Data rich phylodynamics
Trevor Bedford
Fred Hutchinson Cancer Center / Howard Hughes Medical Institute
12 Sep 2024
Cobey Lab Seminar
University of Chicago
Slides at: bedford.io/talks
Genomic epidemiology in emerging outbreaks
Three key insights from genomic epi during COVID-19 pandemic
- Rapid human-to-human spread in Wuhan beyond initial market outbreak
- Extensive local transmission while testing was rare
- Identification of variants of concern and mapping of increased transmission rates
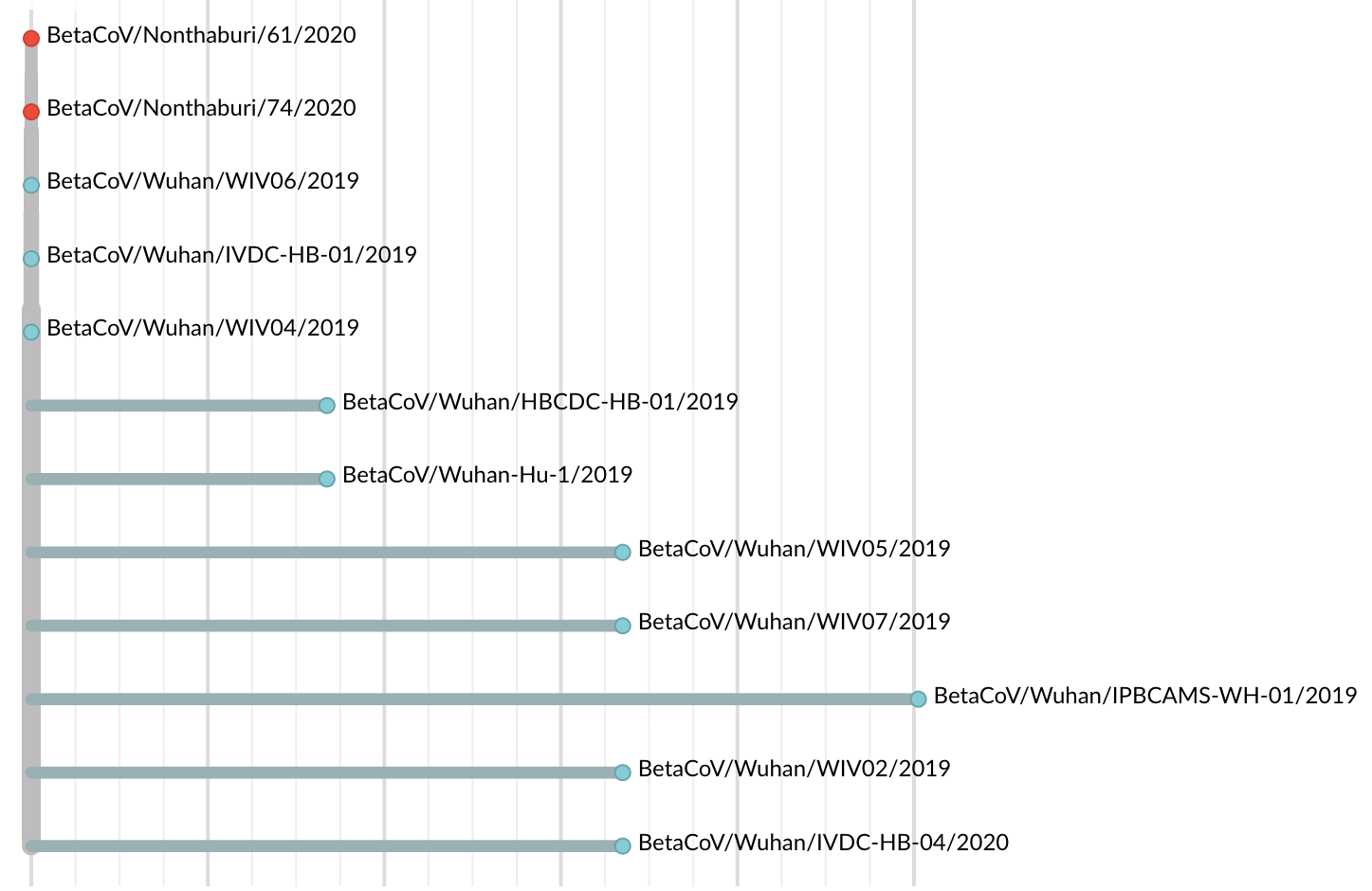
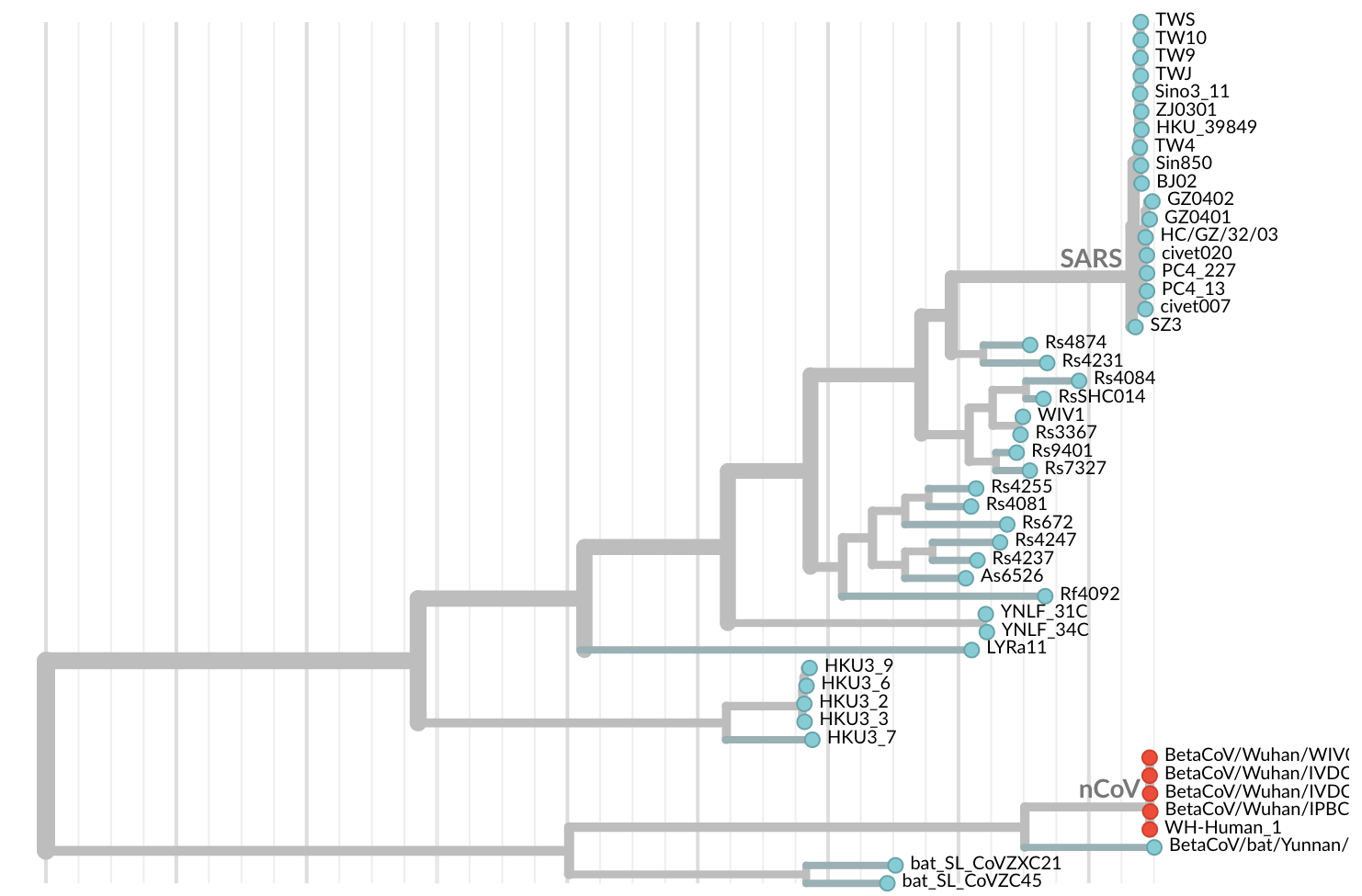
Jan 11: First five genomes showed a novel SARS-like coronavirus
Initially thought clustering due to epi investigation of linked cases at Huanan seafood market
Jan 19: First 12 genomes from Wuhan (blue) and Bangkok (red) showed lack of genetic diversity

Jan 23: Introduction into the human population between Nov 15 and Dec 15 and subsequent rapid human-to-human spread
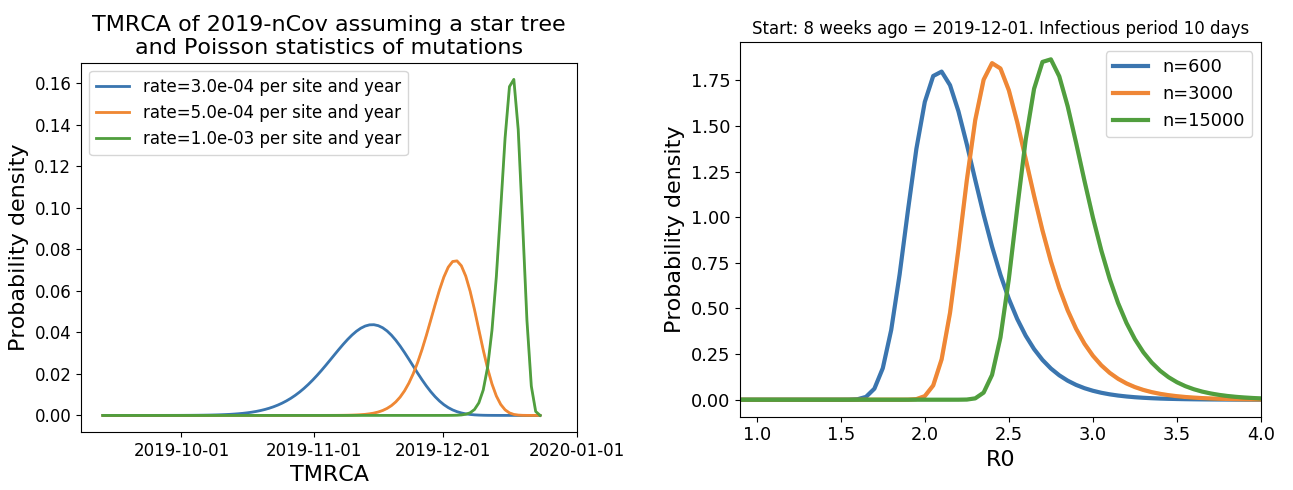
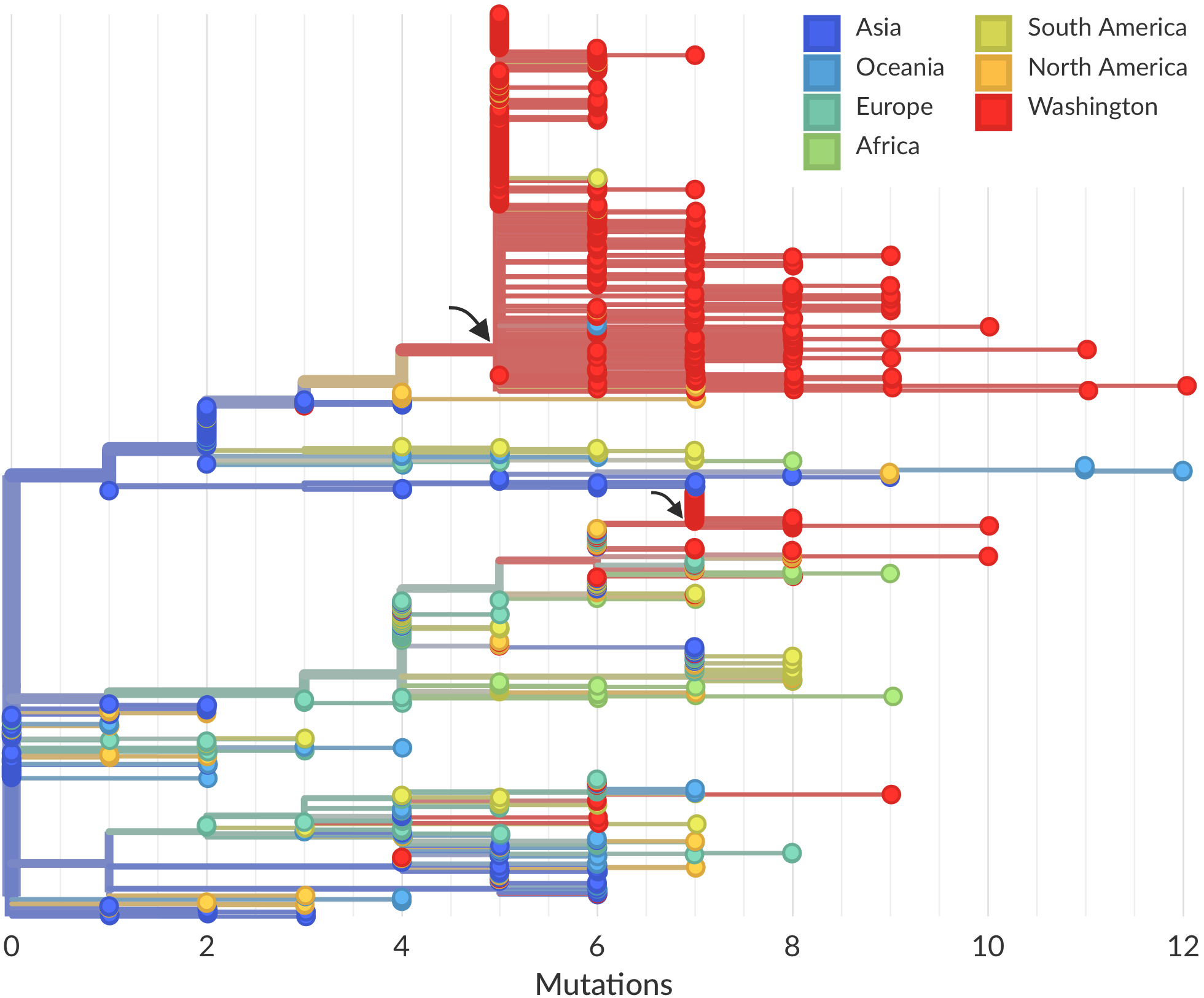
Rapid global epidemic spread from China
