Tracking influenza antigenic evolution and geographic circulation
Trevor Bedford (@trvrb)
27 Jun 2016
Microbiology Department Seminar
Korea University College of Medicine
Slides at bedford.io/talks/
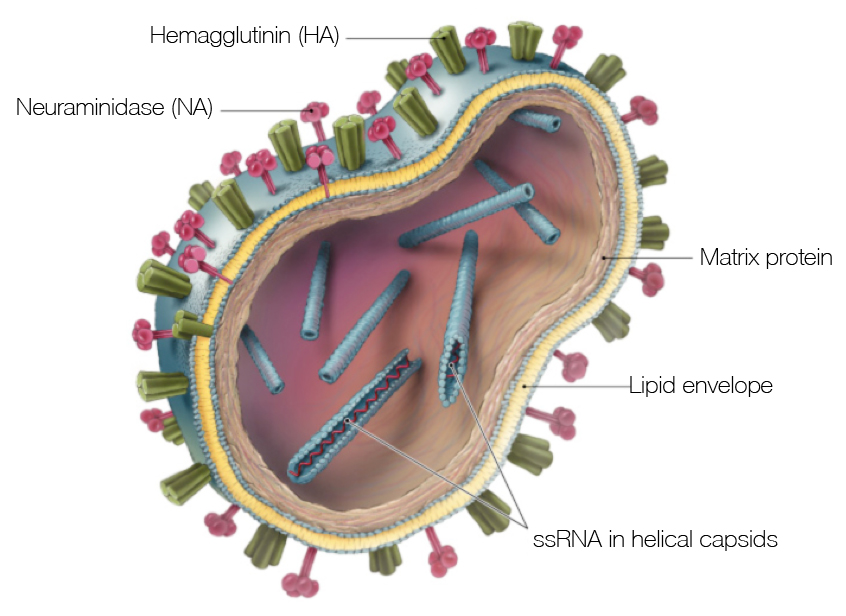
Influenza virus
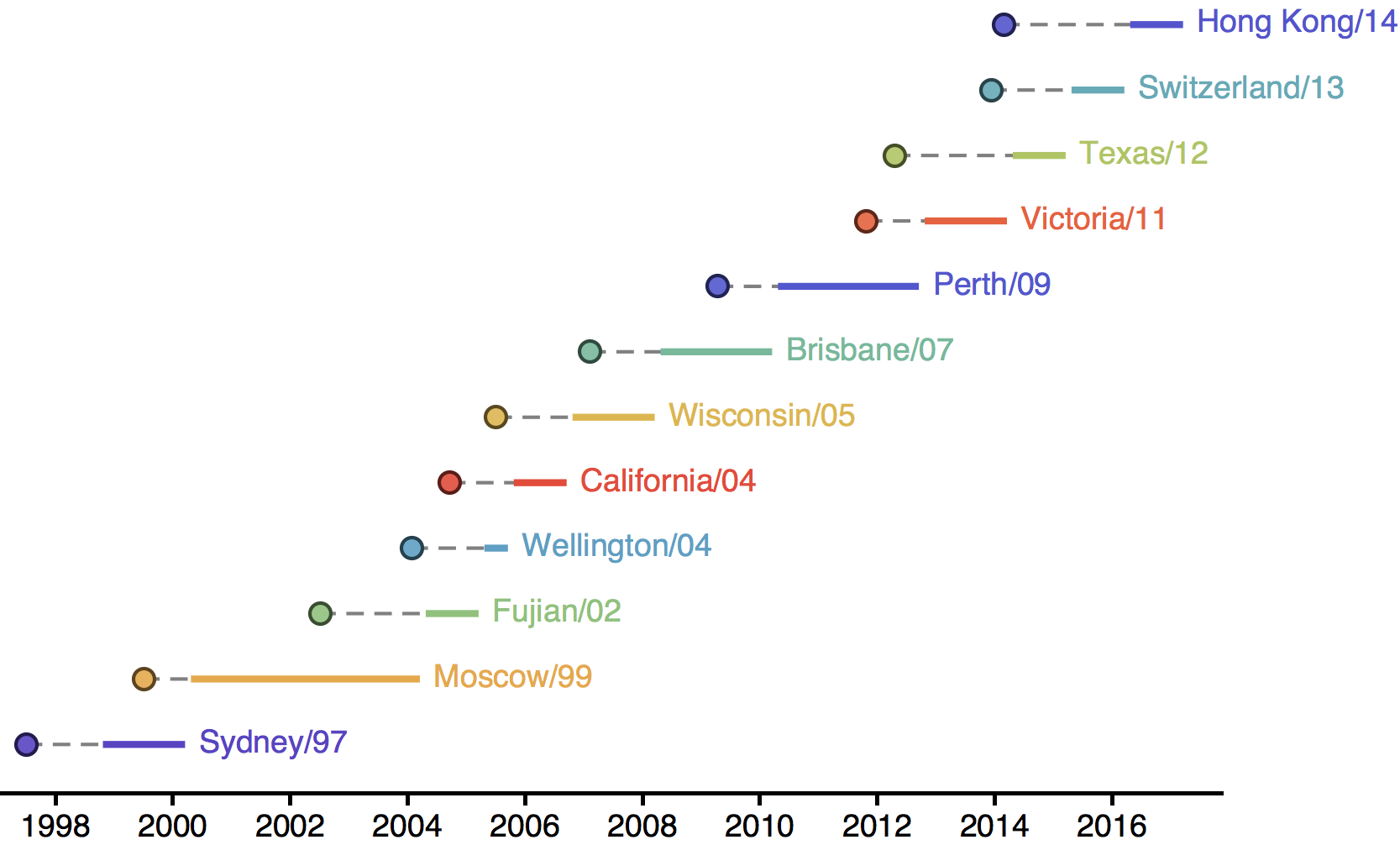
Influenza H3N2 vaccine updates
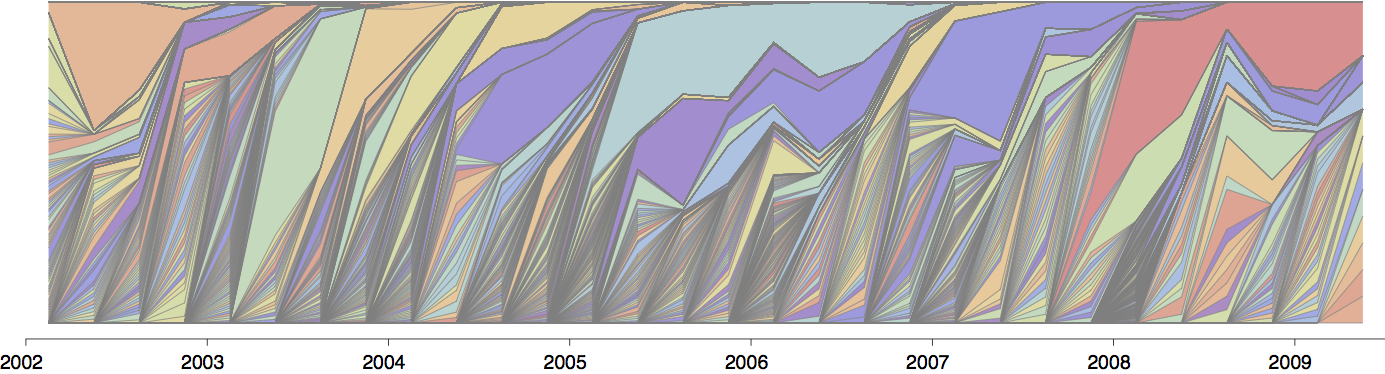
Gene flow and population turnover
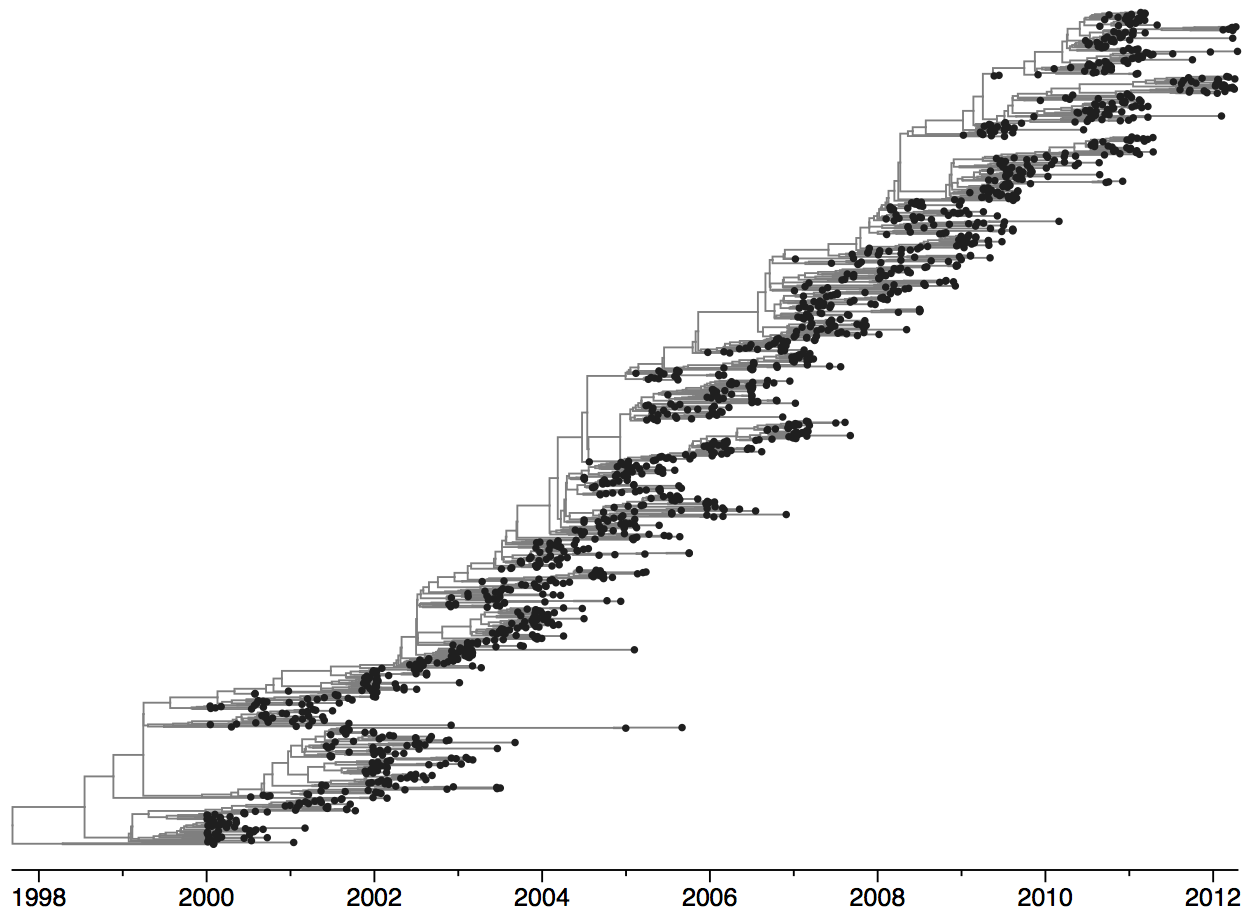
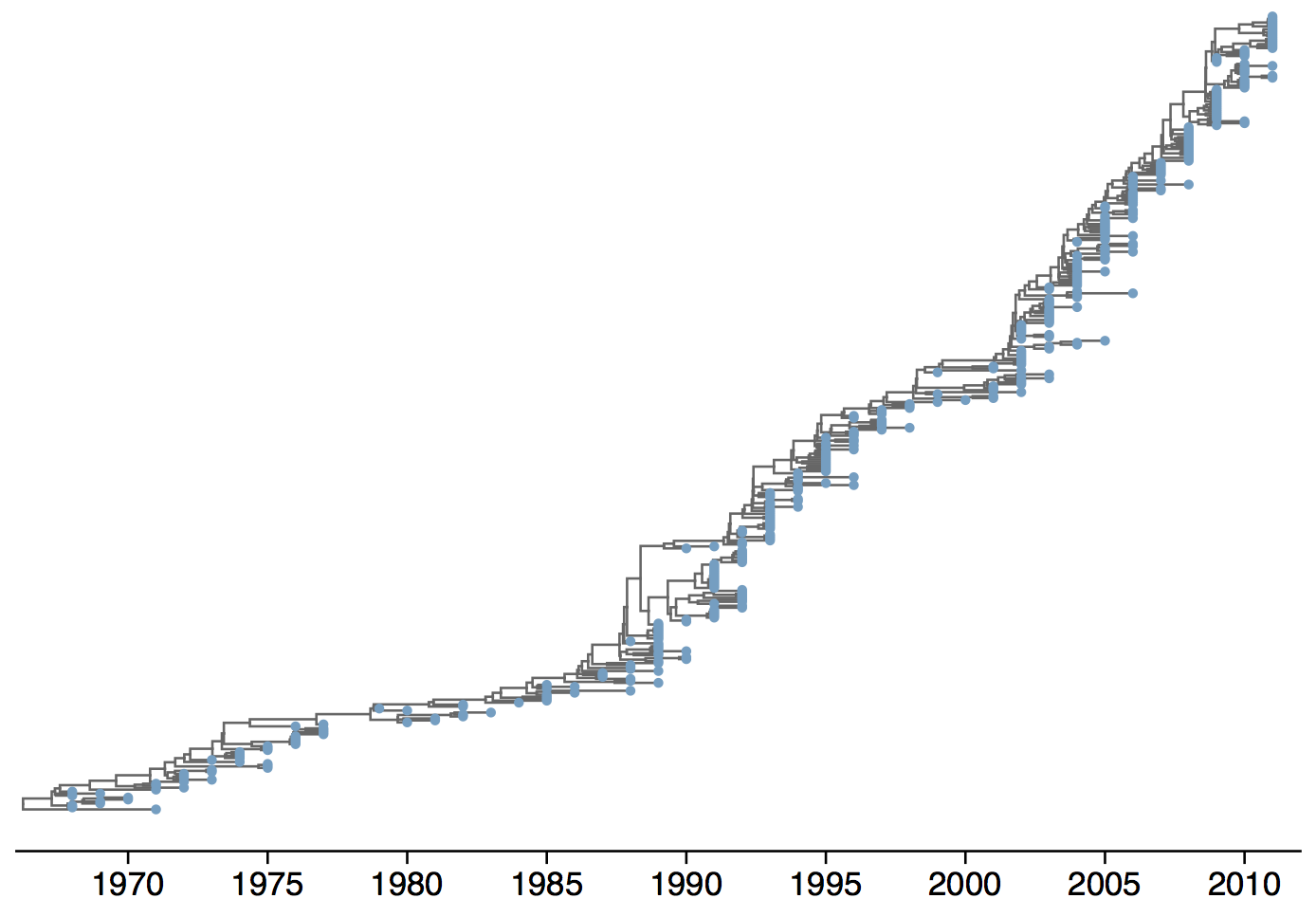
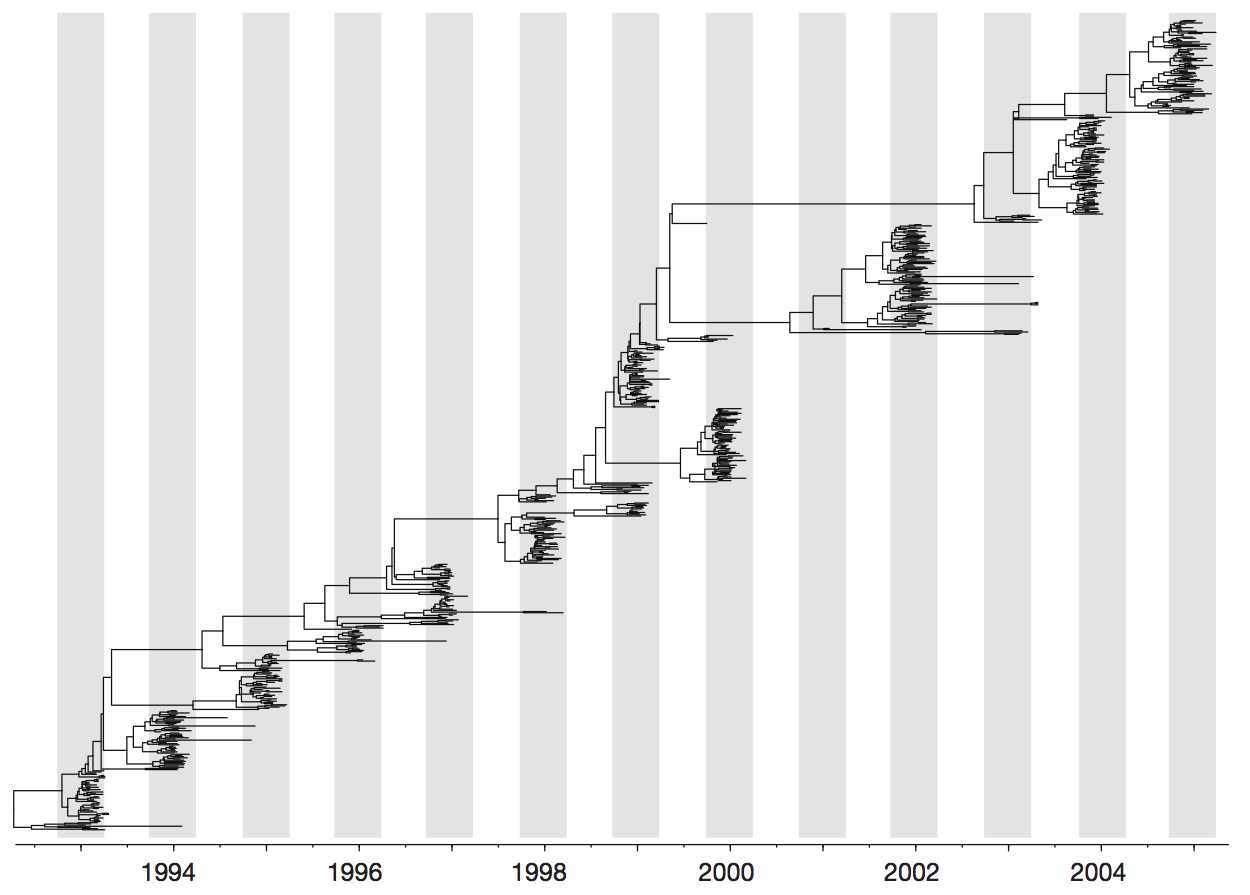
Turnover can be seen in the flu phylogeny
Flu pandemics caused by host switch events
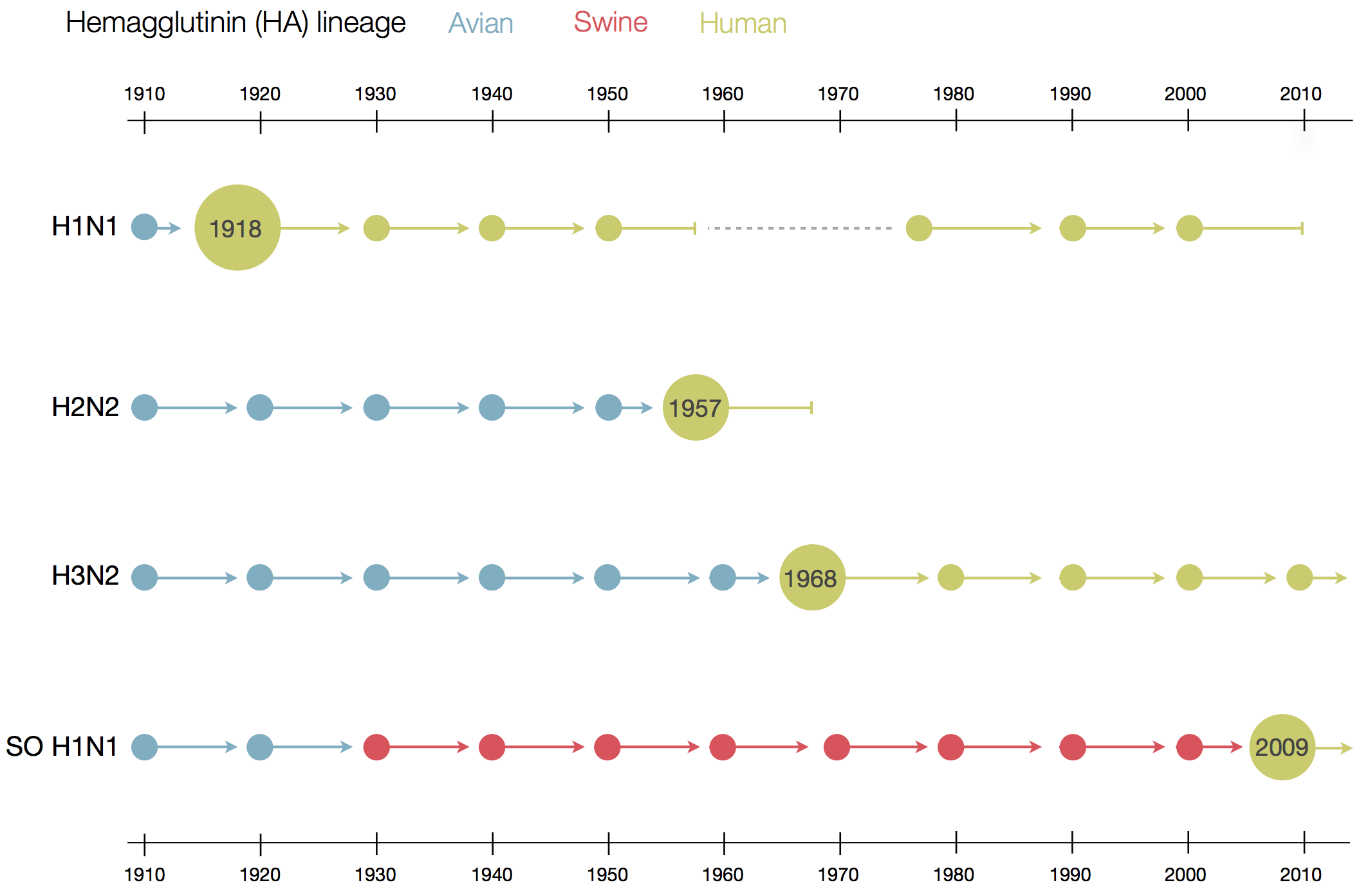
Influenza B does not have pandemic potential
Phylogenetic trees of different influenza lineages
Antigenic evolution drives viral dynamics
Antigenic drift
with Andrew Rambaut, Marc Suchard and others
Bedford et al 2014. Integrating influenza antigenic dynamics
with molecular evolution. eLife.
