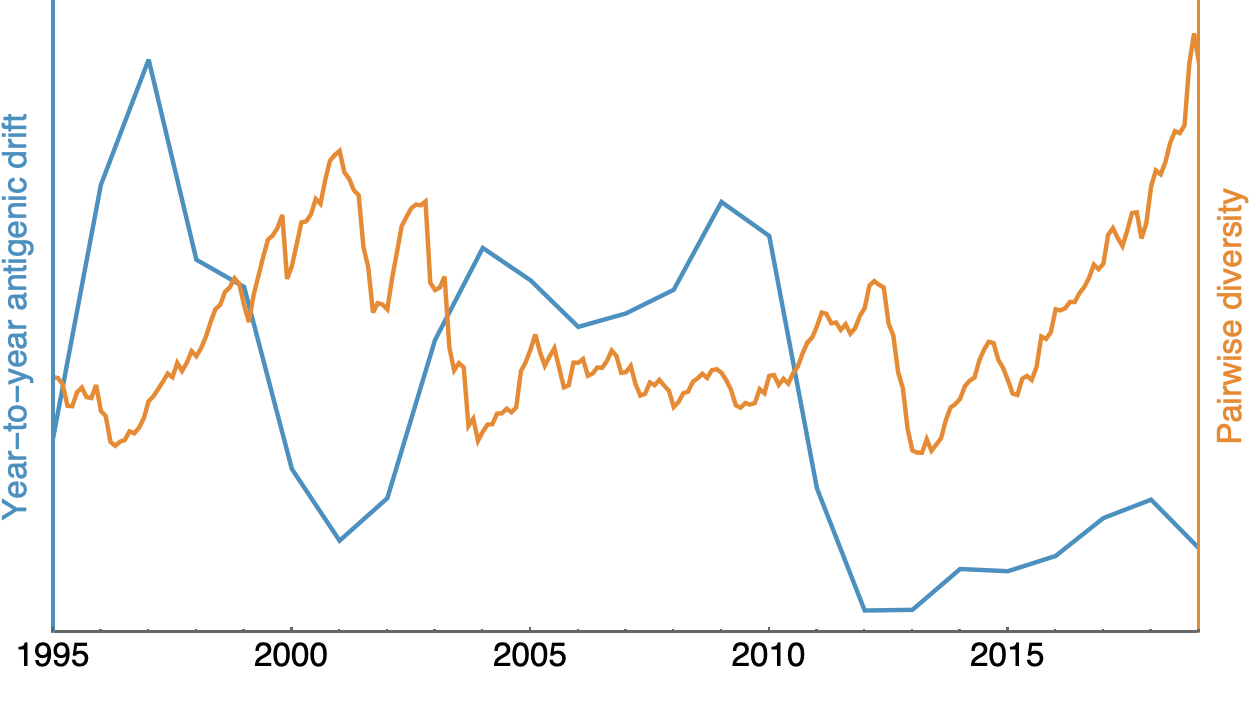
Looking historically shows this to be rare
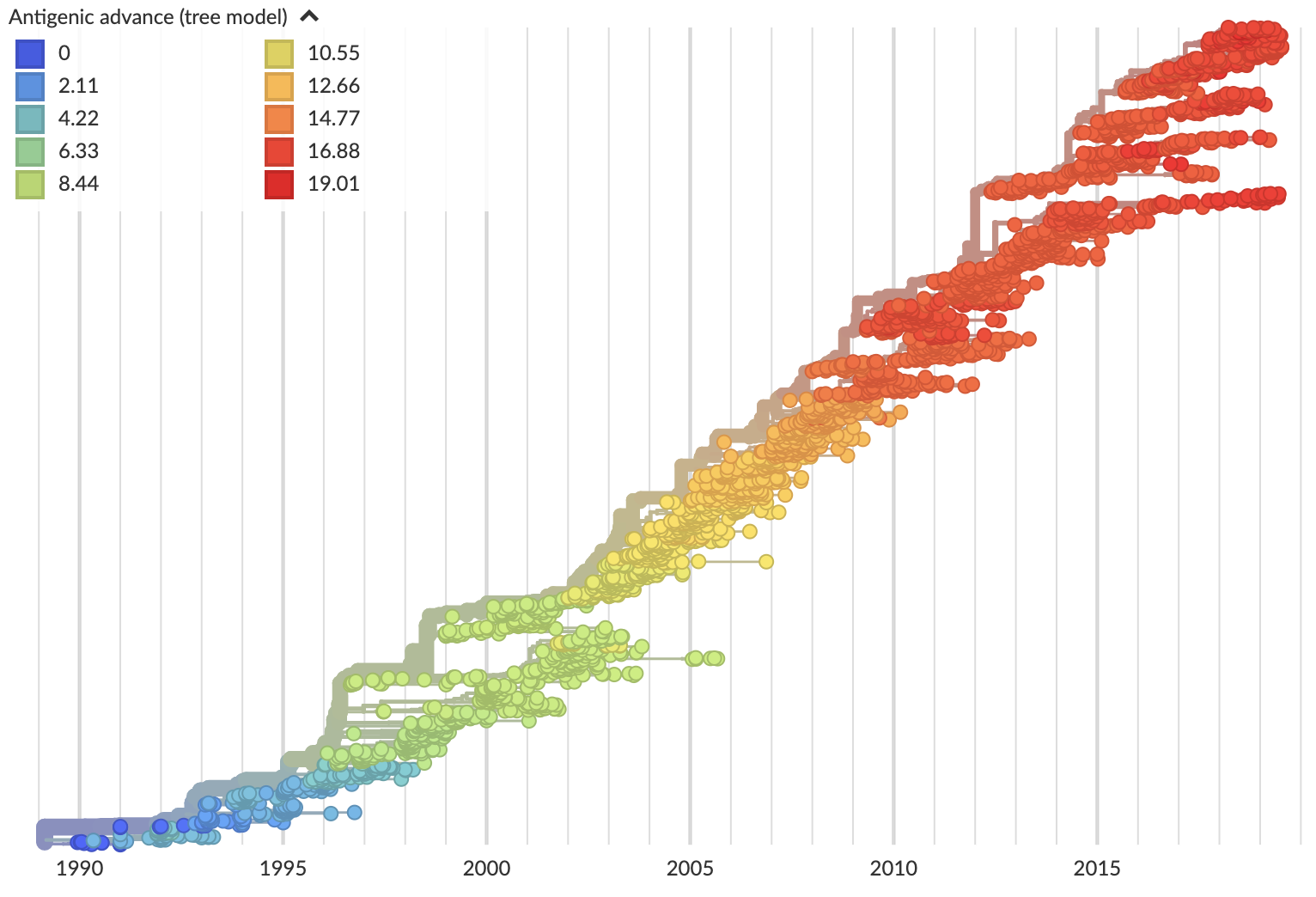
Variation in both diversity and rate of HI drift
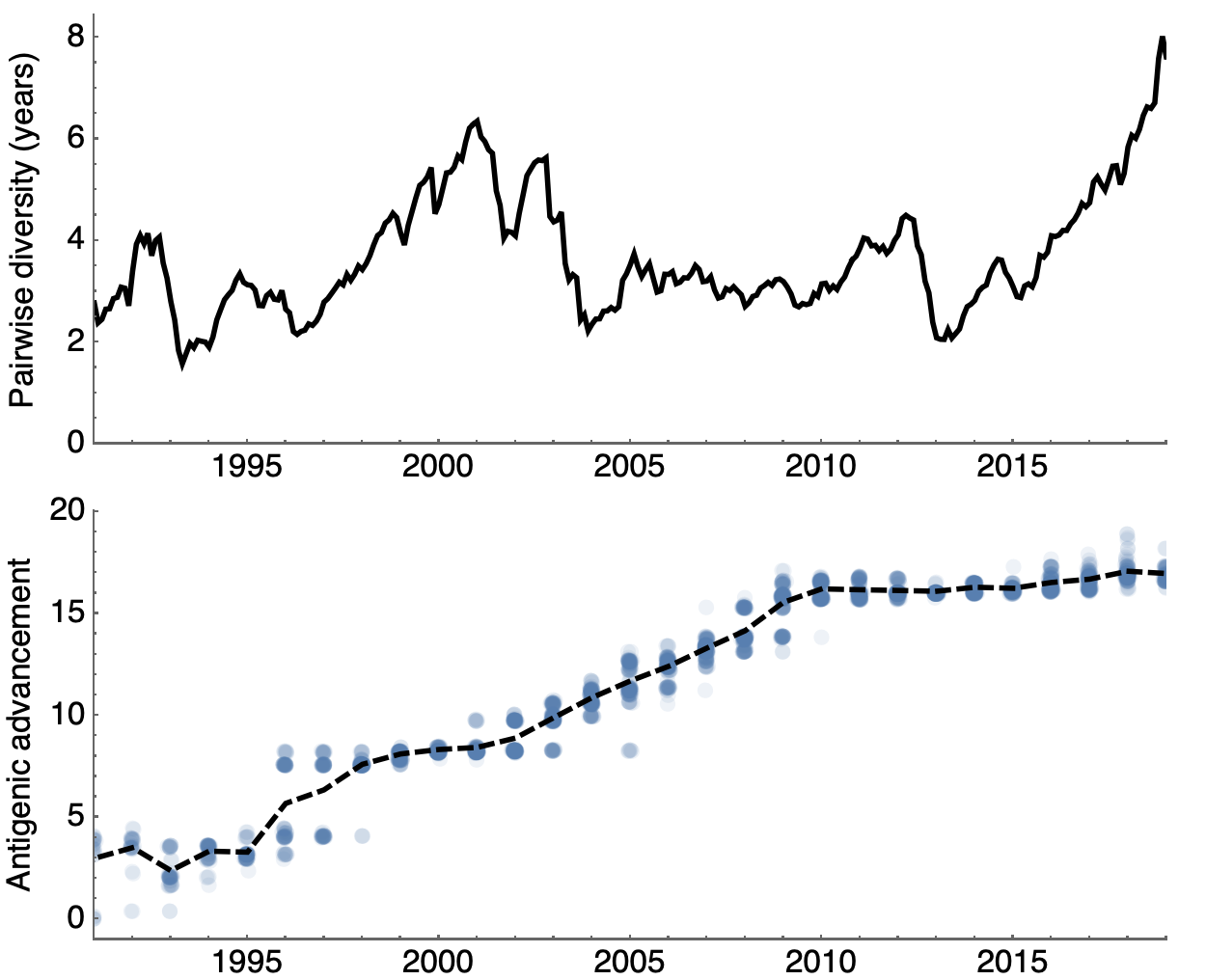
Possible correlation of increased diversity when drift is slow
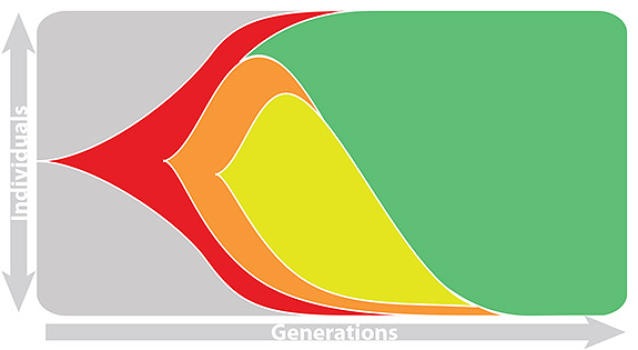
Risk of eventual "speciation"
Forecasting strain turnover
with  John Huddleston
and
John Huddleston
and  Richard Neher
Richard Neher
Fitness models project strain frequencies
Future frequency $x_i(t+\Delta t)$ of strain $i$ derives from strain fitness $f_i$ and present day frequency $x_i(t)$, such that
$$\hat{x}_i(t+\Delta t) = x_i(t) \, \mathrm{exp}(f_i \, \Delta t)$$
Total strain frequencies at each timepoint are normalized. This captures clonal interference between competing lineages.