Epidemic in the USA was introduced from China in late Jan and from Europe during Feb
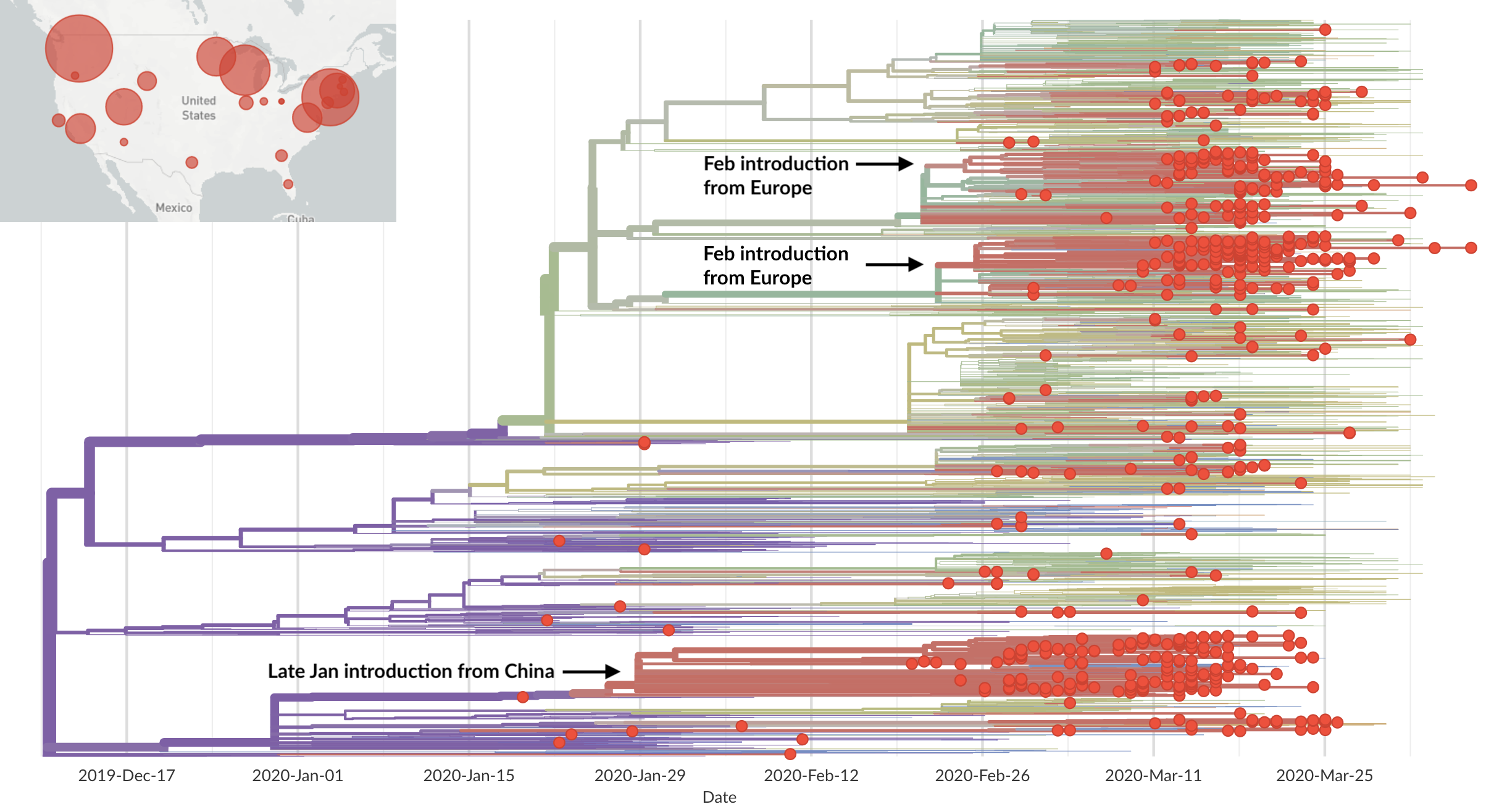
Early sequencing provided best estimate of extent of local outbreak
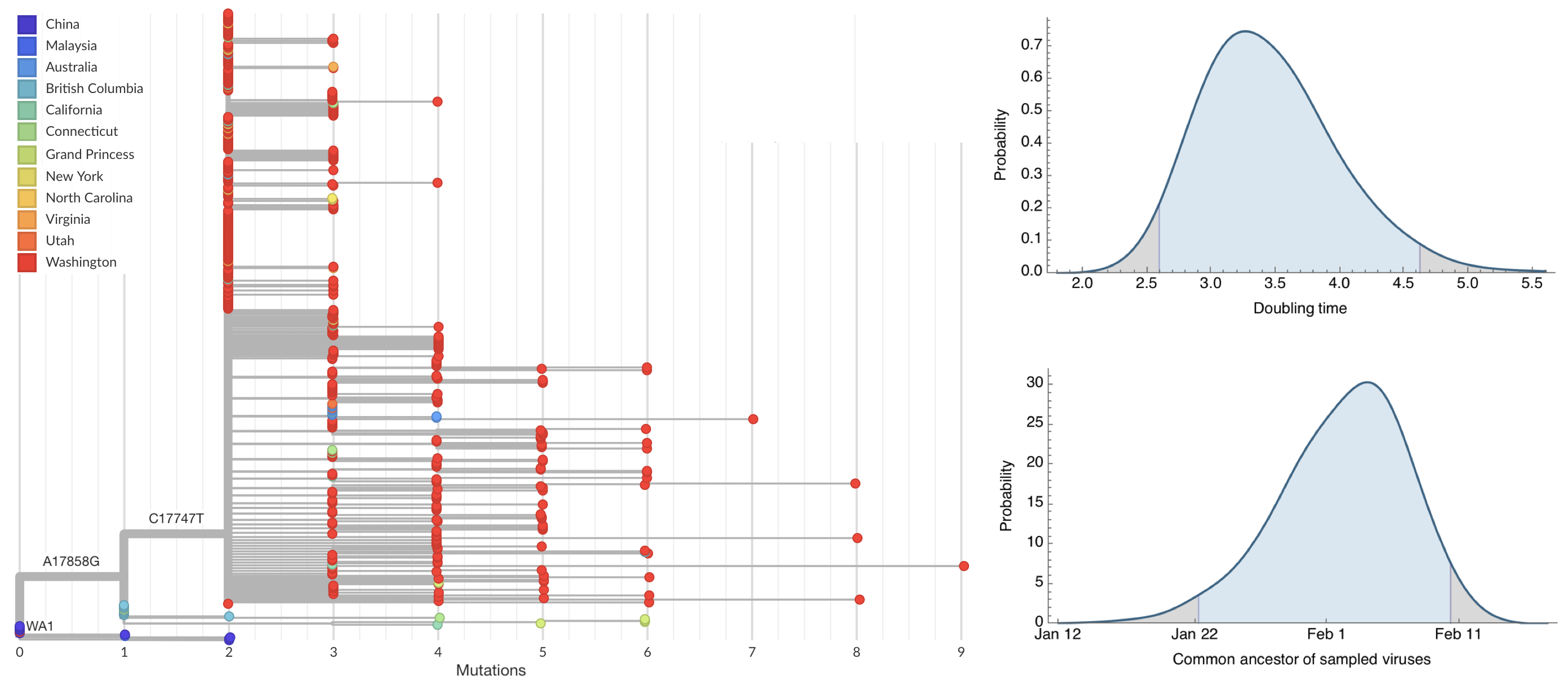
After initial wave, with mitigation
efforts and decreased travel,
regional clades emerge
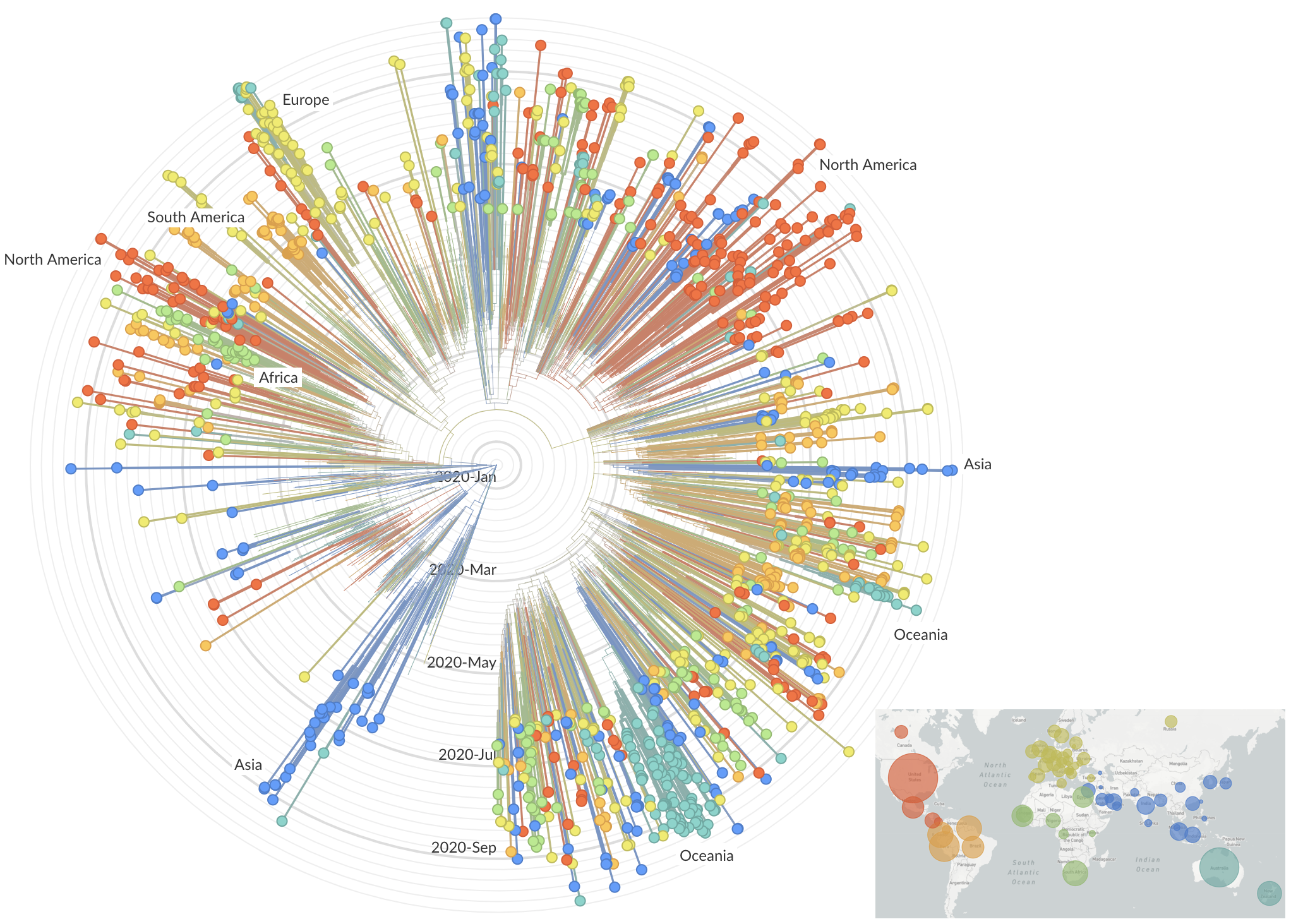
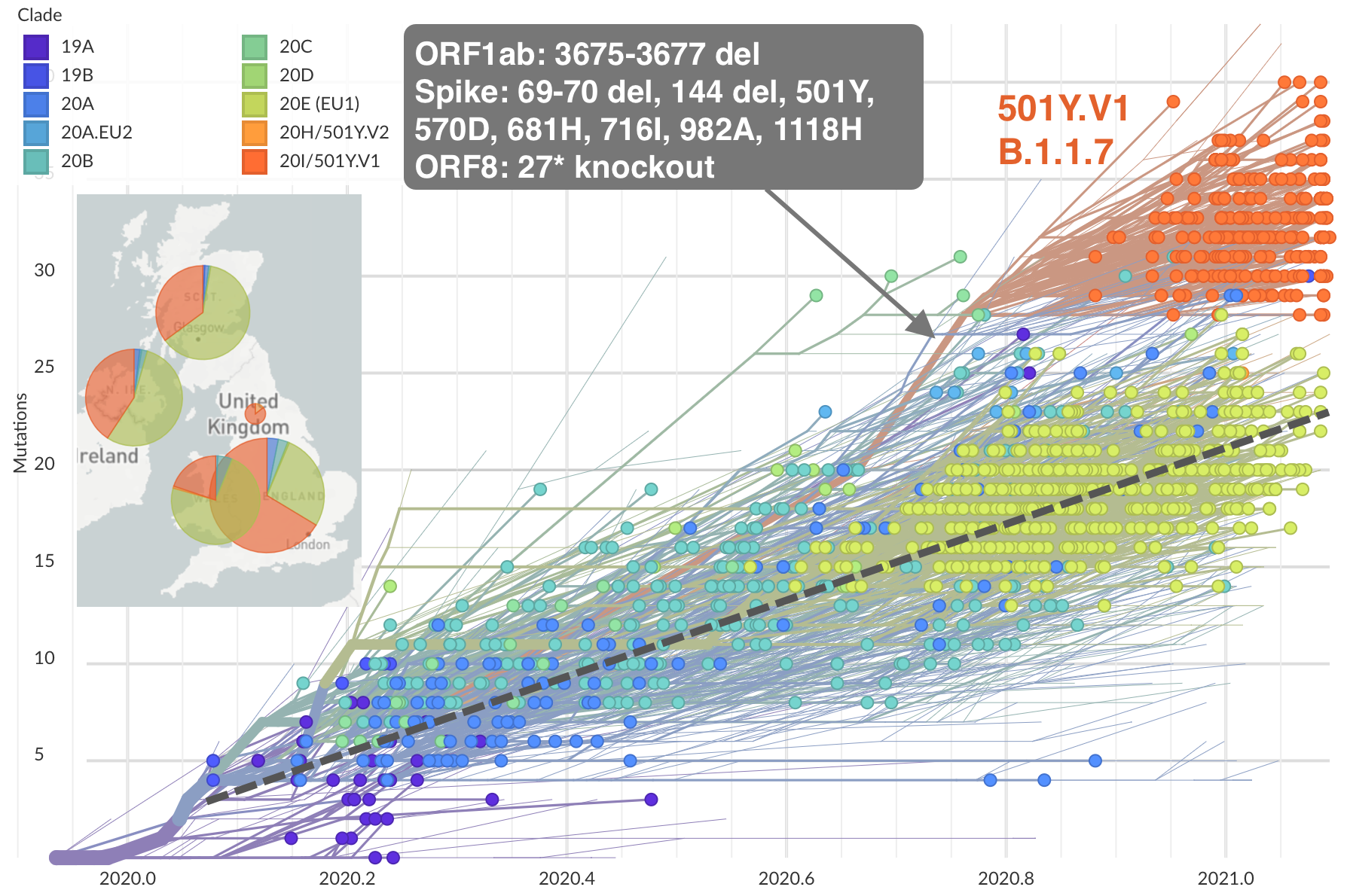
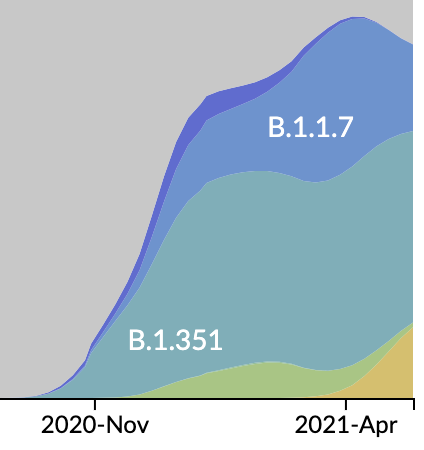
Emergence and spread of initial VOC viruses

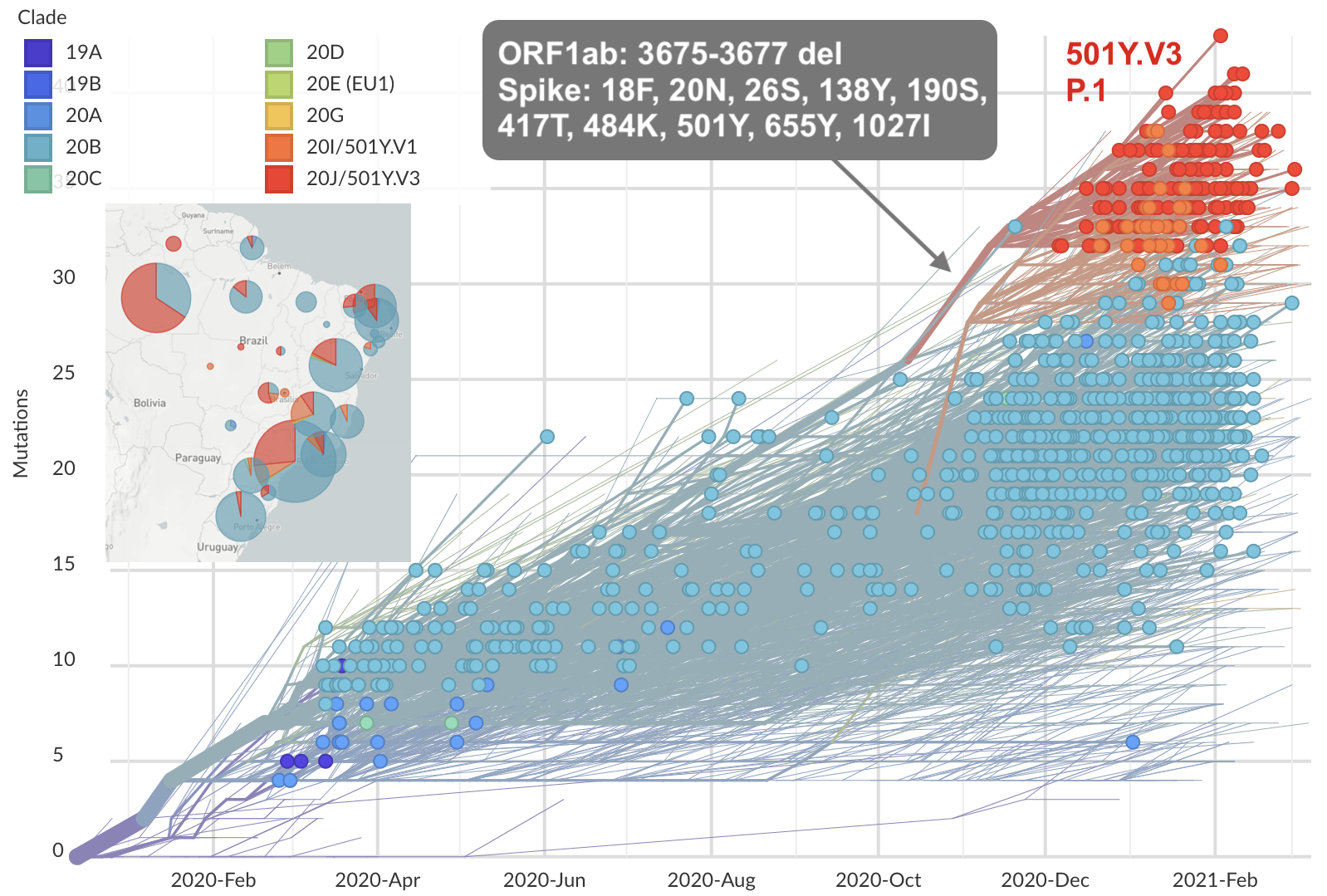
Similar story for other emerging pathogens
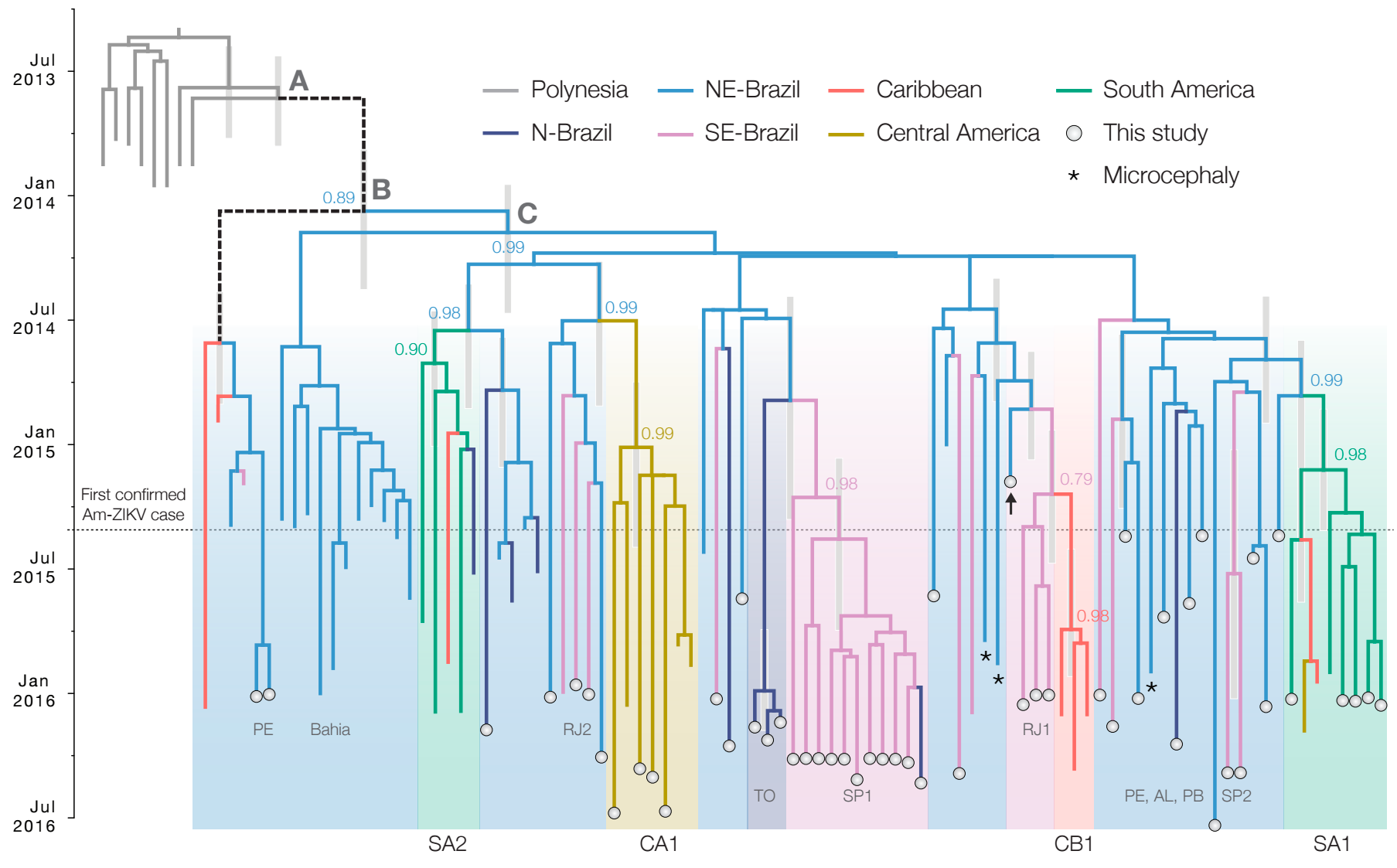
Zika epidemic in the Americas originated ~1.5 years before initial case detection
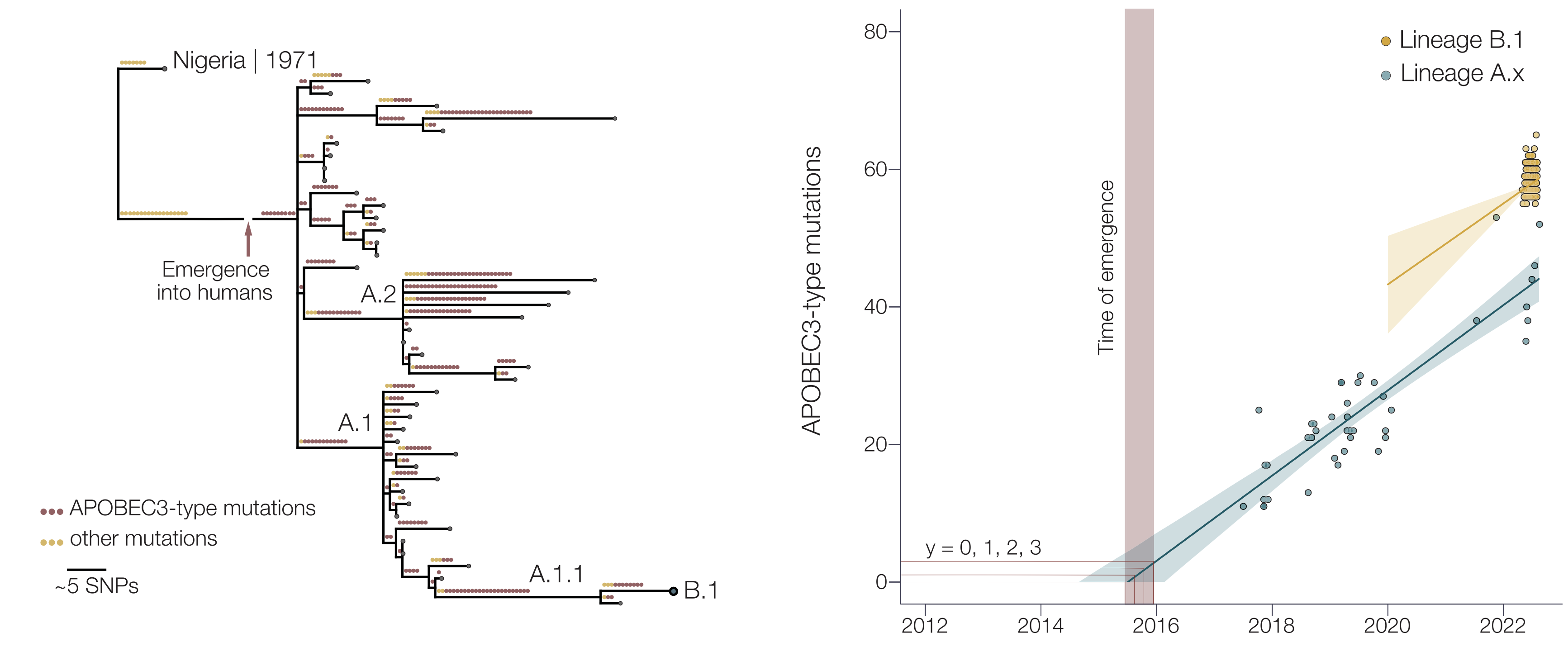
Mpox epidemic of 2022 stems from human-to-human transmission that began in Nigeria in 2016
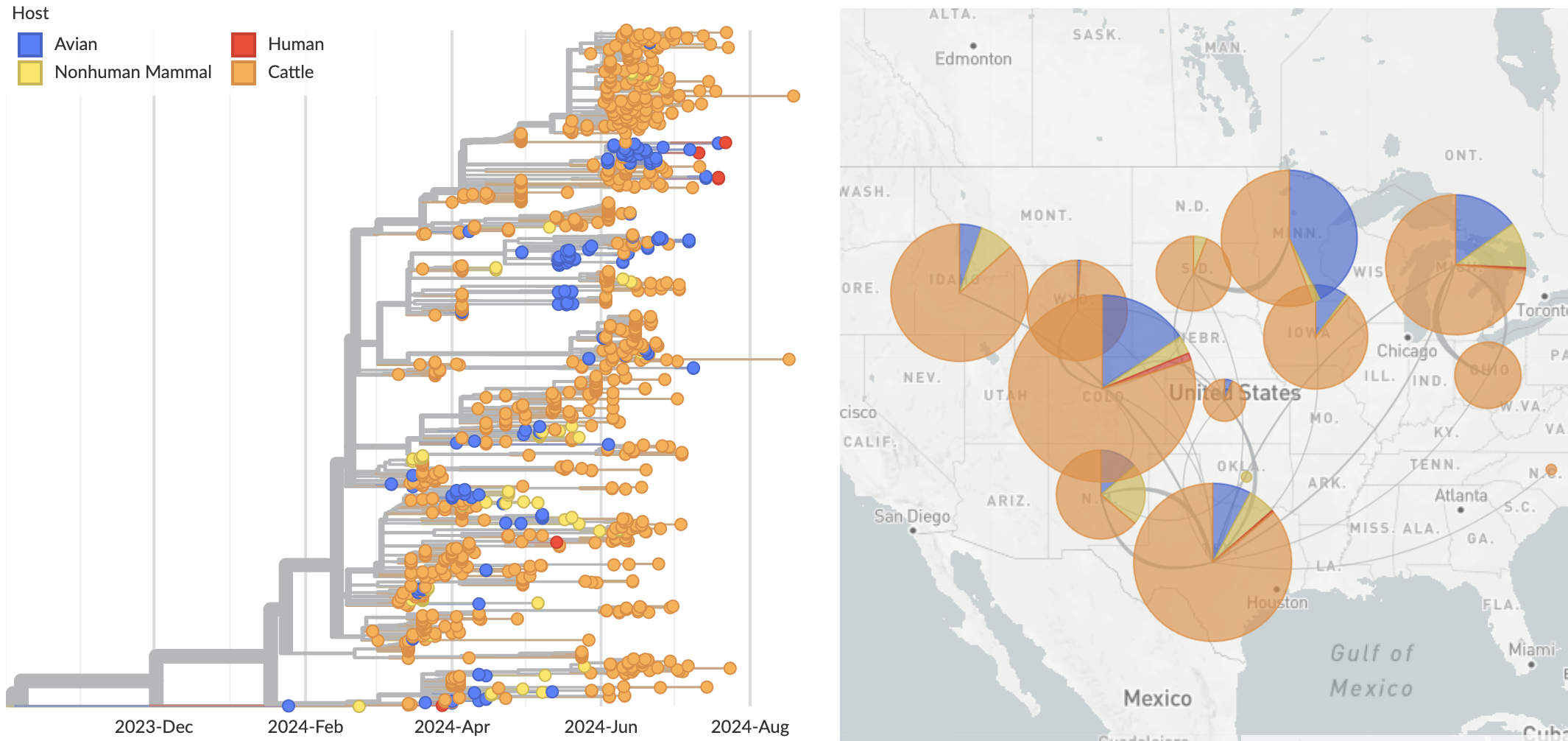
Ongoing H5N1 epizootic in cattle was spreading for months prior to initial case detection in late March
After initial lag, we've seen ramp up of molecular diagnostics and sequencing
The COVID-19 pandemic pushed the field perhaps ~5 years into the future