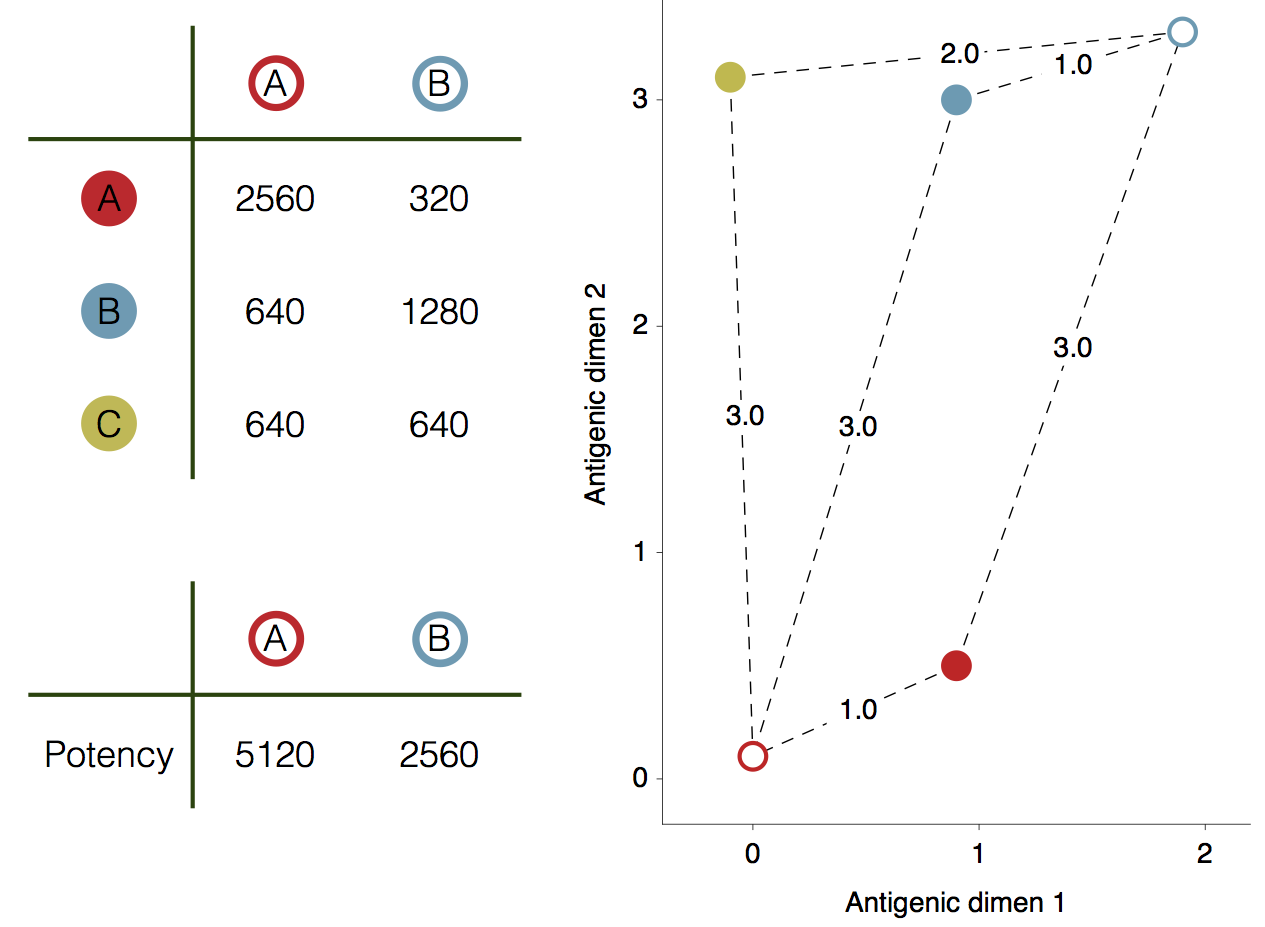
Influenza hemagglutination inhibition (HI) assay
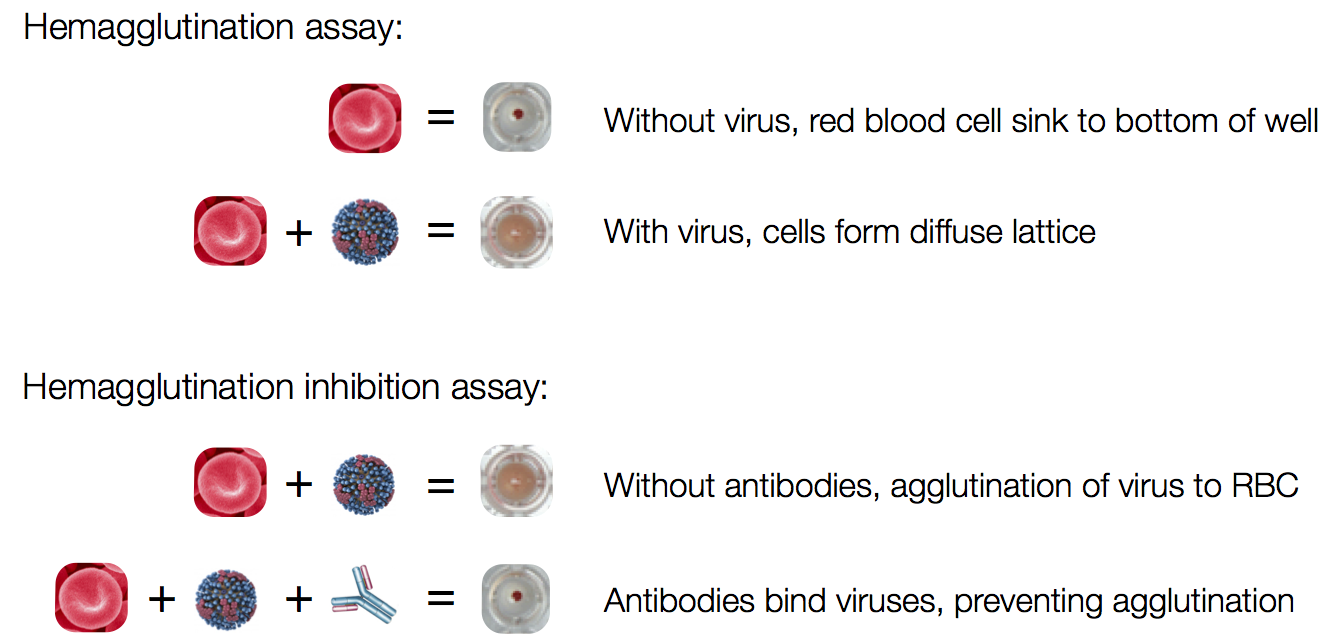
HI measures cross-reactivity across viruses
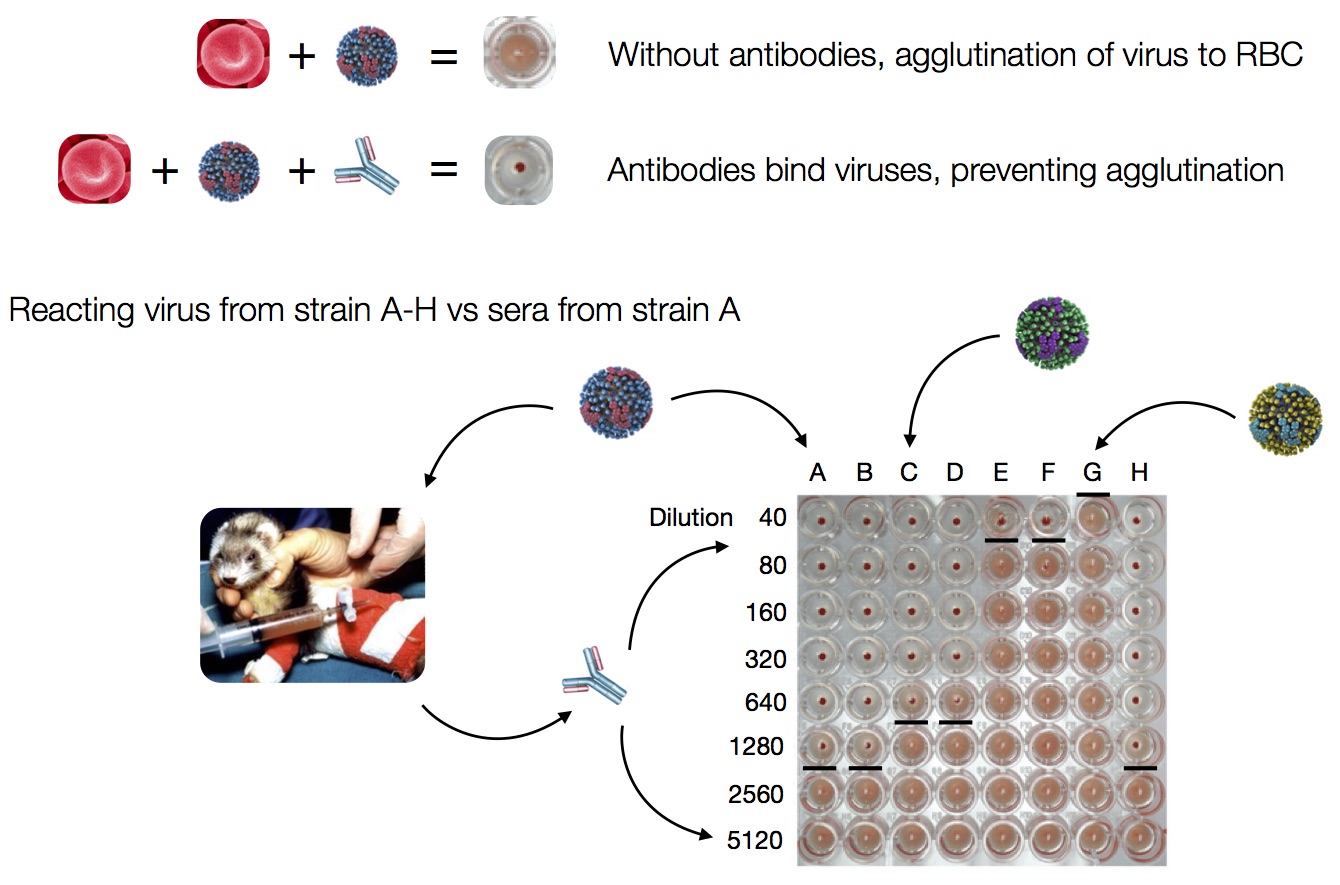
Data in the form of table of maximum inhibitory titers
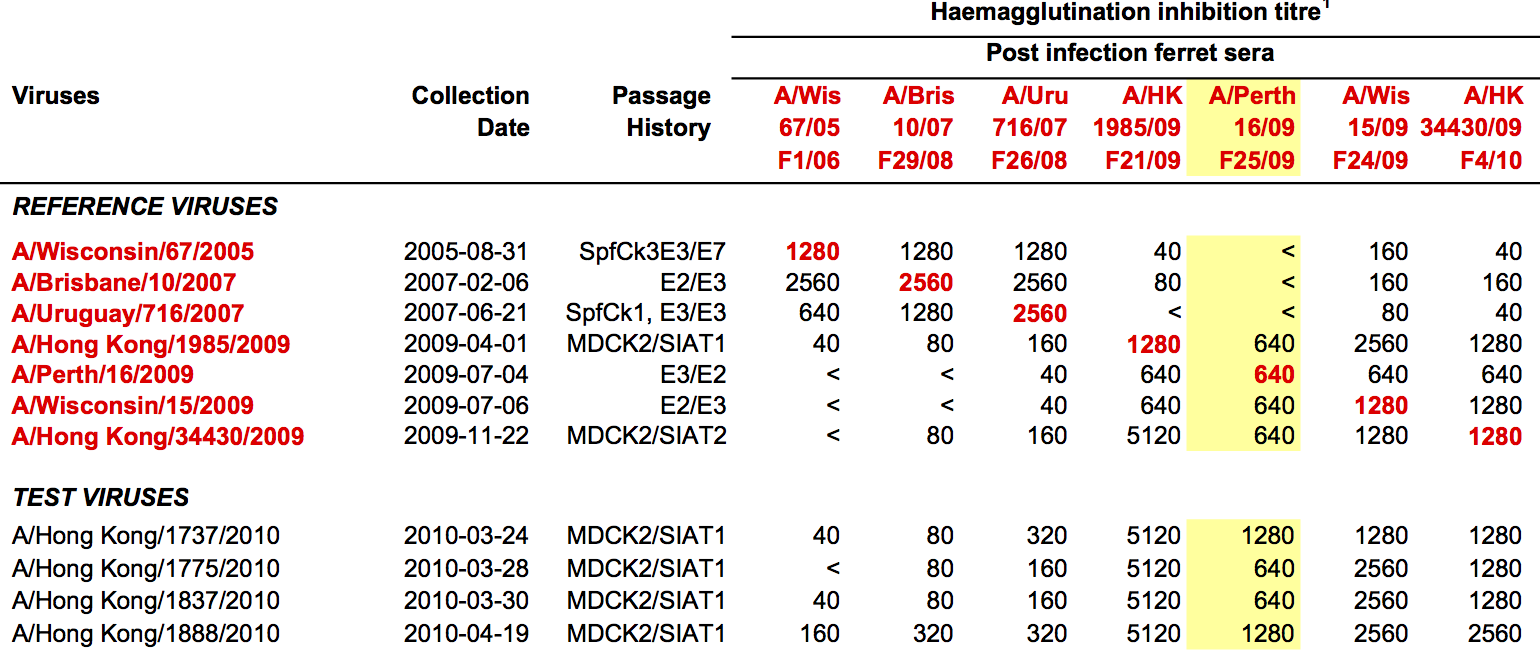
Compiled HI data difficult to work with
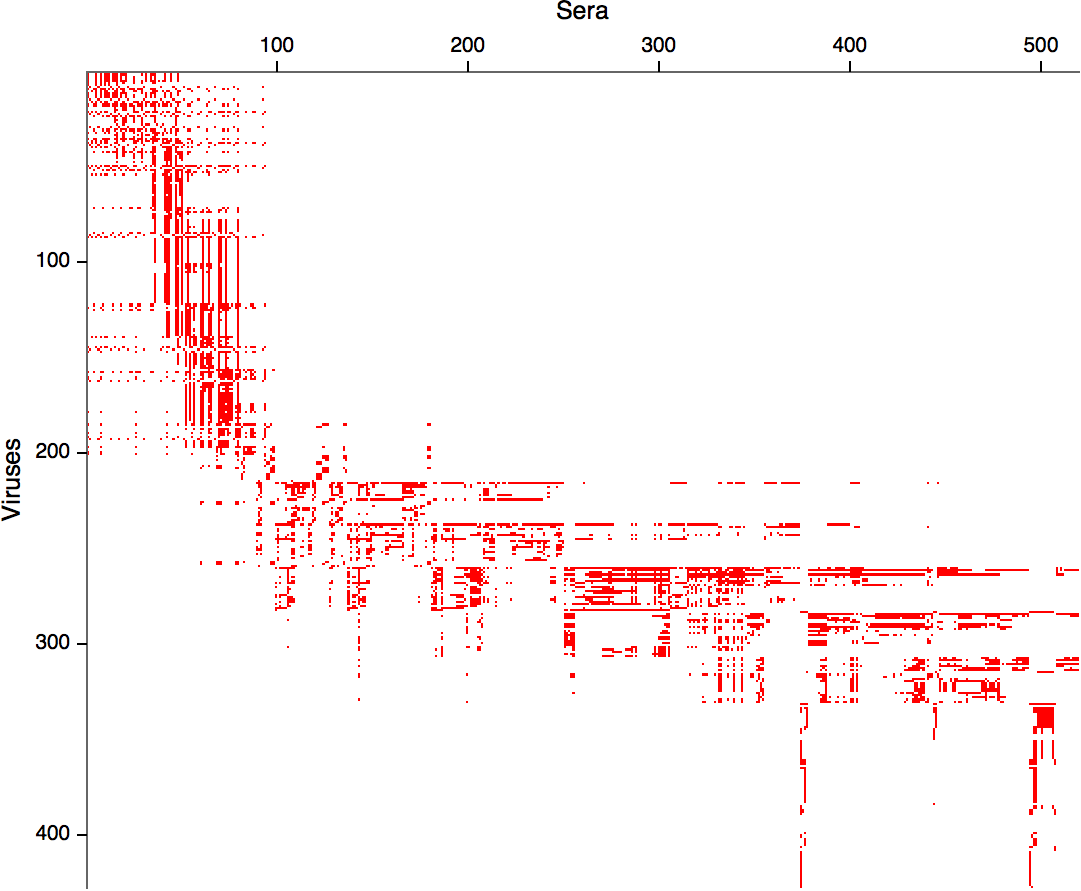
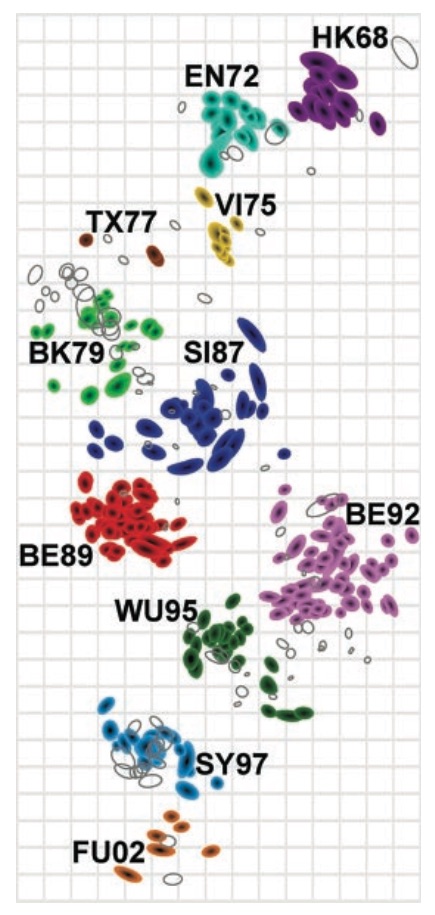
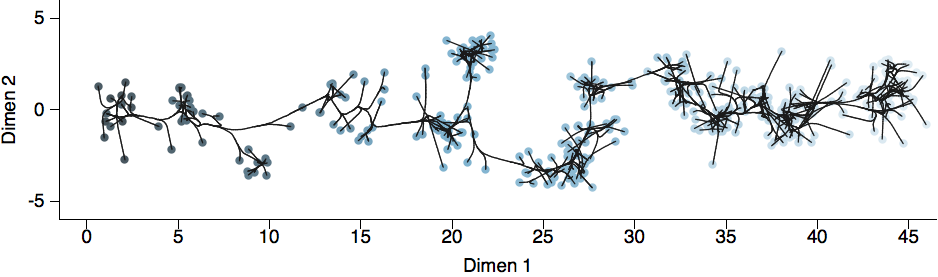
Antigenic cartography positions viruses and sera to recapitulate titer values

Antigenic cartography positions viruses and sera to recapitulate titer values
Schematic HI table and antigenic map
Bayesian multidimensional scaling (BMDS)
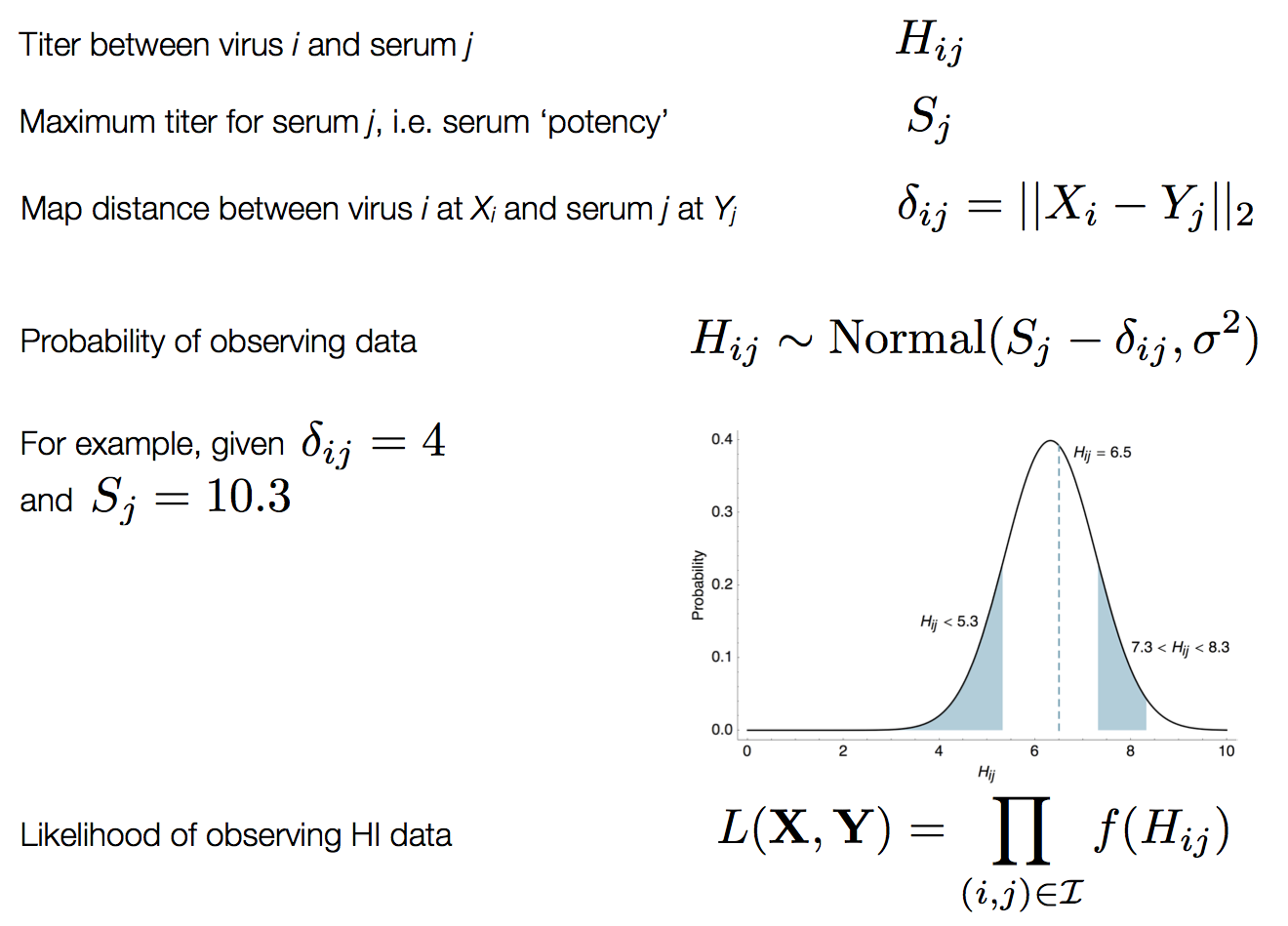
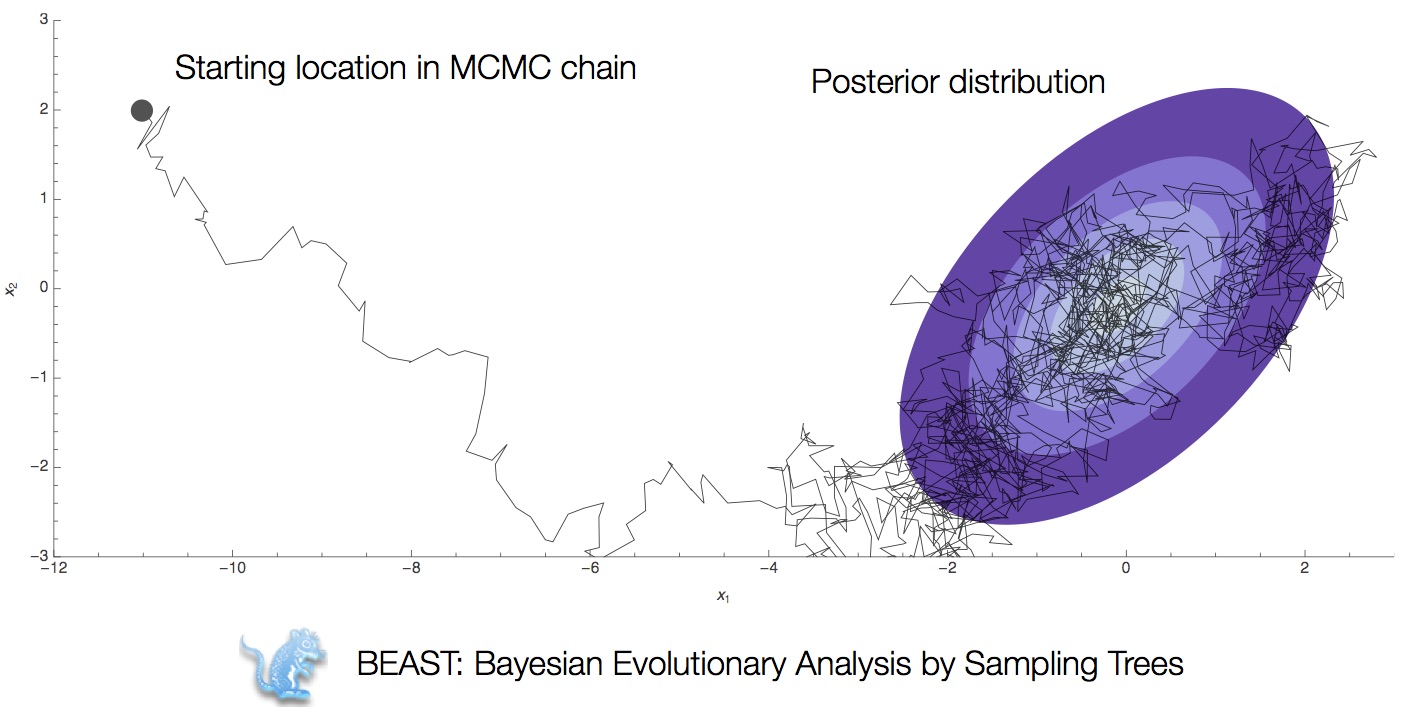
