Phylodynamics to assist in Ebola outbreak response
Trevor Bedford (@trvrb) and Allison Black (@alliblk)
4 Dec 2019
Epidemics7
Charleston, SC
Okay to share on social media
Pathogen genomes can reveal transmission dynamics
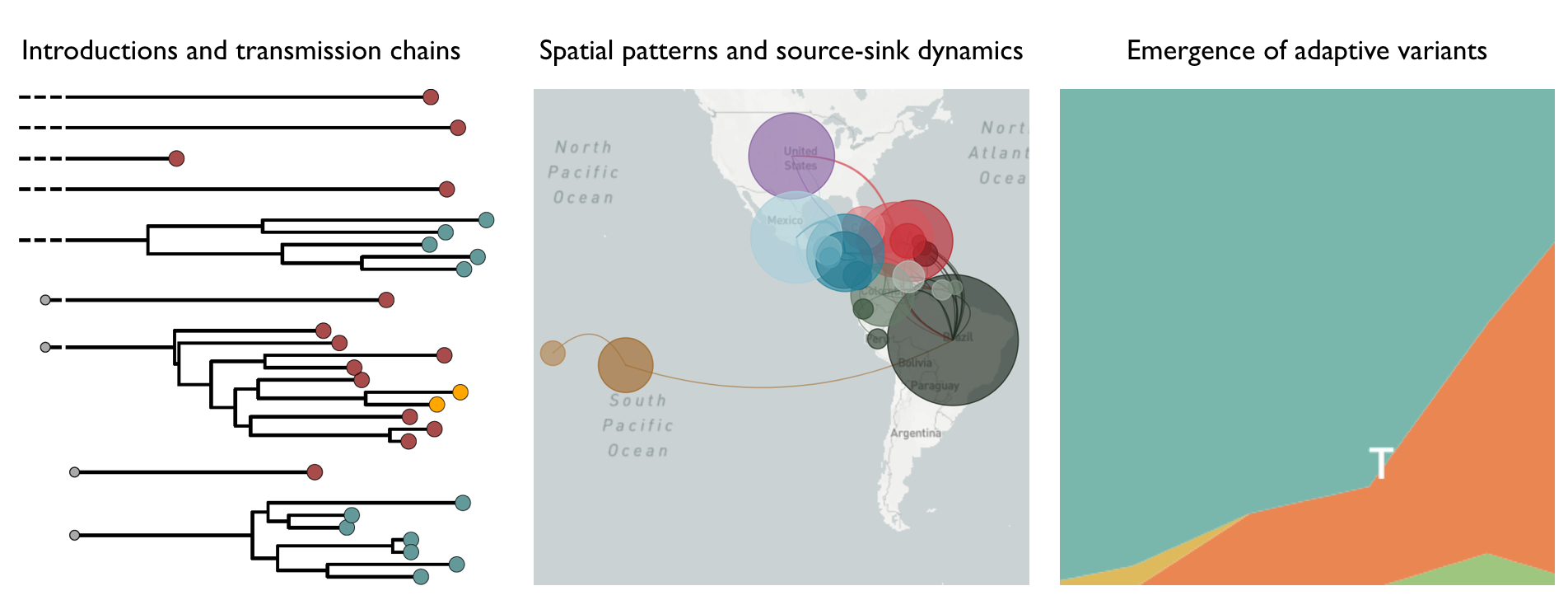
Key challenges to making genomic epidemiology actionable
- Timely analysis and sharing of results critical
- Dissemination must be scalable
- Integrate many data sources
- Results must be easily interpretable and queryable
Nextstrain
Real-time tracking of pathogen evolution
with
![]() Richard Neher,
Richard Neher,
![]() James Hadfield,
James Hadfield,
![]() Emma Hodcroft,
Emma Hodcroft,
![]() Thomas Sibley,
Thomas Sibley,
![]() Colin Megill,
Colin Megill,
![]() John Huddleston,
John Huddleston,
![]() Barney Potter,
Barney Potter,
![]() Sidney Bell,
Sidney Bell,
![]() Louise Moncla,
Louise Moncla,
![]() Charlton Callender,
Charlton Callender,
![]() Misja Ilcisin,
Misja Ilcisin,
![]() Kairsten Fay,
Kairsten Fay,
![]() Jover Lee
Jover Lee
Nextstrain is two things
- a bioinformatics toolkit and visualization app, which can be used for a broad range of datasets
- a collection of real-time pathogen analyses kept up-to-date on the website nextstrain.org
Ongoing DRC outbreak
Genomic epidemiology applied to North Kivu Ebola outbreak
with Placide Mbala-Kingebeni, Eddy Kinganda Lusamaki, Catherine Pratt, Mike Wiley, James Hadfield, Allison Black, Jean-Jacques Muyembe Tamfum, Steve Ahuka-Mundeke, Daniel Mukadi, Gustavo Palacios, Amadou Sall, Ousmane Faye, Eric Delaporte, Martine Peeters and many others
Nextstrain is being used to track North Kivu outbreak
- Institut National de Recherche Biomédicale (INRB) sequencing samples in Kinshasa and Katwa
- Data and pipeline housed at github.com/inrb-drc/ebola-nord-kivu
- Resulting analyses are being deployed live to nextstrain.org/community/inrb-drc/ebola-nord-kivu
Allison Black (PhD student) and James Hadfield (postdoc) working with scientists at the INRB. Goal is to provide training in bioinformatics, Nextstrain and genomic epidemiology.