Two inputs
- Estimate of present-day strain frequencies $x(t)$
- Estimate of present-day strain fitnesses $f$
Strain frequency estimated via region-weighted KDE
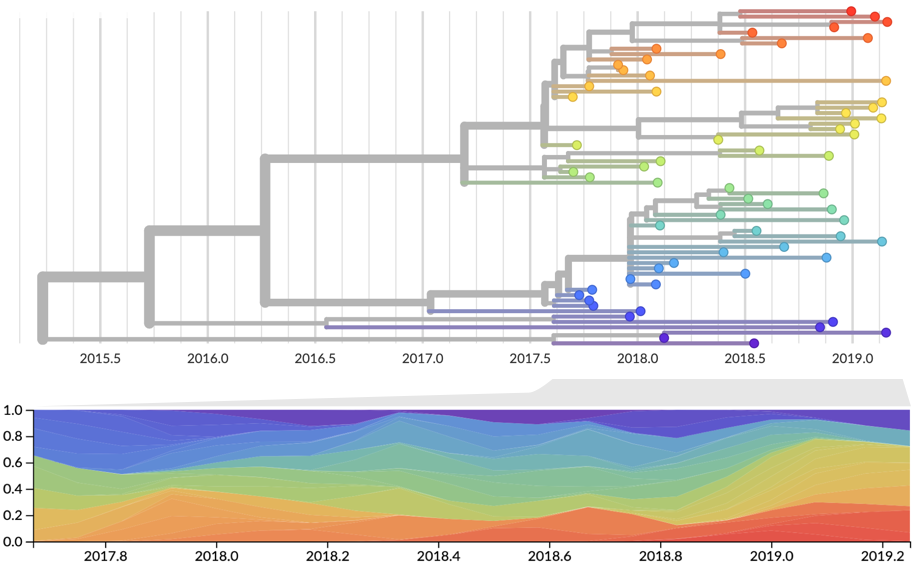
Strain fitness estimated from viral attributes
The fitness $f$ of strain $i$ is estimated as
$$\hat{f}_i = \beta^\mathrm{A} \, f_i^\mathrm{A} + \beta^\mathrm{B} \, f_i^\mathrm{B} + \ldots$$
where $f^A$, $f^B$, etc... are different standardized viral attributes and $\beta^A$, $\beta^B$, etc... coefficients are trained based on historical evolution