Integration through Markov chain Monte Carlo (MCMC)
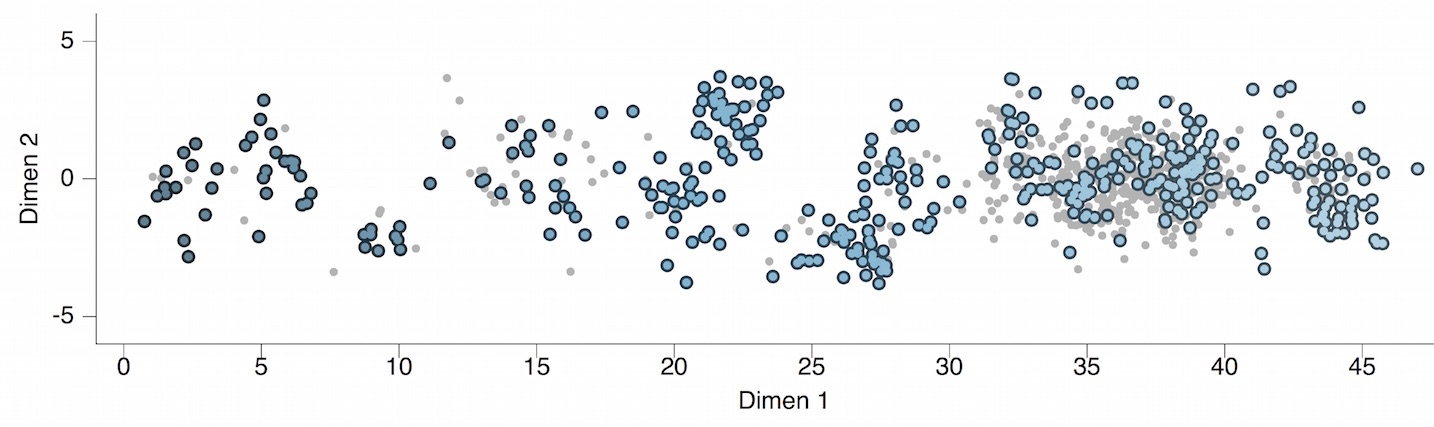
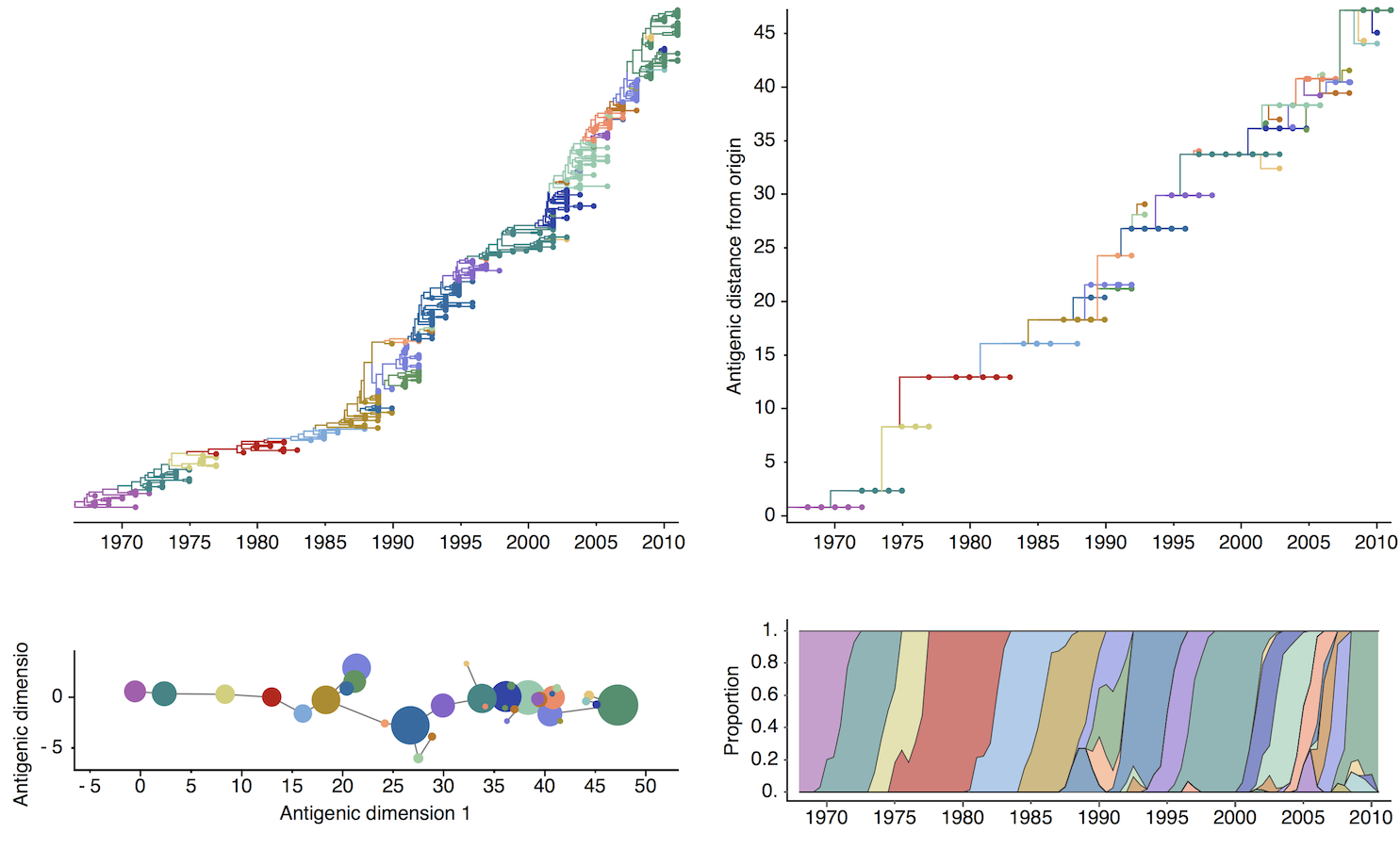
Antigenic map of H3N2 influenza from 1968 to 2011
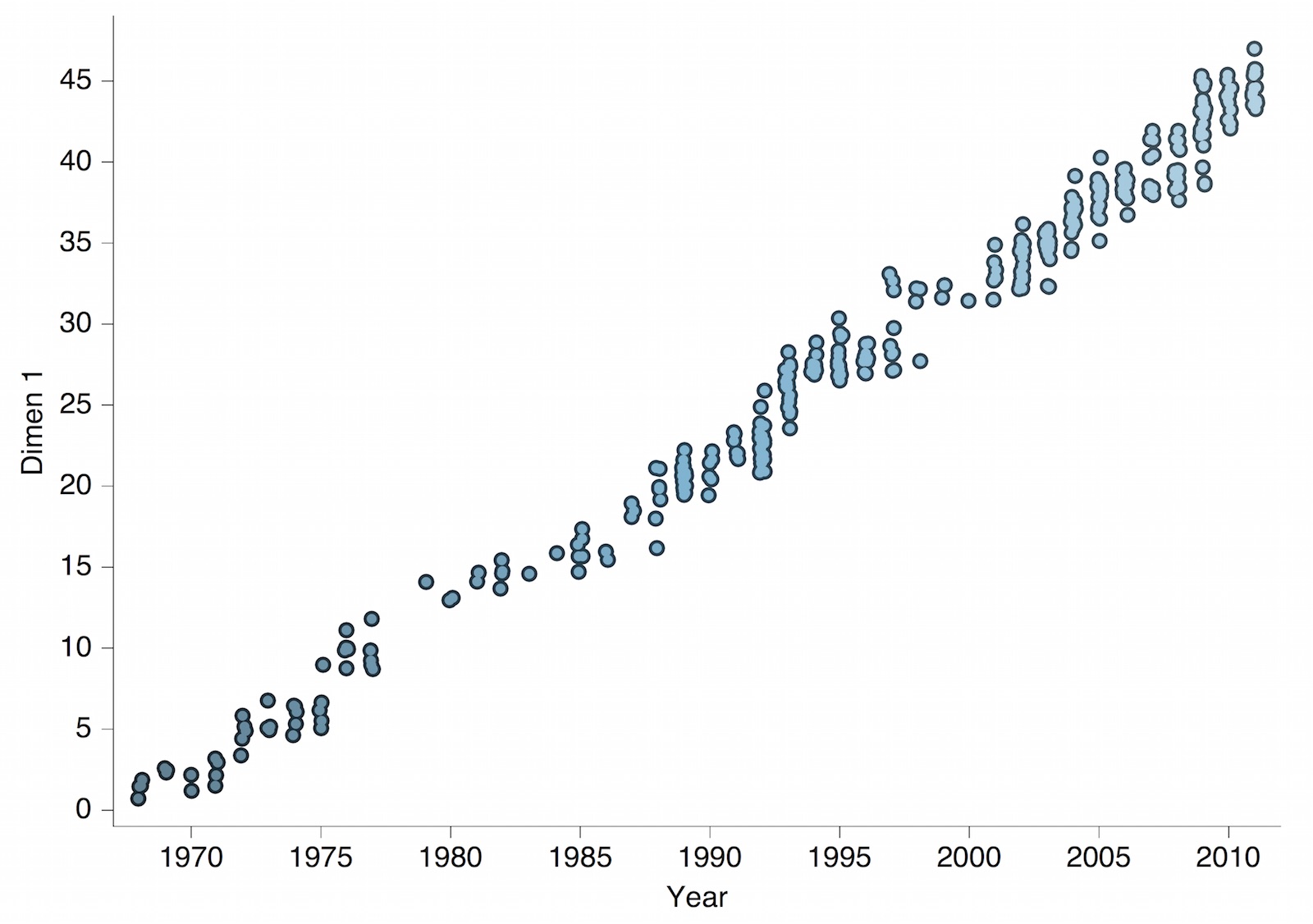
Antigenic drift of H3N2 influenza
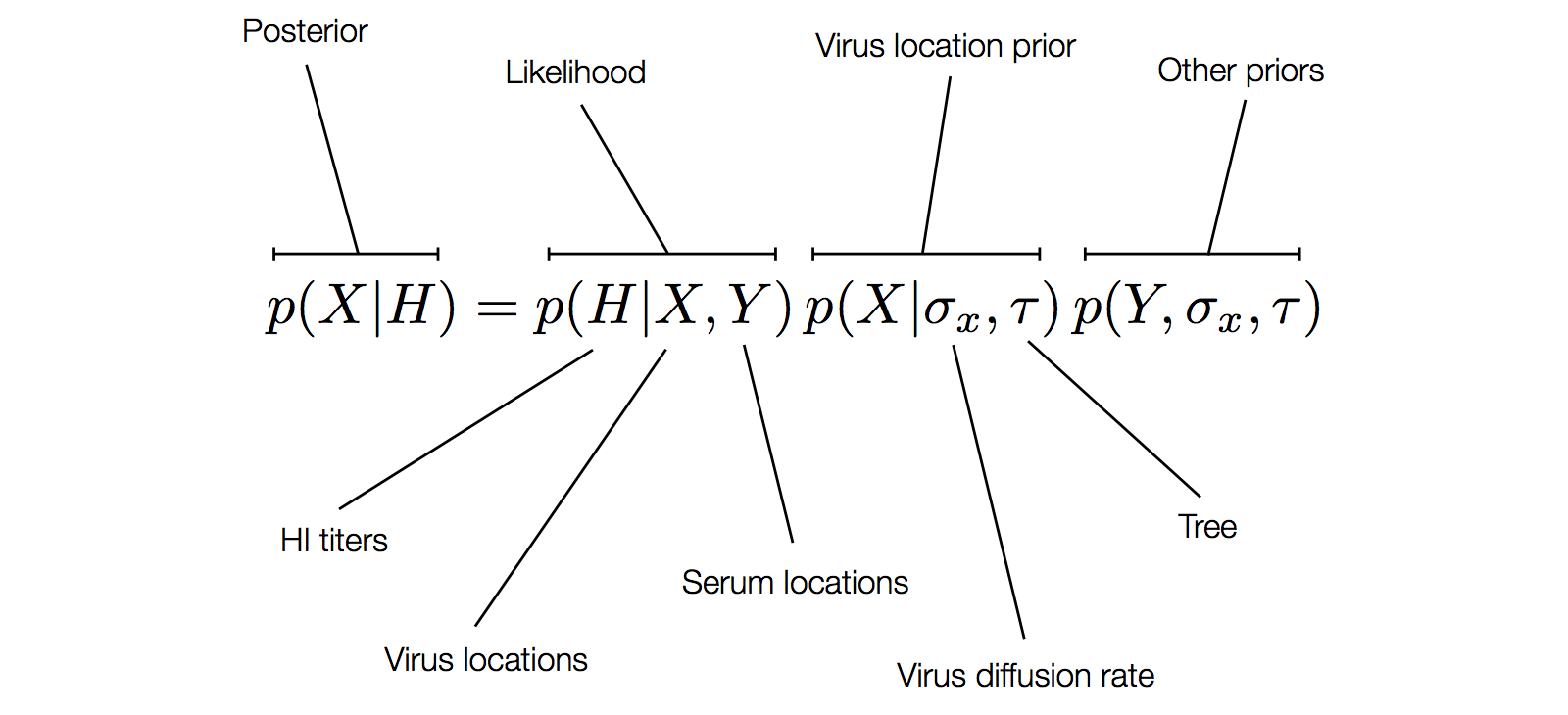
Factoring the BMDS model
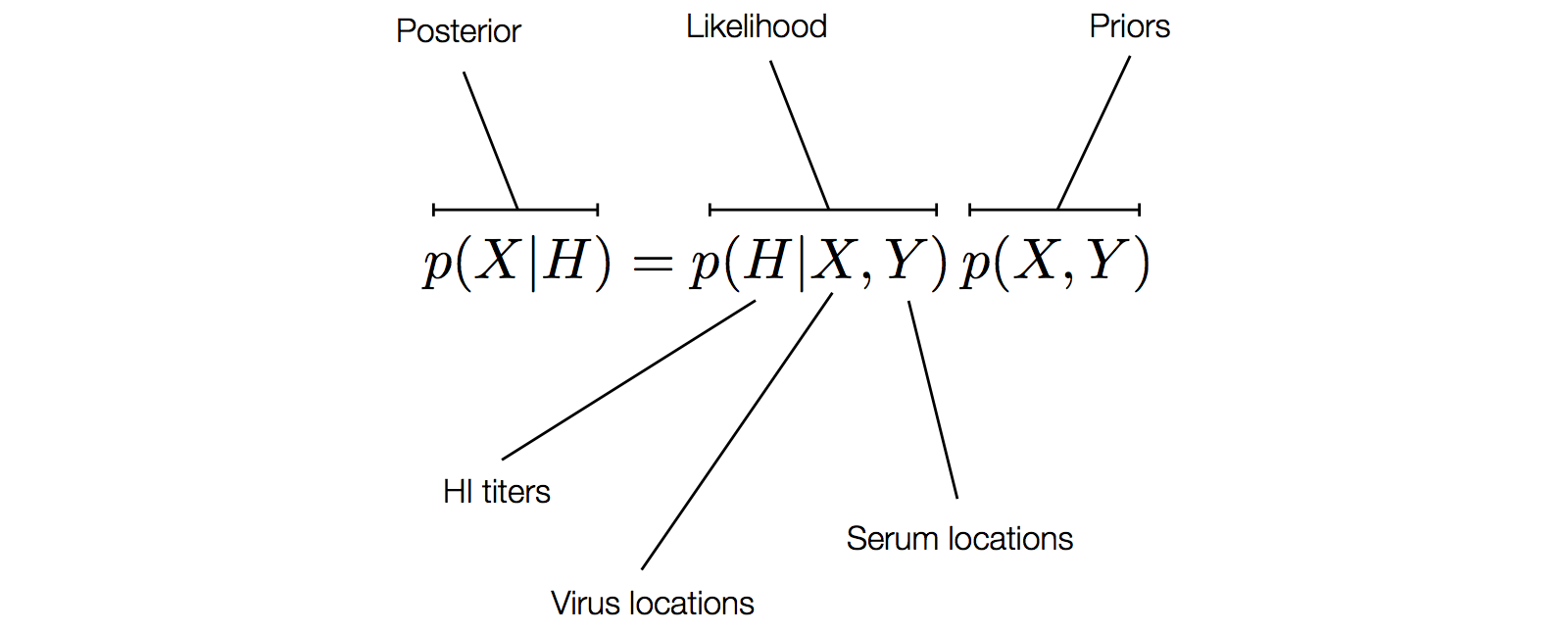
Phylogeny of H3N2 virus sequences
Modeling continuous characters via Brownian motion
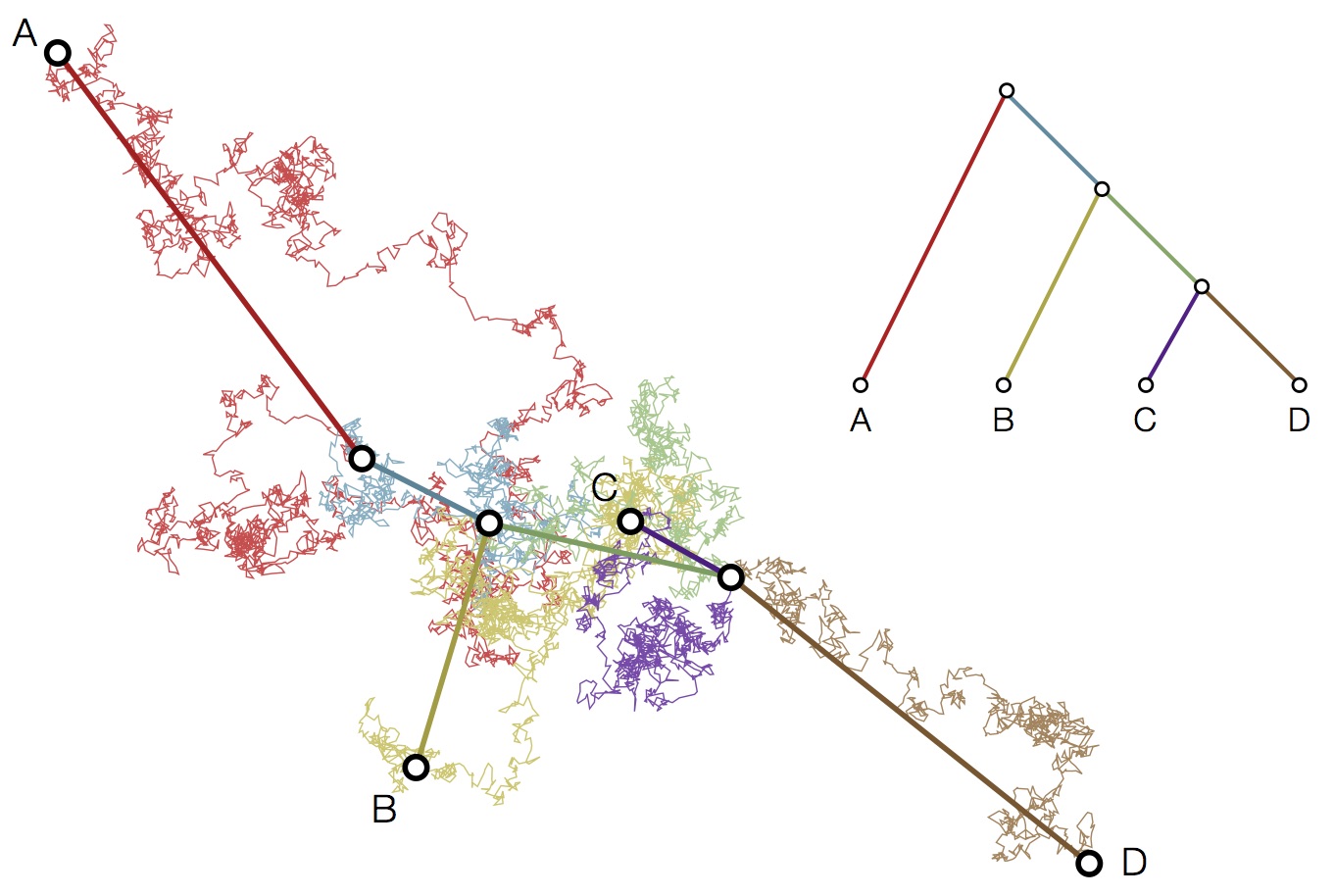
Including diffusion in MDS model

Factoring the BMDS diffusion model
Antigenic cluster transitions
with Charles Cheung, Andrew Rambaut and Marc Suchard
Cheung et al 2015. Detailed antigenic dynamics of influenza virus revealed by Bayesian phylogenetic clustering. In prep.