View of current genomic data
432 genomes most recent from late Oct
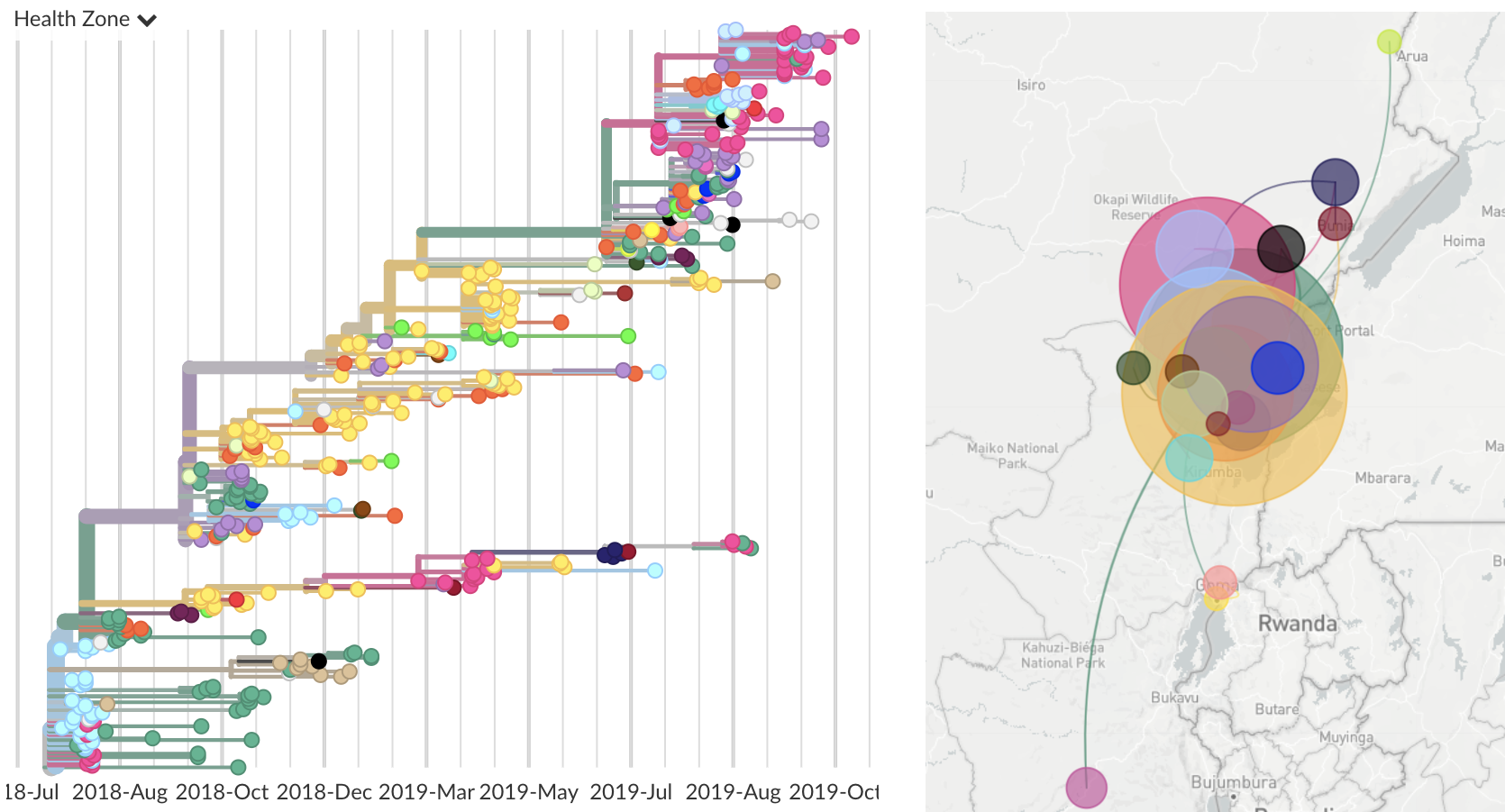
Often (but not always) need specific actionable pieces of information rather than large-scale understanding. For Ebola outbreak response, I believe this needs to revolve around contact tracing.