| Antigenic drift | Intrinsic fitness | Recent growth |
|---|---|---|
| epitope mutations | non-epitope mutations | local branching index |
| HI titers | DMS data (via Bloom lab) | delta frequency |
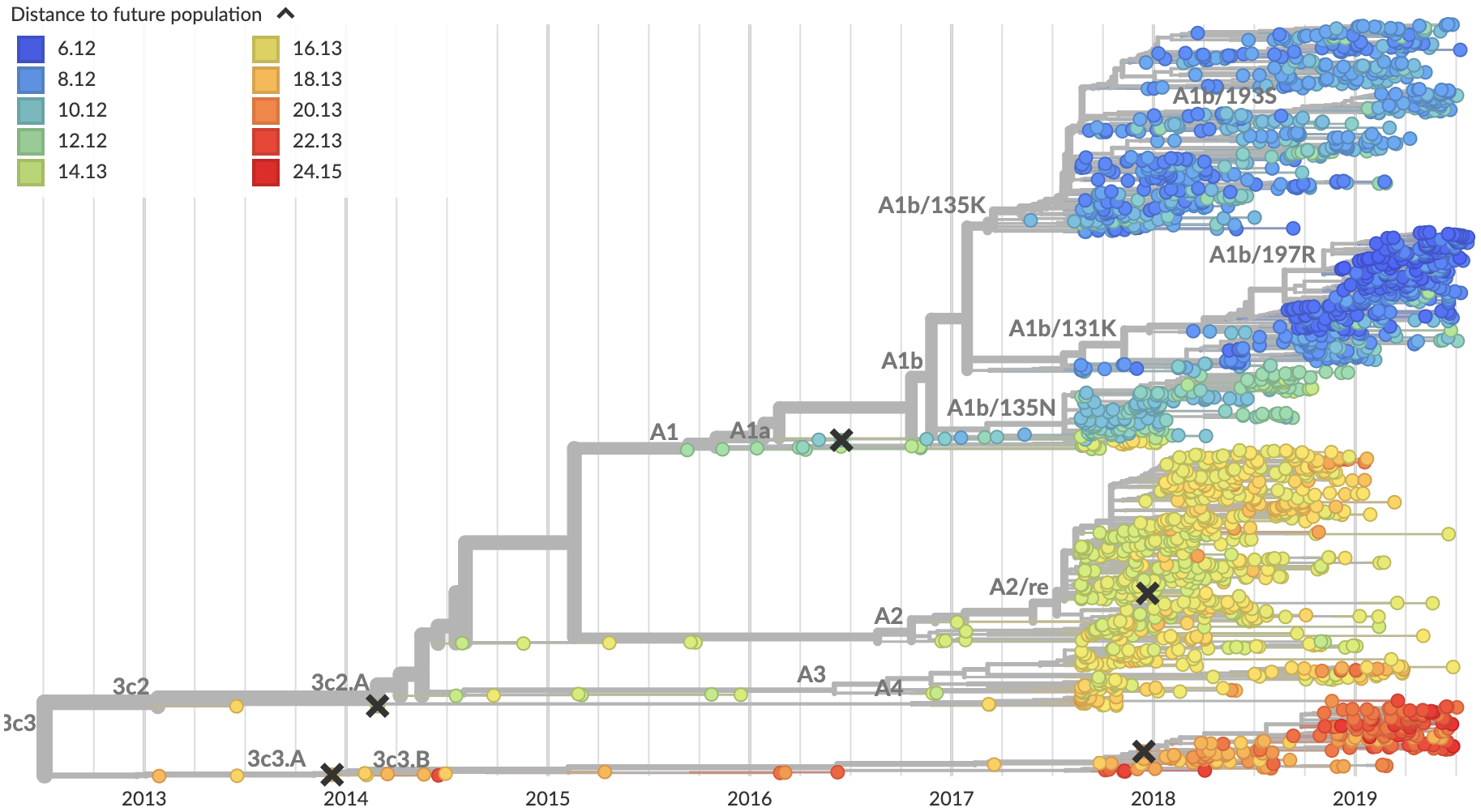
Future population depends on frequency and fitness
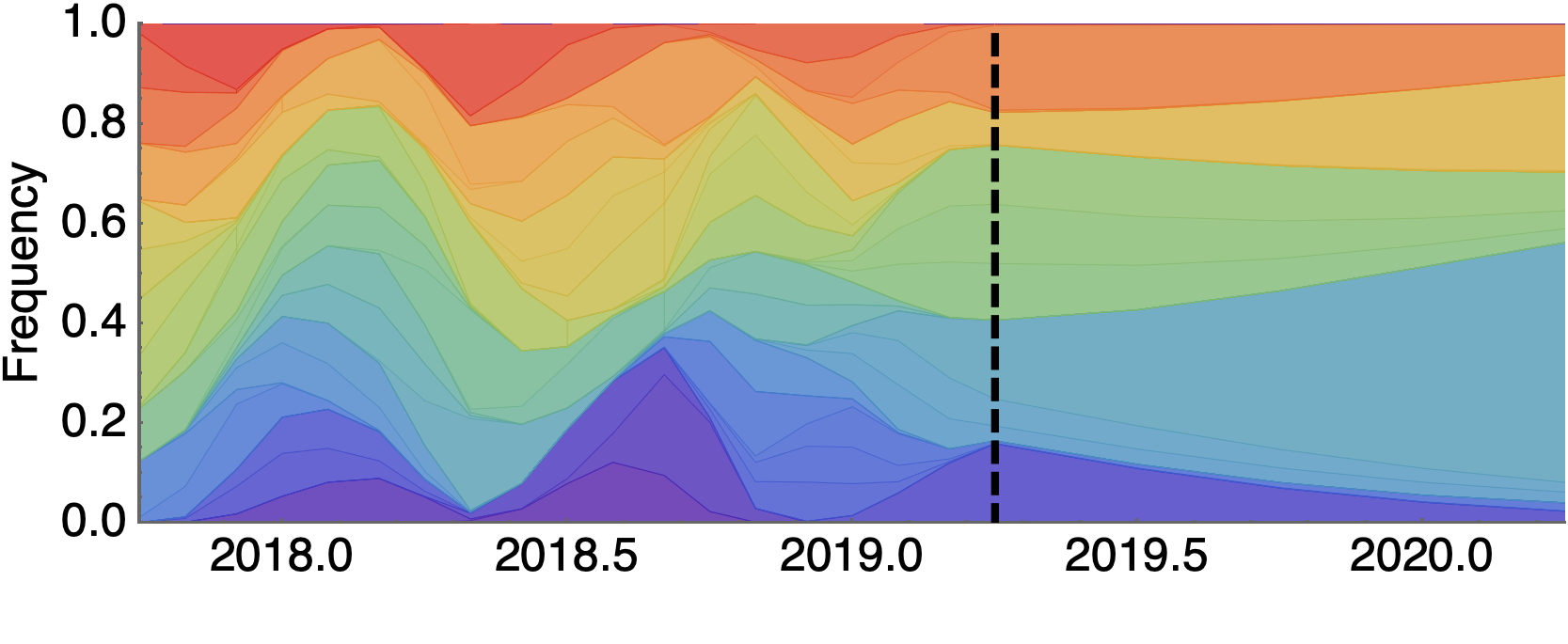
Forecast assessed based on weighted distance match to observed future population
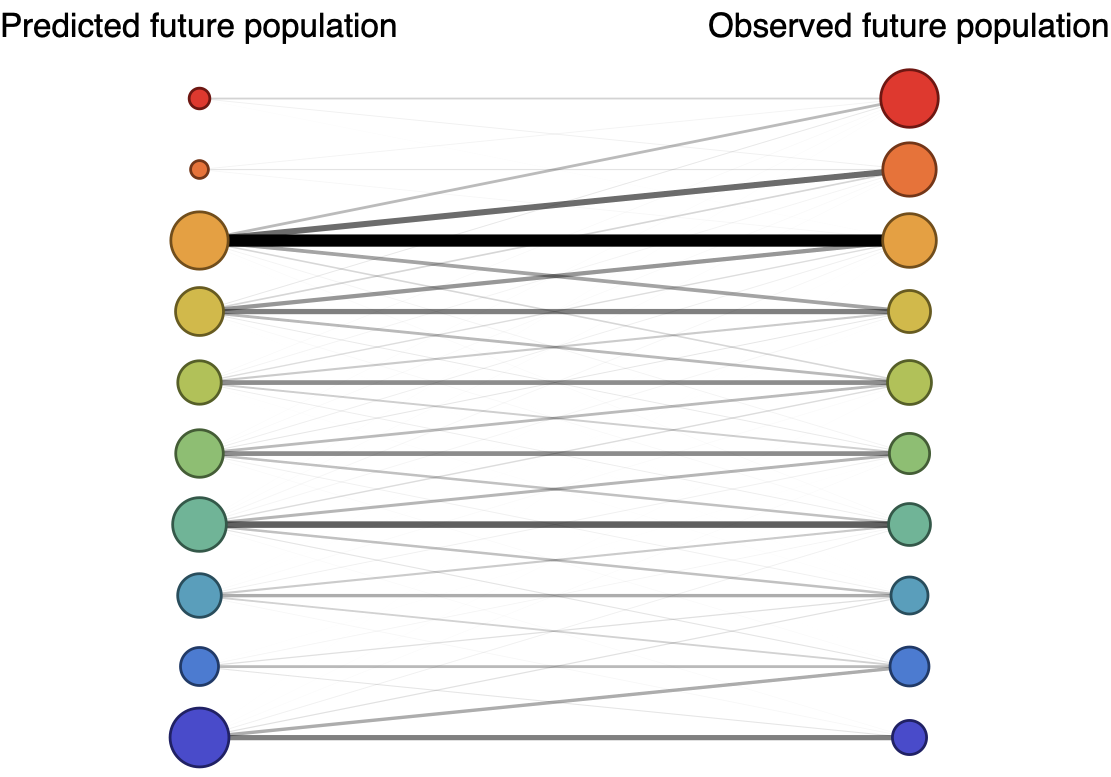