Factoring the BMDS discrete transition model
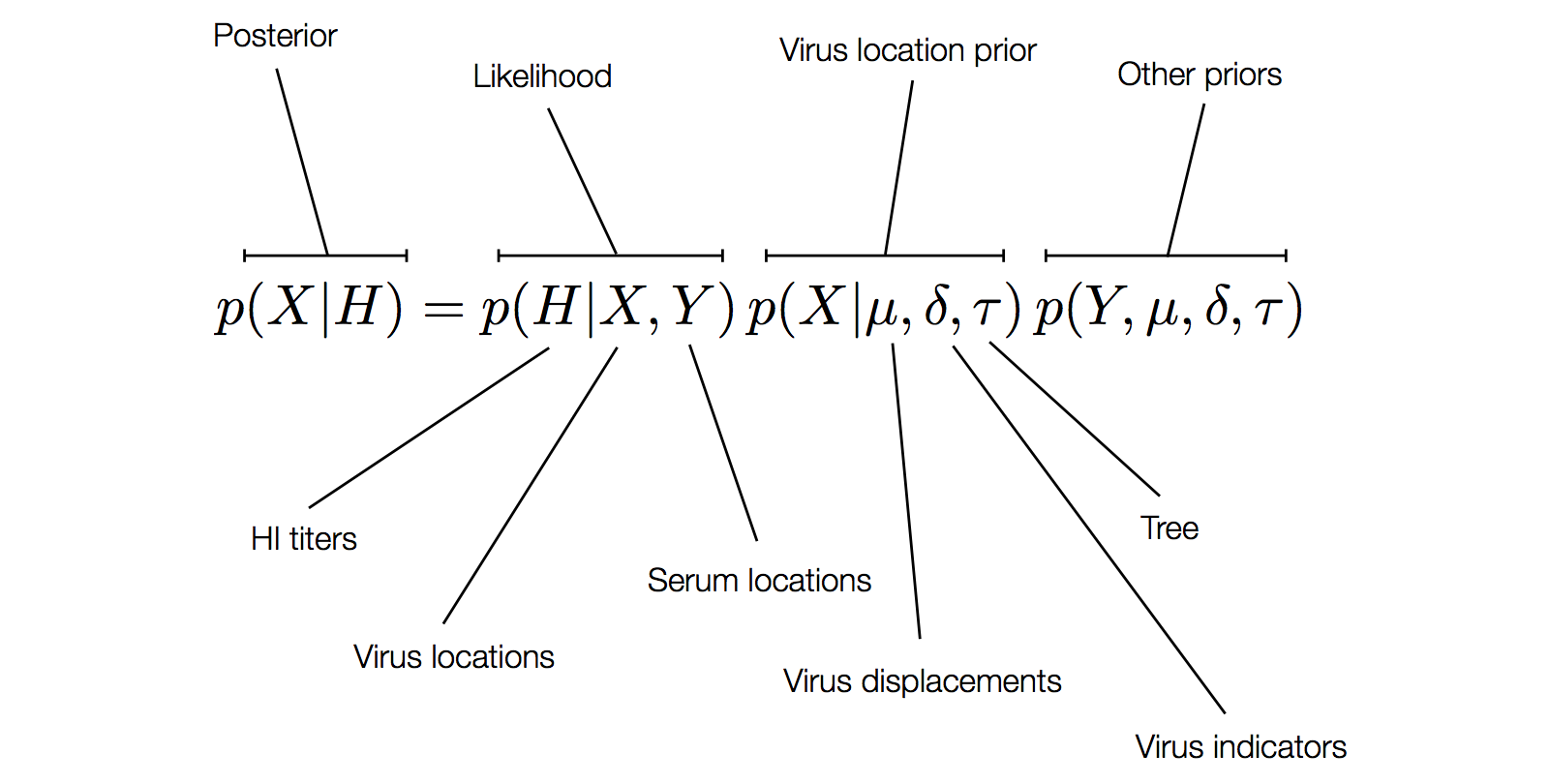
H3N2 discrete antigenic dynamics
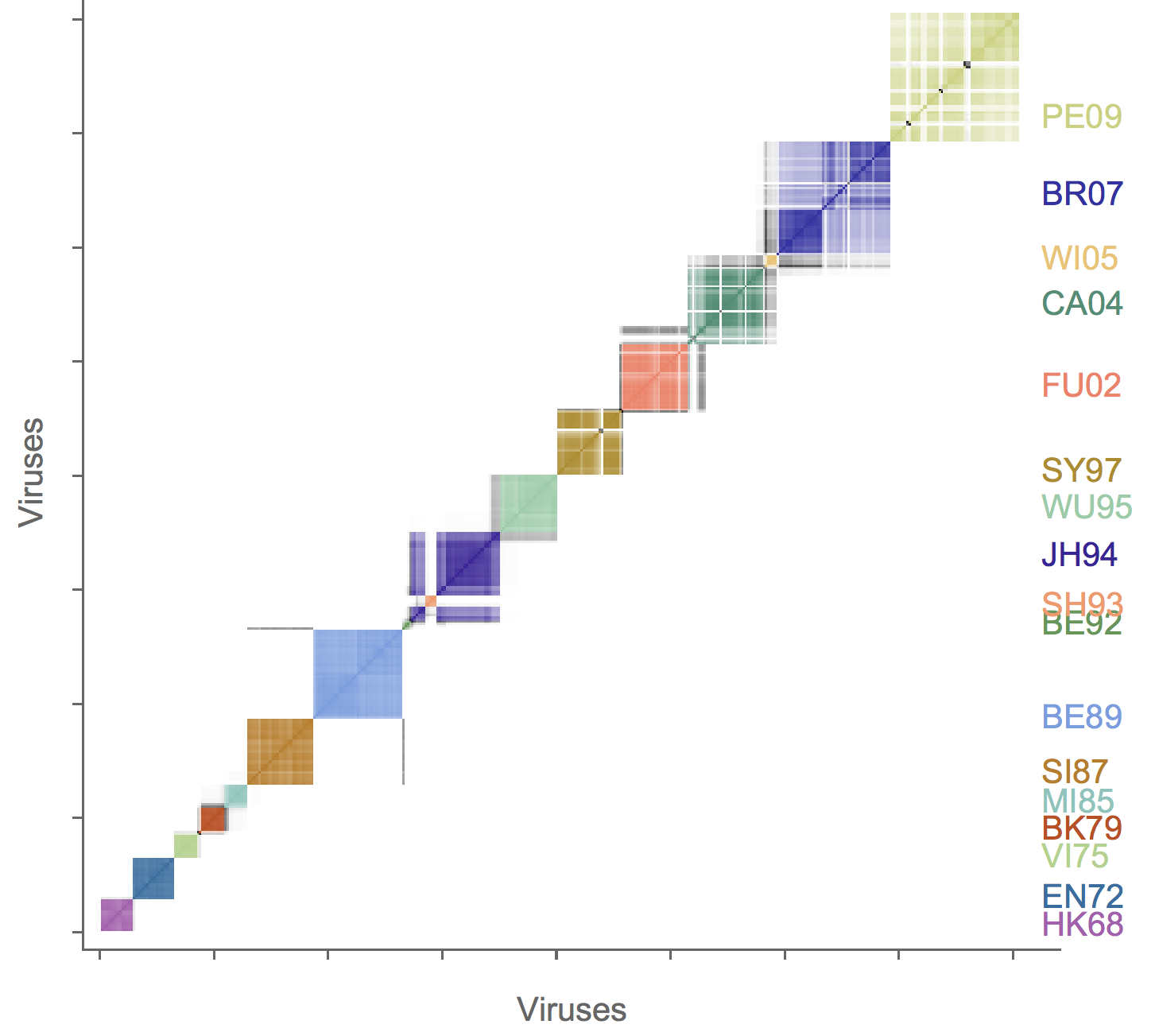
Association map of H3N2 antigenic clustering
H3N2 driver mutations
Sites: 135, 144, 145, 155, 156, 157, 158, 159, 189, 223 , 225, 226
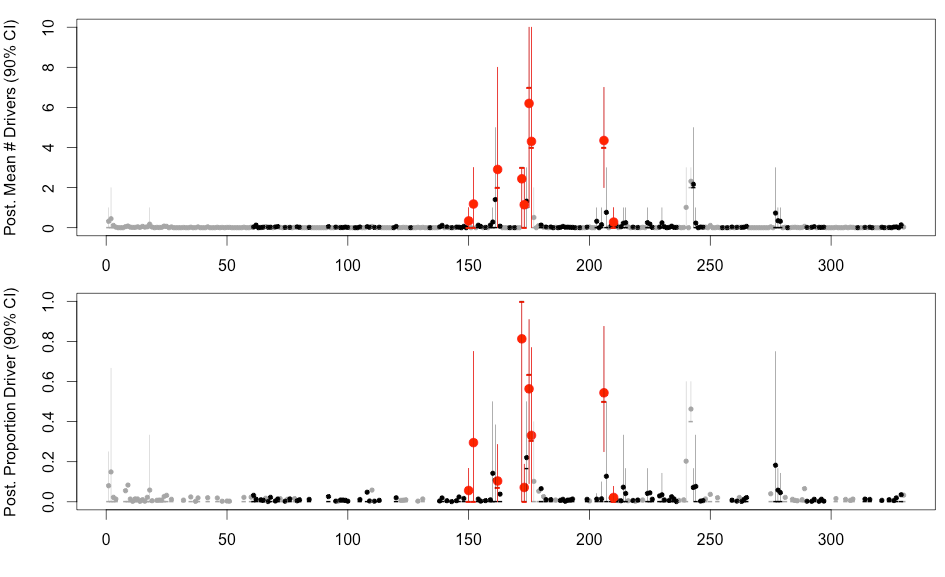
H3N2 clustering with driver mutations
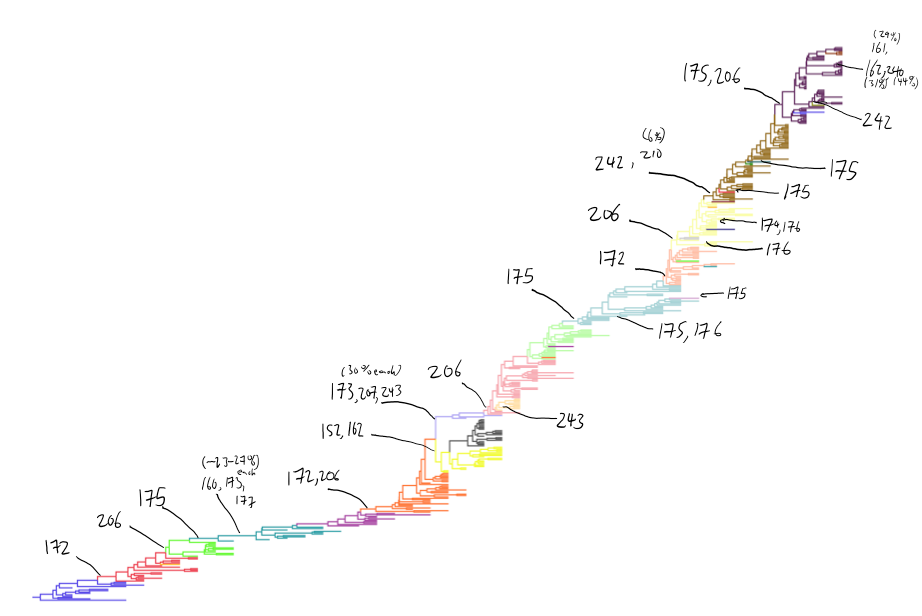
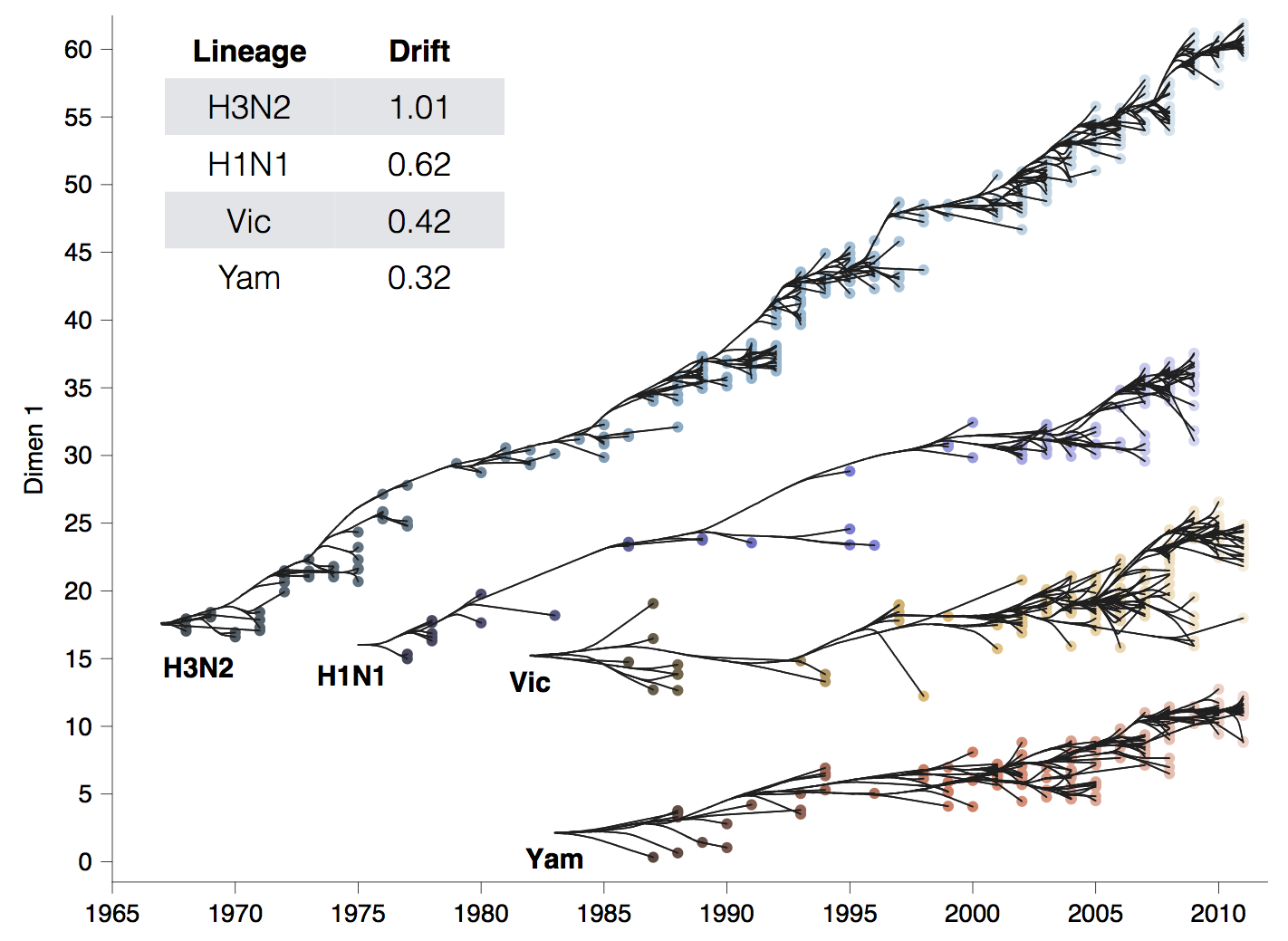
Drift across viruses
Bedford et al 2014. Integrating influenza antigenic dynamics
with molecular evolution. eLife.
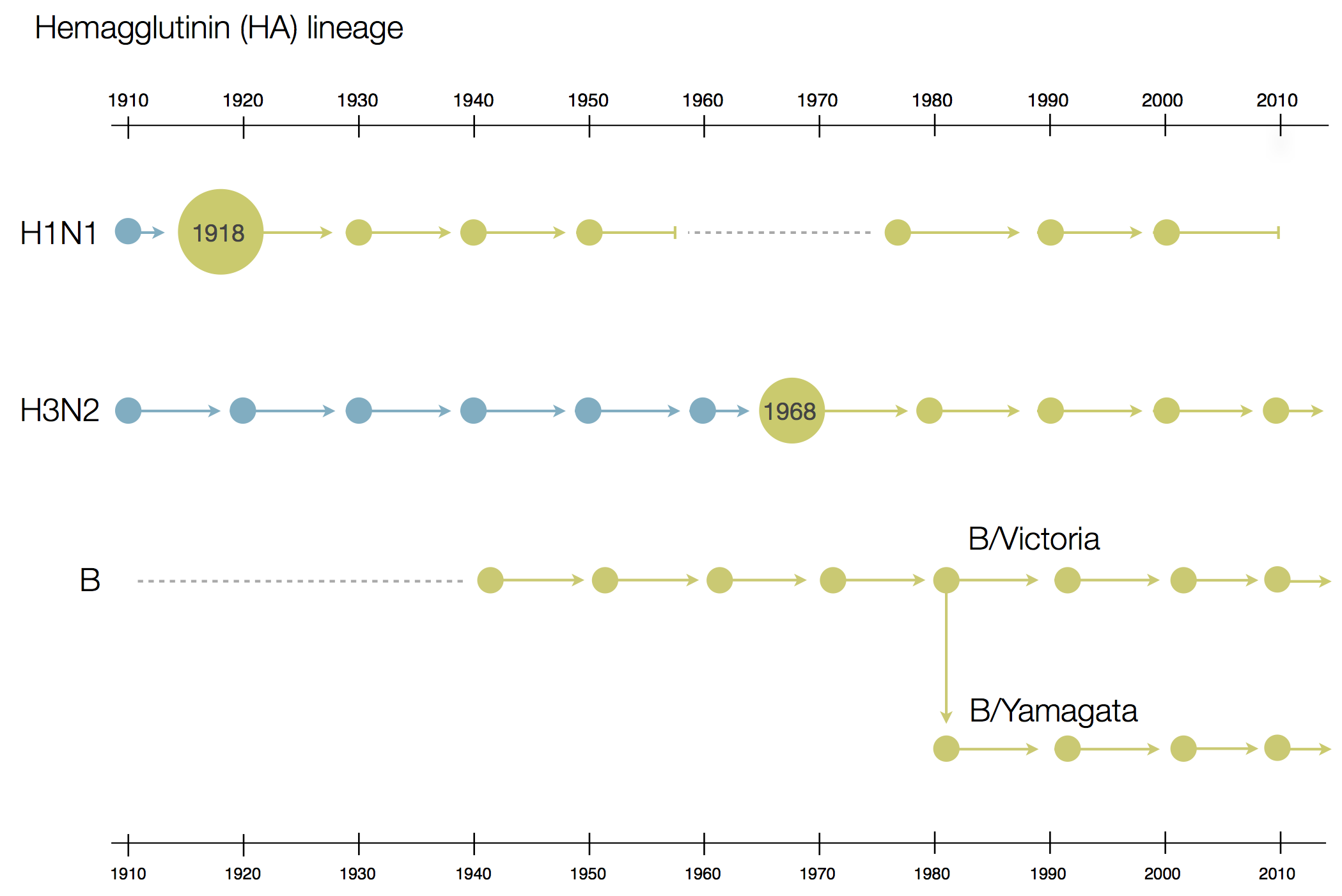
Phylogenetic trees of different influenza lineages

Antigenic phenotype across lineages
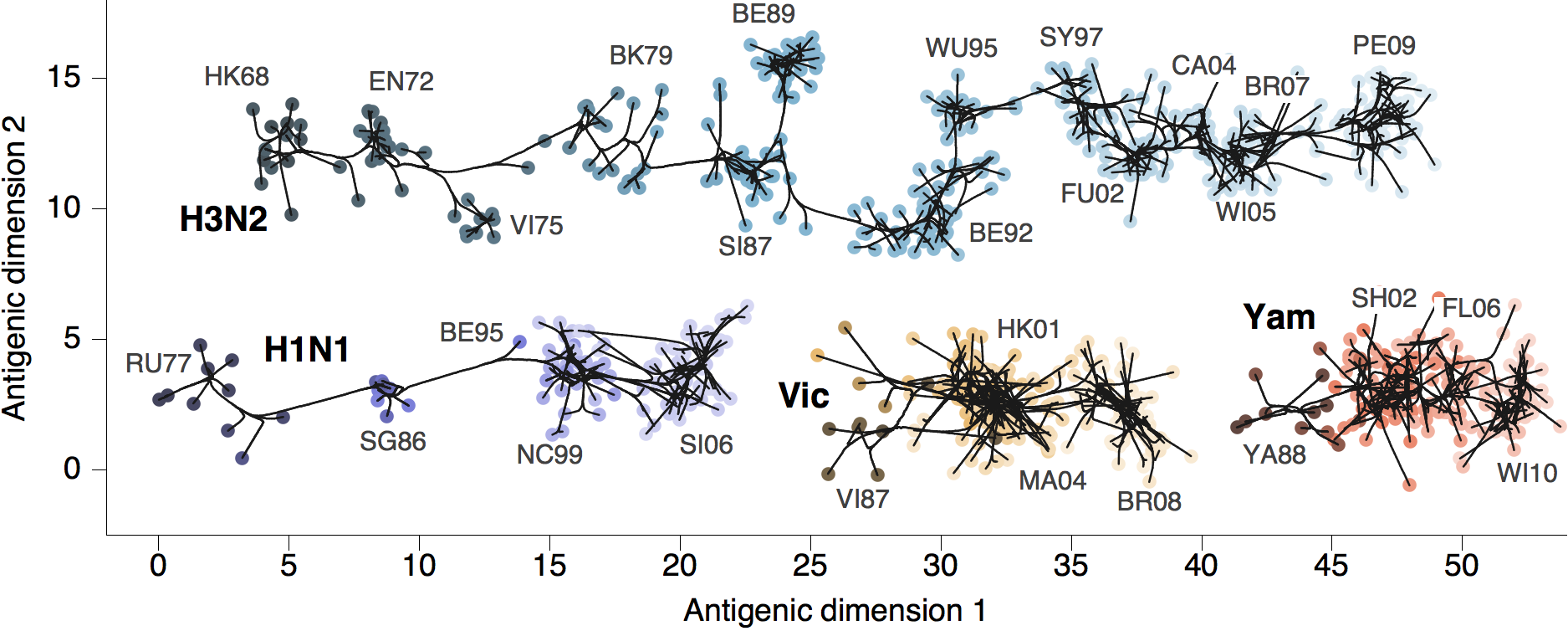
Antigenic drift across lineages
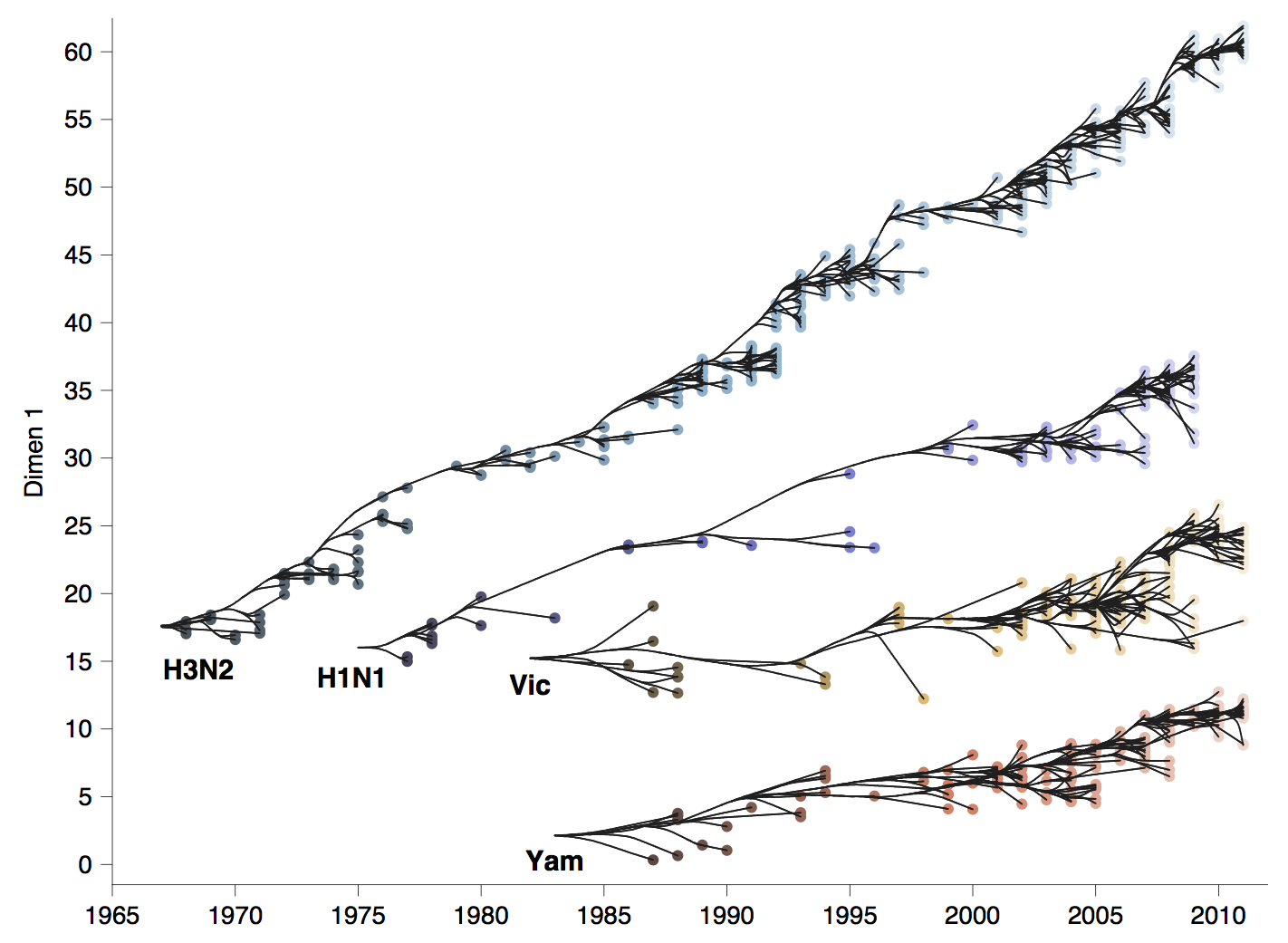
Antigenic drift across lineages
Geographic circulation
with Colin Russell, Philippe Lemey and many others
