Pathogen diversity
A pathogen is a disease-causing biological entity capable of replication within a host and transmission between hosts
Broad categories
- RNA viruses
- DNA viruses
- Bacteria
- Parasites and fungi
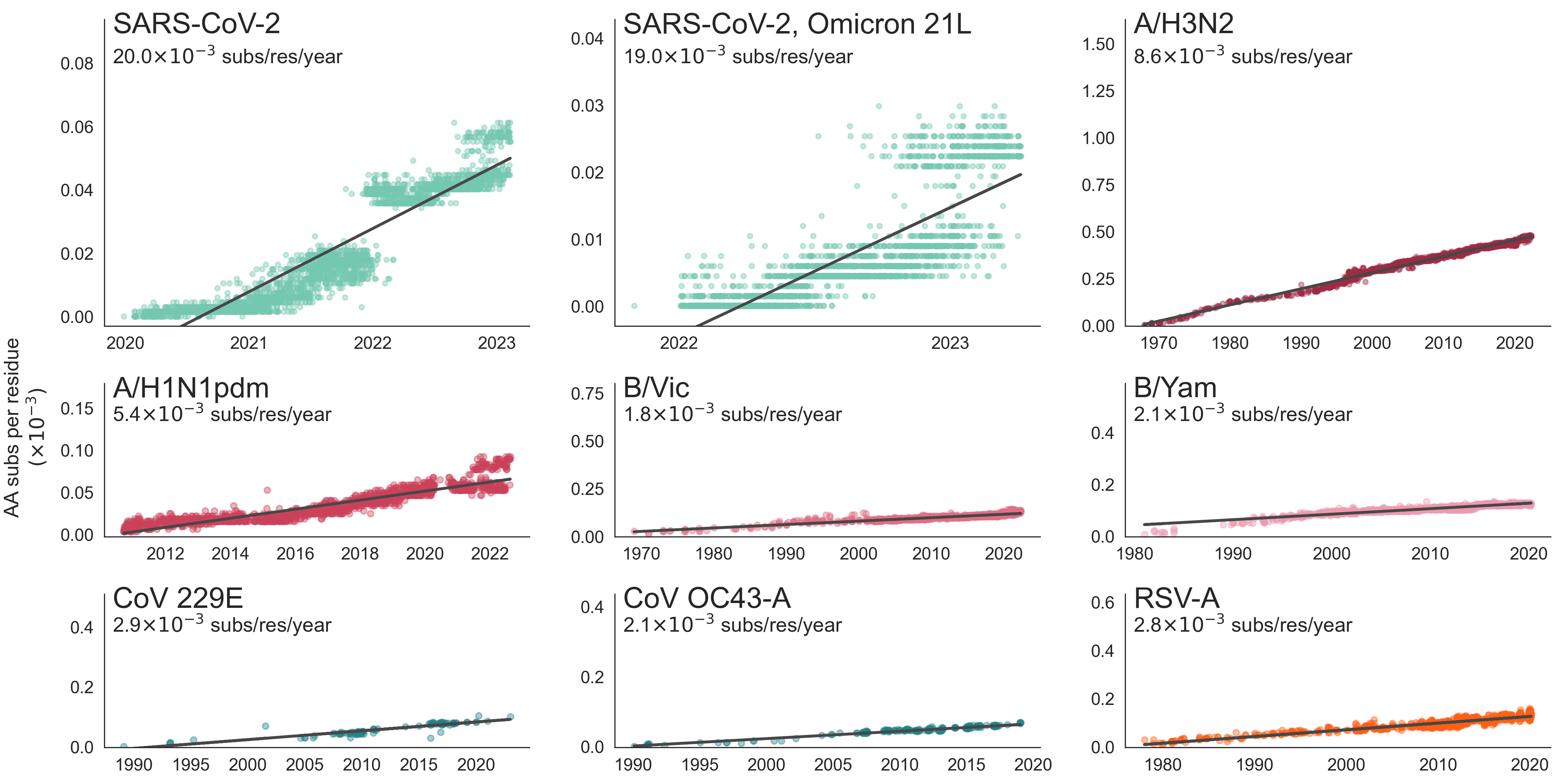
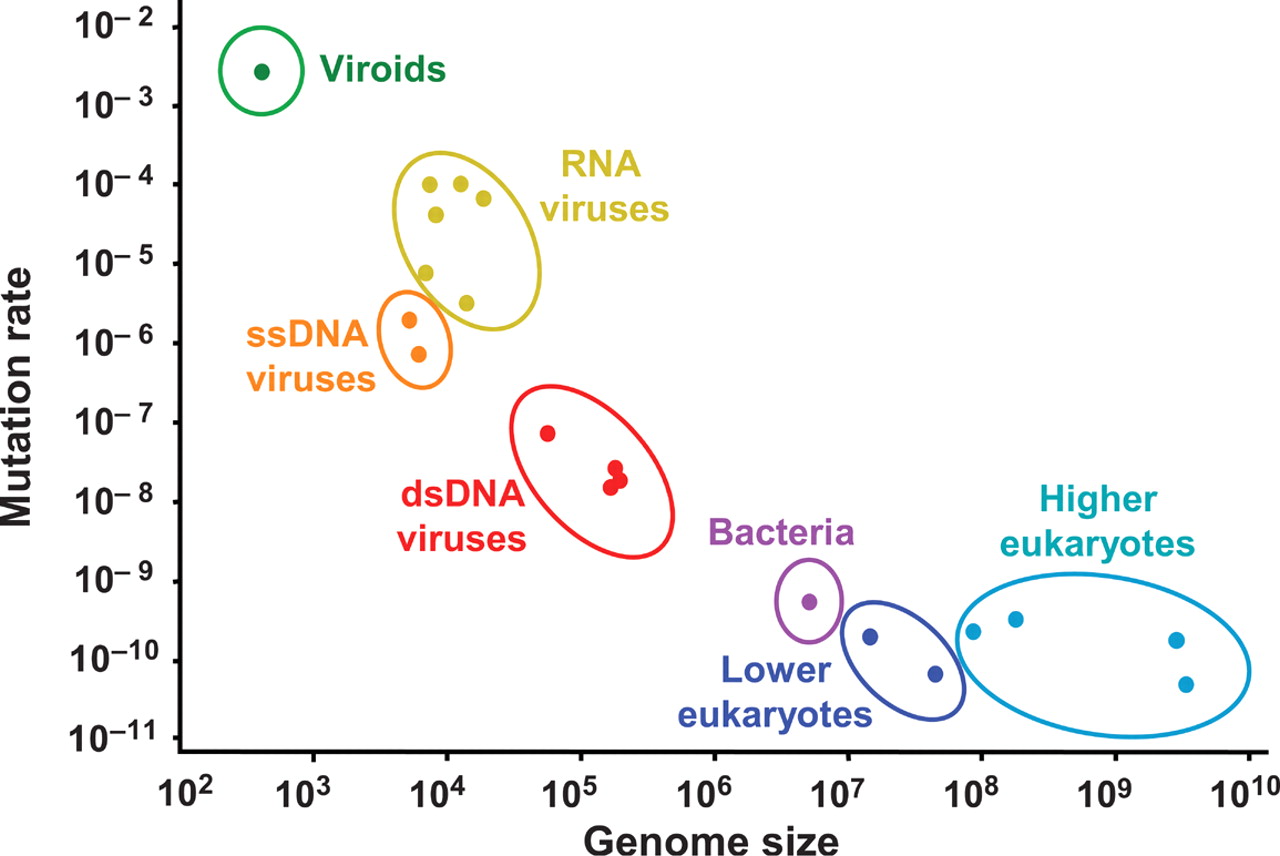
Mutation rate varies with genome size
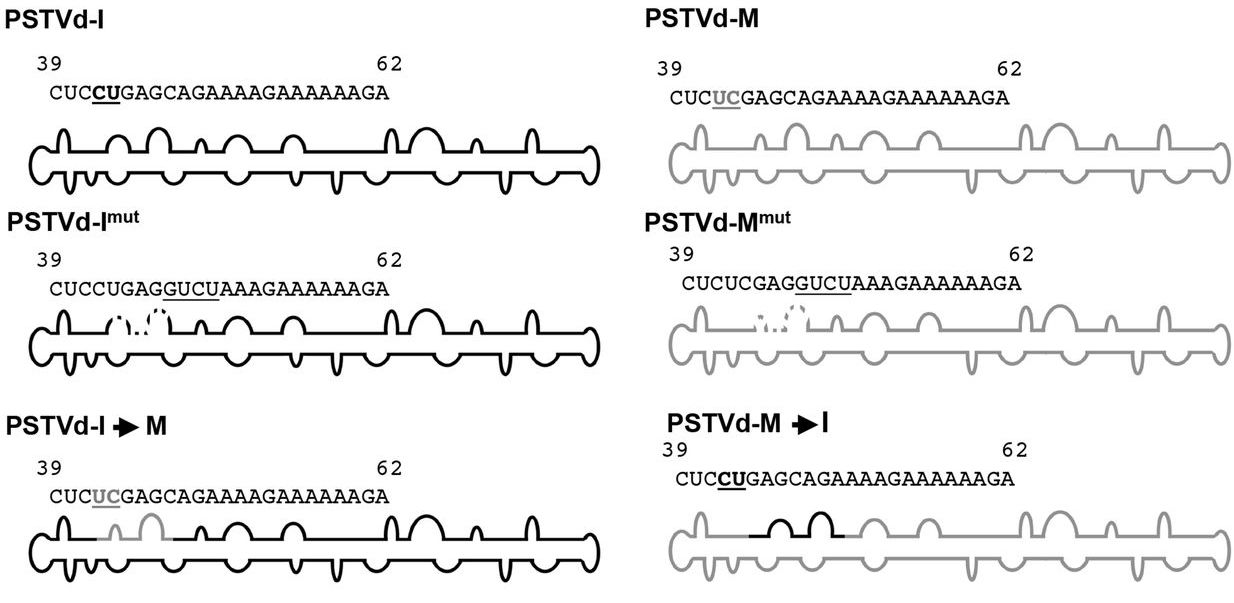