Forecast assessed based on weighted distance match to observed future population
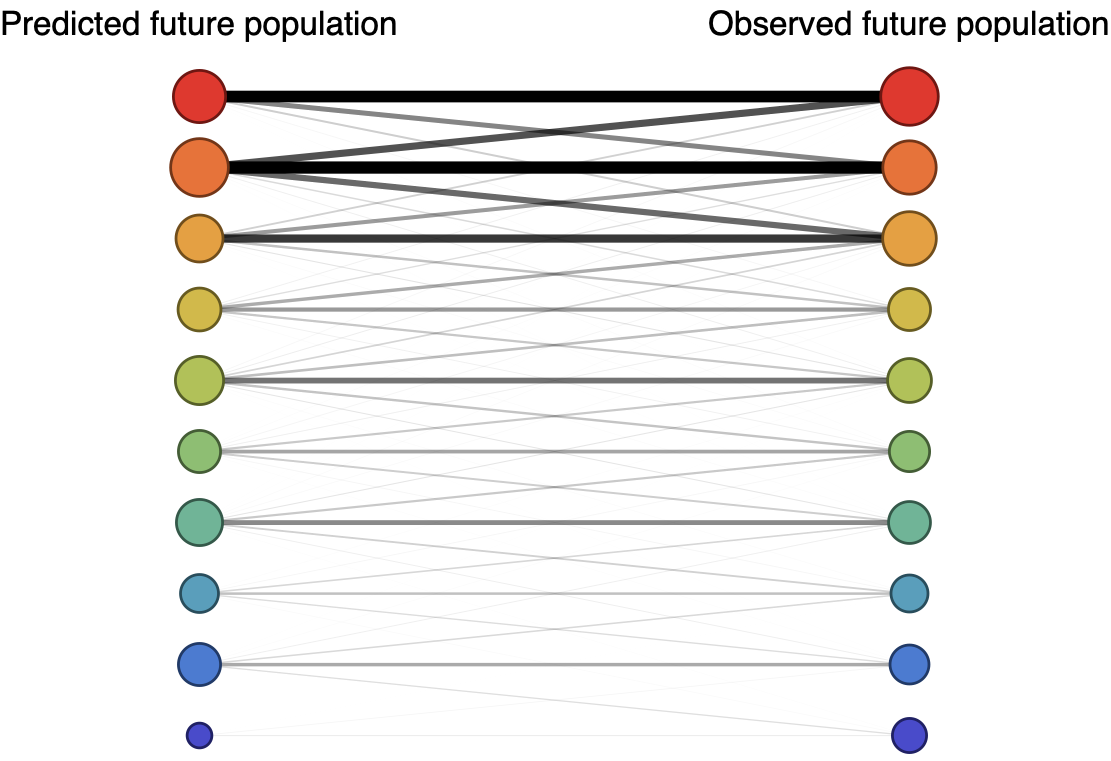
Train in 6-year sliding windows from 1995 to 2015 with most recent years held out as test
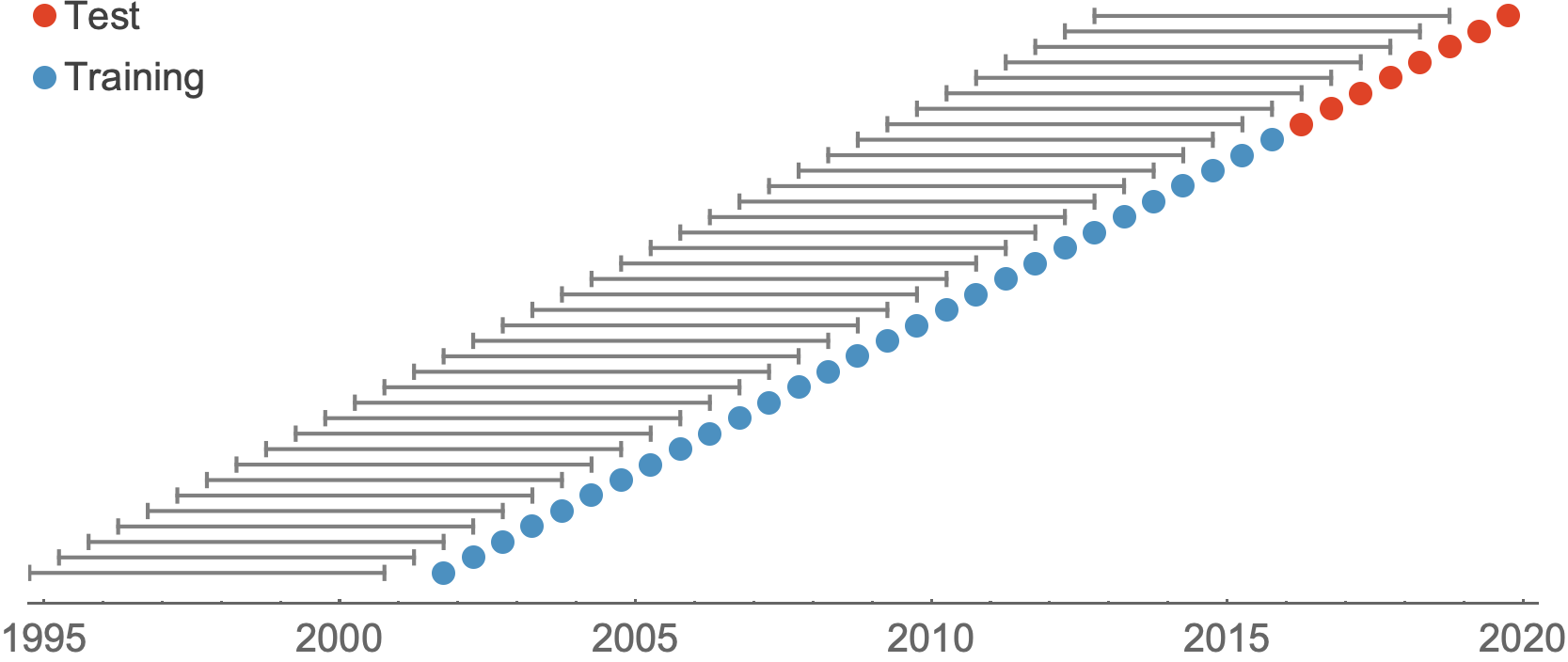
Within-category performance favors HI drift, non-epitope fitness and local branching index
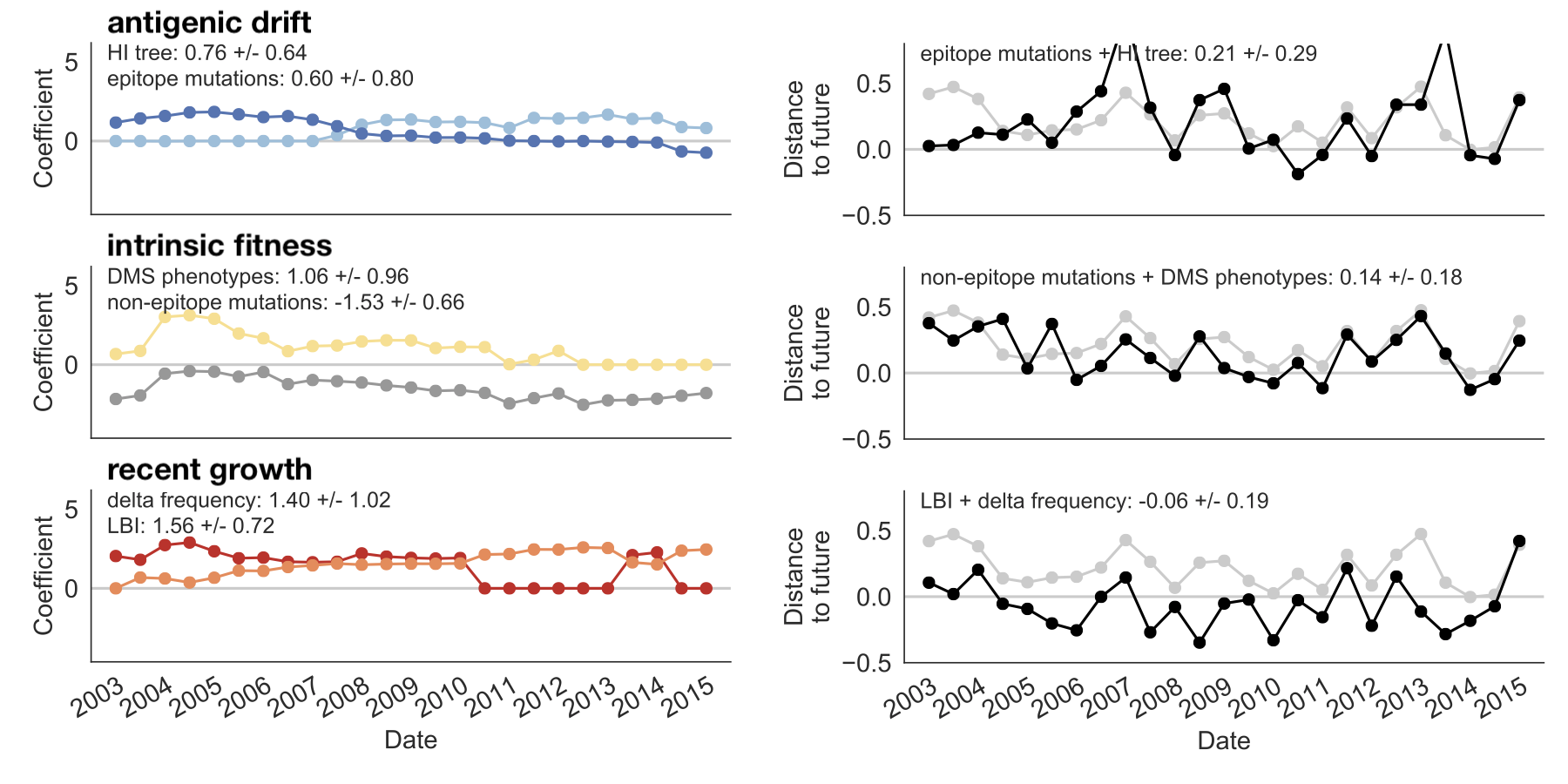
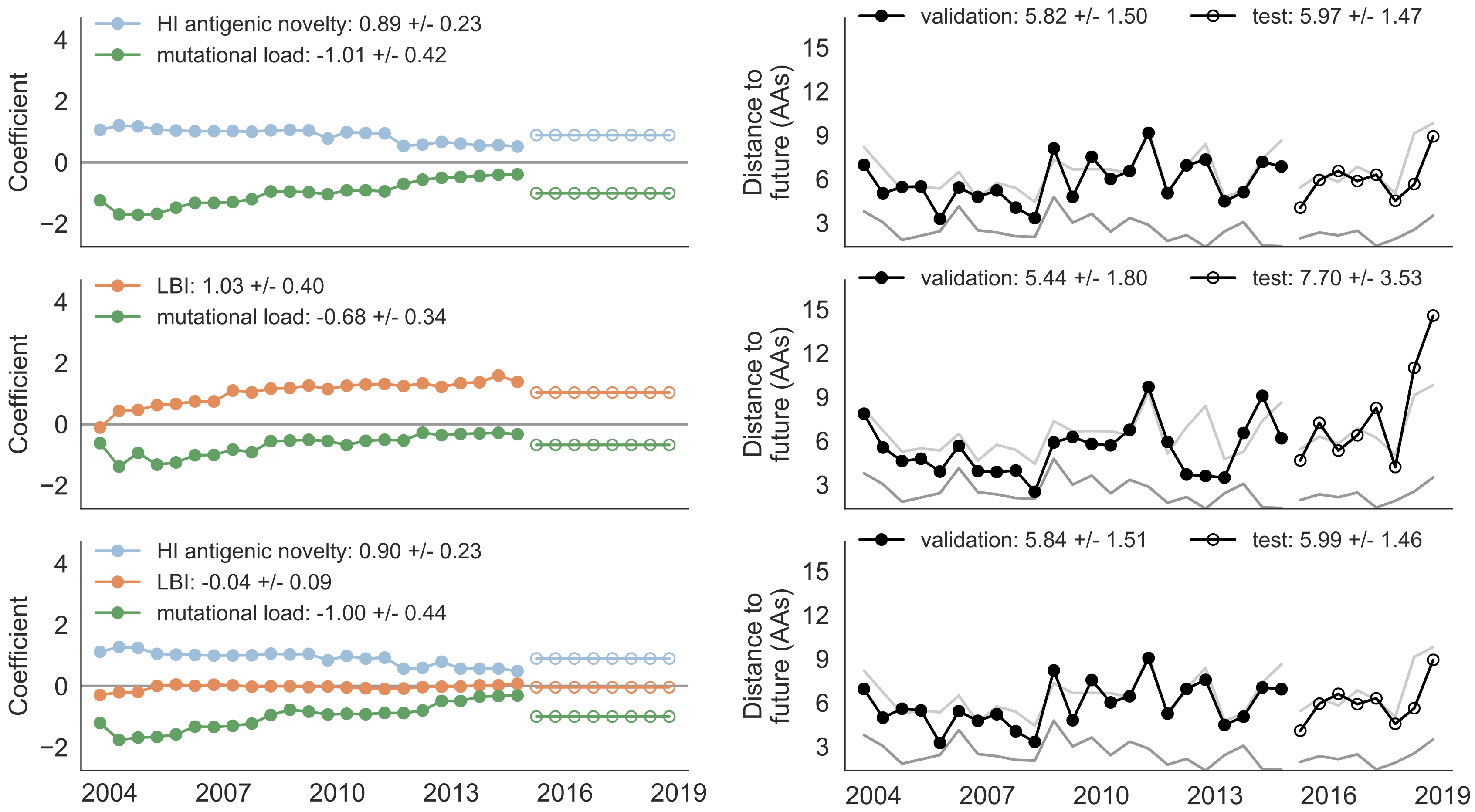
Composite model relies on local branching index and non-epitope fitness