| 2013-16 Ebola in West Africa | 29k confirmed cases | 1610 genomes |
| 2015-17 Zika in the Americas | 223k confirmed cases | 942 genomes |
| 2018-19 seasonal flu in US | 290k confirmed cases | 8864 genomes |
| 2020-22 COVID-19 pandemic | 732M confirmed cases | 14.5M genomes |

Scalable approaches to genomic epidemiology
- Determinants of transmission: Use high volume sequencing datasets to reconstruct transmission dynamics across (spatial) compartments
- Evolutionary forecasting: Use high volume sequencing datasets to track fine-scale lineage frequencies and infer selective dynamics
Determinants of transmission
Phylogeography infers migration matrix via phylogeny
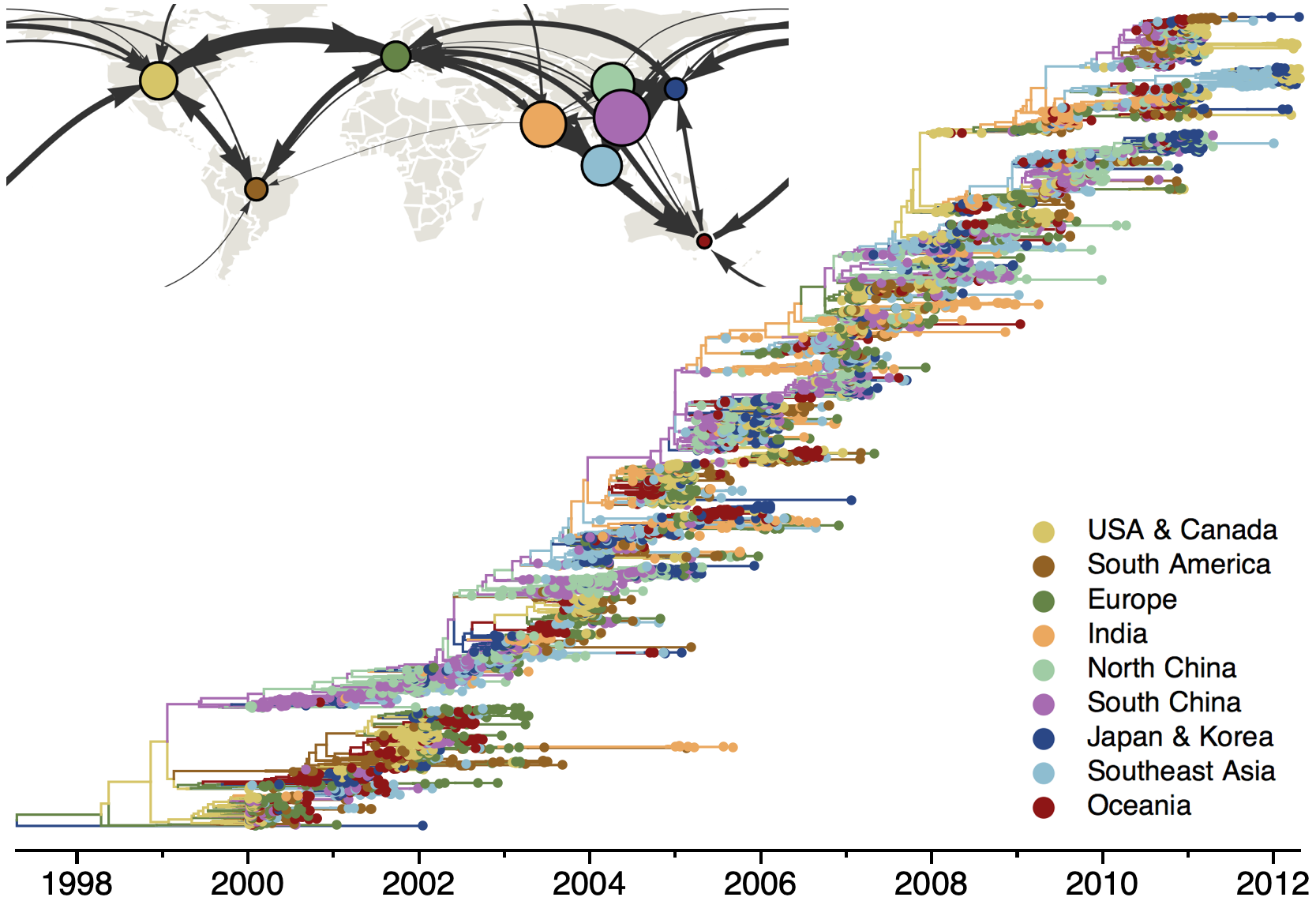
Phylogeography infers migration matrix via phylogeny
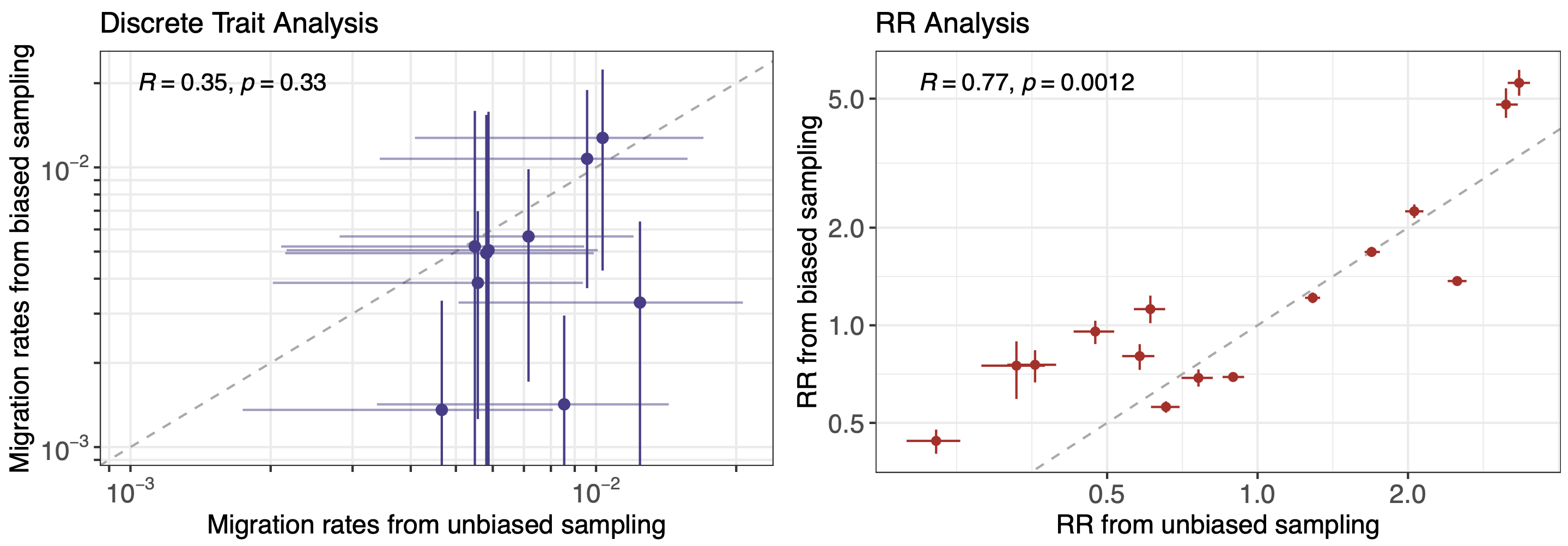
However, this approach faces significant issues with scalability and sampling bias


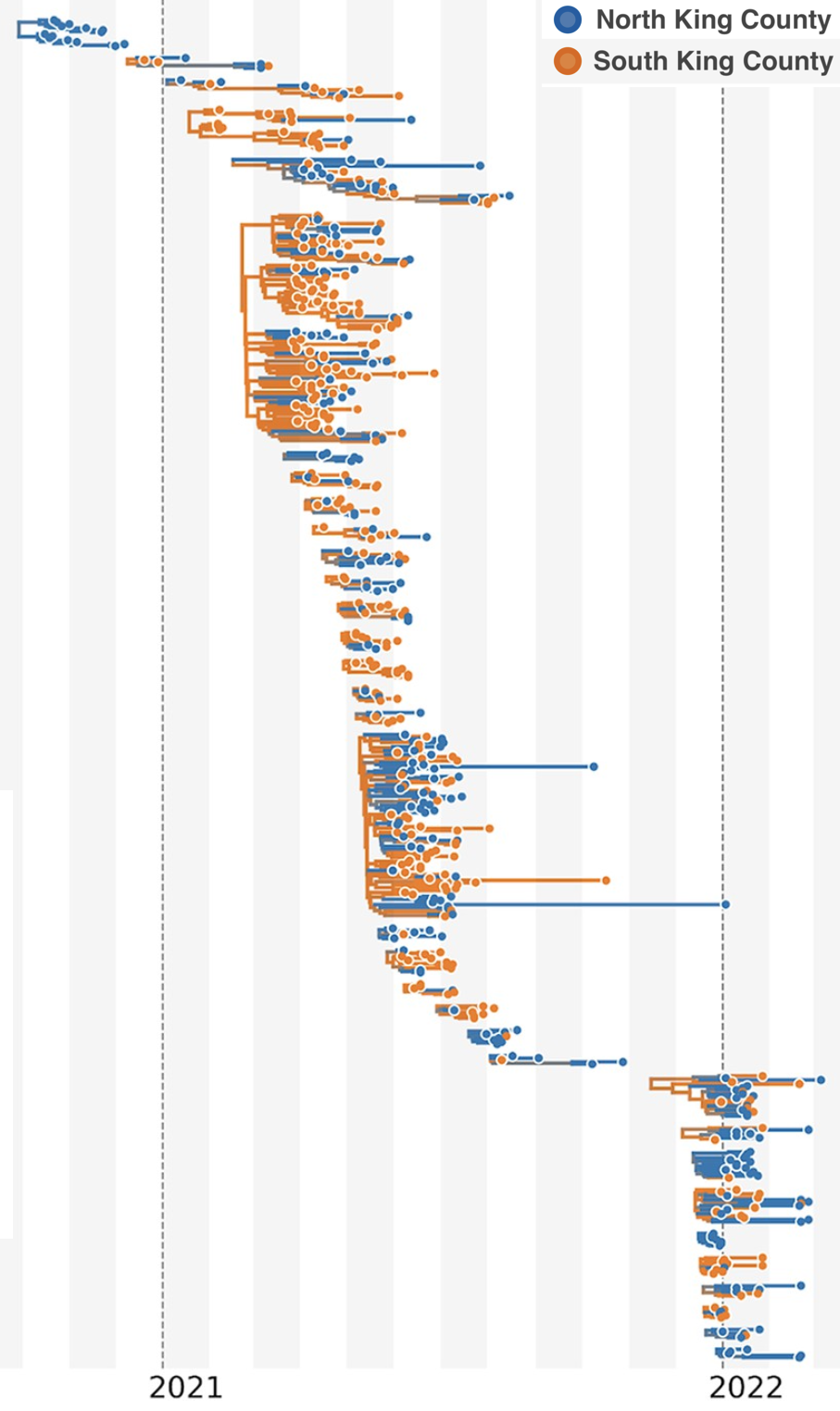
Model emergence, spread and dissolution of clusters of identical sequences
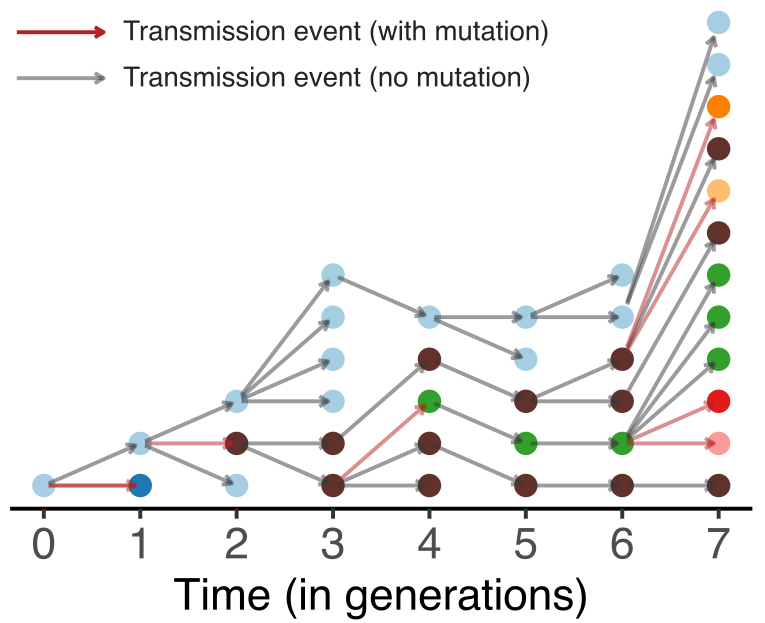