Viroids are parasitic functionless RNA
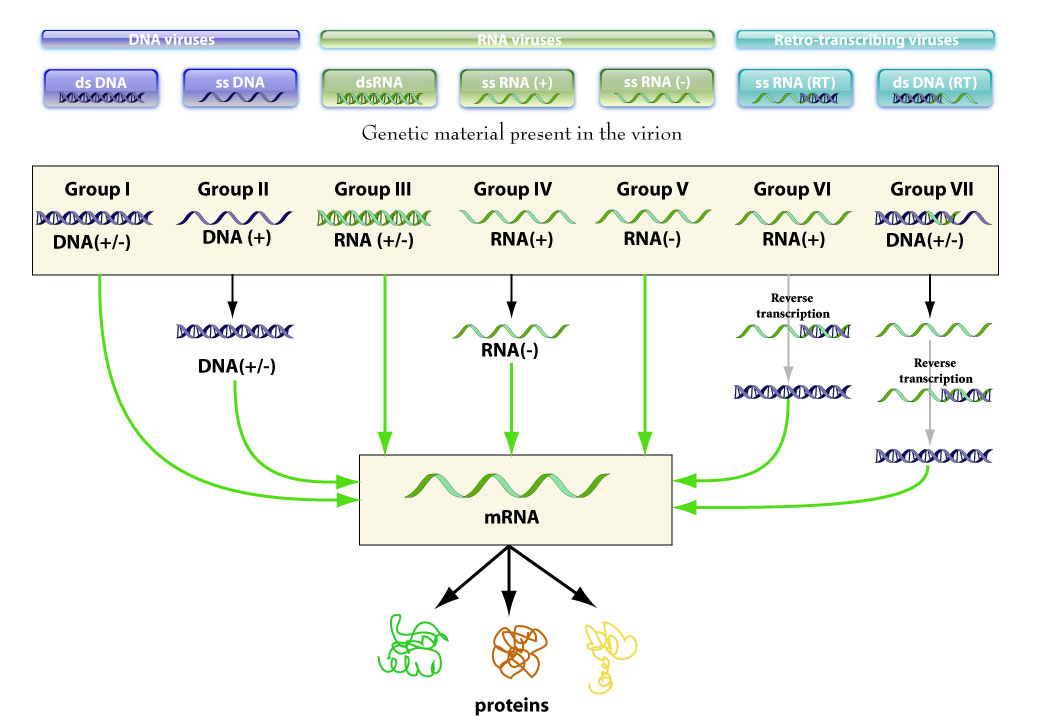
Baltimore classification system describes virus groups
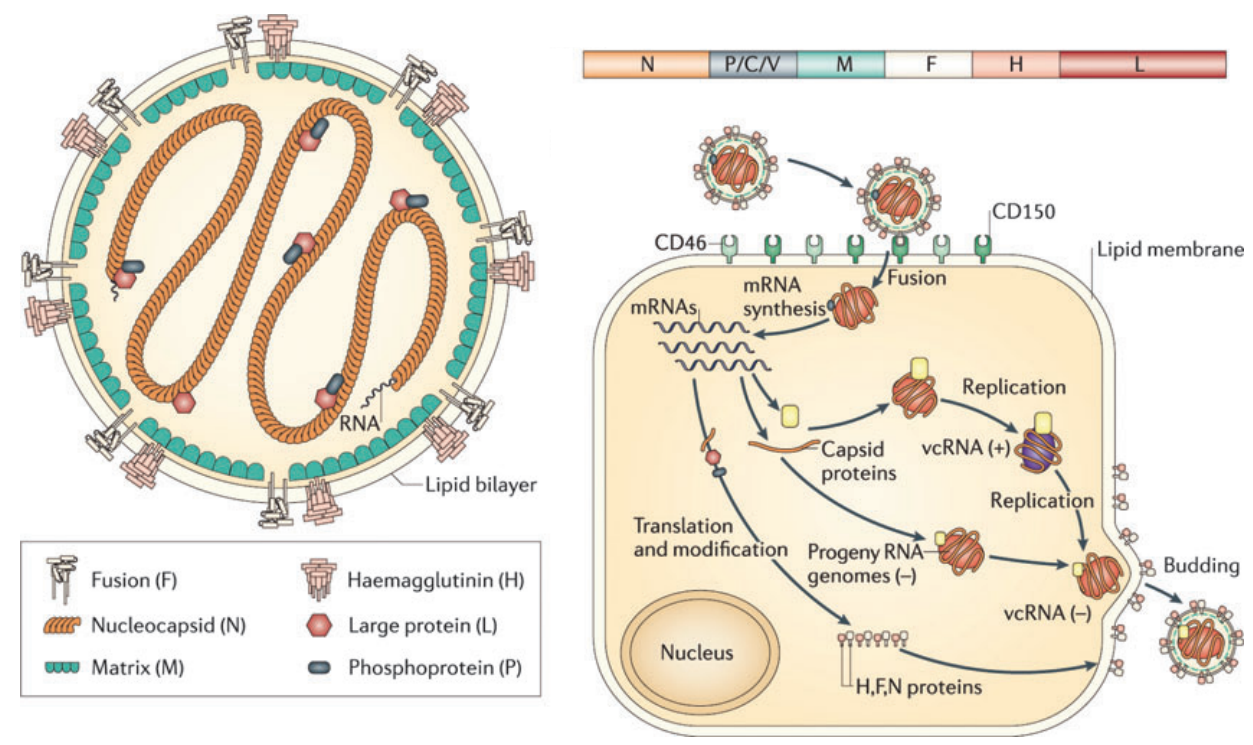