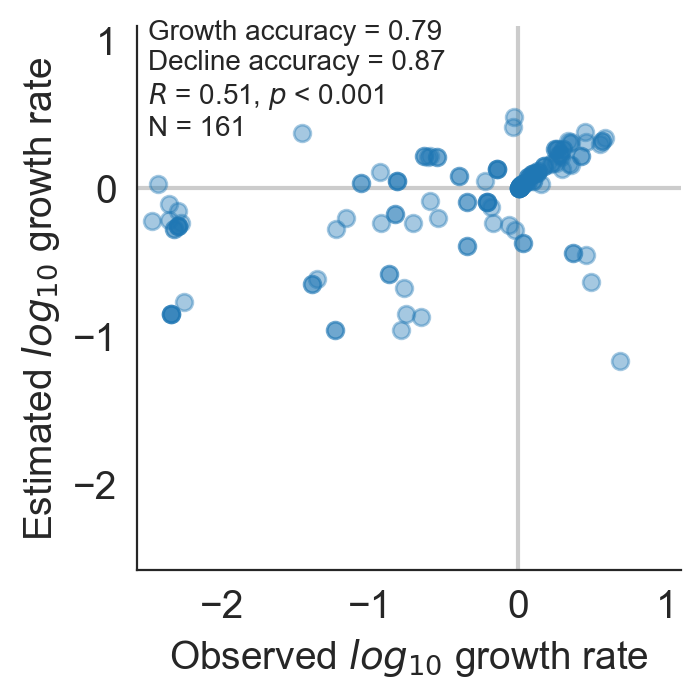
Model successfully predicts clade growth
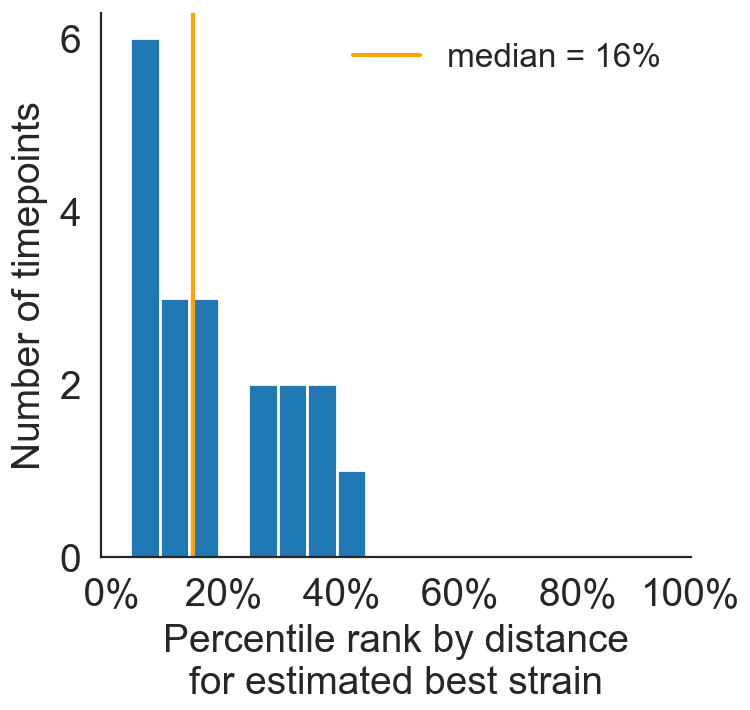
Best pick from model is generally close to best possible retrospective pick
Forecast from current virus population
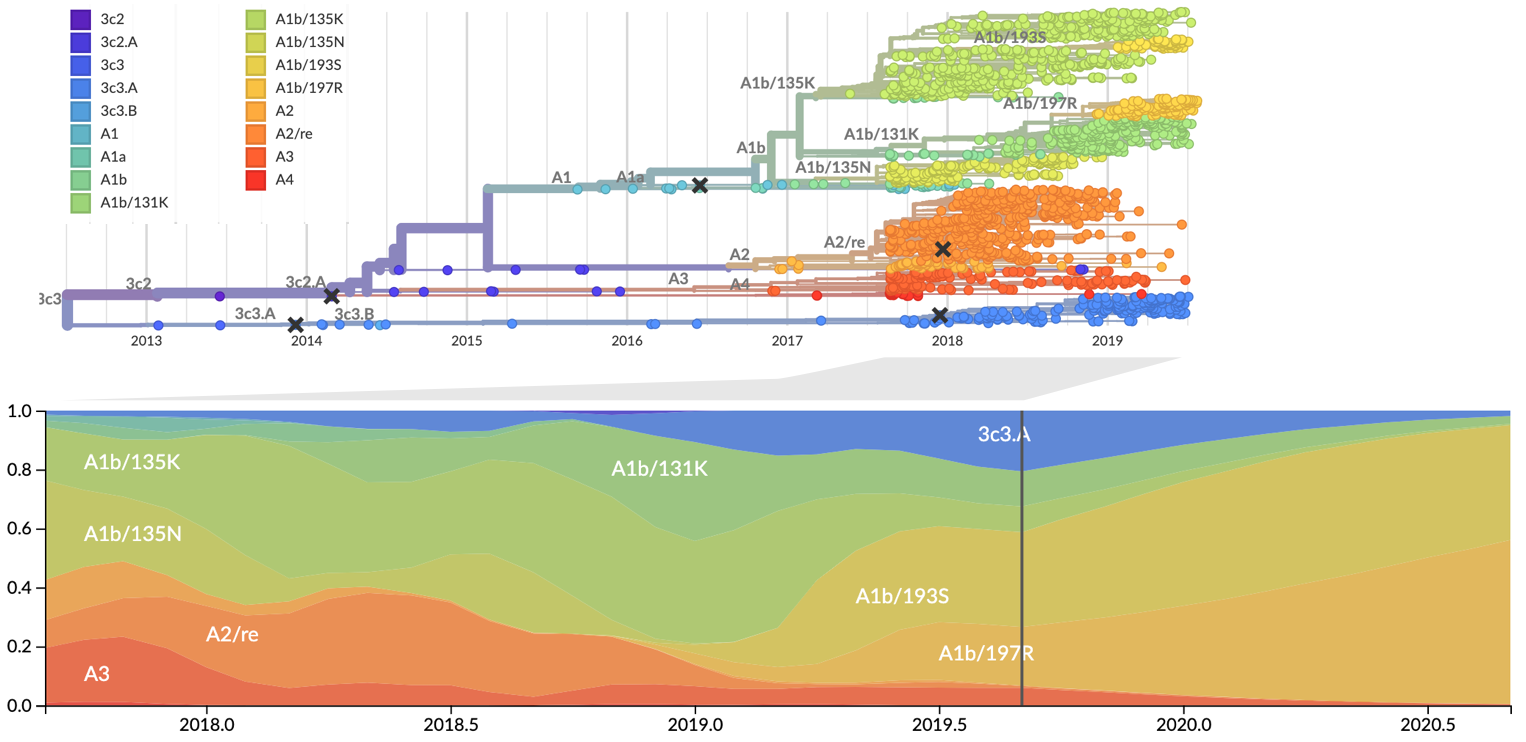
Since March these two clades have begun to spread throughout the world
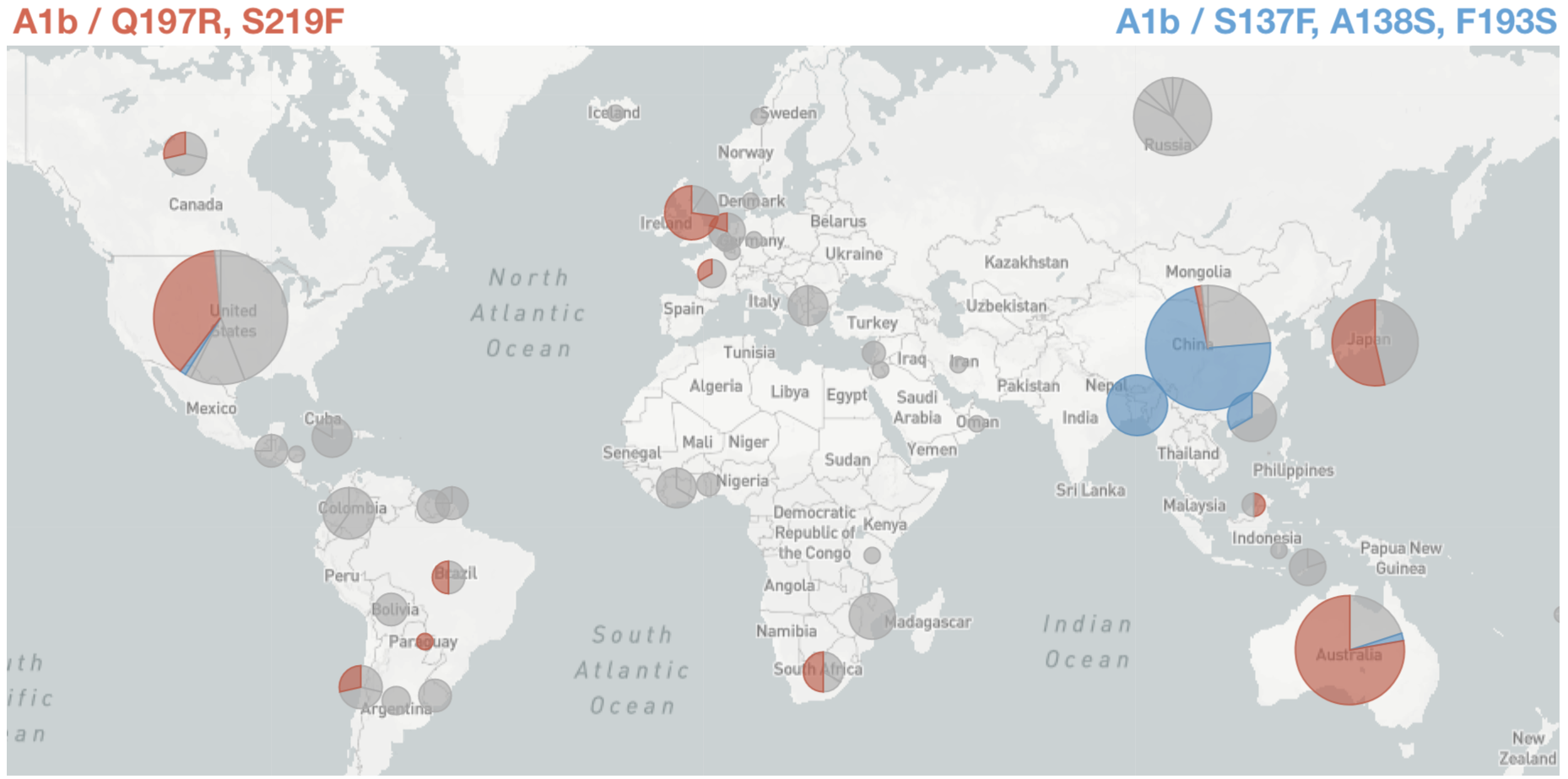
Predicted sequence match of circulating strains to future population
Goal is to start rolling these forecasts out live to nextstrain.org/flu shortly
Future work
- Better predictors for antigenic drift
- Incorporate NA evolution
- Incorporate geography
- Forecast H1N1pdm, Vic and Yam
Acknowledgements
Bedford Lab:
![]() Alli Black,
Alli Black,
![]() John Huddleston,
John Huddleston,
![]() Barney Potter,
Barney Potter,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Louise Moncla,
Louise Moncla,
![]() Maya Lewinsohn,
Maya Lewinsohn,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Kairsten Fay,
Kairsten Fay,
![]() Misja Ilcisin
Misja Ilcisin
This work: WHO Global Influenza Surveillance Network, GISAID, John Huddleston, Richard Neher, Barney Potter, James Hadfield, Dave Wentworth, Becky Garten, Marta Łuksza, Michael Lässig, Richard Reeve, Jackie Katz, Colin Russell, John McCauley, Rod Daniels, Kanta Subbarao, Ian Barr, Aeron Hurt, Tomoko Kuwahara, Takato Odagiri






Contact
- Lab website: bedford.io
- Flu tracking: nextstrain.org
- Twitter: @trvrb
- Slides: bedford.io/talks/flu-forecasting-options-2019/