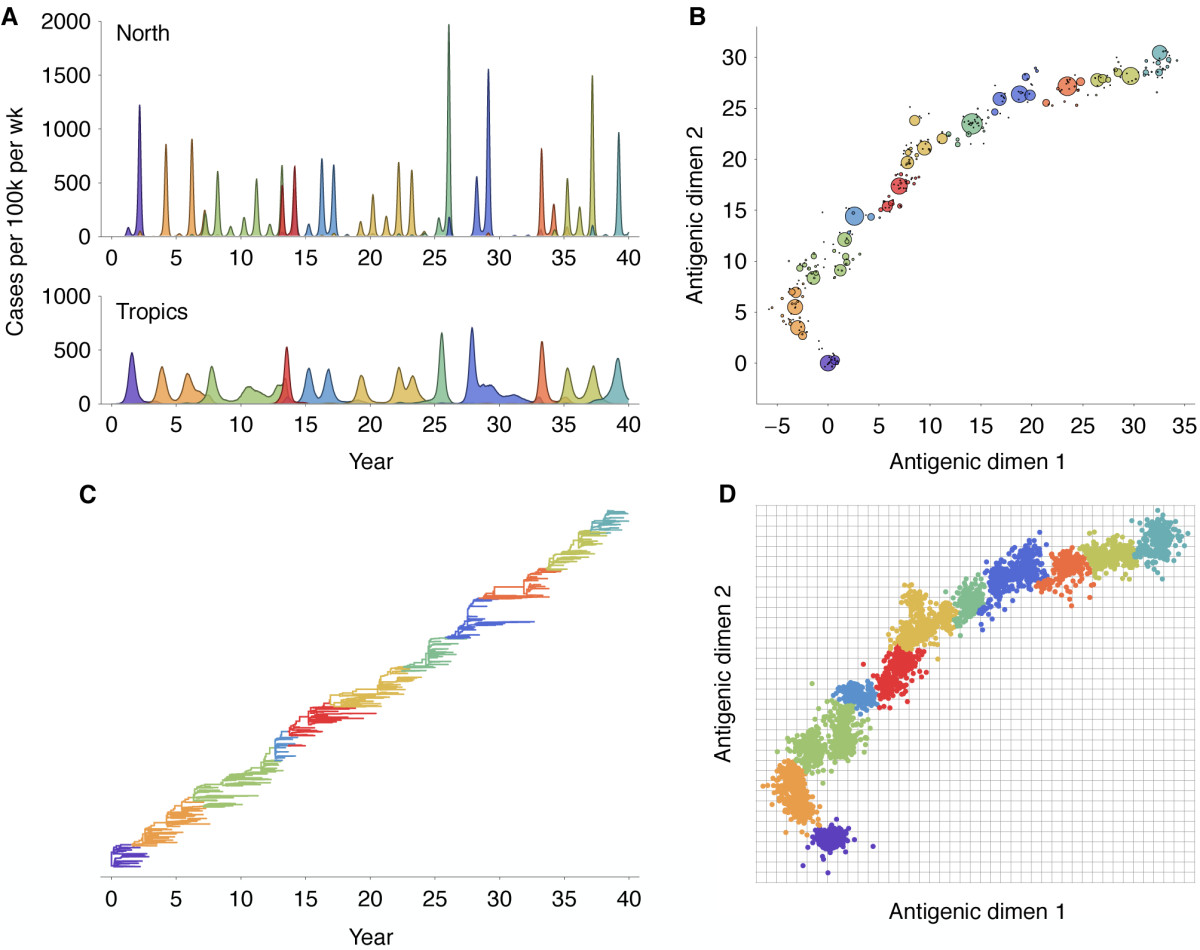
Seasonality in influenza
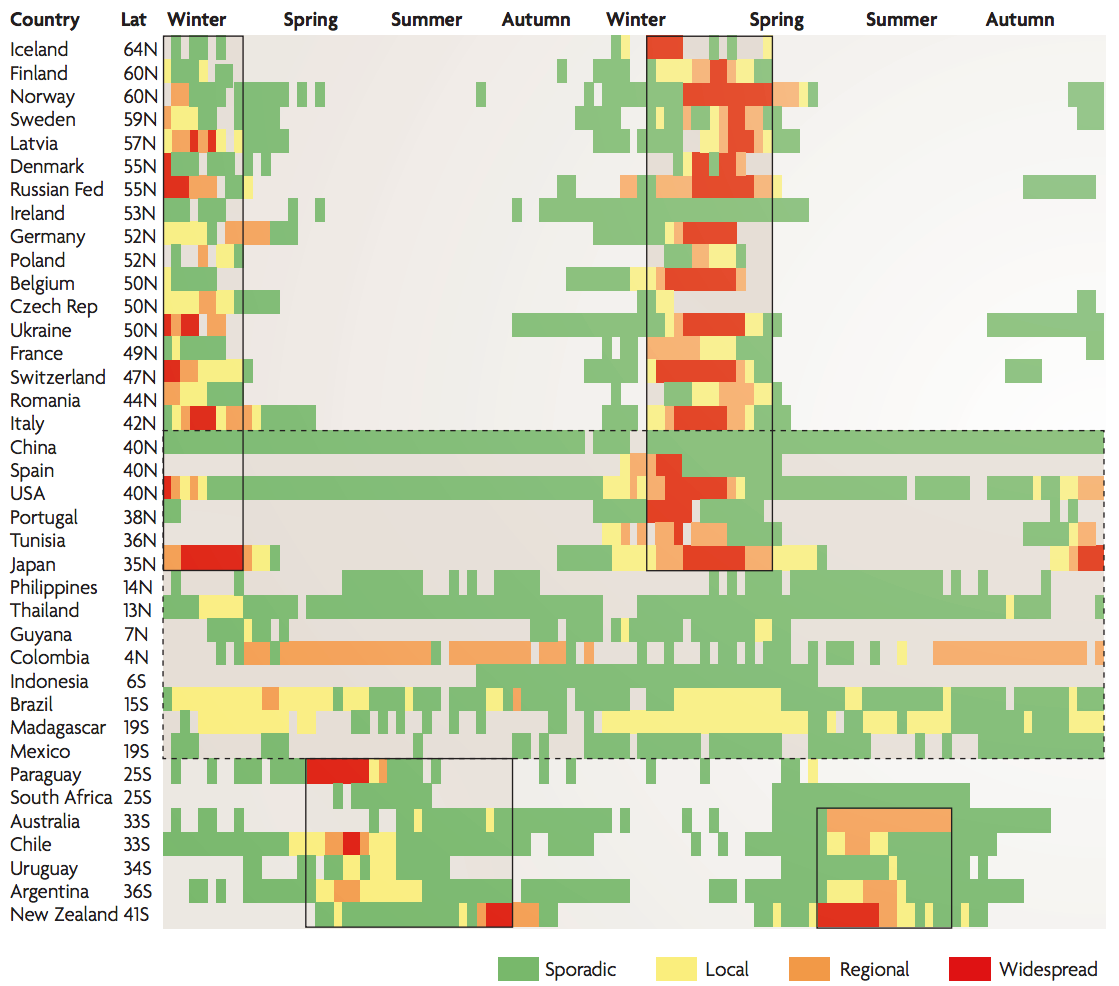
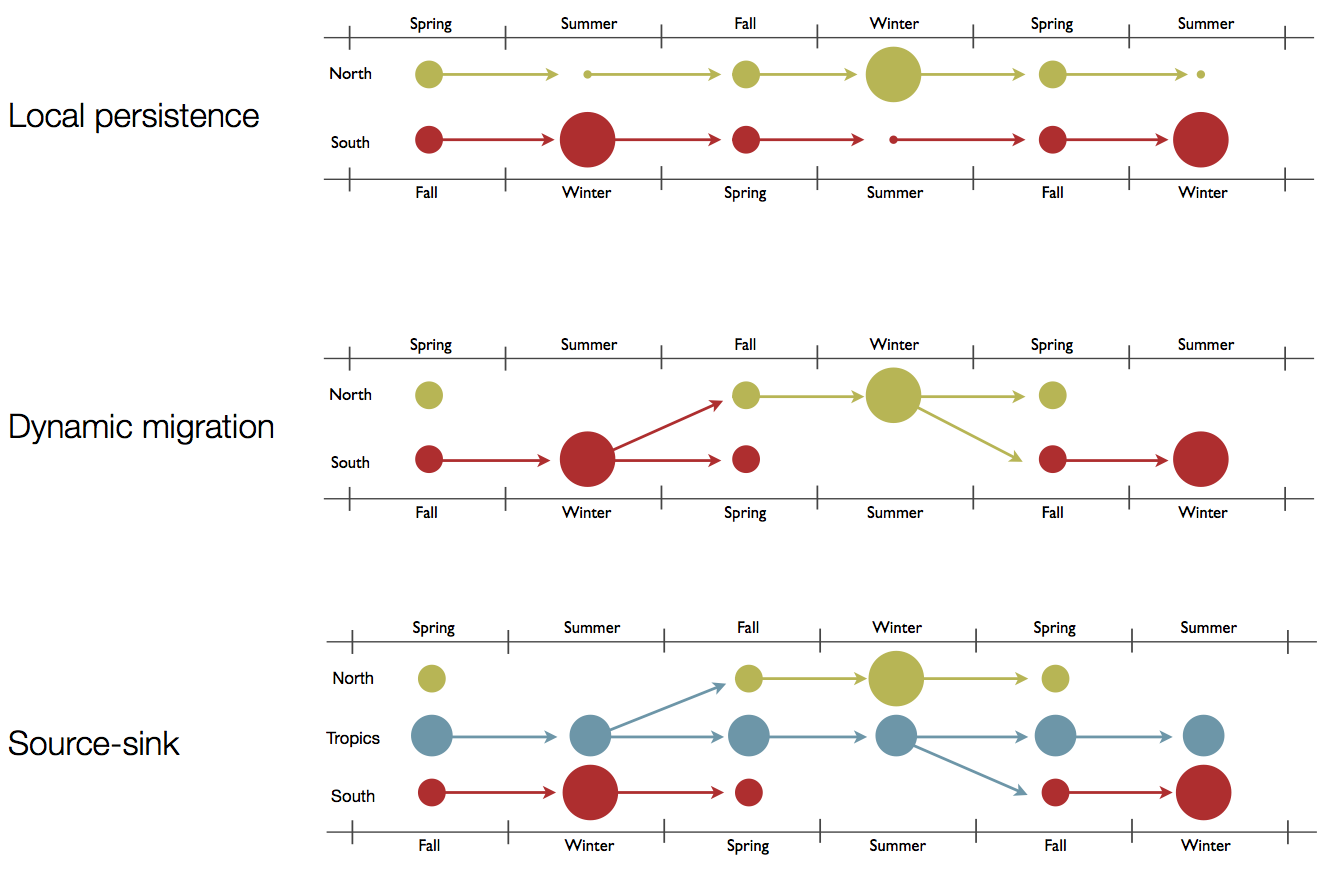
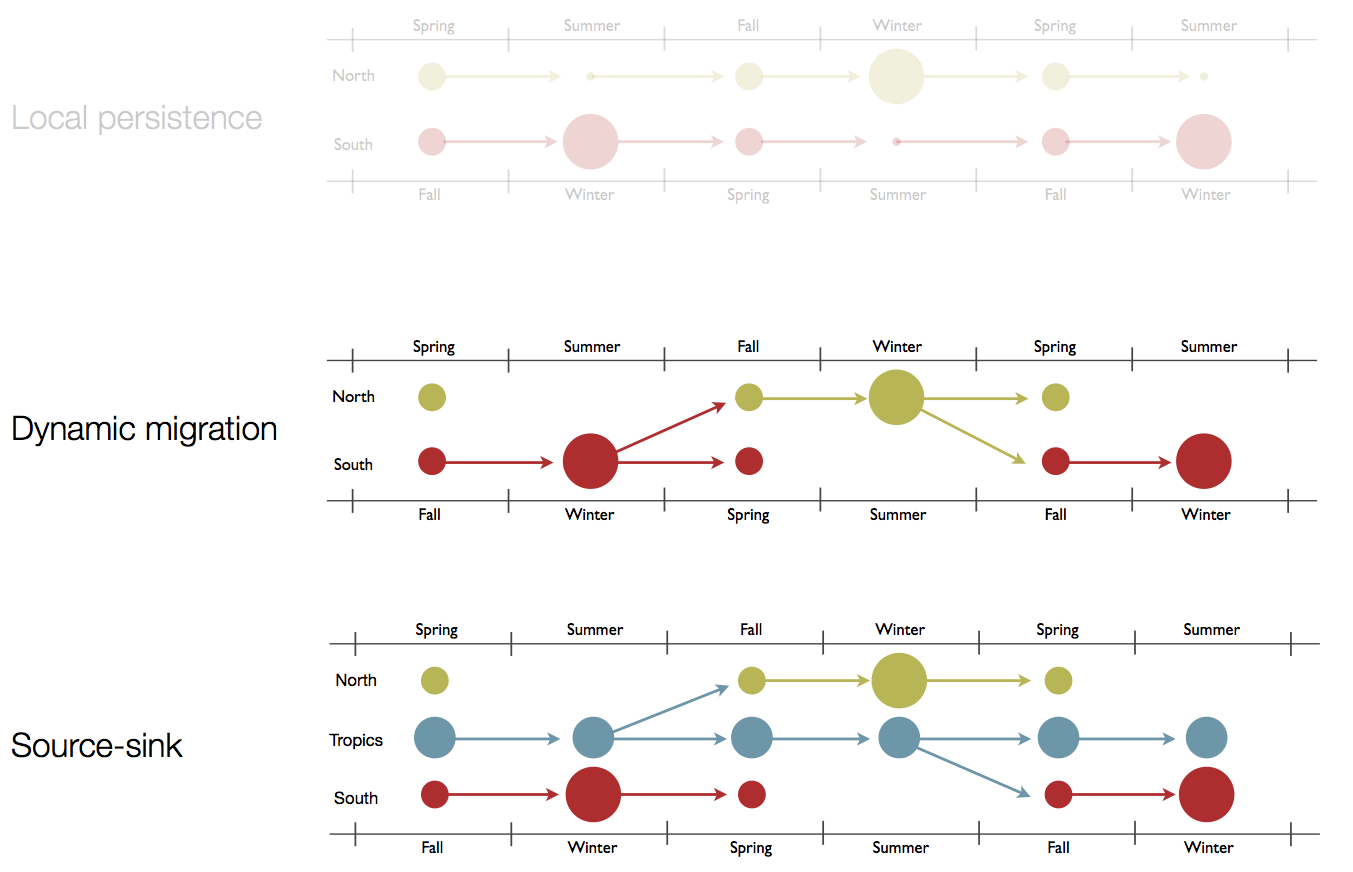
Hypotheses of influenza circulation patterns
Influenza H3 genealogy for NY state viruses
Hypotheses of influenza circulation patterns
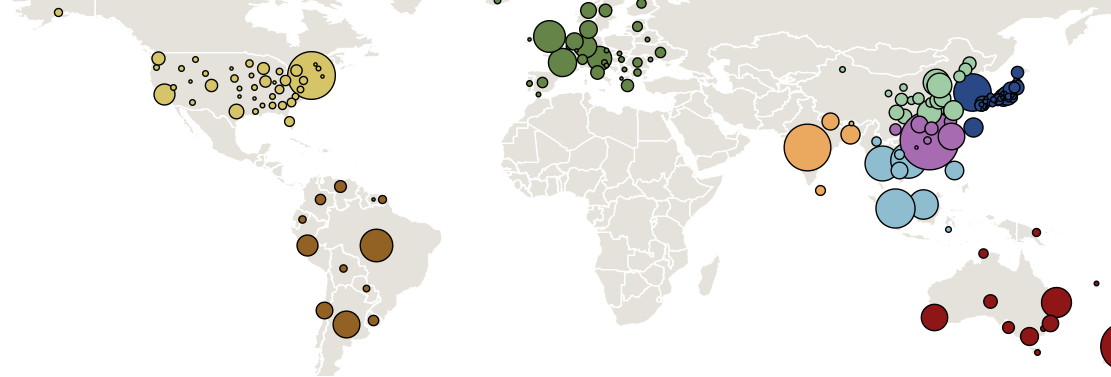
Sample H3N2 from around the world
Treating geographic state as an evolving character
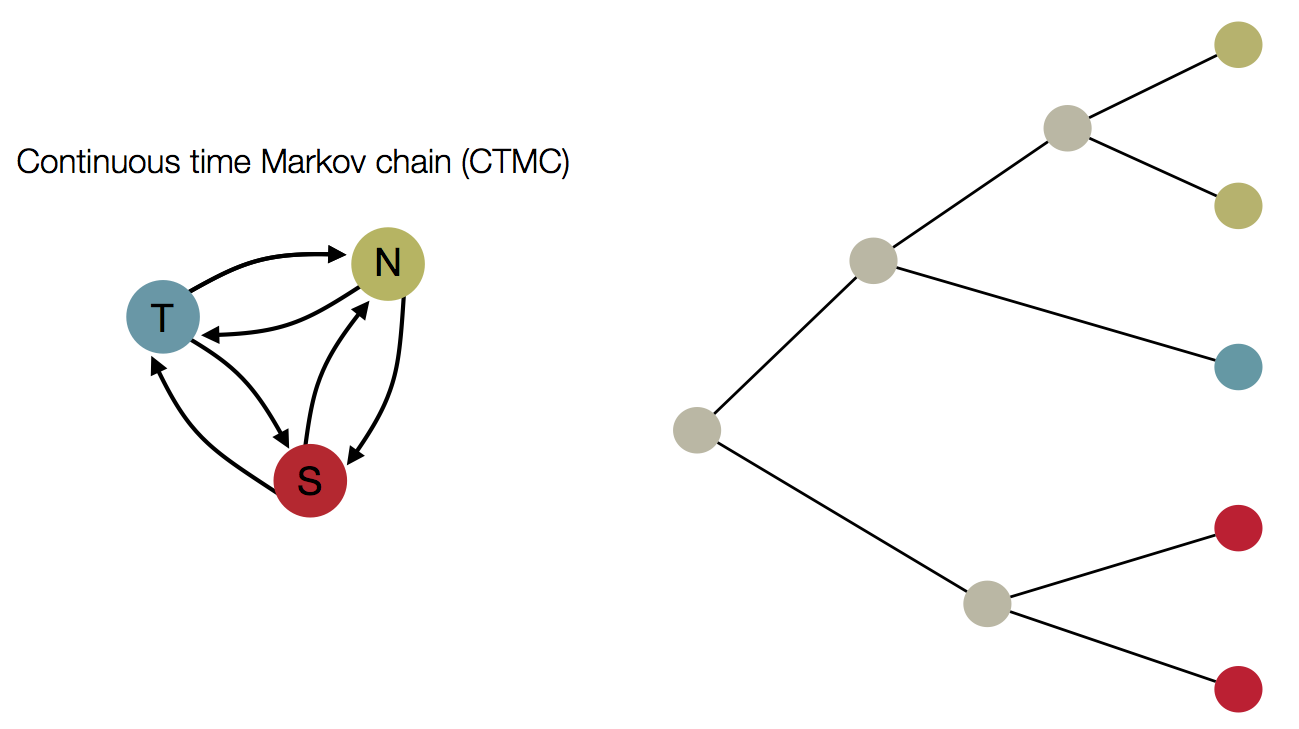
Phylogeny of H3 with geographic history
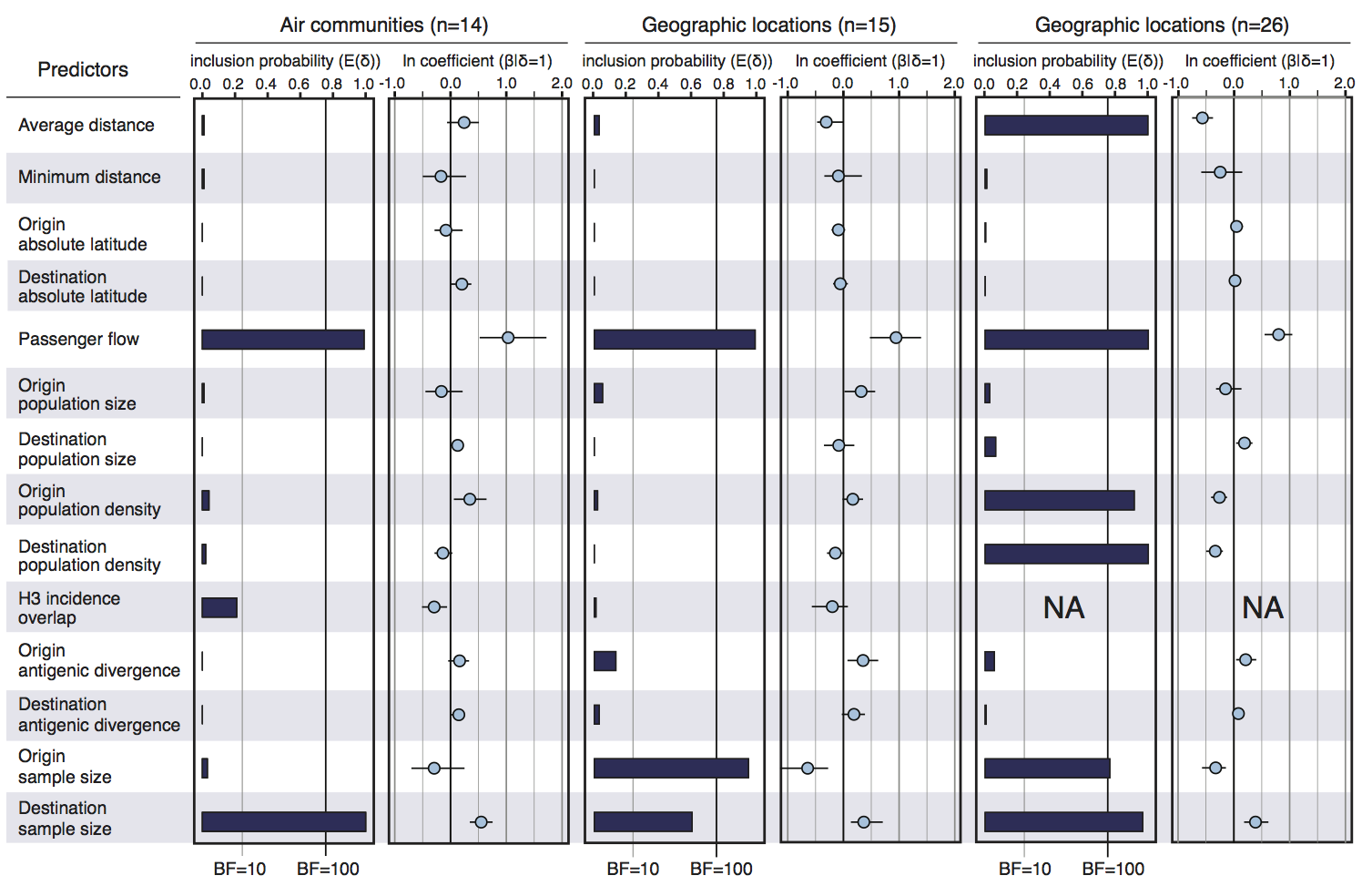
Infer geographic transition matrix
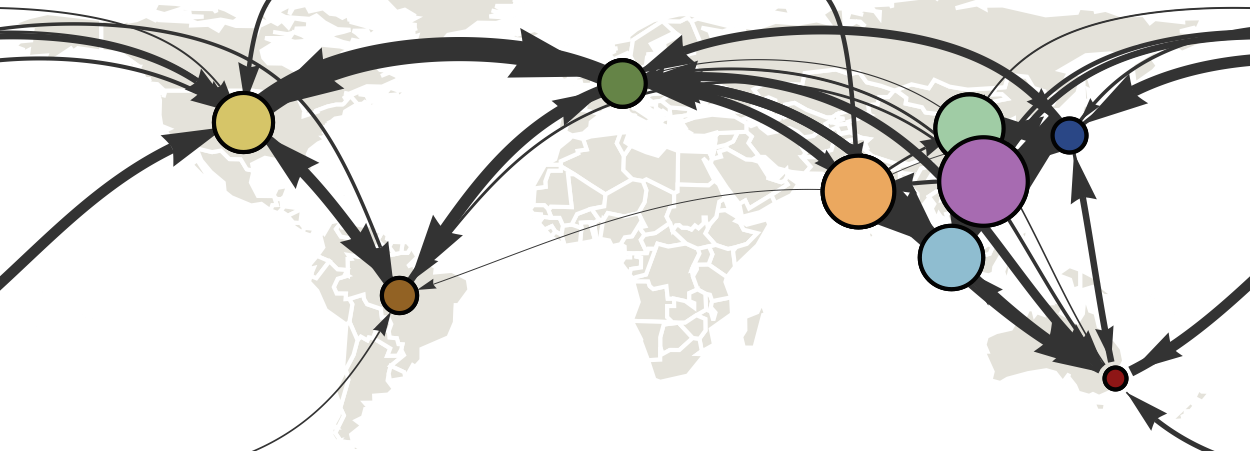
Air travel predicts migration rates
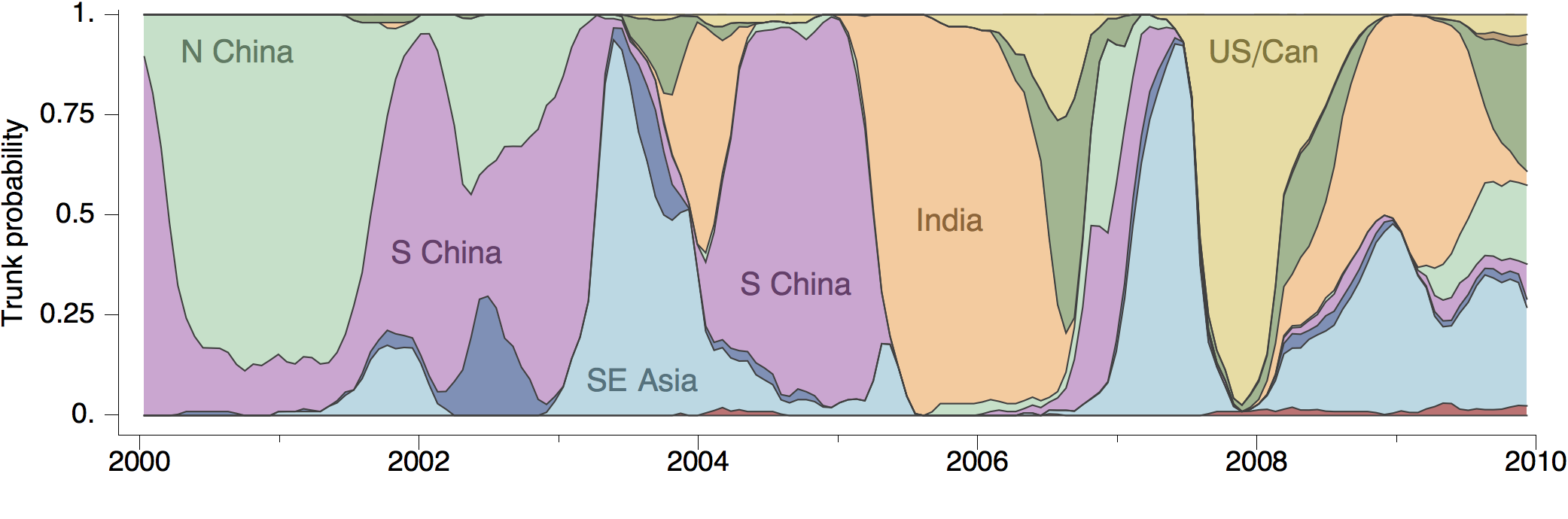
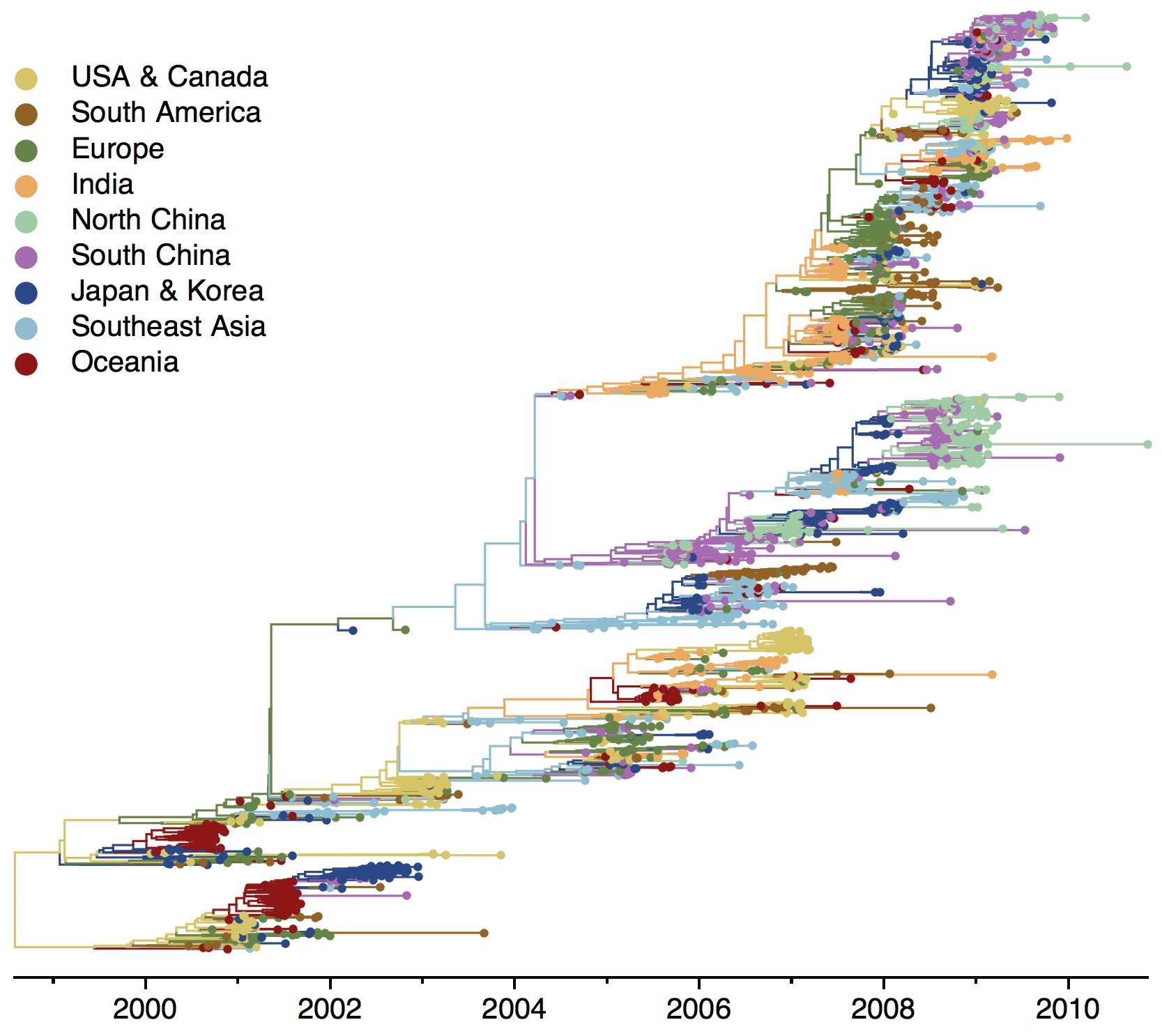
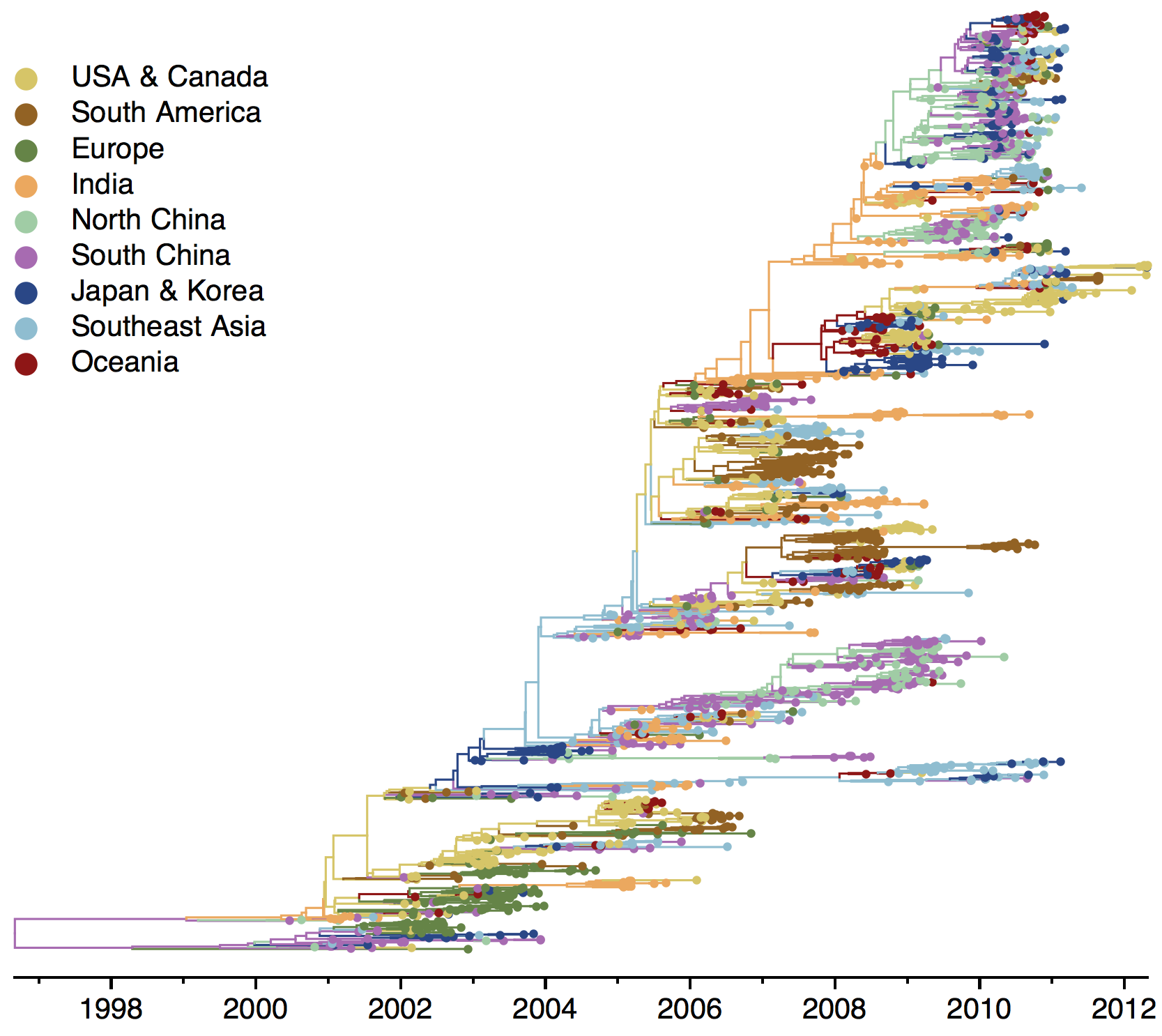
Geographic location of phylogeny trunk
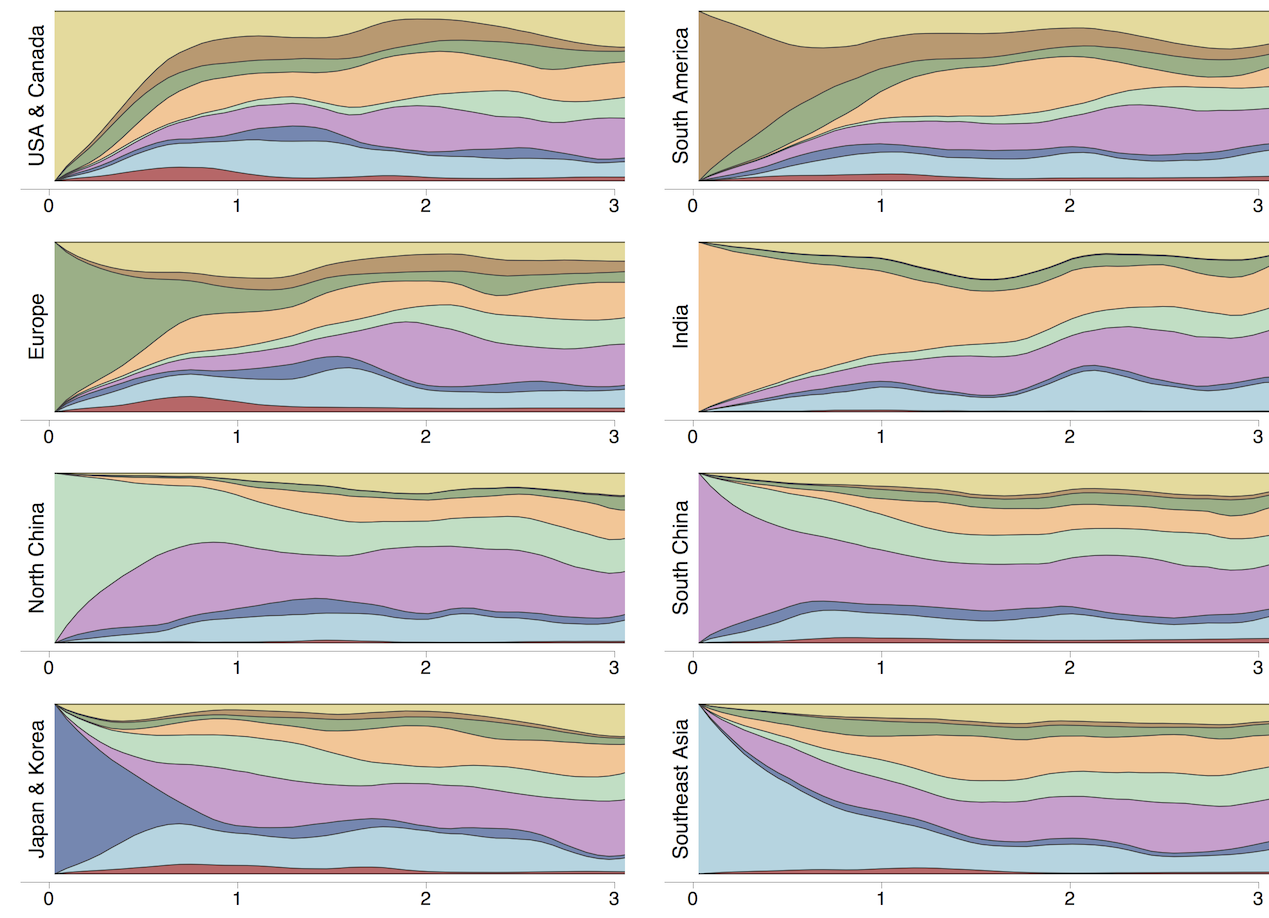