Superspreader event in June
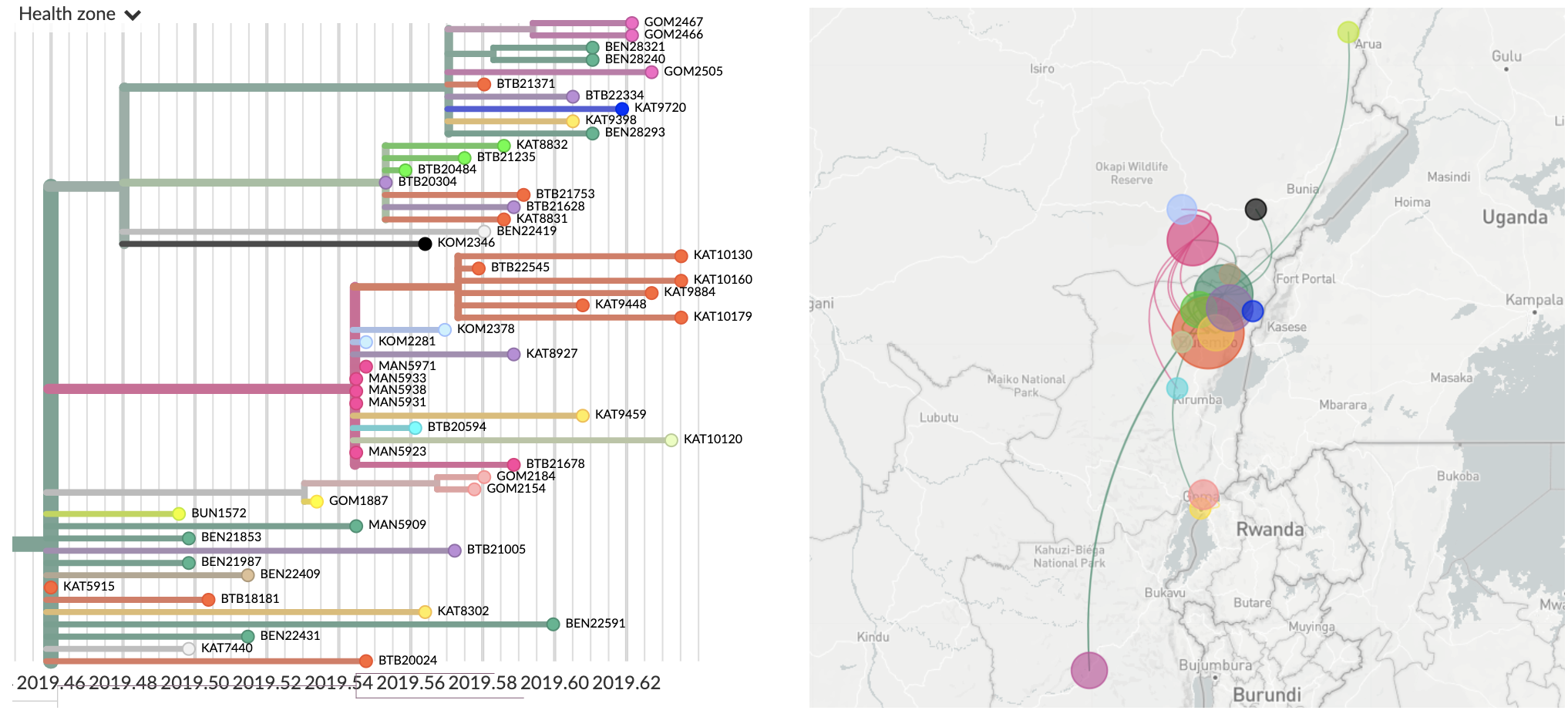
To increase public health impact, we need good ways to communicate genomic findings to field epidemiologists
We think interpretation can be improved by providing information tied to specific views of the tree
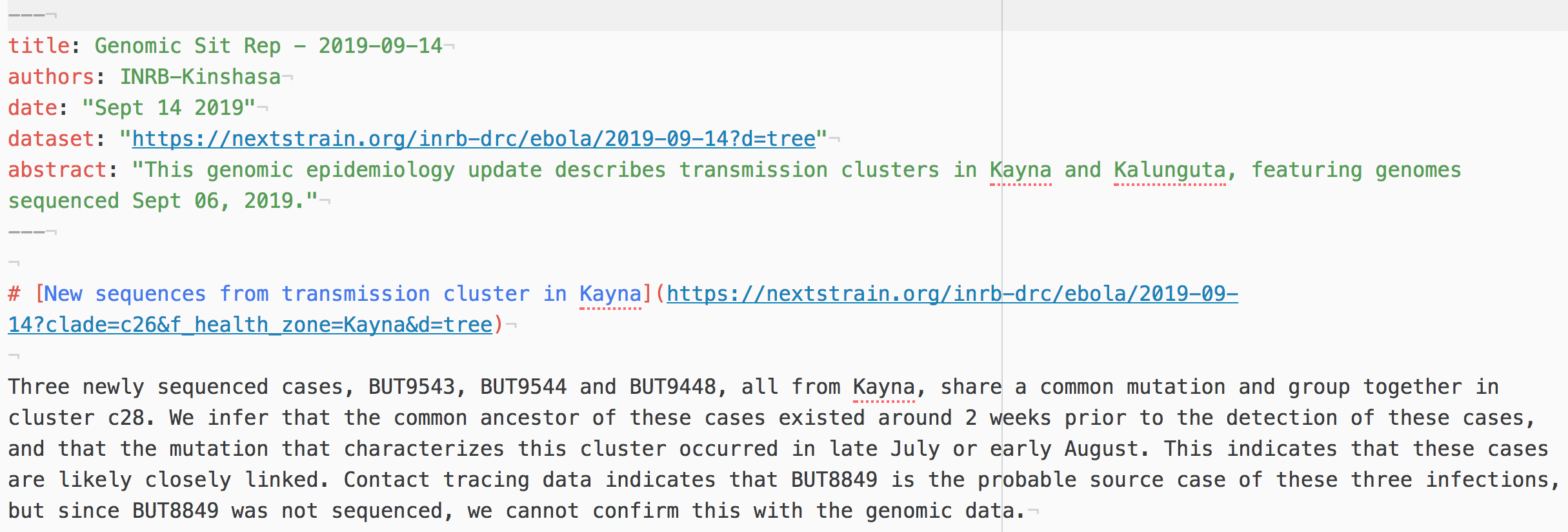
Nextstrain now provides this type of view through support of "Nextstrain Narratives"

Narratives are written as Markdown files that link visualization with descriptive text
What are we looking for in the phylogenies?