Identical sequences generally closely linked
One mutation every ~13 days vs duration of infection of ~5 days
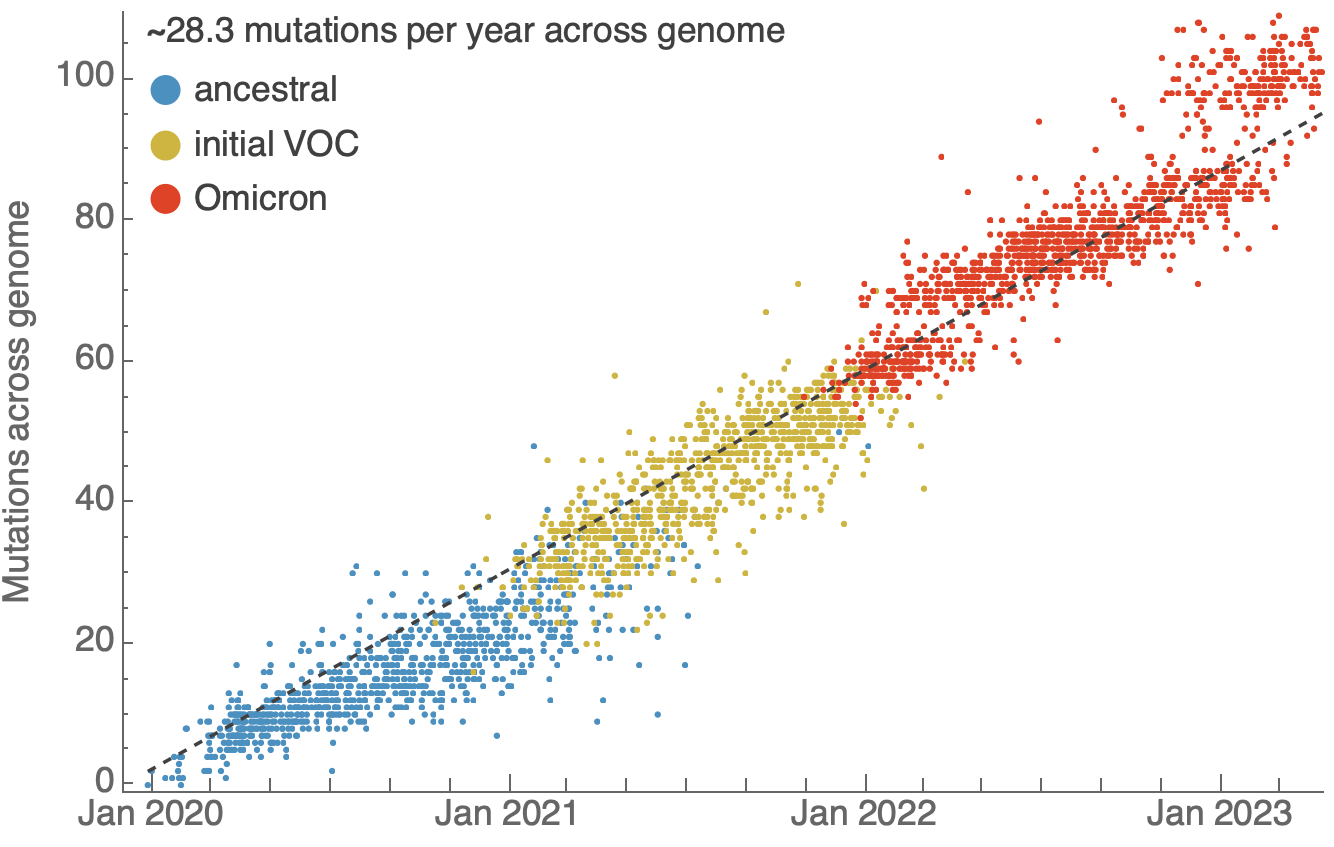
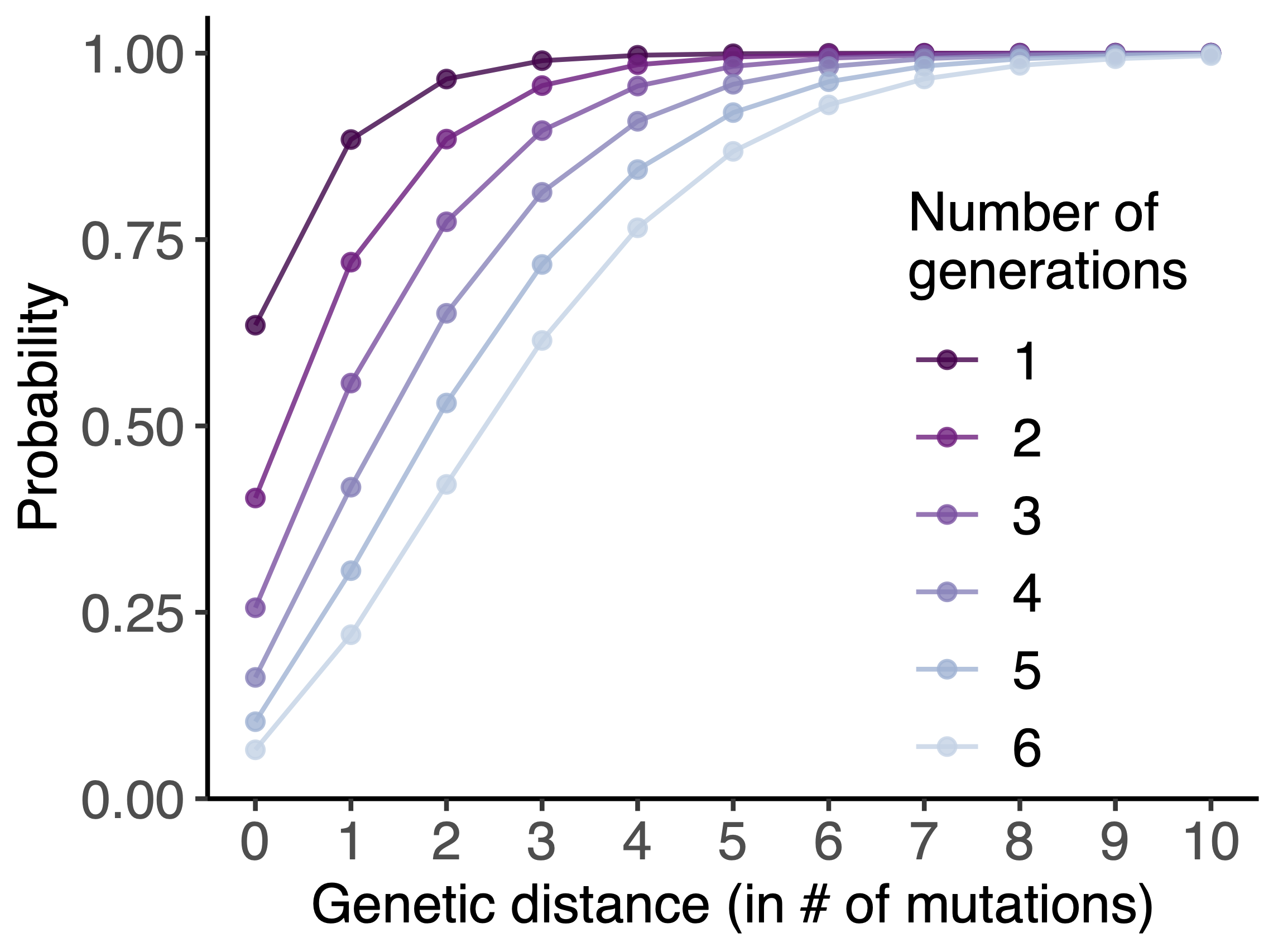
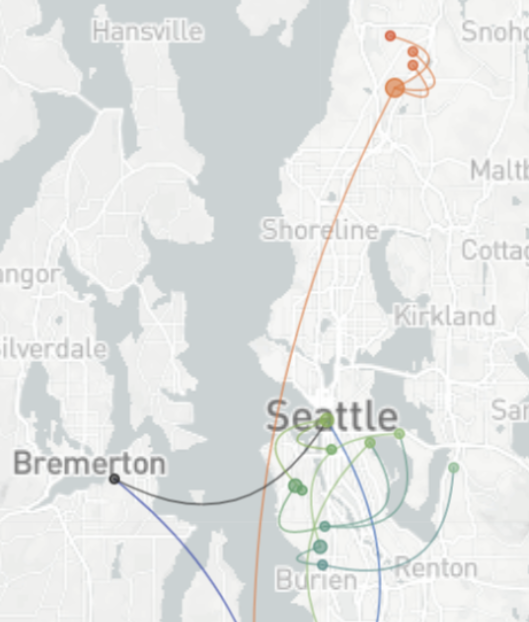
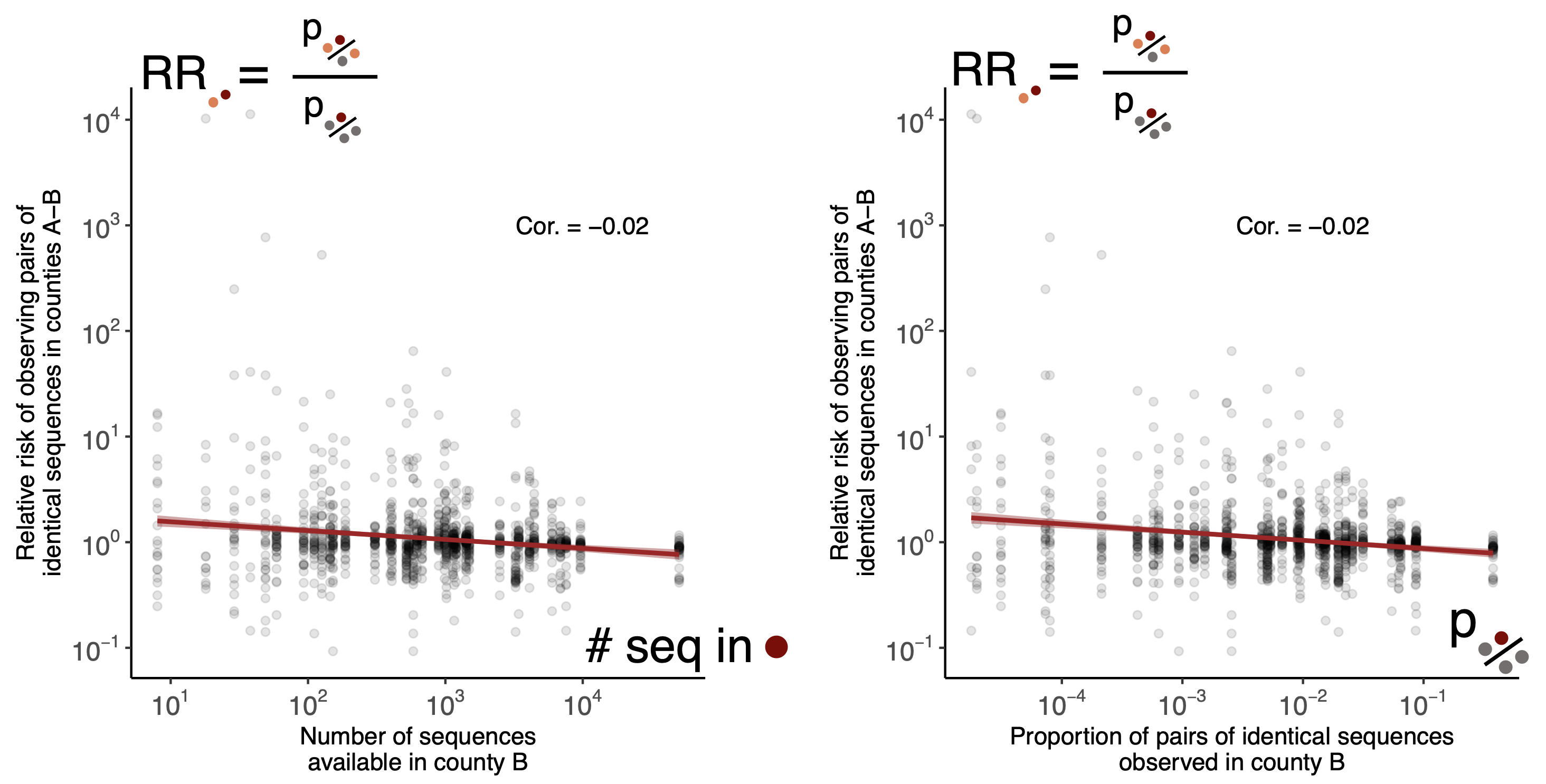
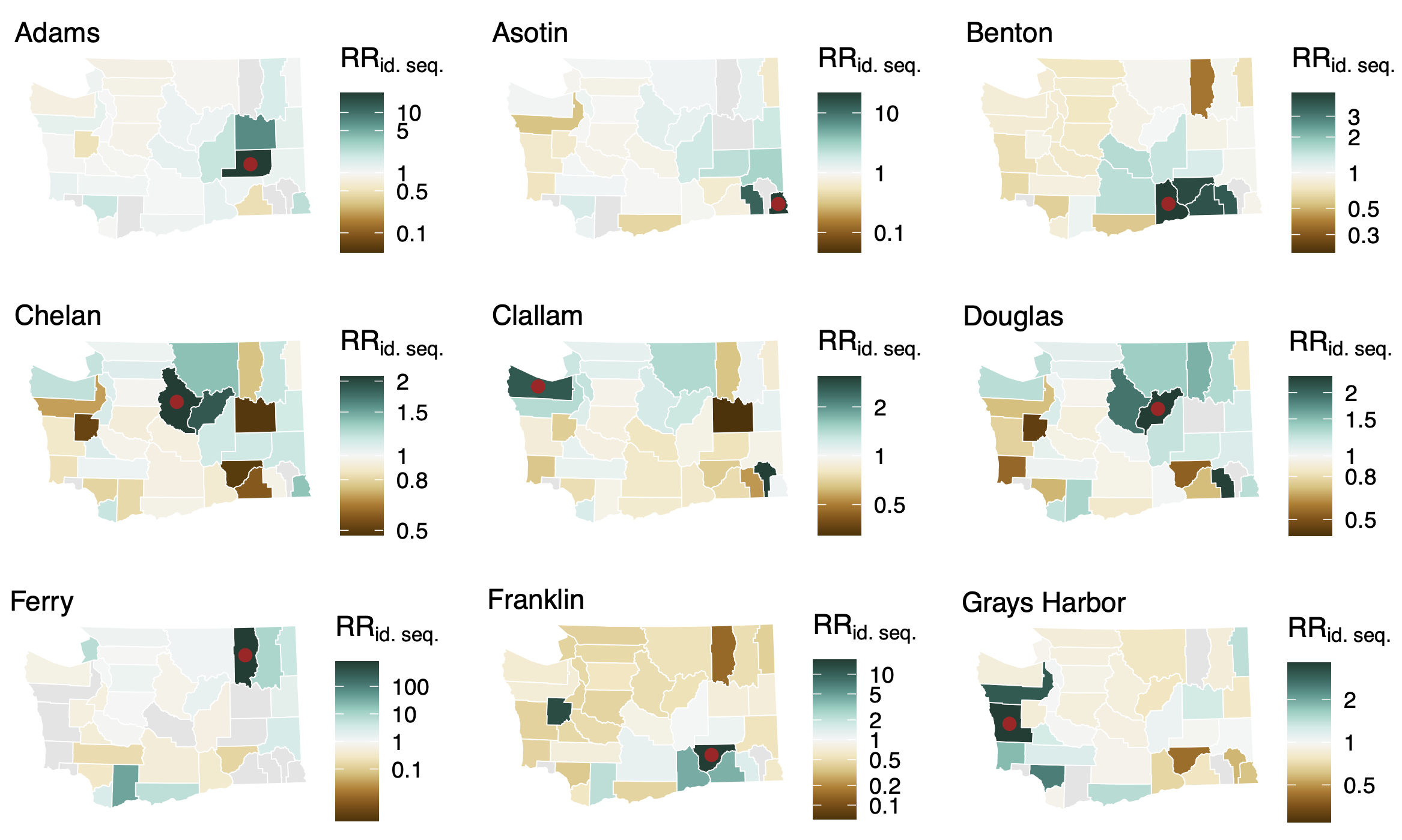
Spatial and social determinants of transmission from clusters of identical sequences
114k SARS-CoV-2 genomes from Washington State sentinel surveillance annotated with
geographic location and age
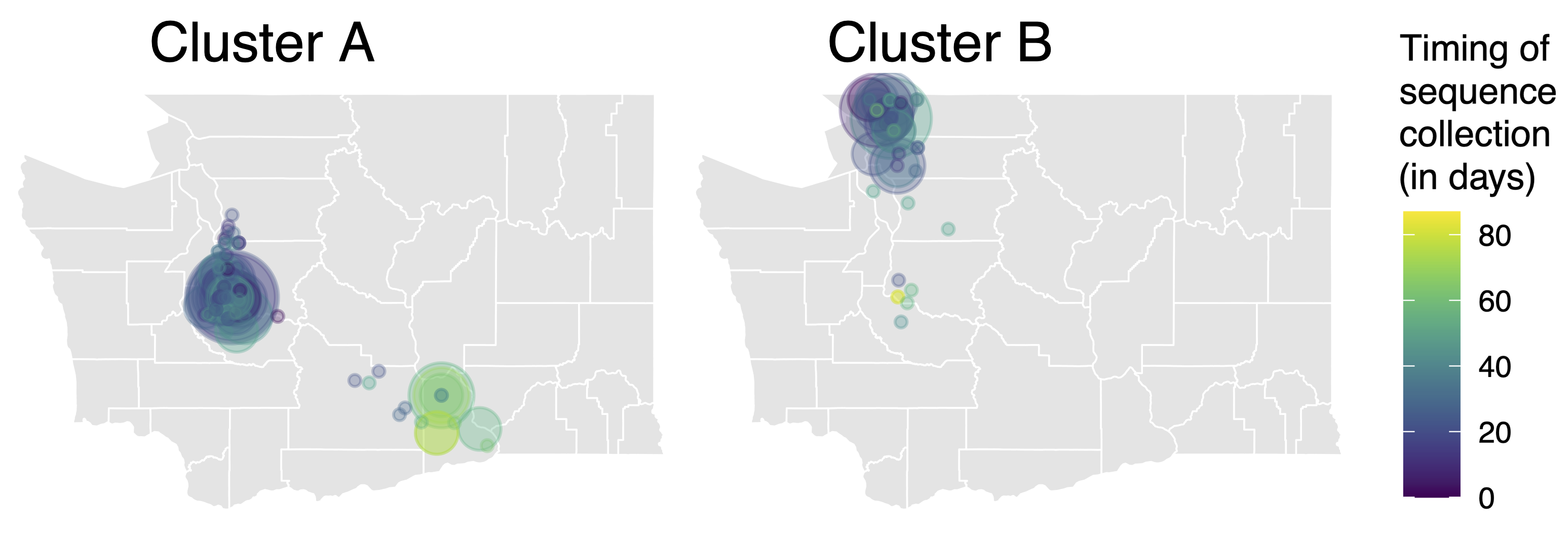
Pairs of identical sequences 5.1 times more likely to come from same county
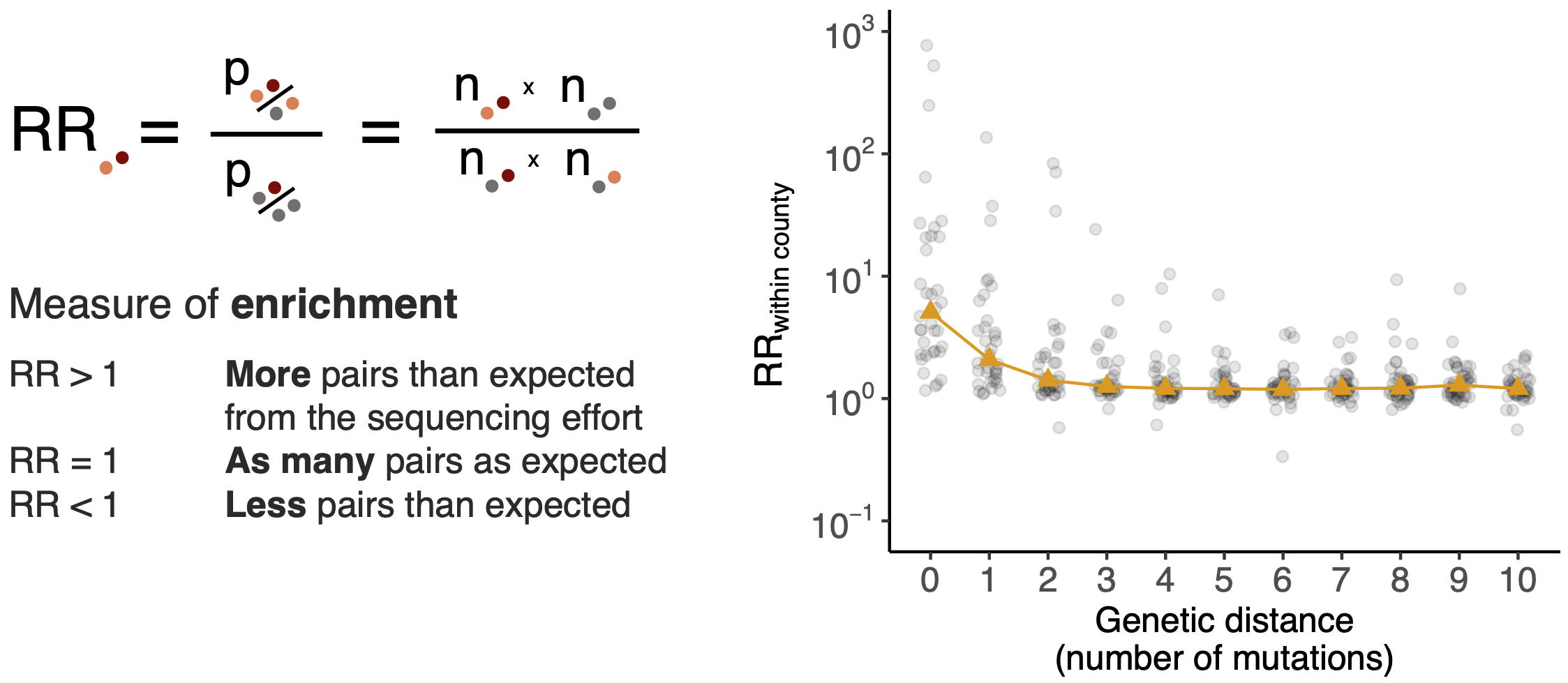
Relative risk score robust to sampling intensity
Between county enrichment corresponds well with geography
East-vs-west split due to geographic barrier in Washington State
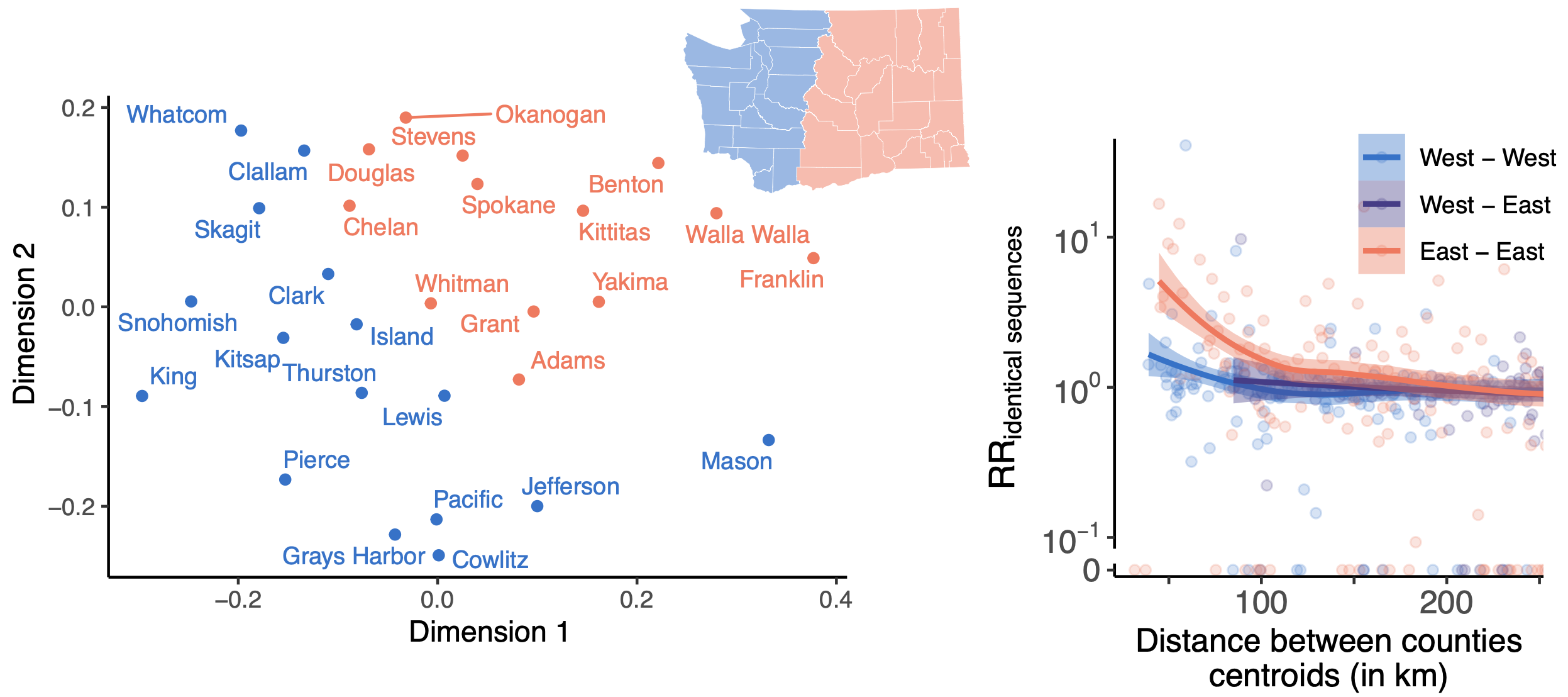
Sequence-derived mixing patterns correlate with cell-phone mobility data
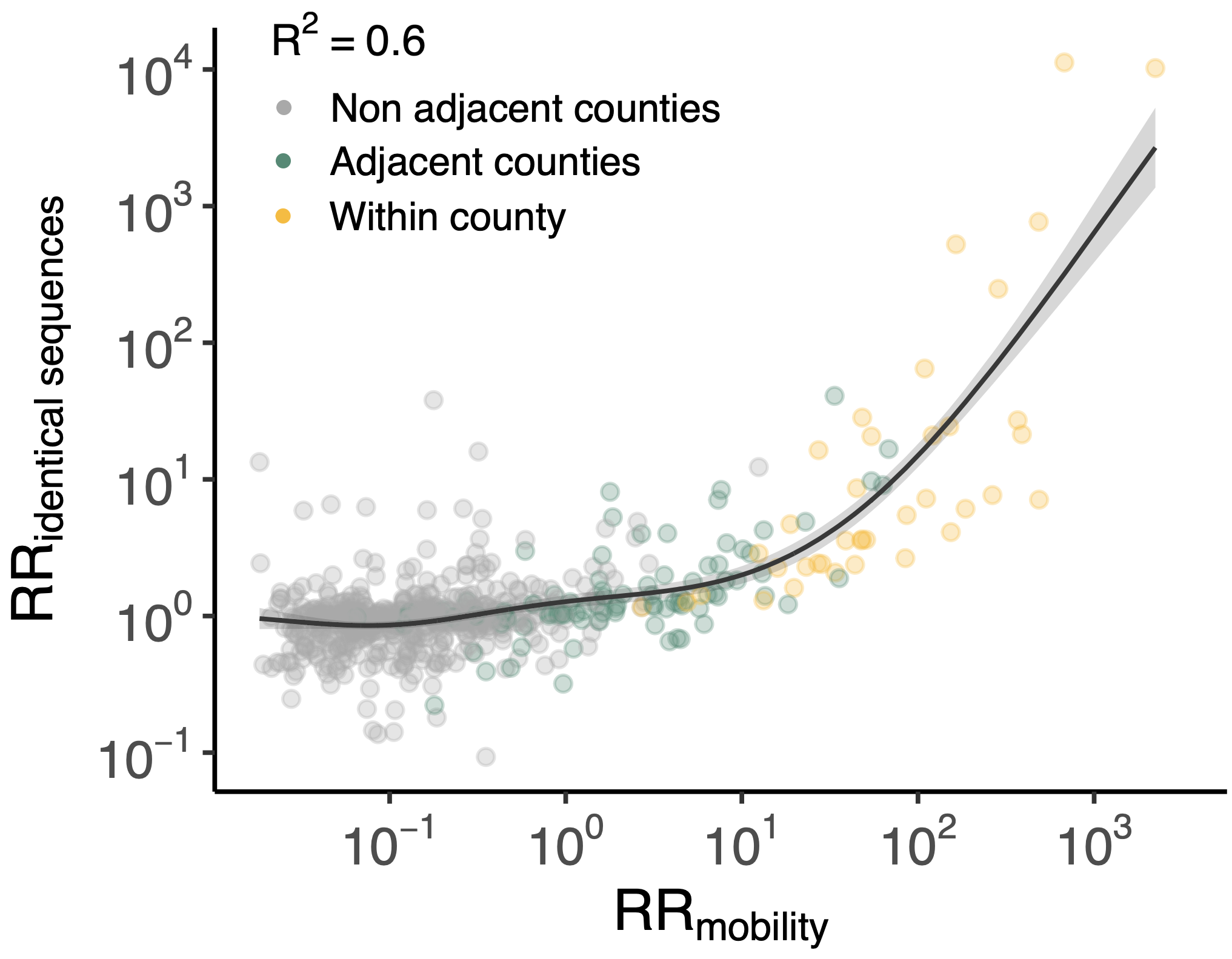
Between age group enrichment shows social mixing patterns
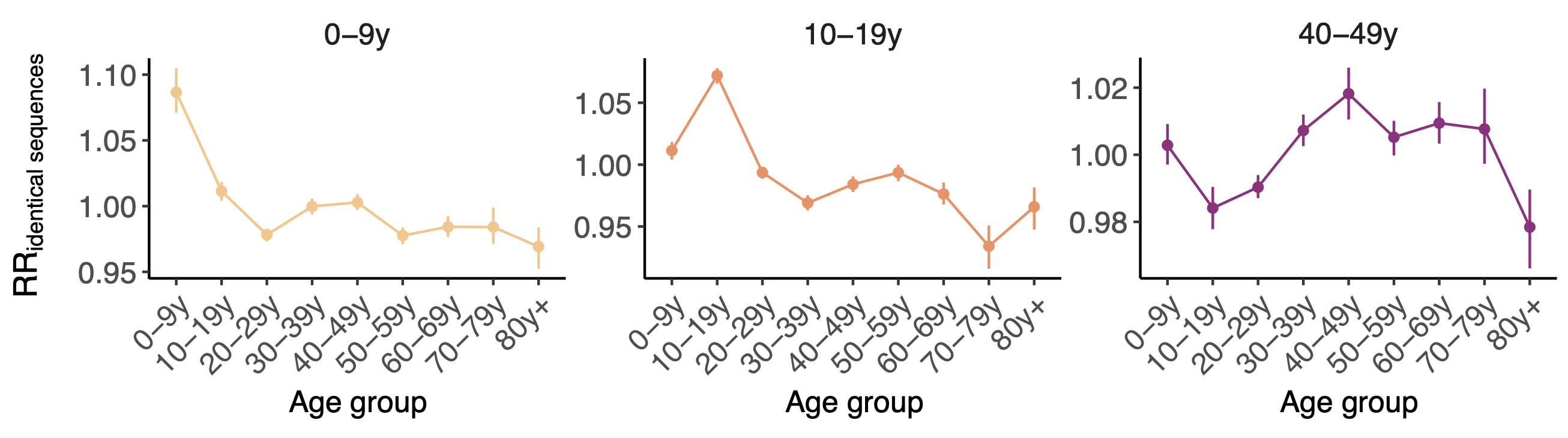