Region-specific ancestry
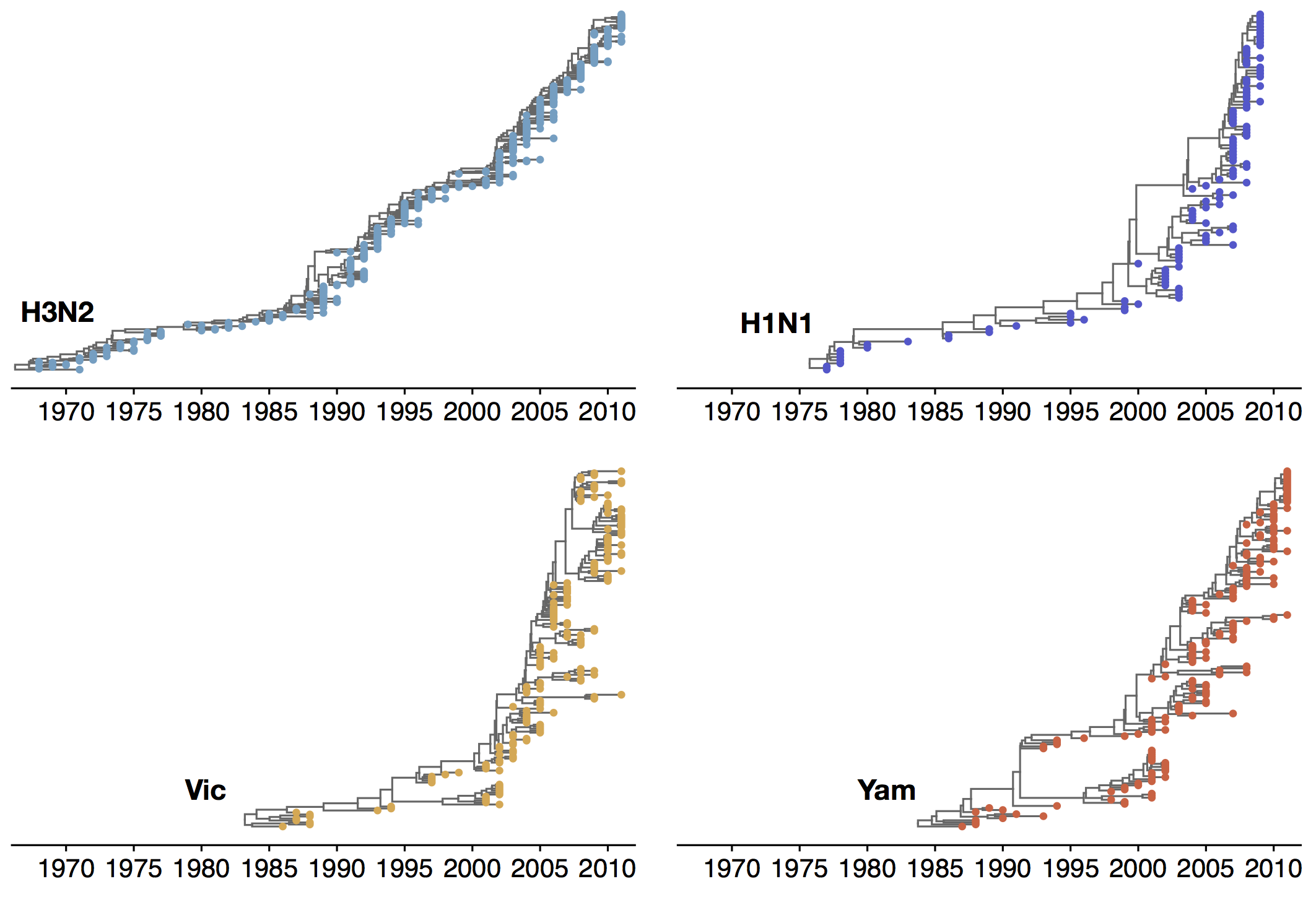
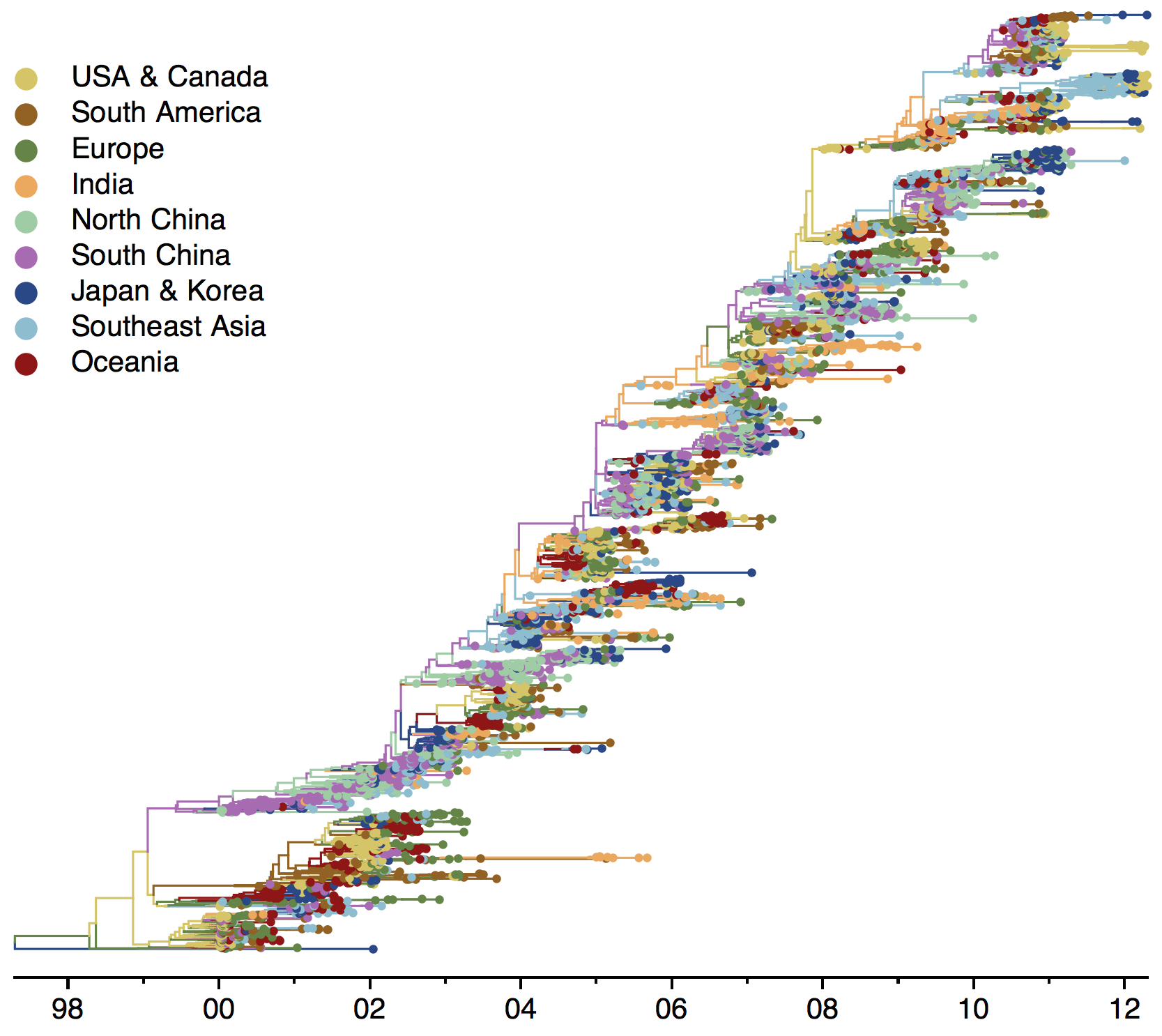
Phylogenies across subtypes / lineages
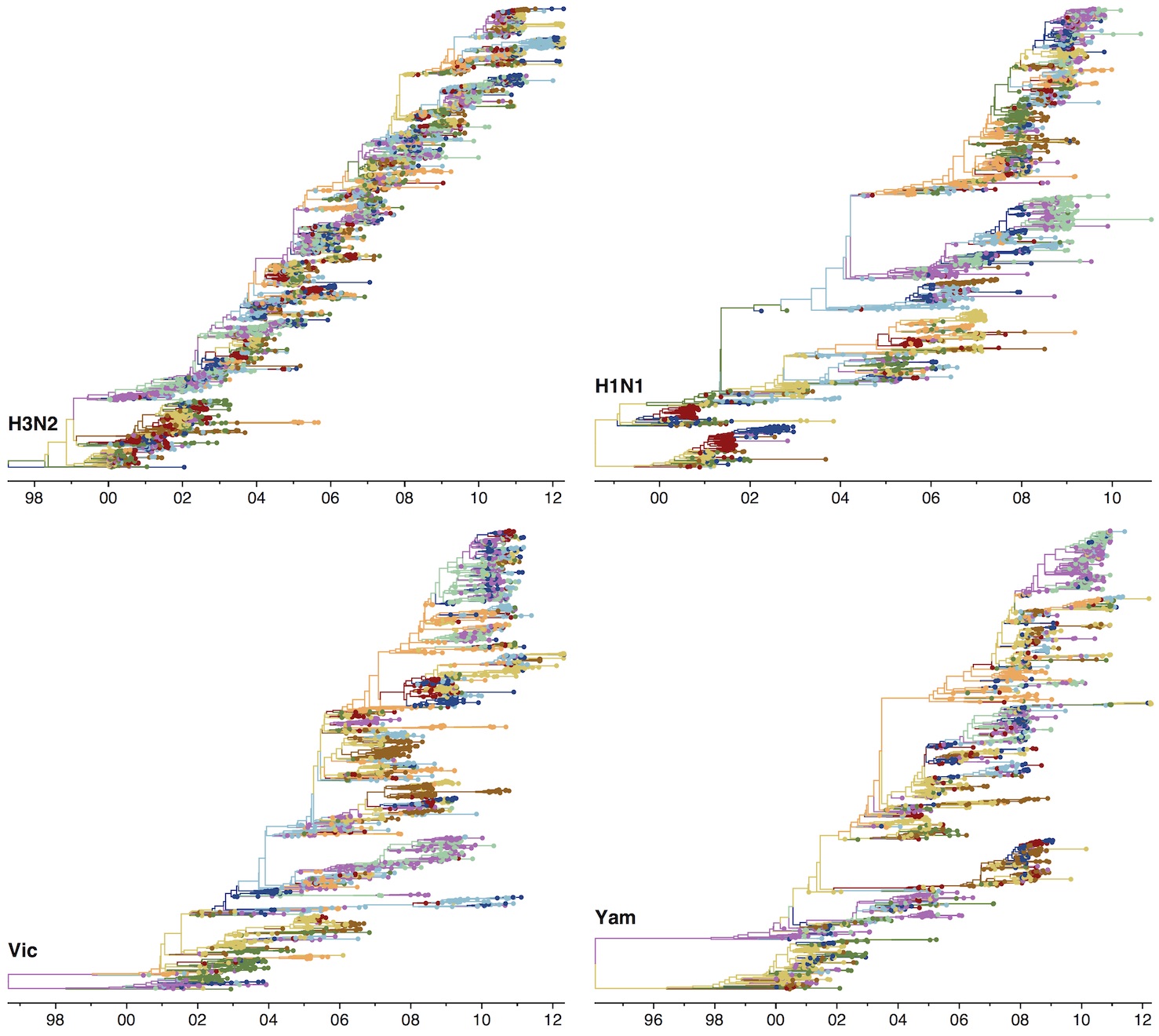
H3N2 phylogeny

H1N1 phylogeny
B/Vic phylogeny
B/Yam phylogeny
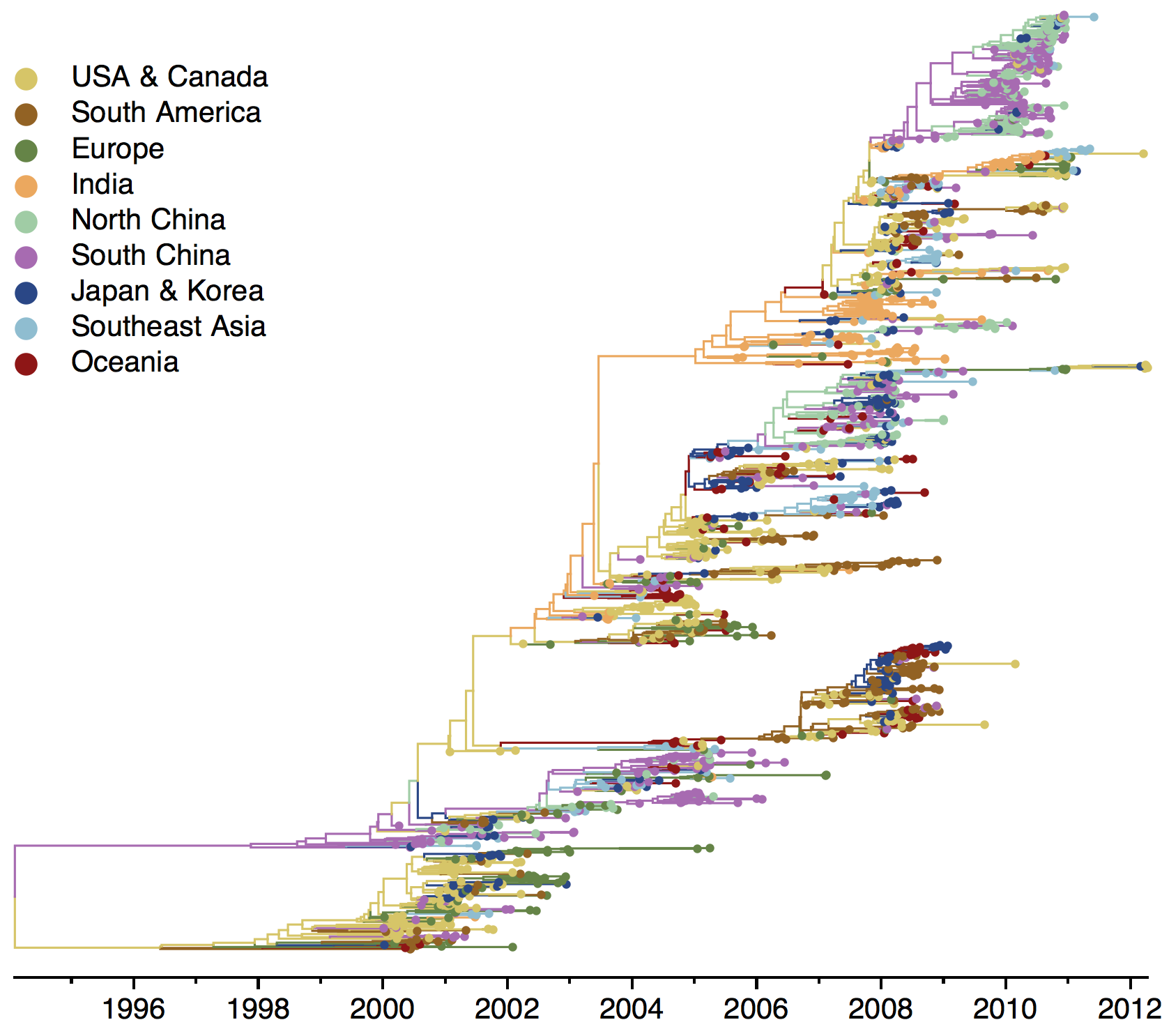
Ancestry patterns across lineages
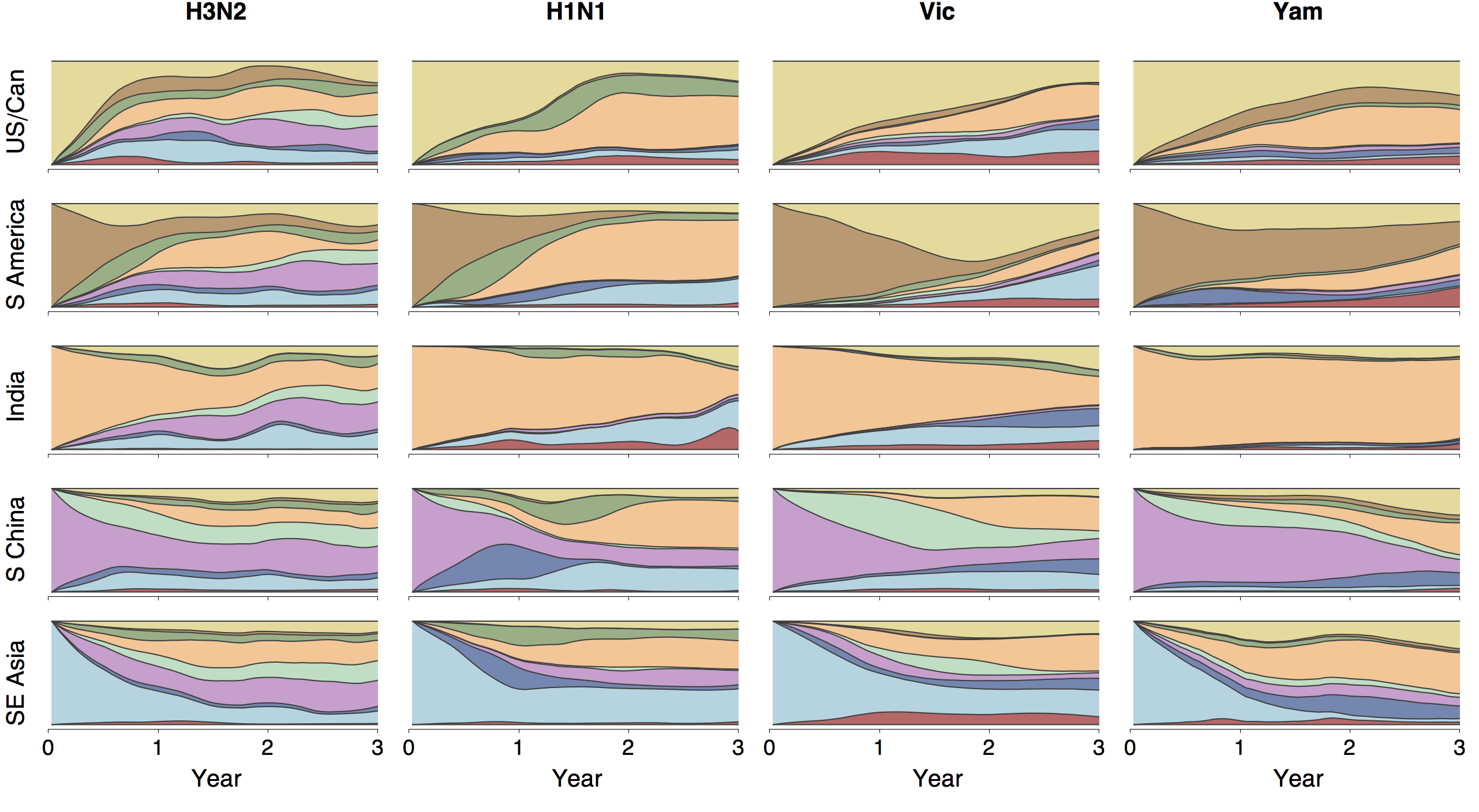
Regional persistence patterns
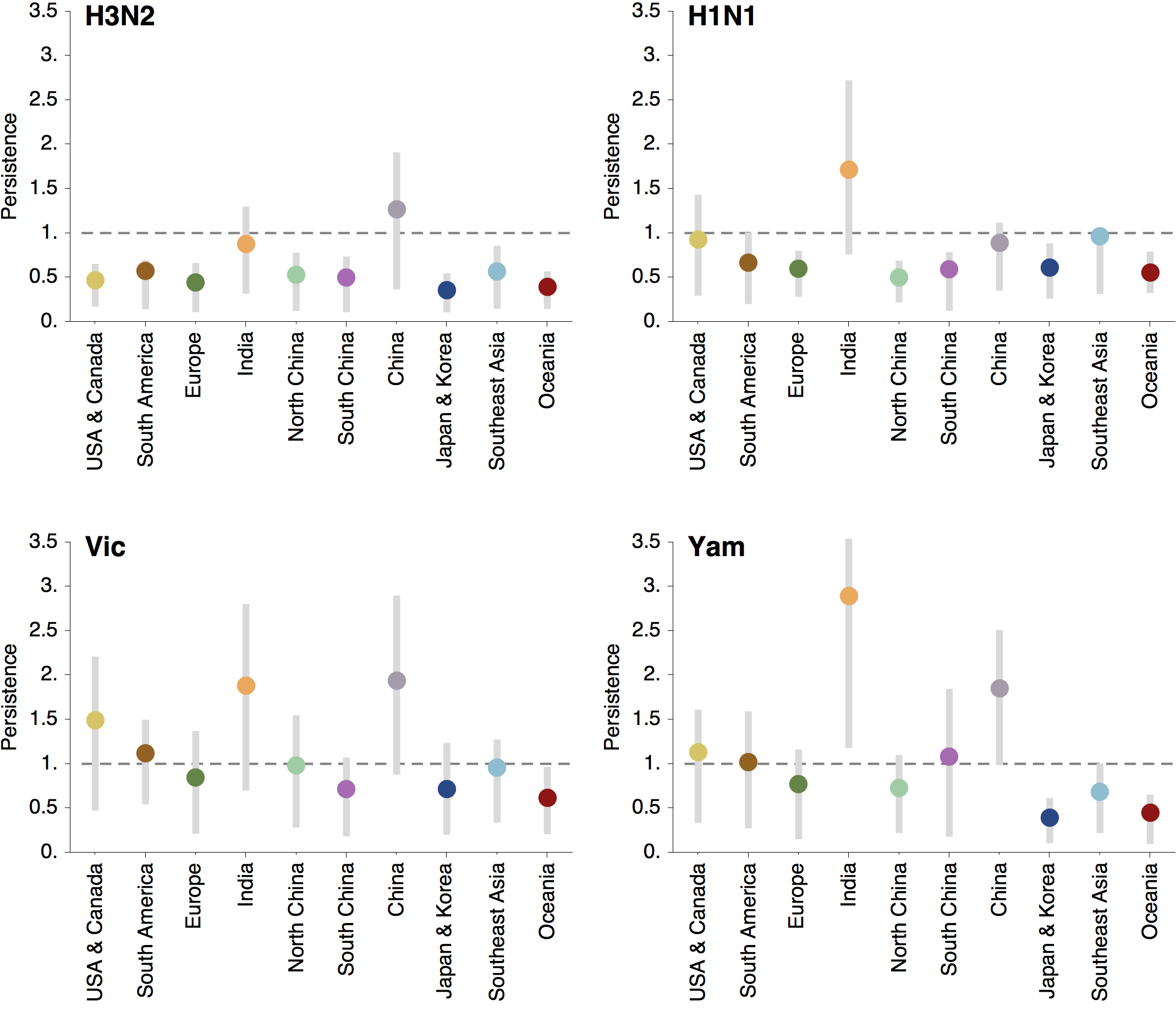
How to explain these differences?
Age distribution across viruses
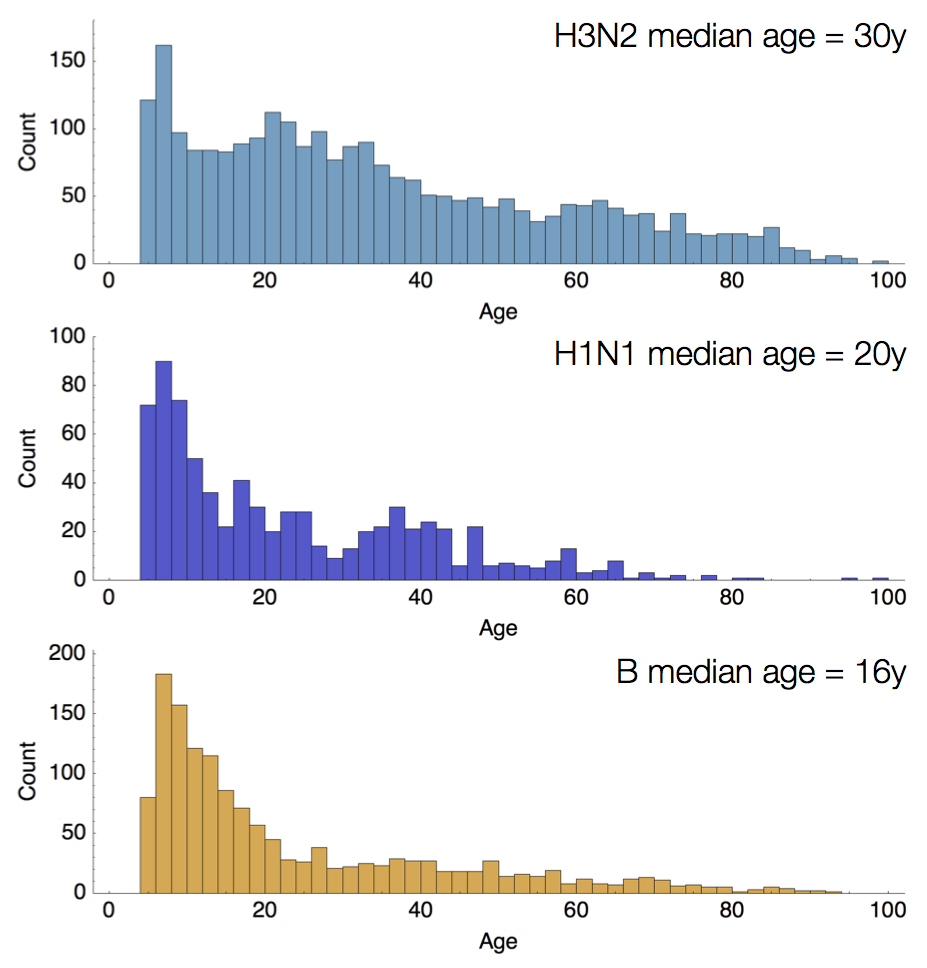
Air travel differences between adults and children
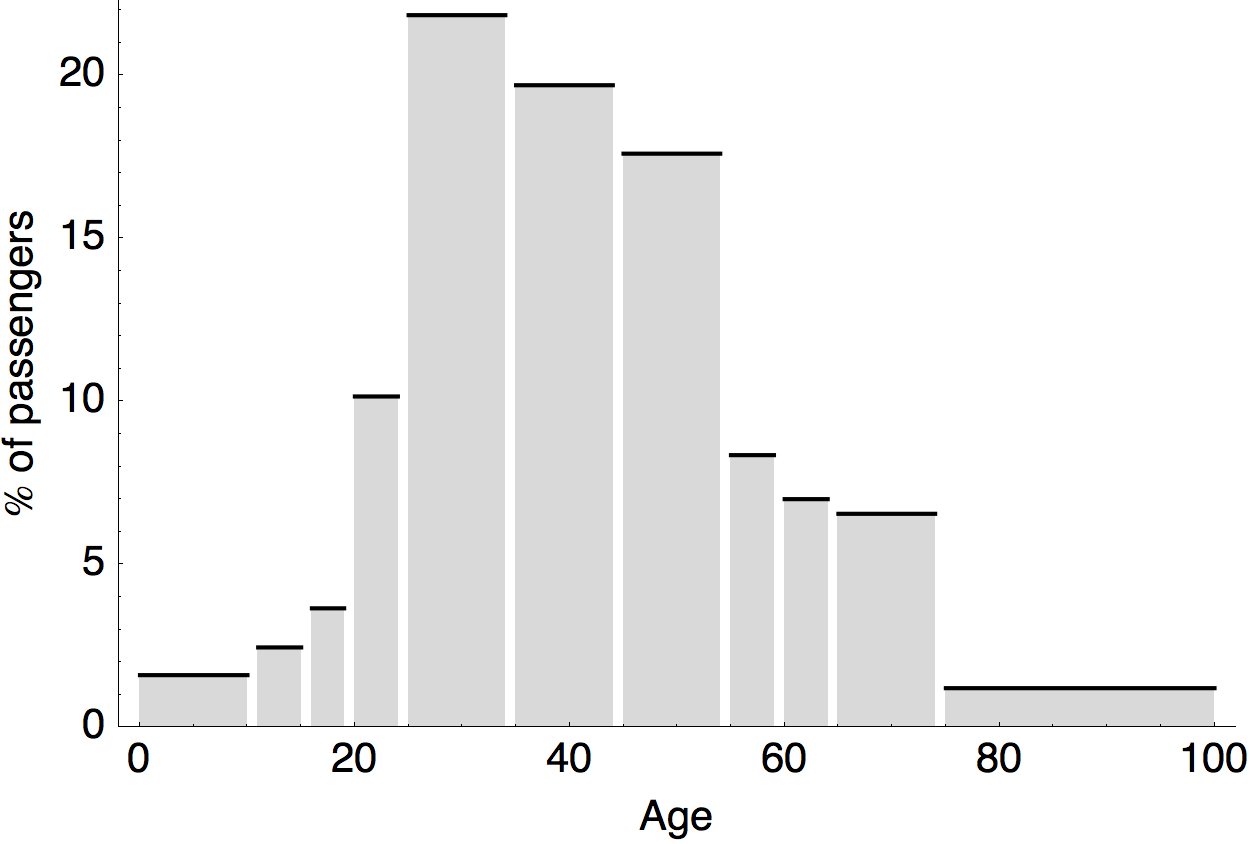
Epidemiological model of varying rates of antigenic drift
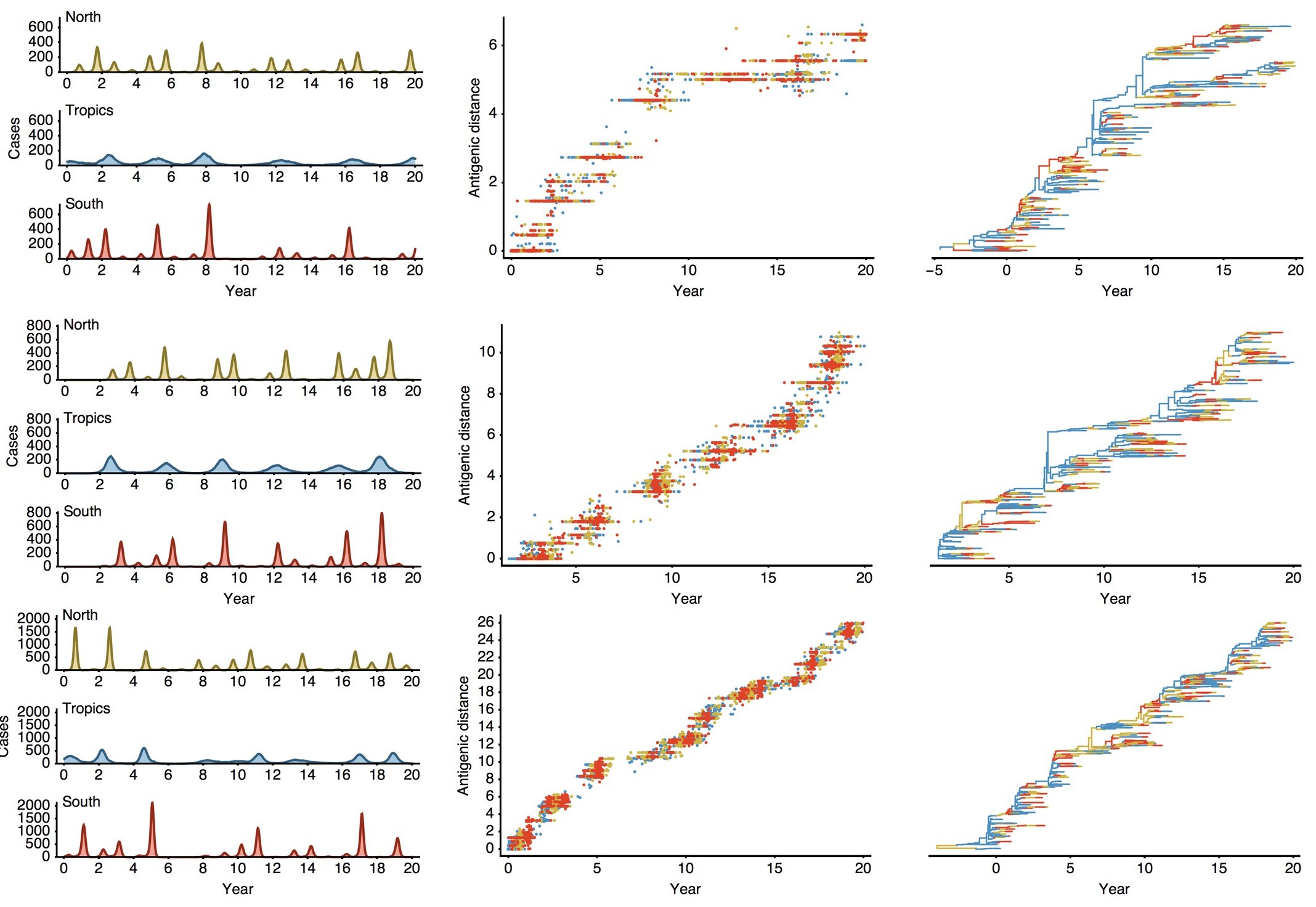
Results of varying antigenic drift
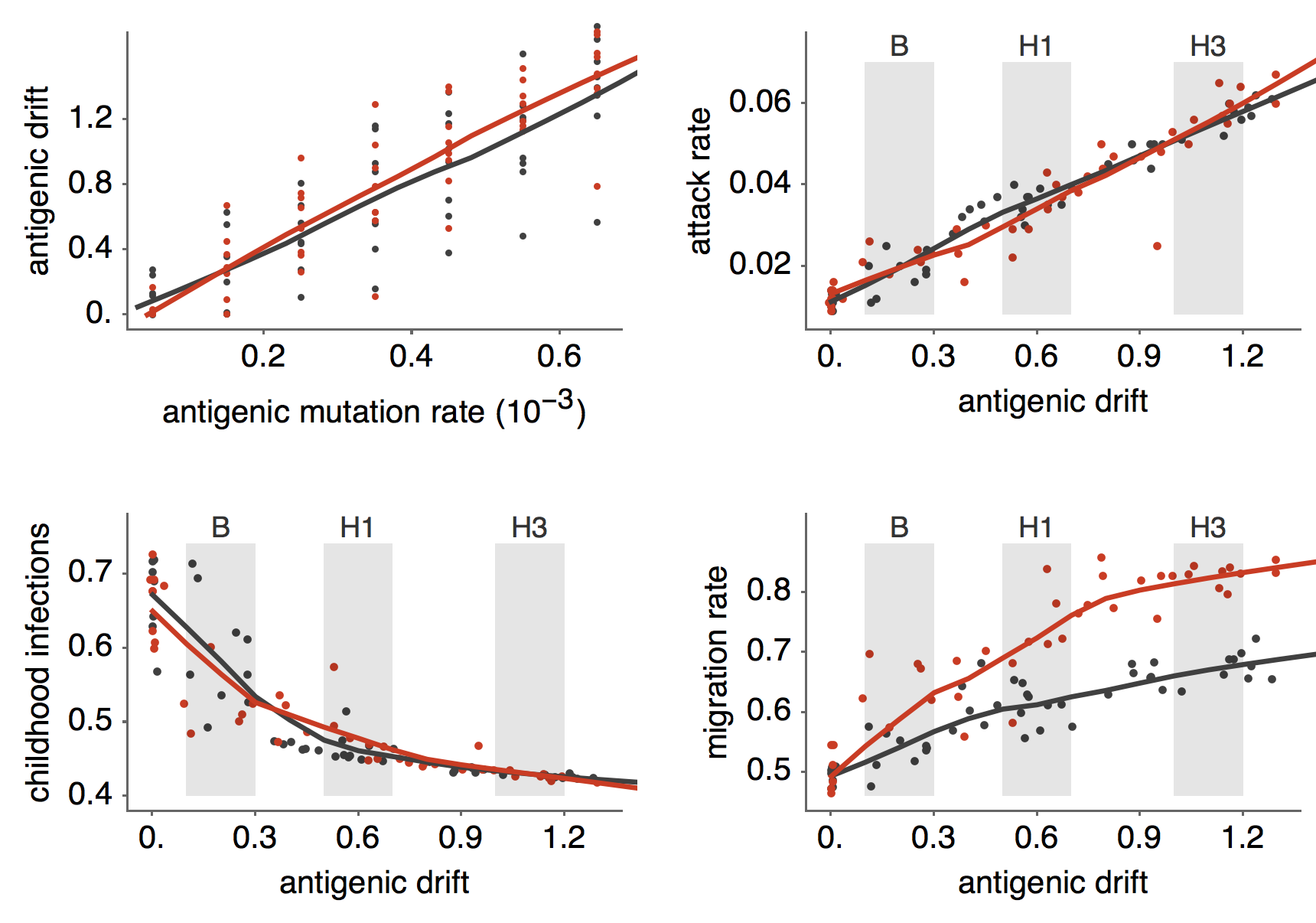
Interaction between virus evolution, epidemiology and human behavior drives migration rate differences