Spatial patterns, sources, and sinks
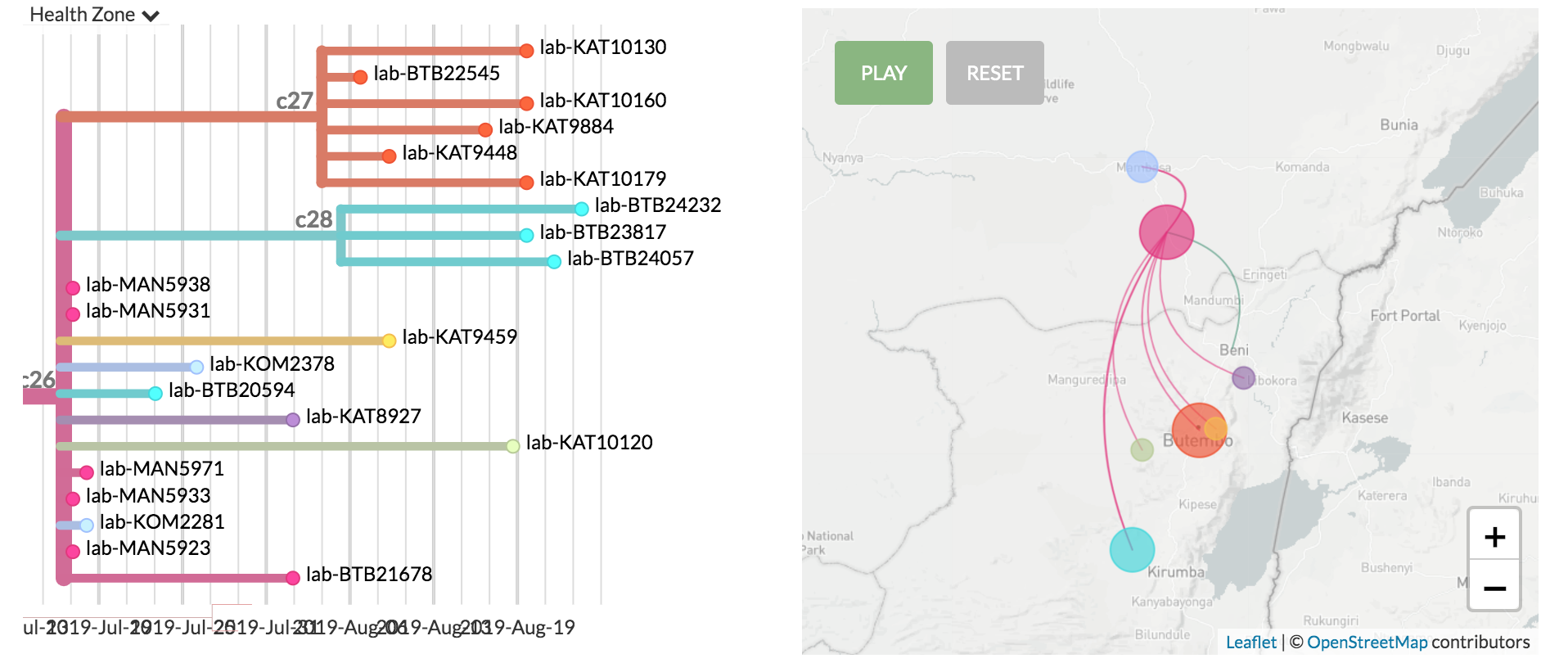
How are cases connected?