Sequence-derived mixing patterns correlate well with expectations from journaling studies
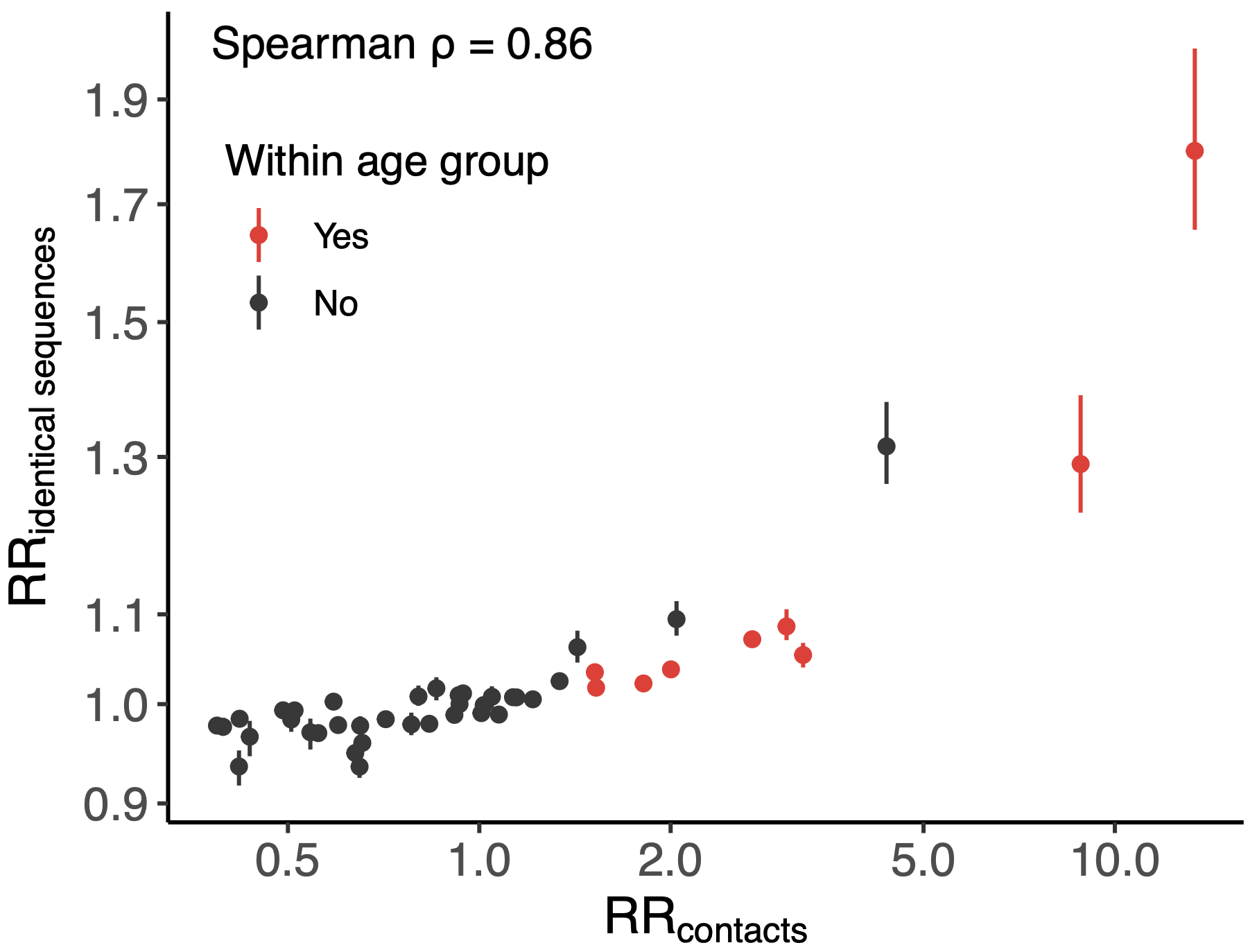
Signal for differential local vs long-distance age mixing
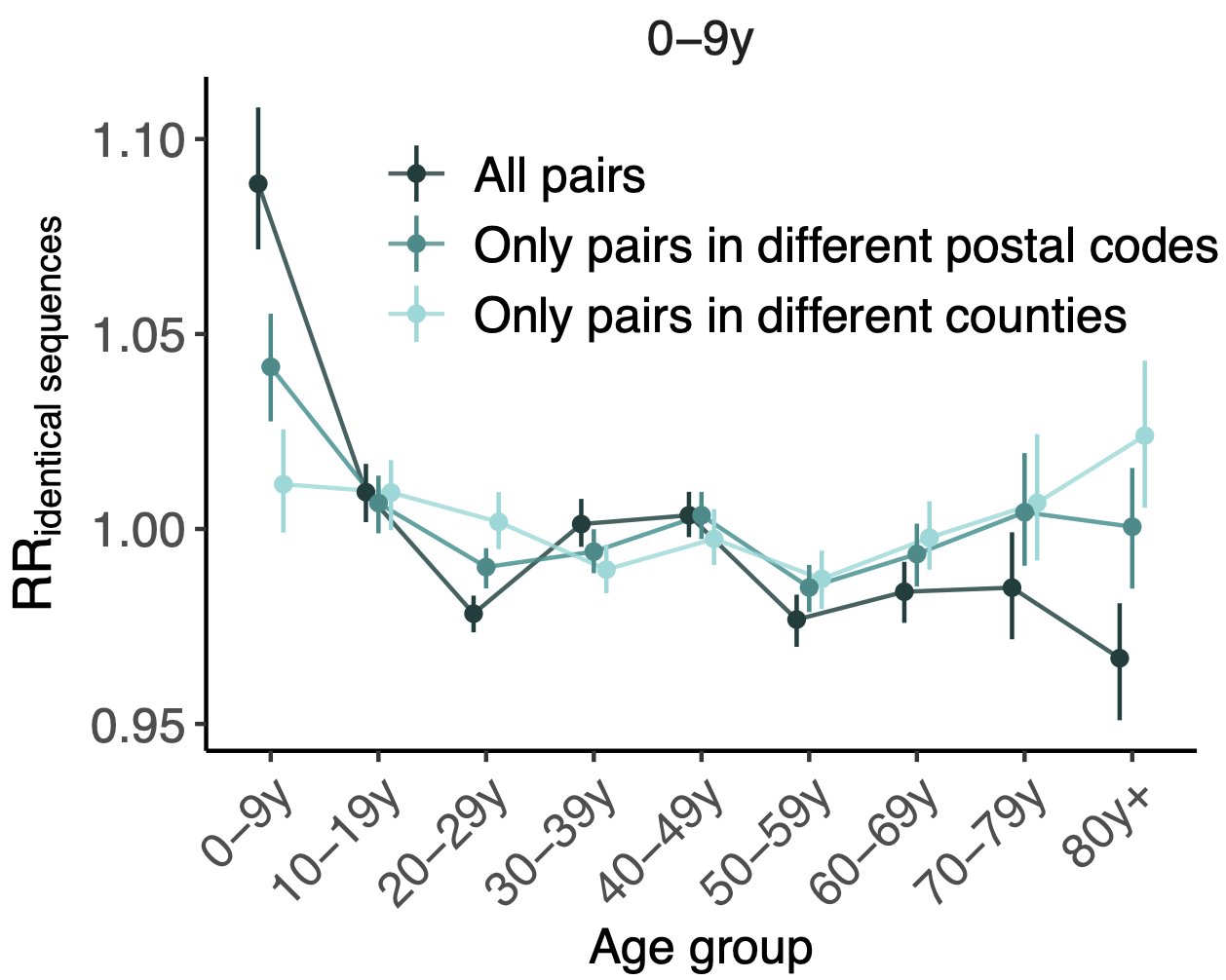
More methods development to be done, but accuracy compared to traditional discrete traits phylogeography is promising
Future work
- Incorporate assymetry in transmission direction between groups
- Estimate group-level contributions to epidemic growth
- Apply more broadly
Evolutionary forecasting
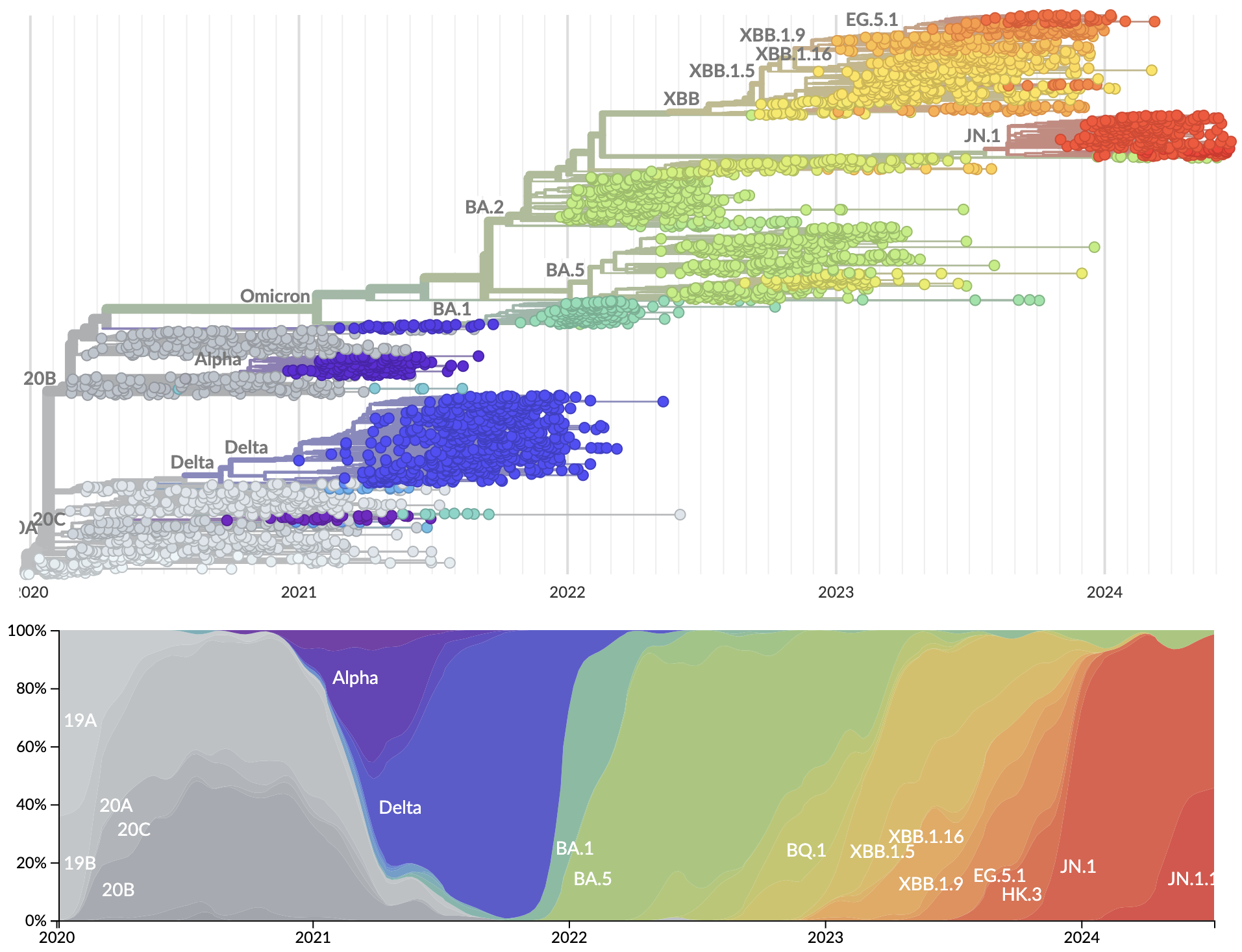
SARS-CoV-2 evolution since 2020
Recent advances
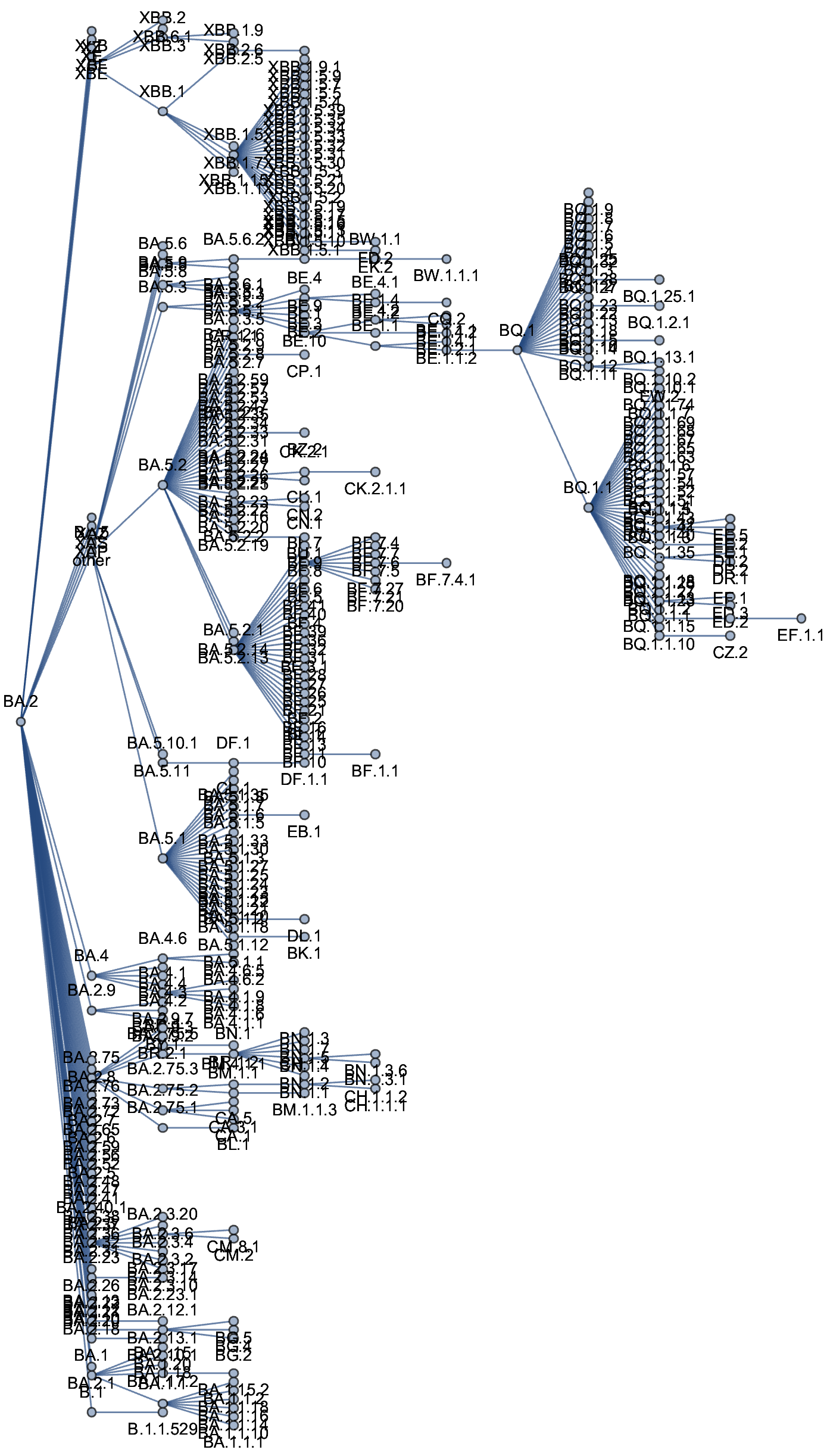
- Granular nomenclature and rapid classification
- Detailed frequency data allowing direct models of selection
Currently 3328 Pango lineages with rapid assignment of samples to lineages by Nextclade or UShER
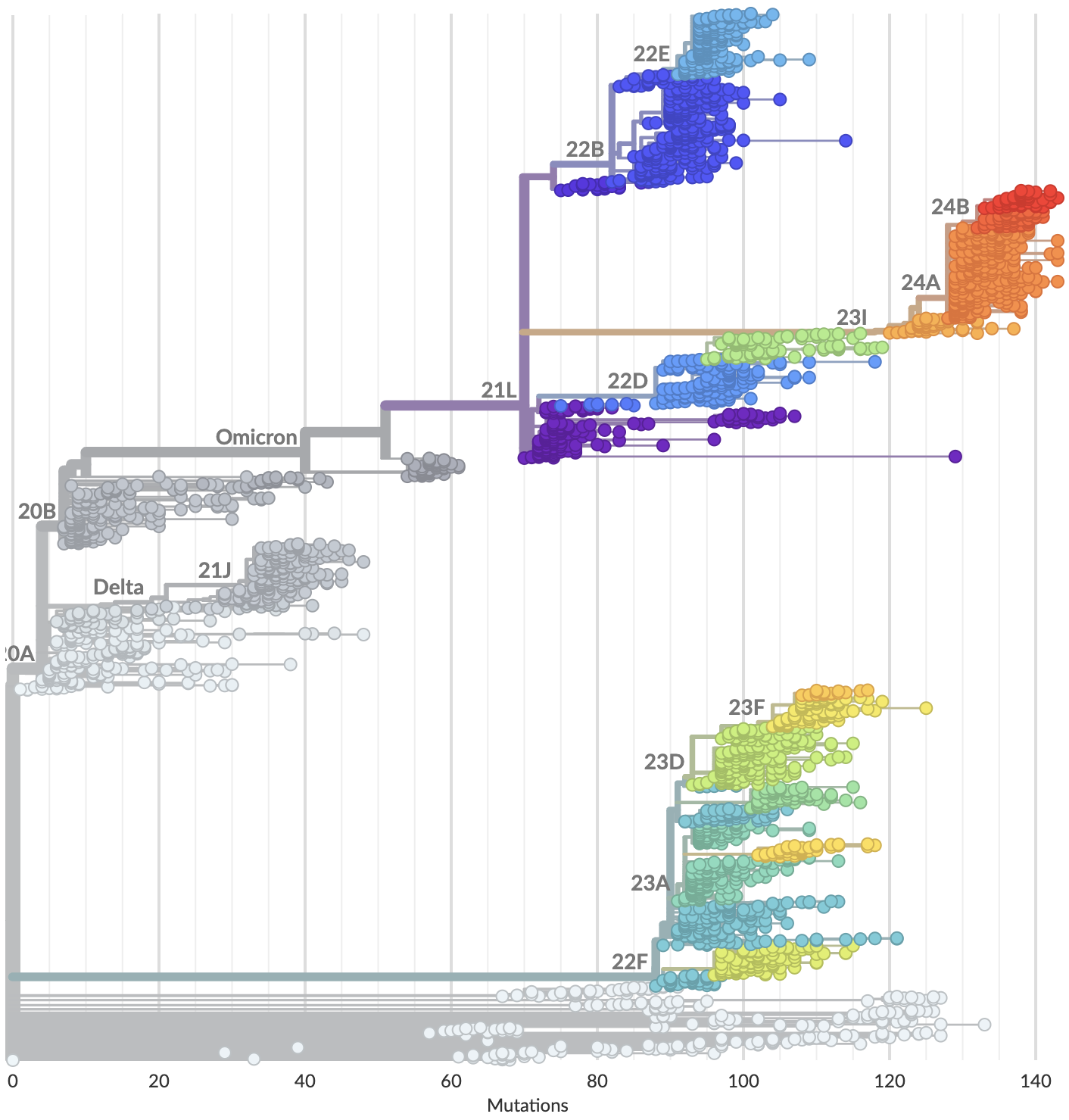
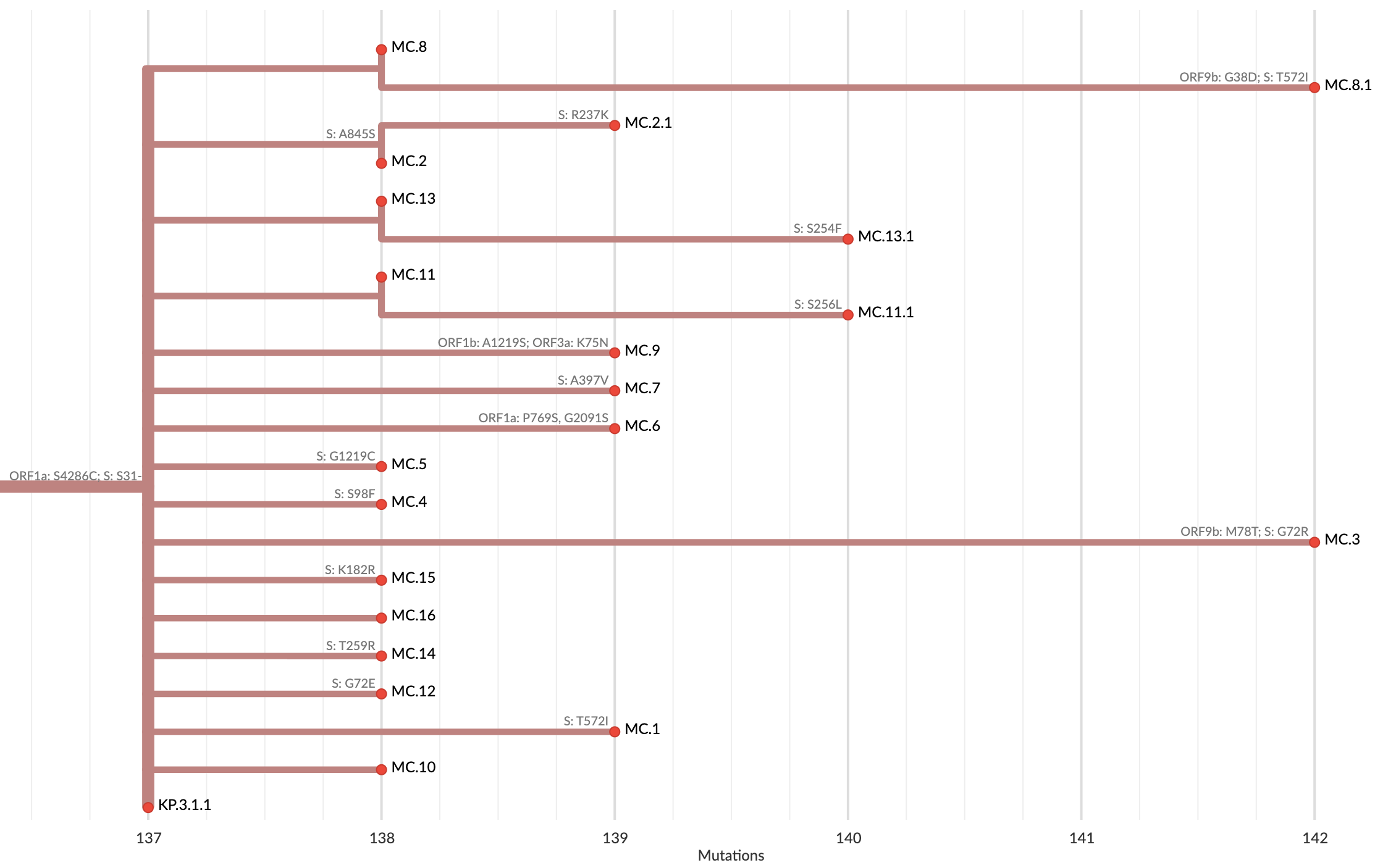
Variants that are just 1-2 mutations different will get a label
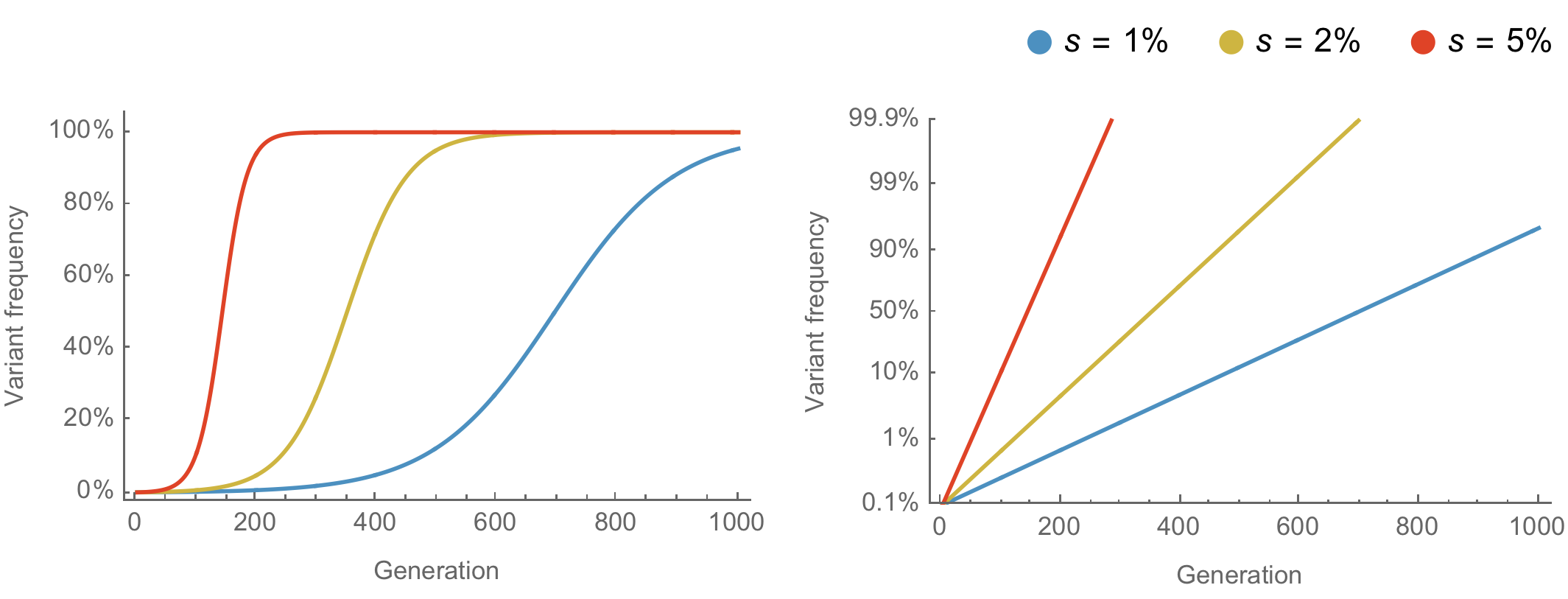