Circulating antigenic variants
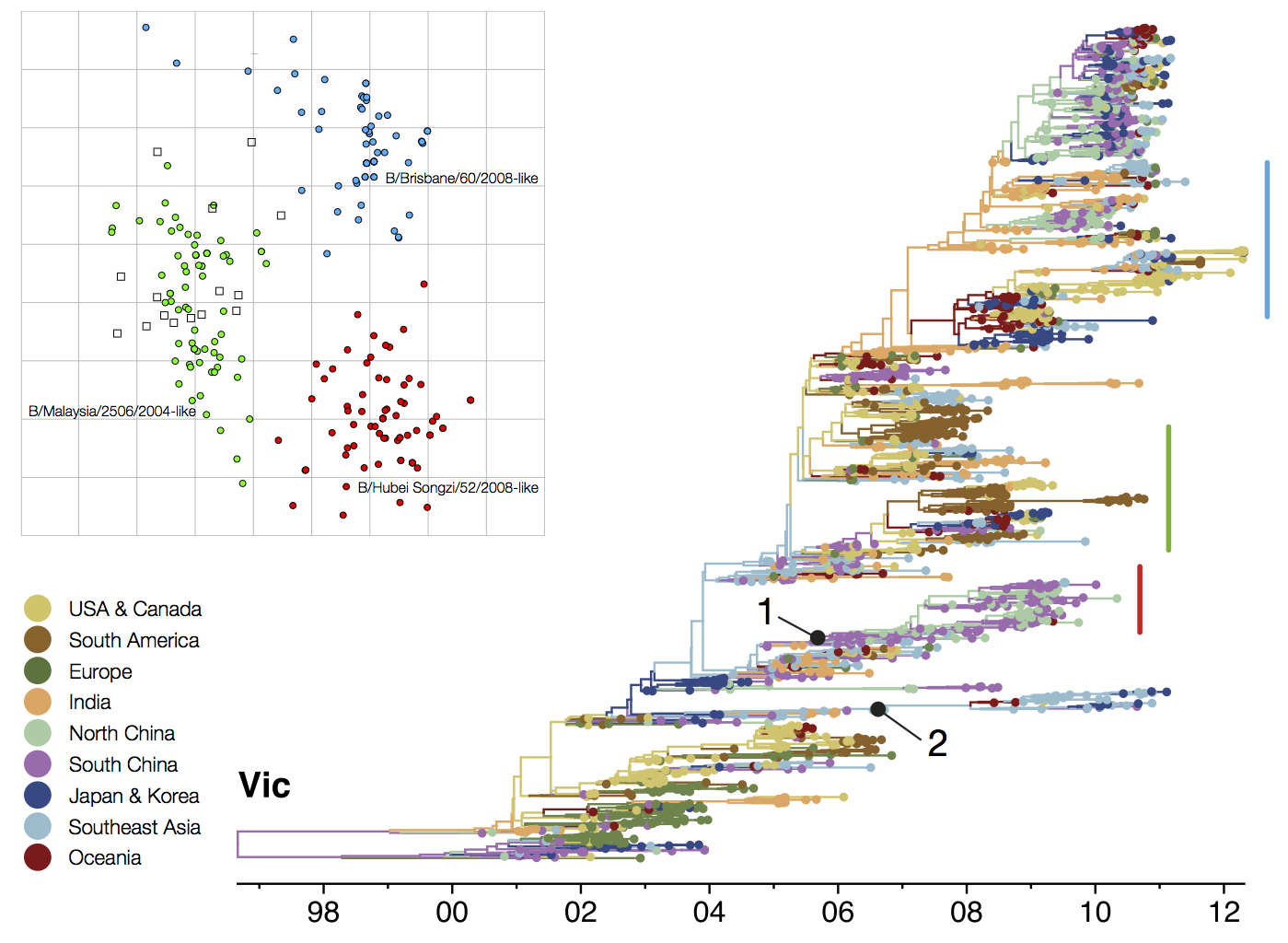
Vaccine strain prediction
Antigenic evolution in H3N2
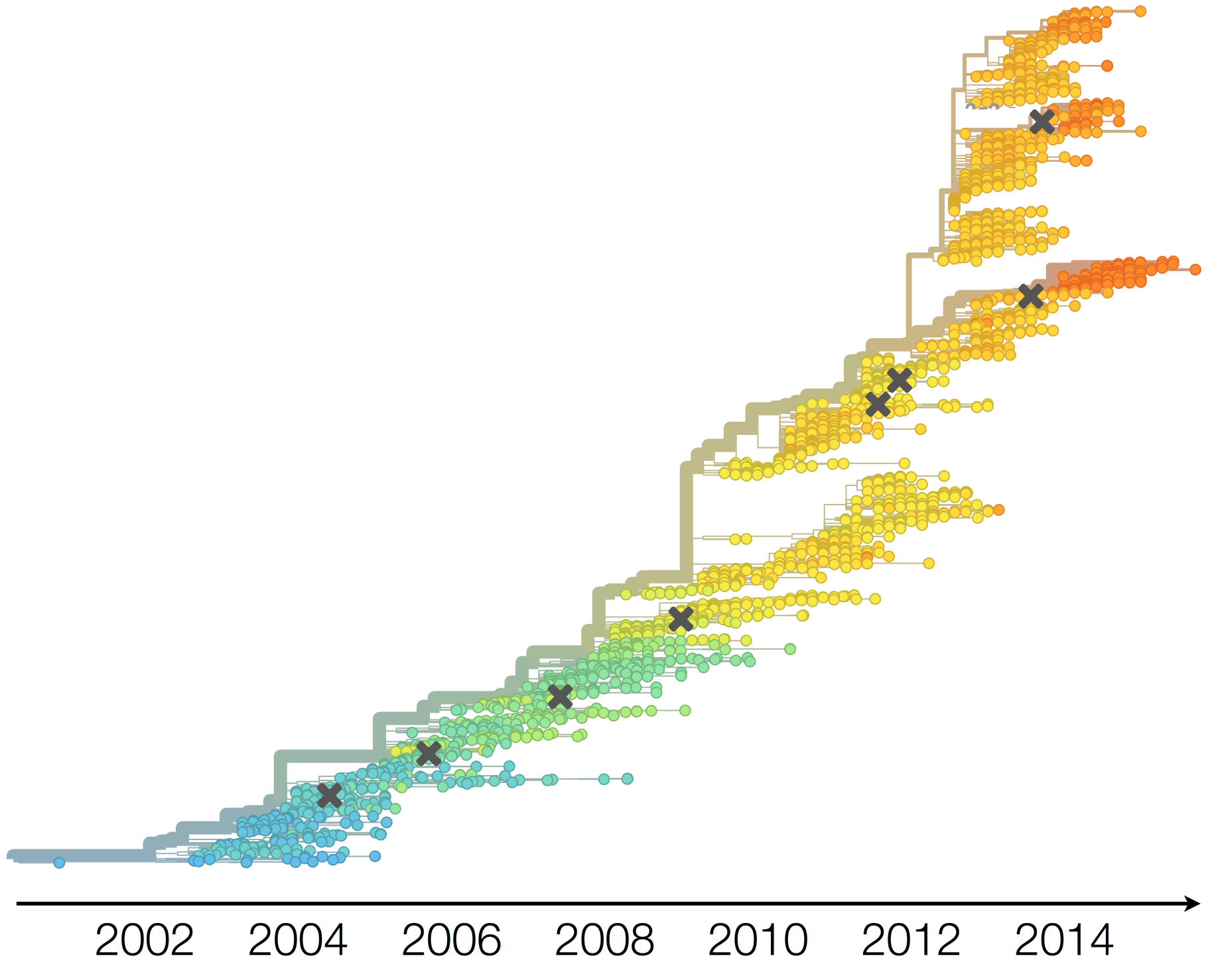
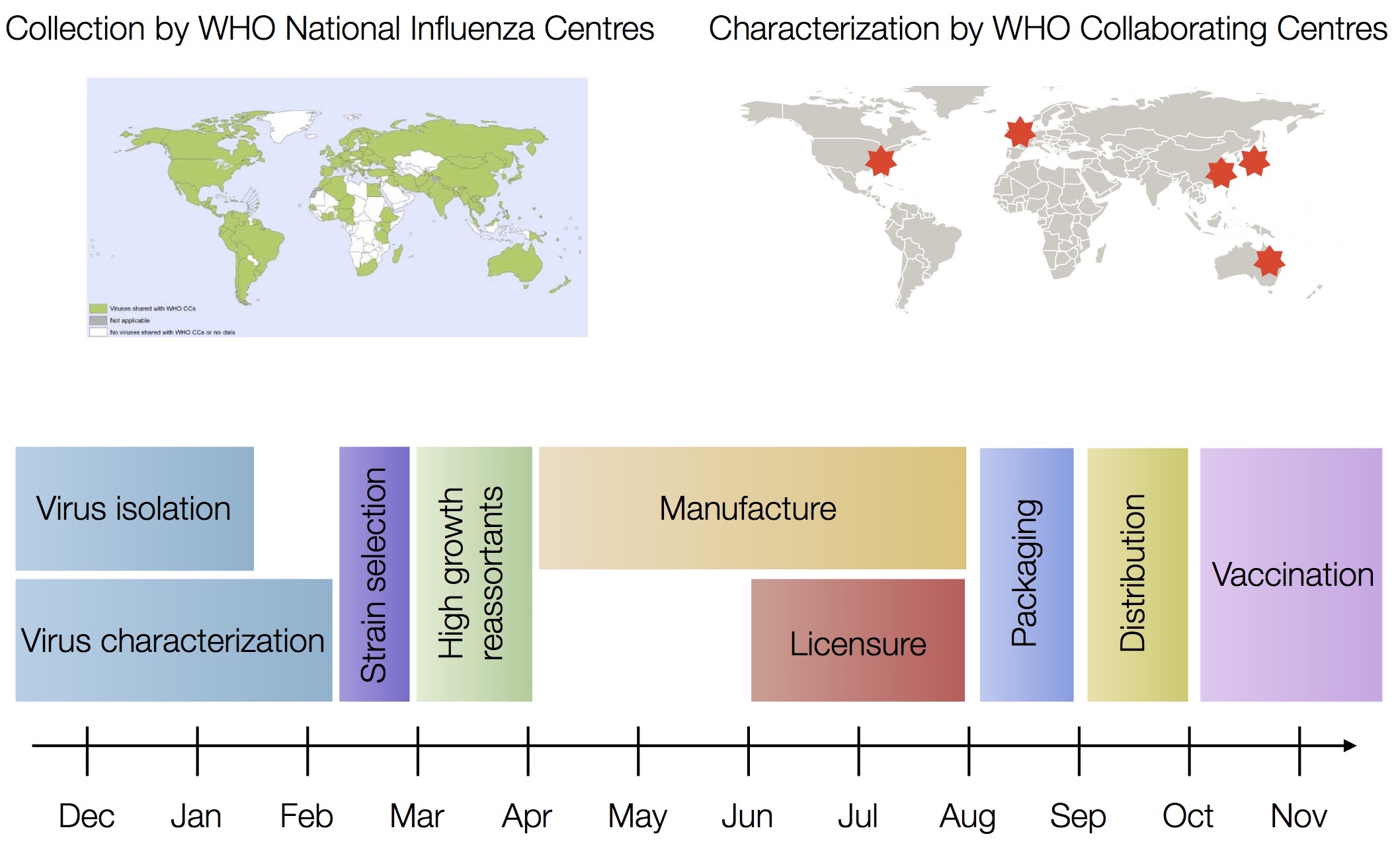
Vaccine strain selection timeline
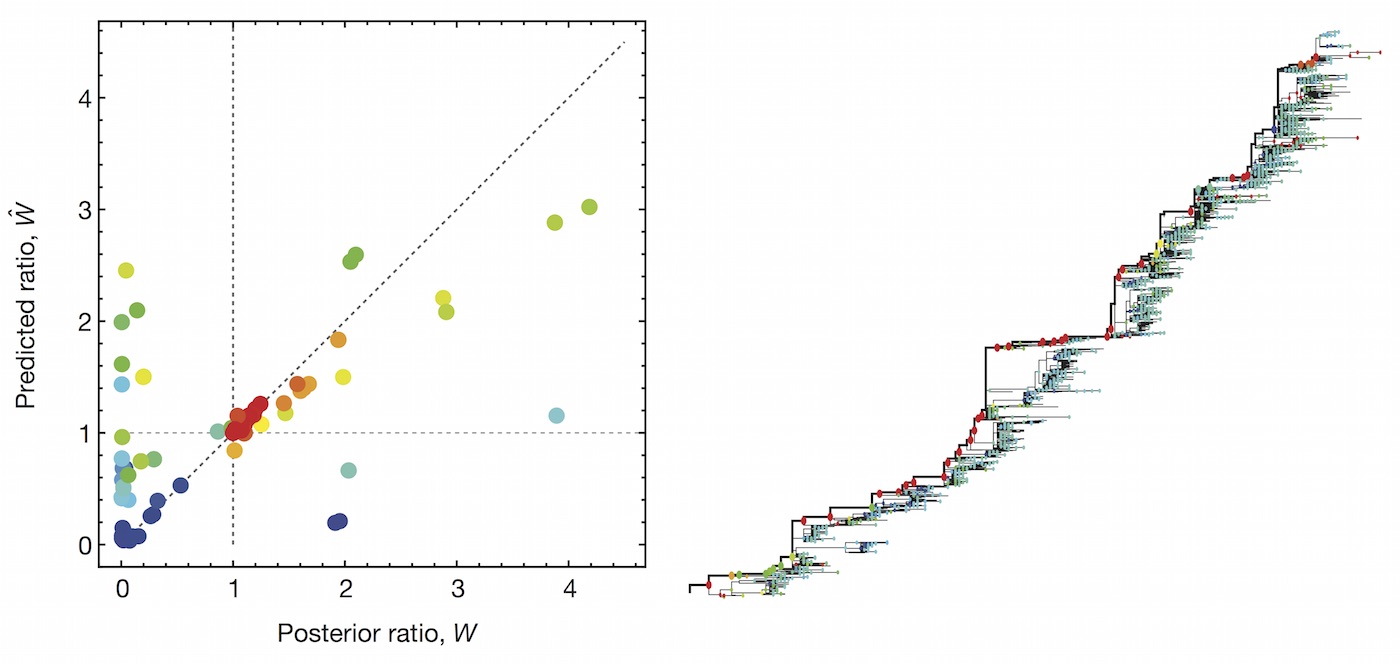
Predictive models
A simple predictive model estimates the fitness $f$ of virus $i$ as
$$\hat{f}_i = \beta^\mathrm{ep} \, f_i^\mathrm{ep} + \beta^\mathrm{ne} \, f_i^\mathrm{ne}$$
where $f_i^\mathrm{ep}$ measures cross-immunity via substitutions at epitope sites and $f_i^\mathrm{ep}$ measures mutational load via substitutions at non-epitope sites.
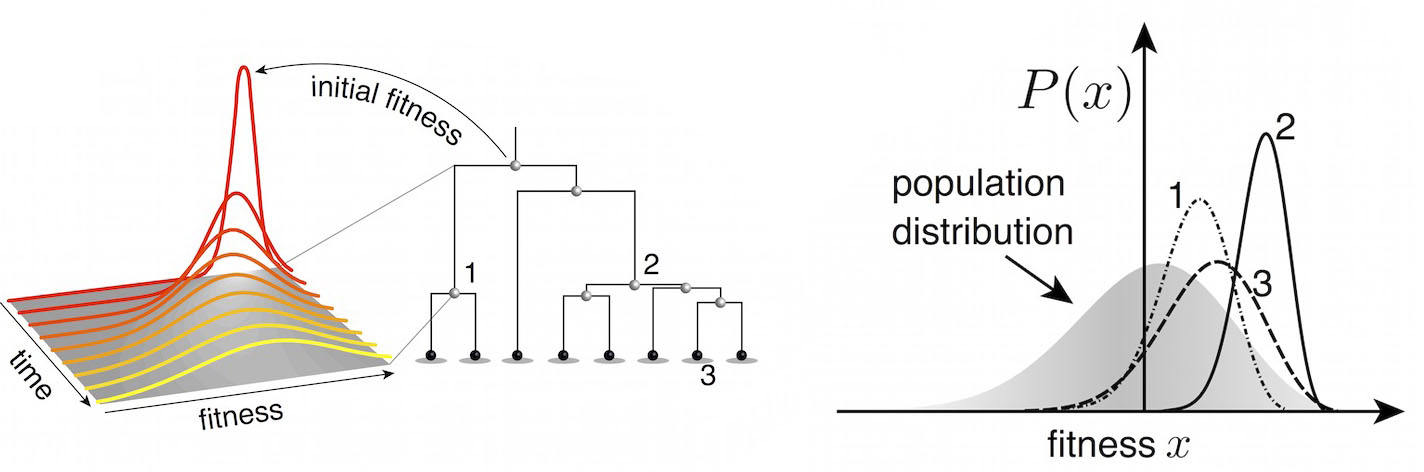
Predictive models
Another approach quantifies phylogenetic branching patterns
nextflu
Project to provide a real-time view of the evolving influenza population
All in collaboration with Richard Neher

nextflu pipeline
- Download all recent HA sequences from GISAID
- Filter to remove outliers
- Align sequences
- More filtering
- Build tree
- Estimate frequencies
- Export JSON for visualization
nextflu.org
Acknowledgements
Richard Neher (Max Planck Tübingen), Andrew Rambaut (University of Edinburgh),
Colin Russell (Cambridge University), Charles Cheung (Fred Hutch),
Marc Suchard (UCLA), Steven Riley (Imperial College),
Philippe Lemey (Philippe Lemey (KU Leuven), Gytis Dudas (University of Edinburgh)
WHO Global Influenza Surveillance Network / GISAID: Ian Barr, Shobha Broor, Mandeep Chadha, Nancy Cox, Rod Daniels, Palani
Gunasekaran, Aeron Hurt, Anne Kelso, Alexander Klimov, Nicola Lewis, Xiyan Li, John McCauley, Takato Odagiri, Varsha Potdar, Yuelong Shu, Eugene Skepner, Masato Tashiro, Dayan Wang, Xiyan Xu


Contact
- Website: bedford.io
- Twitter: @trvrb
- Slides: bedford.io/talks/flu-dynamics-korea-university/