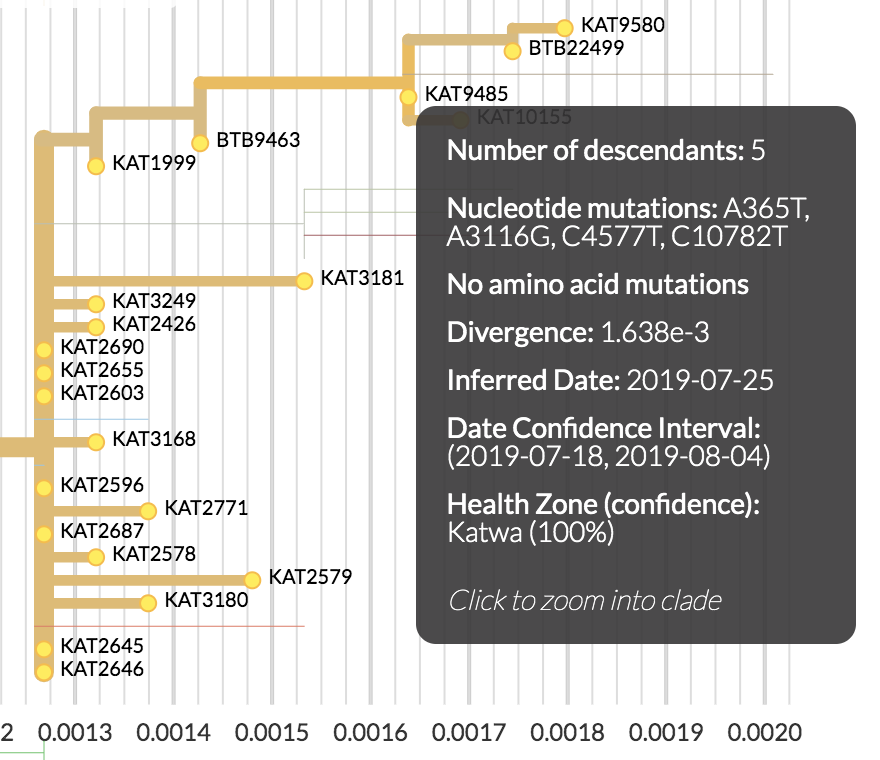
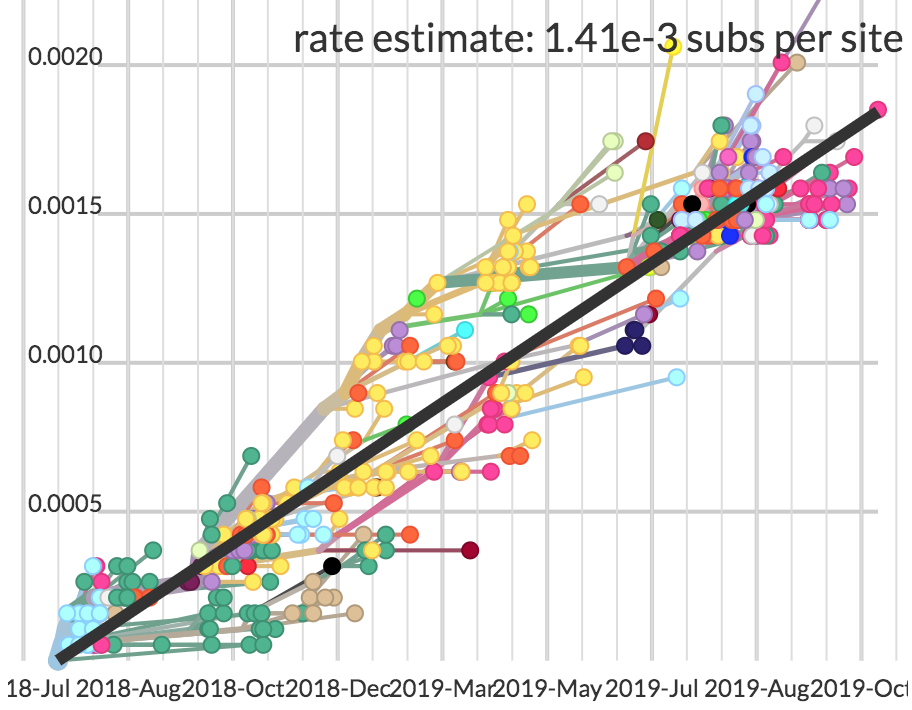
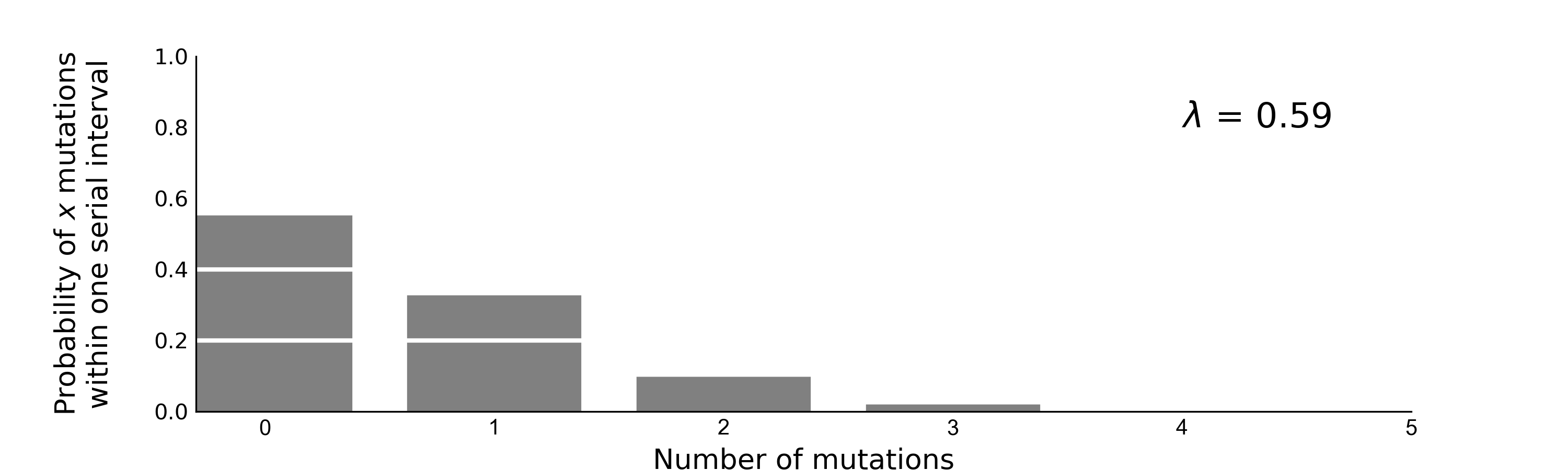
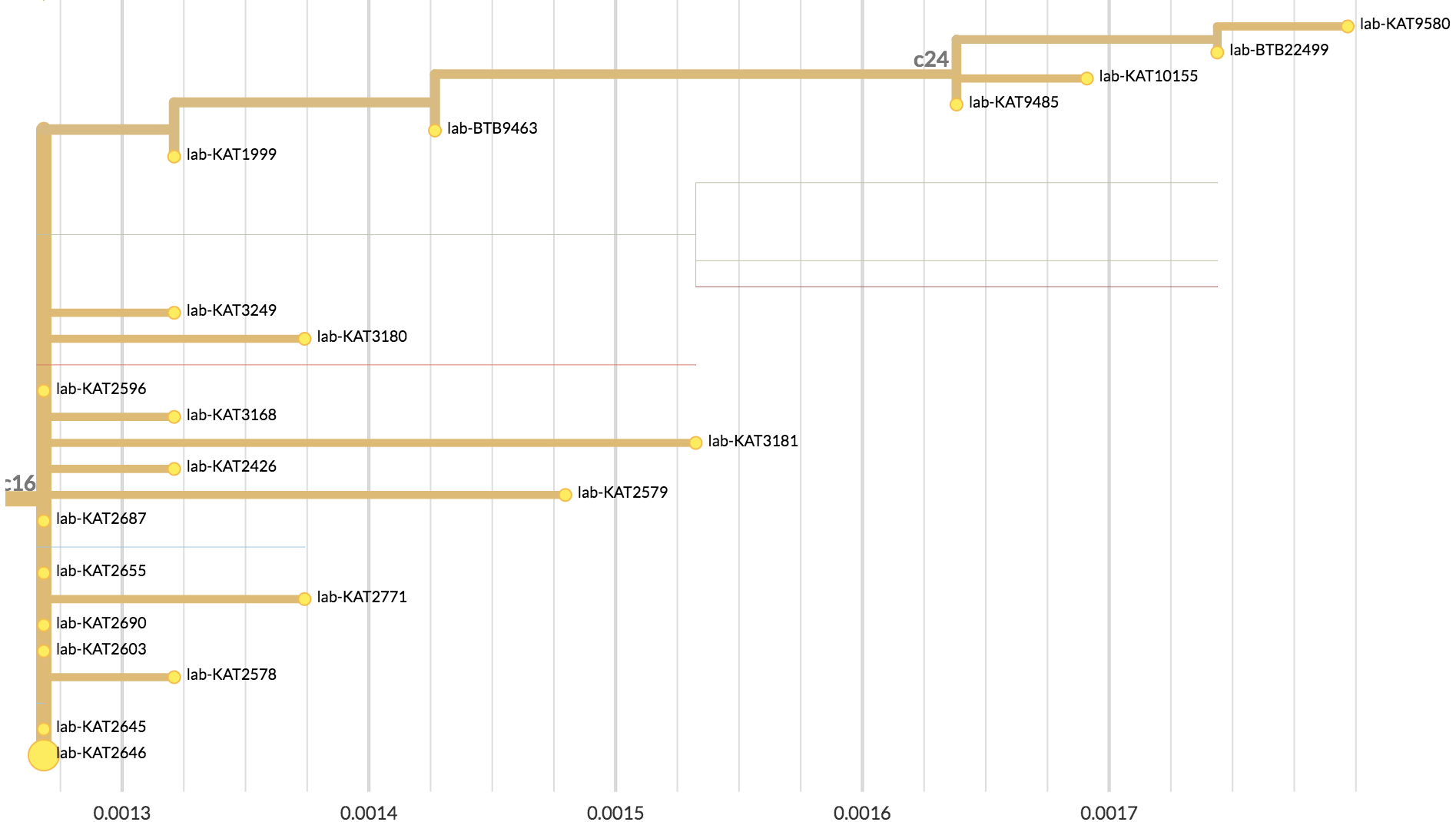
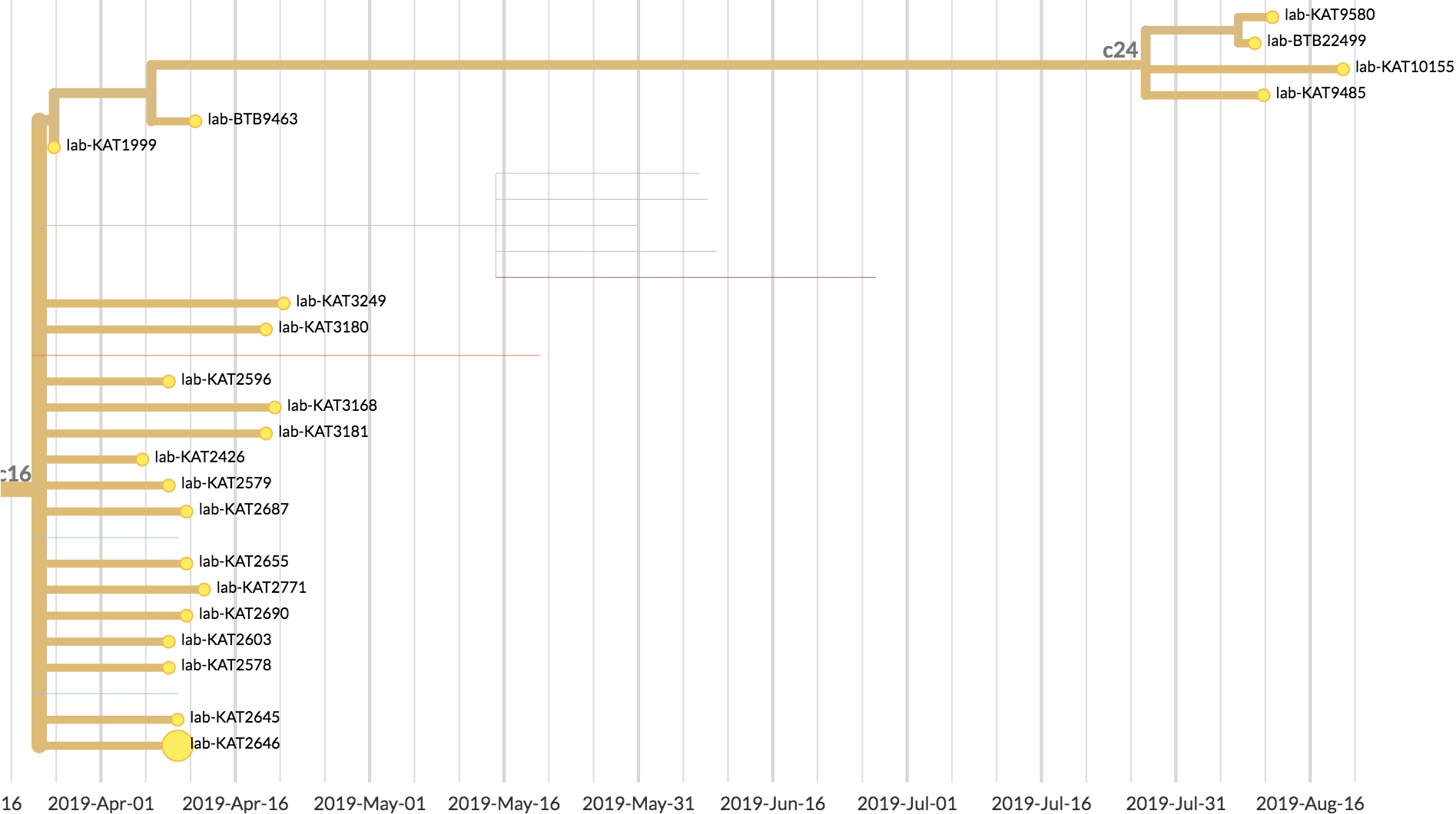
| Mutations | Time |
|---|---|
| 4 mutations | 108 days |
| ~0.59 mutations per serial interval | ~13 days per serial interval |
| 4/0.59 = ~7 serial intervals | 108/13 = ~8 serial intervals |
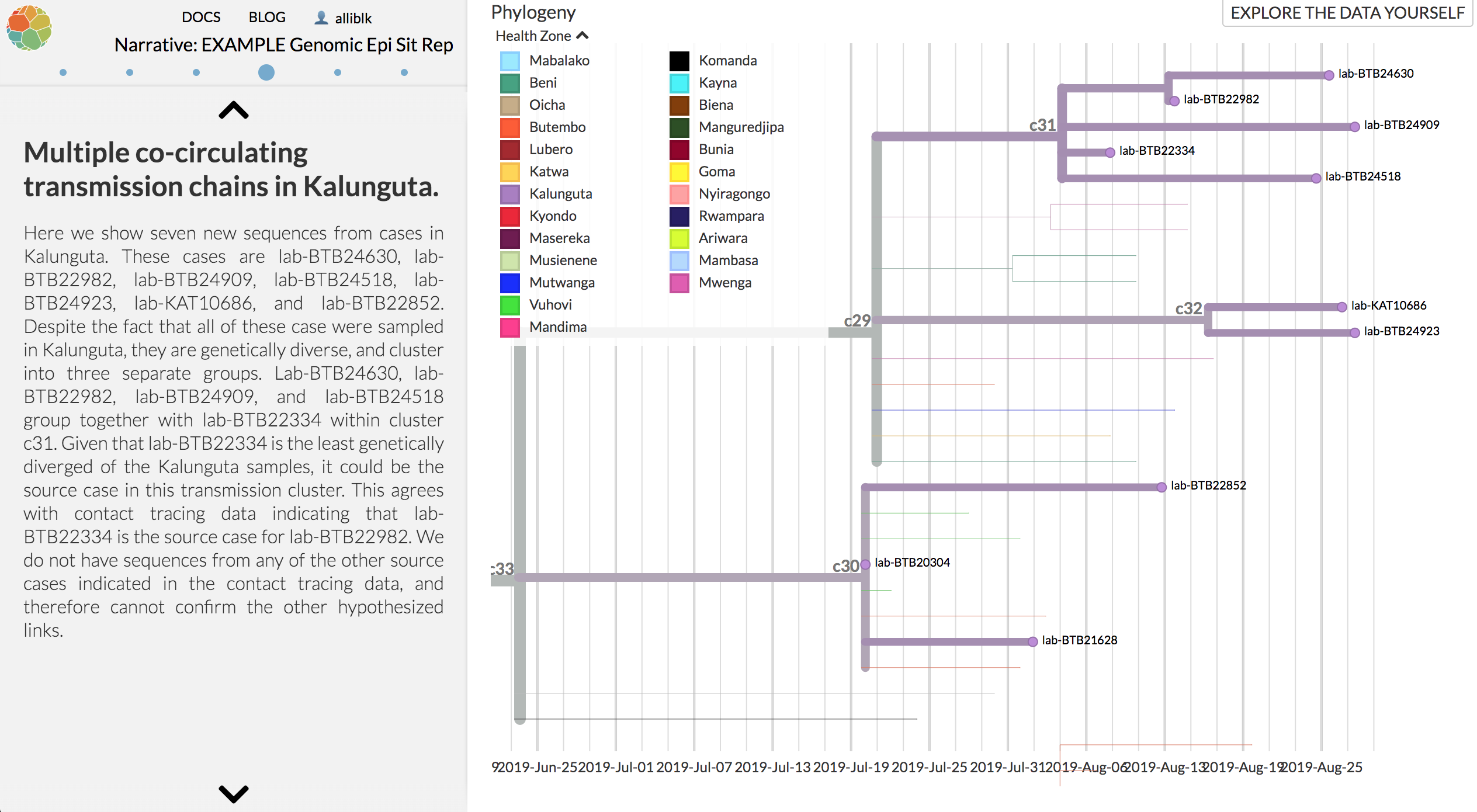