Population genetic expectation of variant frequency under selection
$x' = \frac{x \, (1+s)}{x \, (1+s) + (1-x)}$ for frequency $x$ over one generation with selective advantage $s$
$x(t) = \frac{x_0 \, (1+s)^t}{x_0 \, (1+s)^t + (1-x_0)}$ for initial frequency $x_0$ over $t$ generations
Trajectories are linear once logit transformed via $\mathrm{log}(\frac{x}{1 - x})$
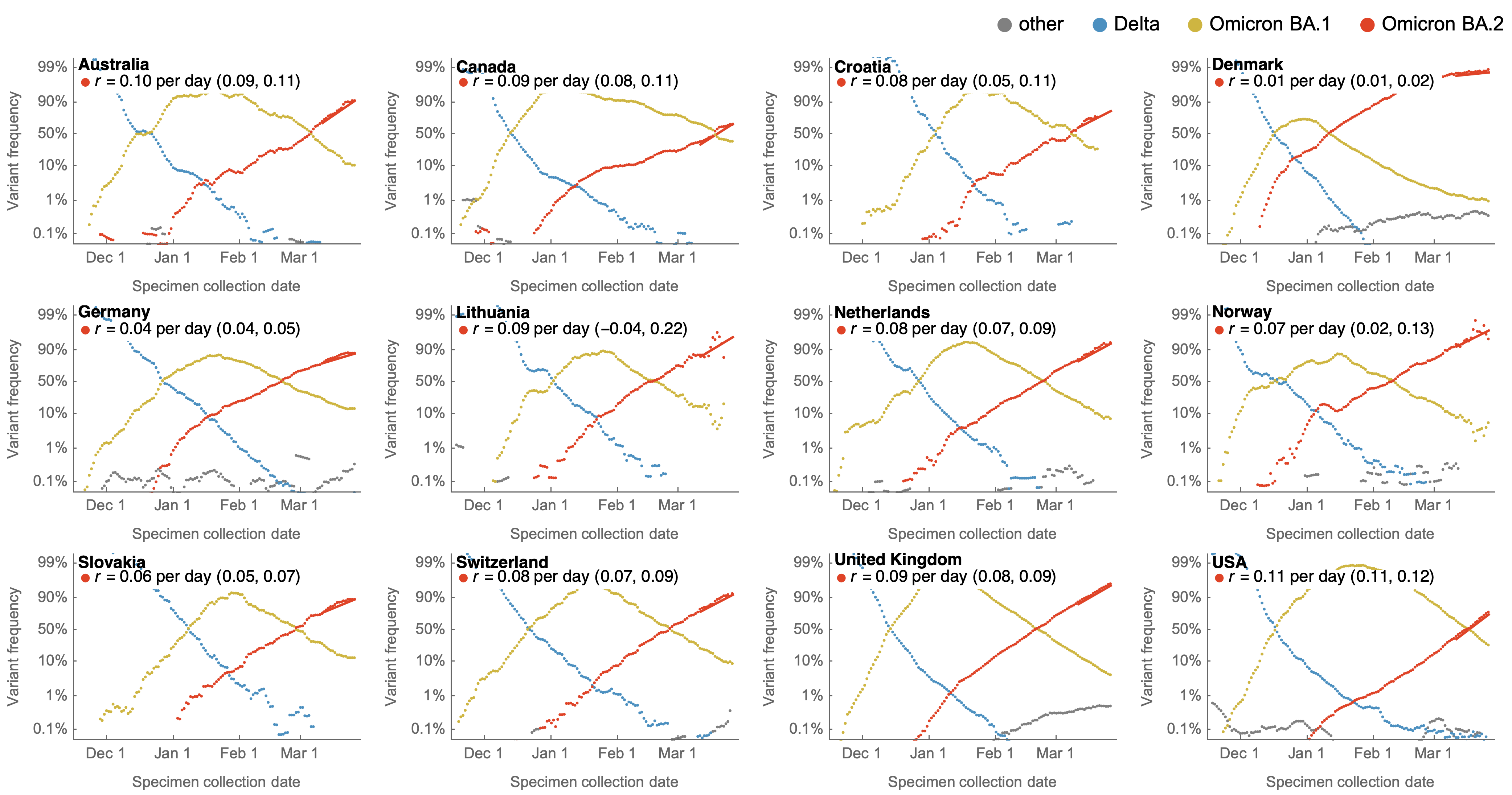
Consistent frequency dynamics in logit space (BA.2 Mar 2022)
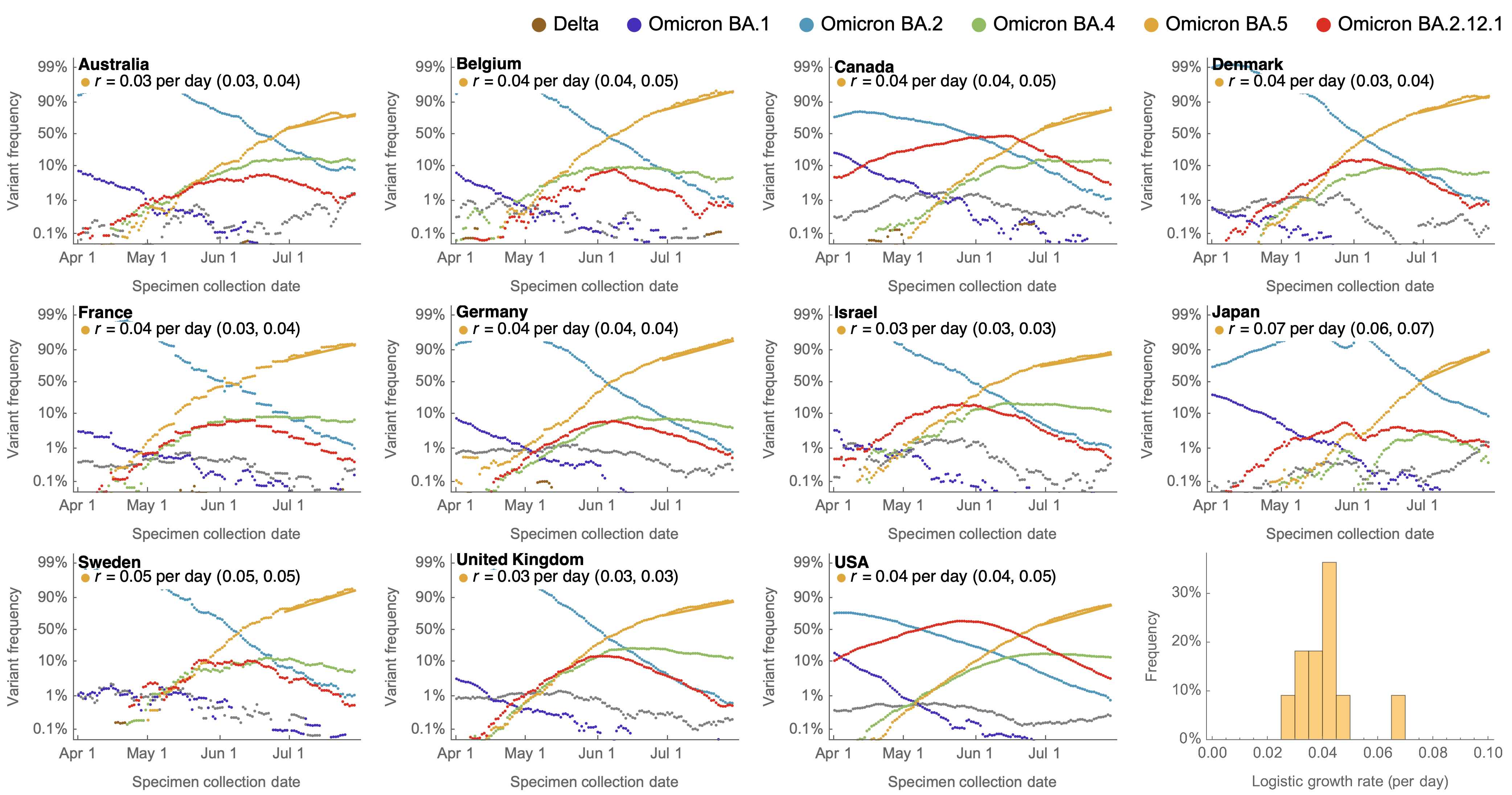
Consistent frequency dynamics in logit space (BA.5 Jul 2022)
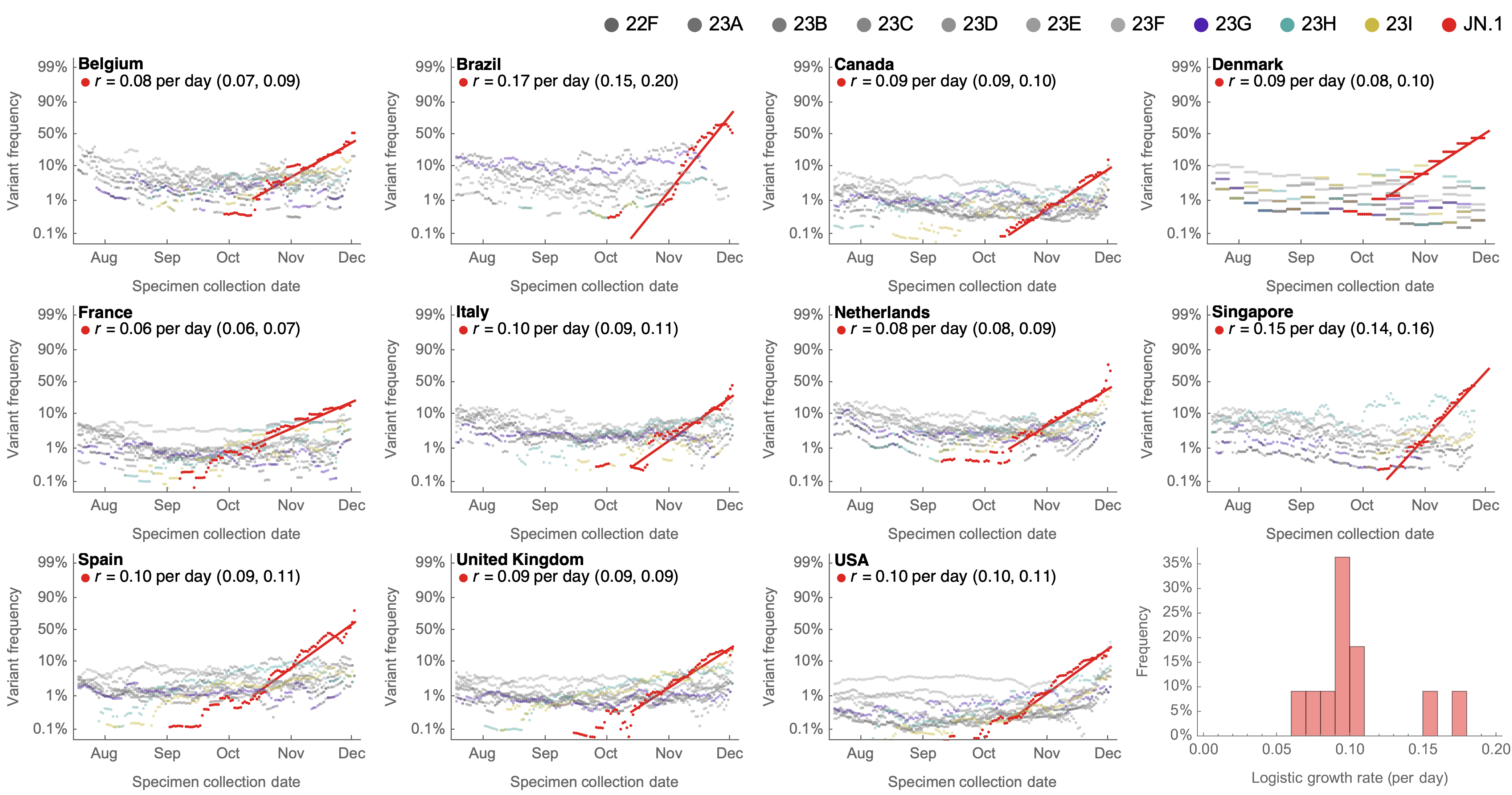
Consistent frequency dynamics in logit space (JN.1 Dec 2023)
Multinomial logistic regression
Multinomial logistic regression across $n$ variants models the probability of a virus sampled at time $t$ belonging to variant $i$ as
$$\mathrm{Pr}(X = i) = x_i(t) = \frac{p_i \, \mathrm{exp}(f_i \, t)}{\sum_{1 \le j \le n} p_j \, \mathrm{exp}(f_j \, t) }$$
with $2n$ parameters consisting of $p_i$ the frequency of variant $i$ at initial timepoint and $f_i$ the growth rate or fitness of variant $i$.
Original VOC viruses had substantially increased transmissibility
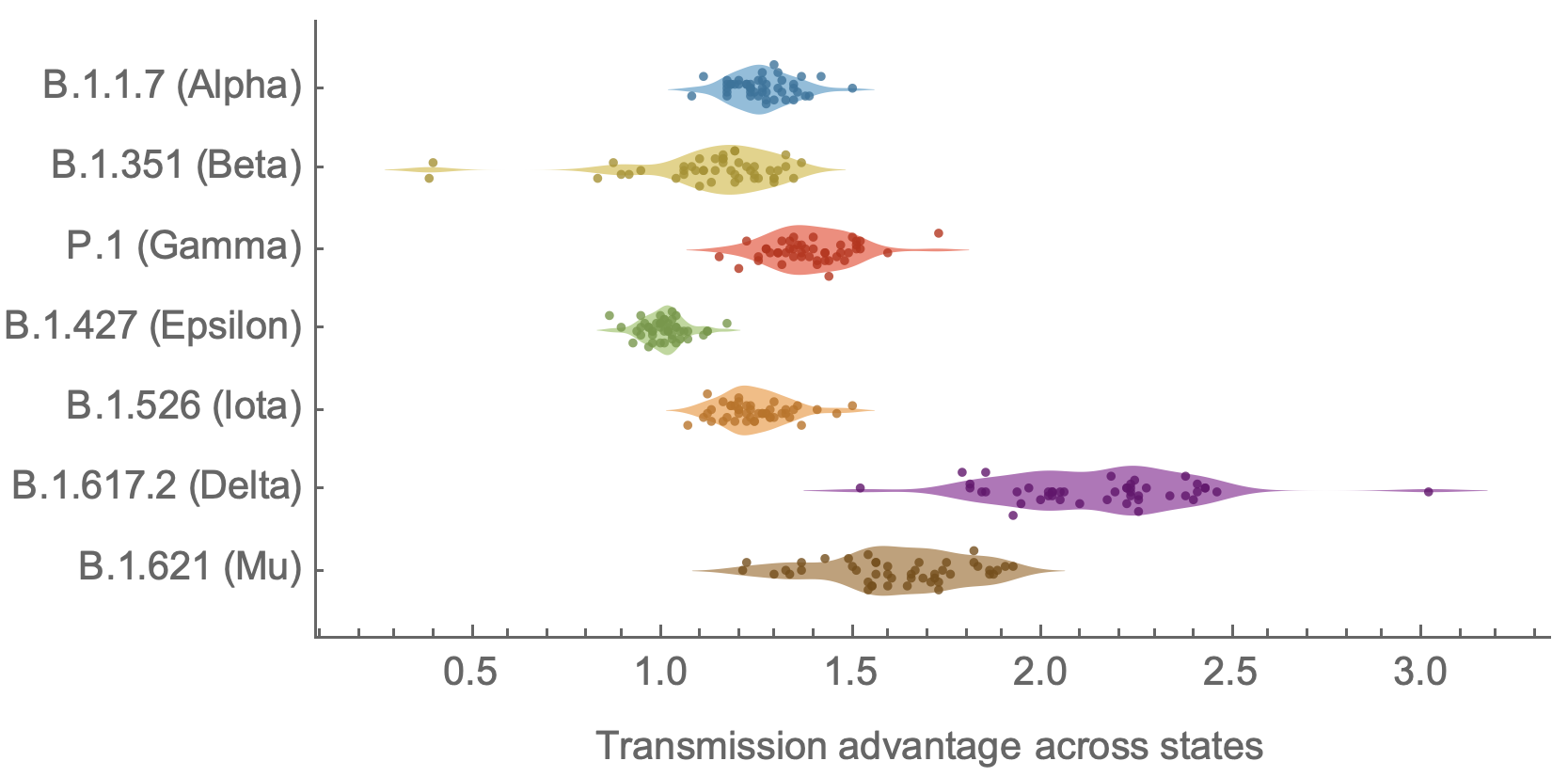
Clade success from 2022 to 2024 driven by antigenic evolution
We find that recent variants are ~250% fitter than original Omicron BA.1

Assessing MLR models for short-term frequency forecasting
Retrospective projections twice monthly during 2022
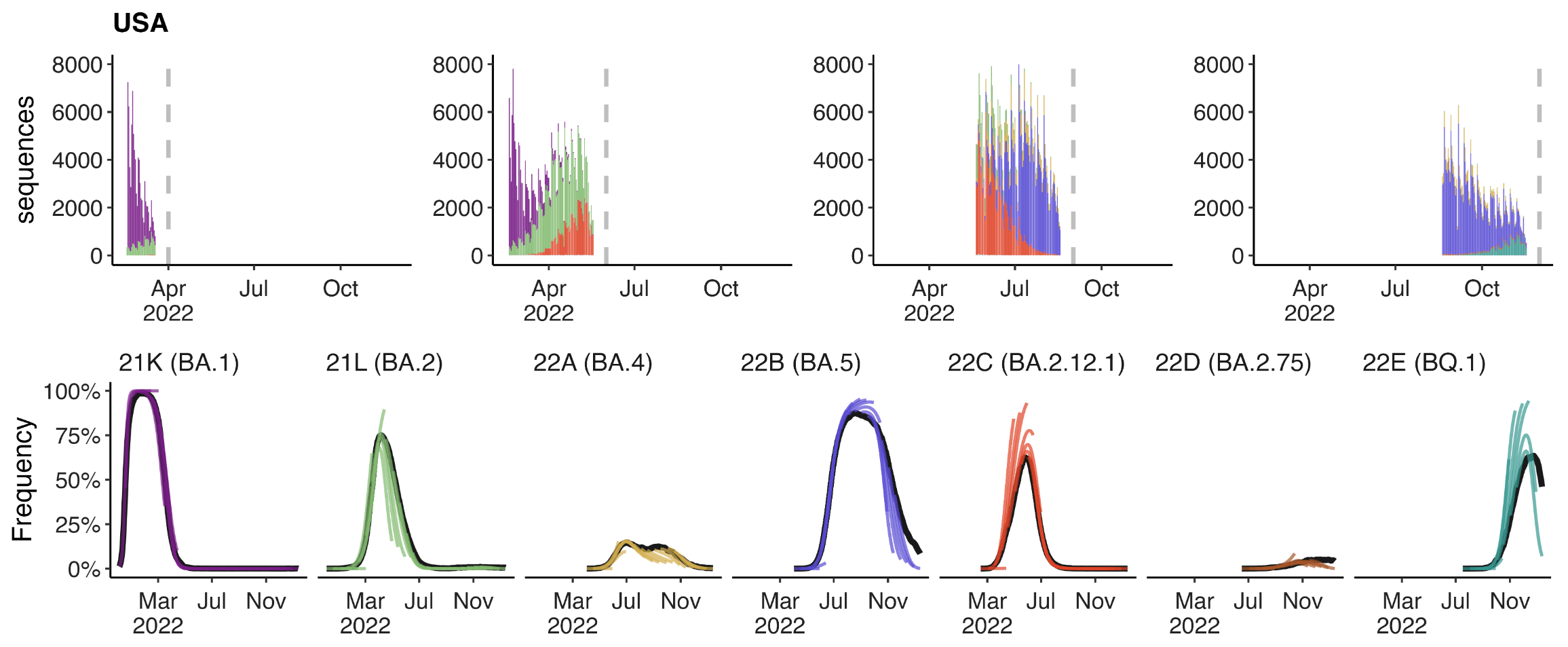
+30 day short-term forecasts across different countries
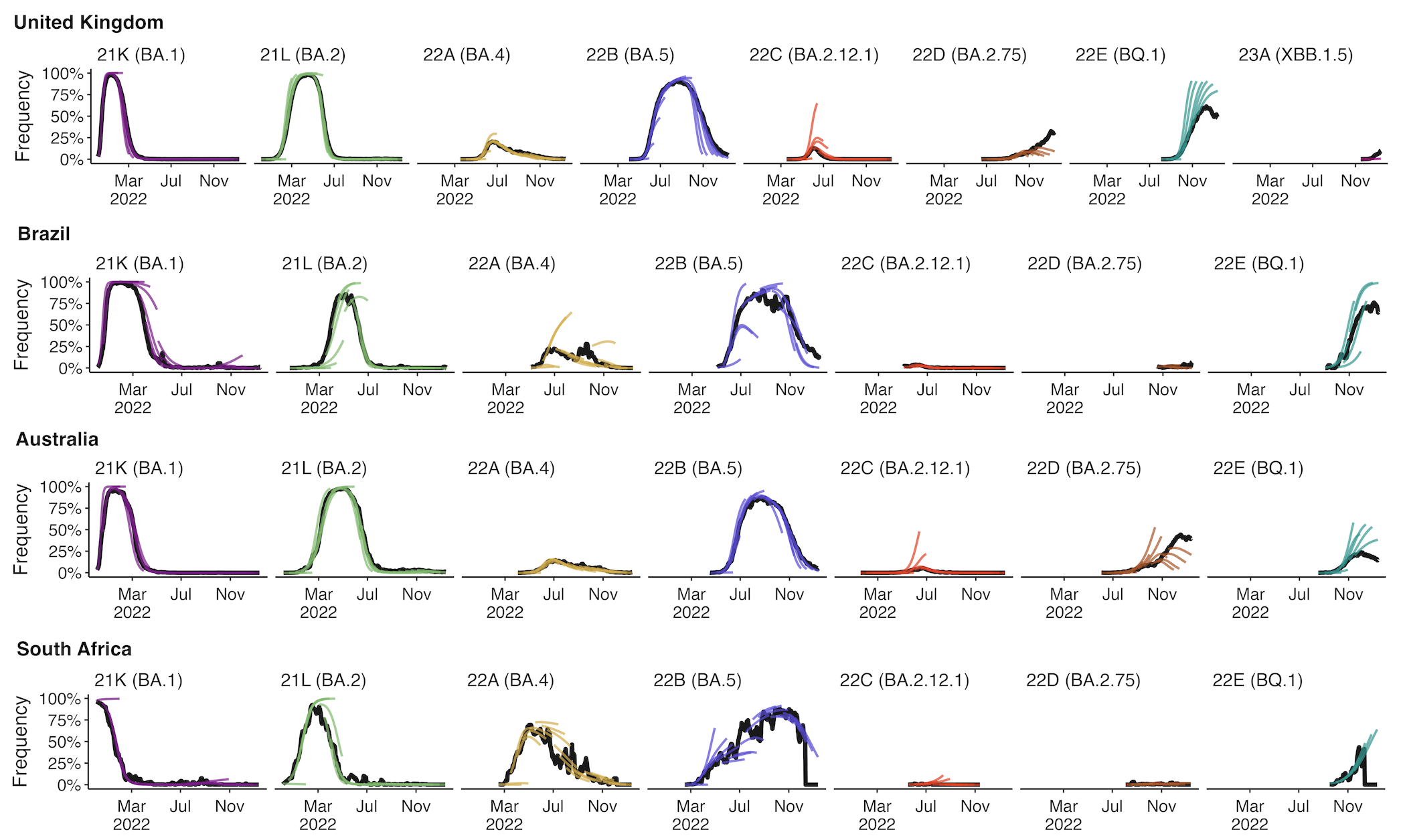
MLR models generate accurate short-term forecasts
30 days out, countries range from 5 to 15% mean absolute error
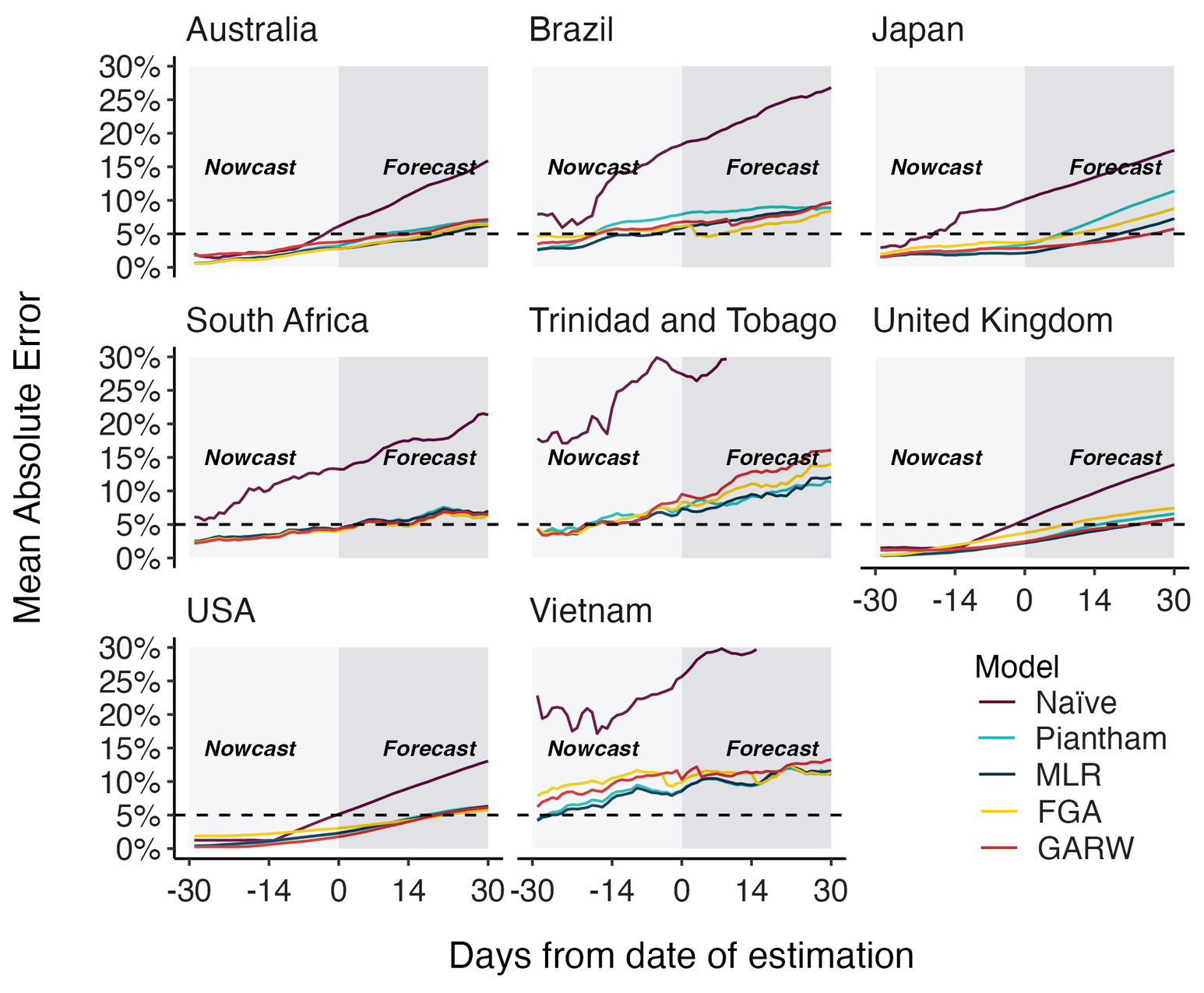
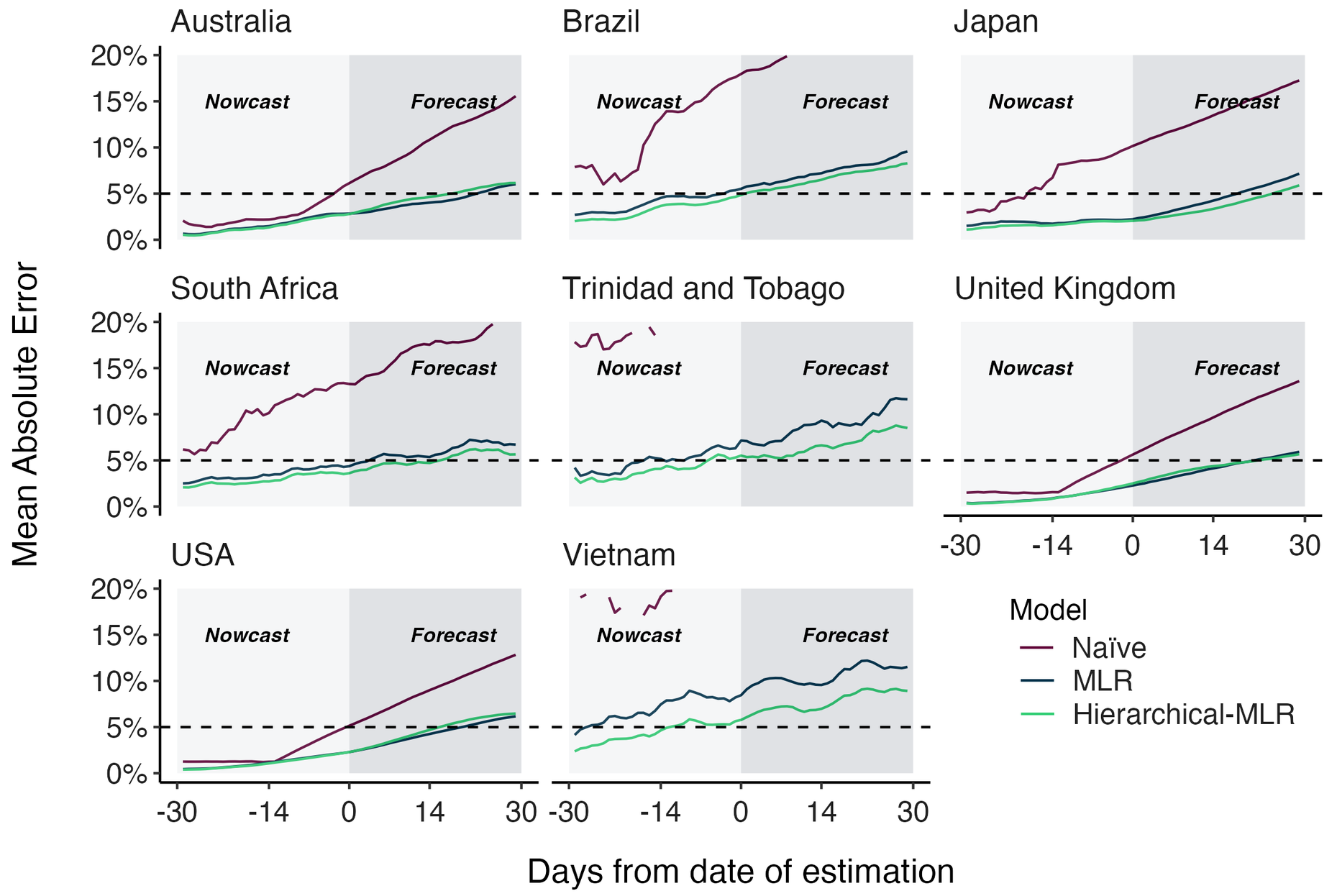
Hierarchical MLR model pools variant fitness estimates across countries
This approach improves poor model accuracy in countries with less intensive genomic surveillance
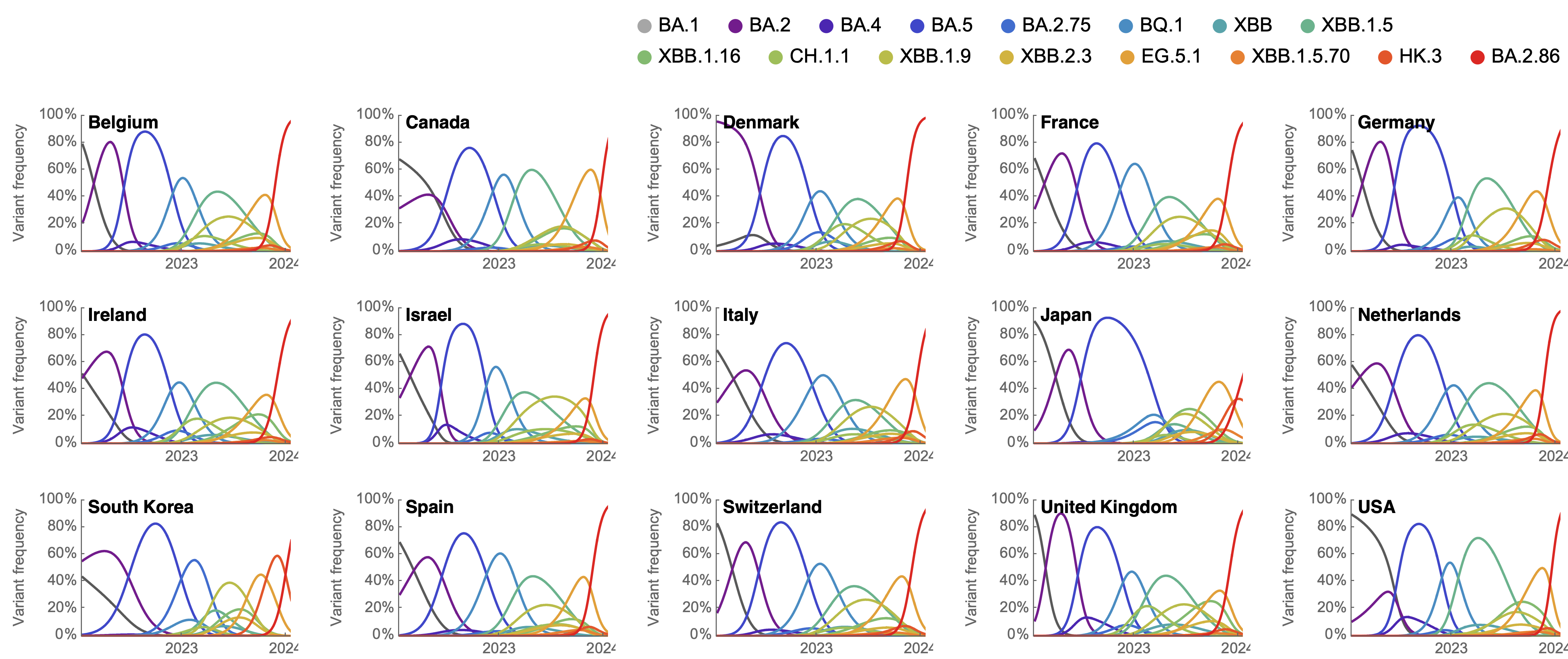
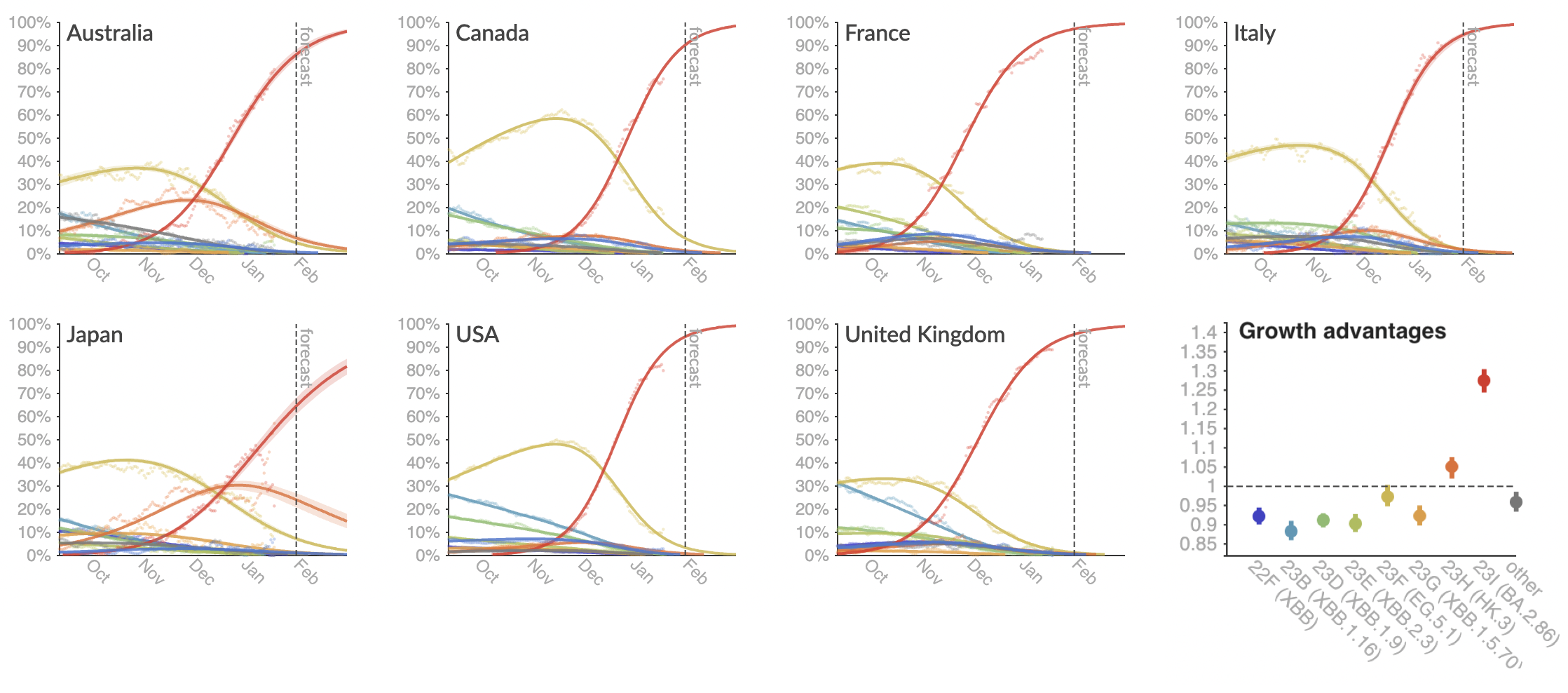
Clade and lineage forecasts continuously updated at nextstrain.org
Rapid sweep of JN.1 over Dec to Jan 2024
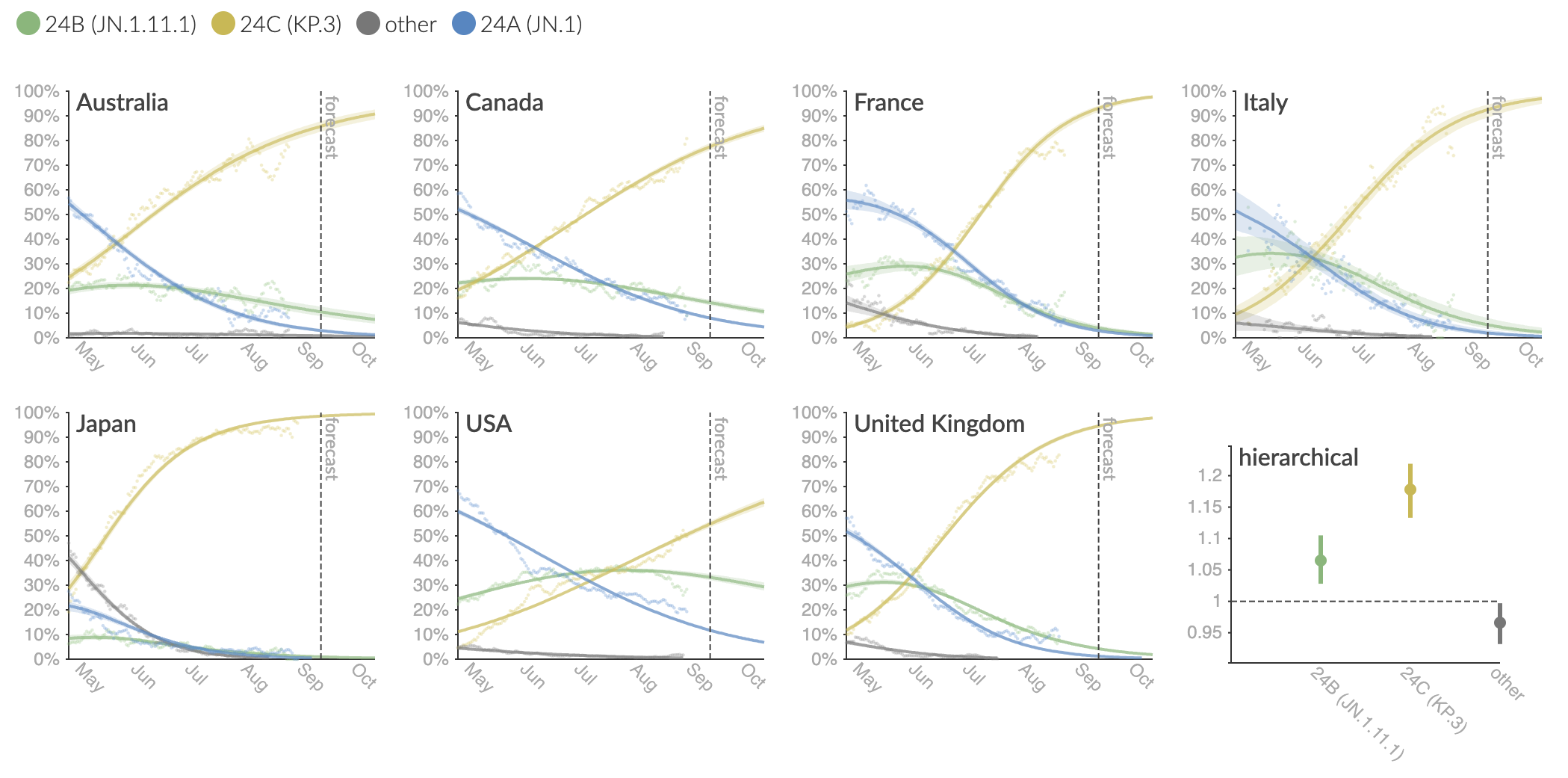
Clade and lineage forecasts continuously updated at nextstrain.org
Serial replacement of descendants of JN.1 with KP.3 the current winner
Assess currently circulating lineages by comparing frequency to population weighted growth advantage
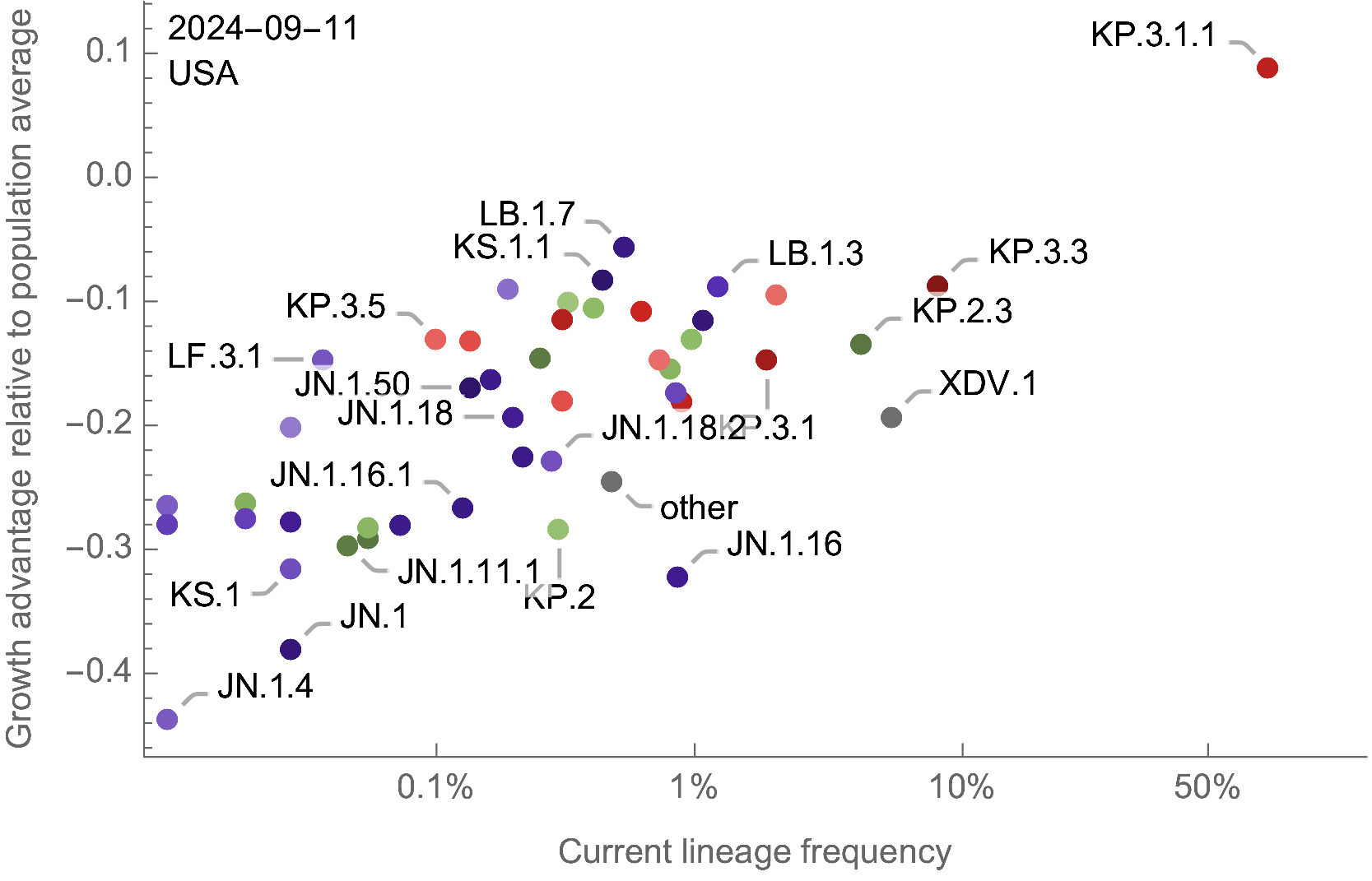
Eventual lineage success determined by initial fitness

Eventual lineage success determined by initial fitness

Eventual lineage success determined by initial fitness
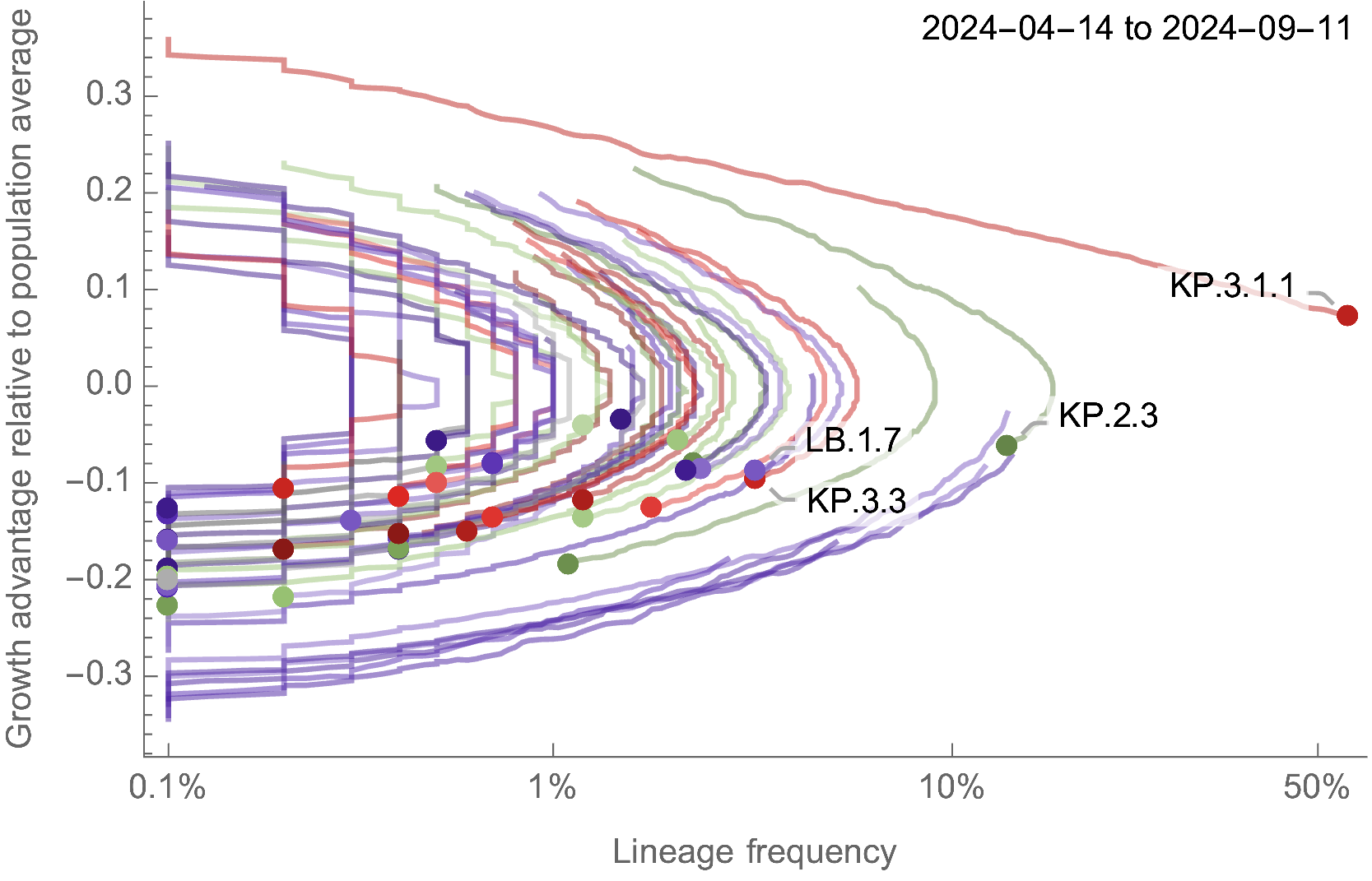
Multinomial logistic regression should work well for SARS-CoV-2 prediction, except new variants have been emerging fast enough that the prediction horizon is limited to 2-3 months
Ongoing work to lengthen prediction horizon by incorporating high-throughput experimental measurements of ACE2 binding and immune escape
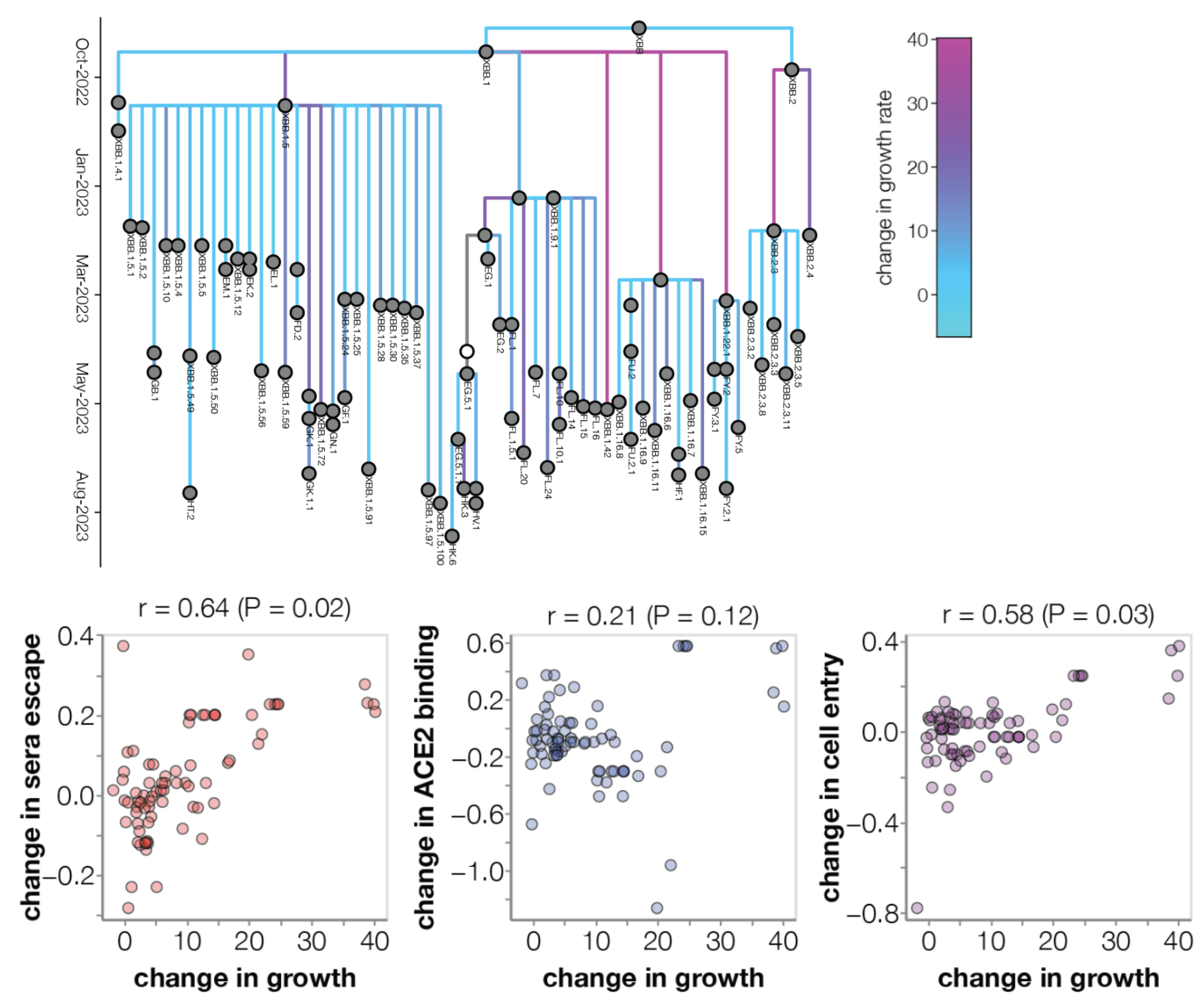
Prediction of variant fitness from empirical priors