Example: Evidence for multiple concurrently circulating transmission chains in Kalunguta
Extraordinarily challenging outbreak, but some good things going on here
- Samples sequenced entirely in country by local scientists
- Bioinformatics for genome assembly and for phylodynamics run entirely in country
- Nextstrain narratives to aid epidemiological interpretation
- Best turnaround time so far is 7 days from sample collection to genomic situation report
Acknowledgements
Bedford Lab:
![]() Alli Black,
Alli Black,
![]() John Huddleston,
John Huddleston,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Louise Moncla,
Louise Moncla,
![]() Maya Lewinsohn,
Maya Lewinsohn,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Kairsten Fay,
Kairsten Fay,
![]() Misja Ilcisin,
Misja Ilcisin,
![]() Nicola Müller
Nicola Müller
Ebola in DRC: Allison Black, James Hadfield, Eddy Kinganda Lusamaki, Placide Mbala-Kingebeni, Catherine Pratt, Mike Wiley, Jean-Jacques Muyembe Tamfum, Steve Ahuka-Mundeke, Daniel Mukadi, Gustavo Palacios, Amadou Sall, Ousmane Faye, Eric Delaporte, Martine Peeters, David Blazes, Cécile Viboud
Nextstrain: Richard Neher, James Hadfield, Emma Hodcroft, Tom Sibley, John Huddleston, Sidney Bell, Barney Potter, Colin Megill, Charlton Callender