Rather than estimate variant specific fitness $f_i$ directly, we instead parameterize as the "innovation" in fitness in going from parent lineage $p$ to child lineage $i$ as $\psi_i = (f_i - f_p)$.
We then compare a non-informative model of $$\psi_i = (f_i - f_p) \sim \mathrm{Normal}(0, \sigma)$$ to a model where each "innovation" value has an informed prior based on a linear combination of predictors such as ACE2 binding, immune escape and S1 mutations $$\psi_i = (f_i - f_p) \sim \mathrm{Normal}\left(\sum_p \beta_p \, x_p, \sigma\right)$$
Exciting developments in applying protein language models to estimate sequence-level fitness
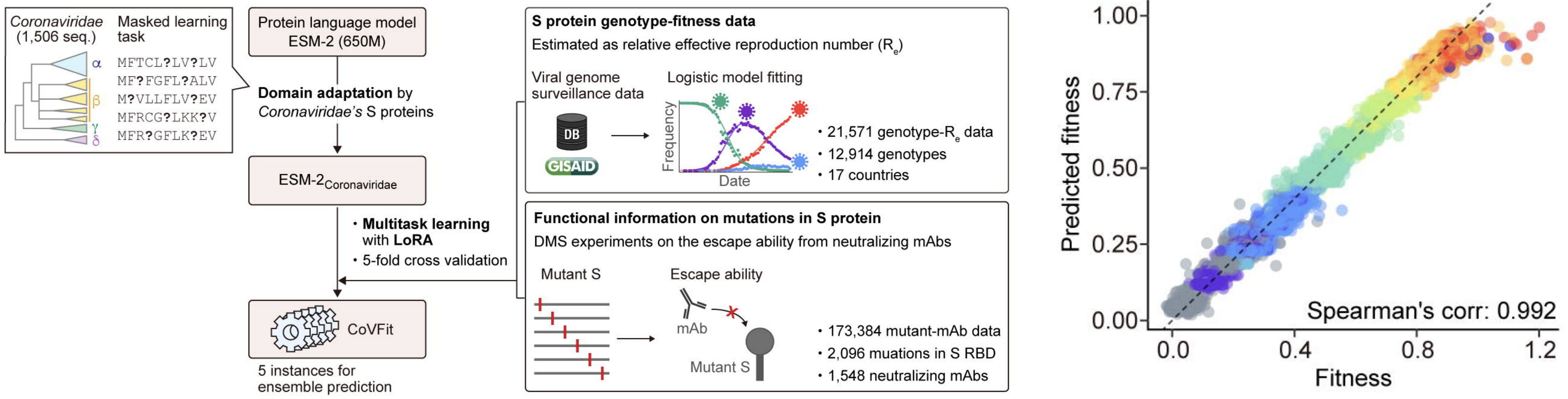
Continued research
- Application of MLR models to seasonal influenza and other pathogens
- Assessing and improving accuracy of "live" models at nextstrain.org/sars-cov-2/forecasts/
- Implementing DMS priors to predict fitness of emerging and yet-to-emerge lineages
- Deep learning approaches to incorporate structural information and avoid classification
Acknowledgements
SARS-CoV-2 genomic epi: Data producers from all over the world, GISAID, UW Virology, BBI, WA PHL
Nextstrain: Richard Neher, Ivan Aksamentov, John SJ Anderson, Kim Andrews, Jennifer Chang, James Hadfield, Emma Hodcroft, John Huddleston, Jover Lee, Victor Lin, Cornelius Roemer, Thomas Sibley
Determinants of transmission: Cécile Tran Kiem, Amanda Perofsky, Miguel Paredes, Lauren Frisbie, Allison Black, Cécile Viboud
Evolutionary forecasting: Marlin Figgins, Jover Lee, James Hadfield, John Huddleston, Eslam Abousamra, Jesse Bloom, Cornelius Roemer, Richard Neher
Bedford Lab:
![]() John Huddleston,
John Huddleston,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Miguel Paredes,
Miguel Paredes,
![]() Marlin Figgins,
Marlin Figgins,
![]() Victor Lin,
Victor Lin,
![]() Jennifer Chang,
Jennifer Chang,
![]() Nashwa Ahmed,
Nashwa Ahmed,
![]() Cécile Tran Kiem,
Cécile Tran Kiem,
![]() Kim Andrews,
Kim Andrews,
![]() Cristian Ovaduic,
Cristian Ovaduic,
![]() Philippa Steinberg,
Philippa Steinberg,
![]() Jacob Dodds,
Jacob Dodds,
![]() John SJ Anderson
John SJ Anderson
![]() Amin Bemanian
Amin Bemanian