RNA virus lifecycle, internal vs surface proteins
Negative-sense RNA virus phylogeny
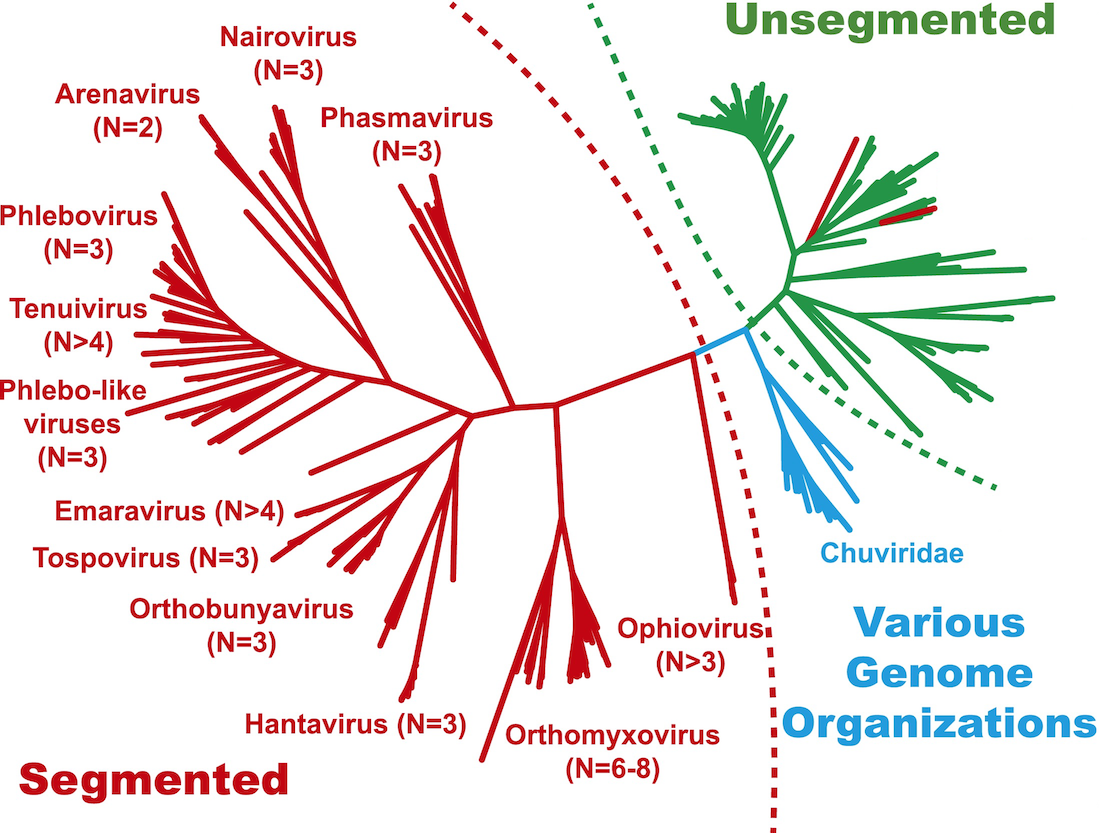
Negative-sense RNA virus phylogeny
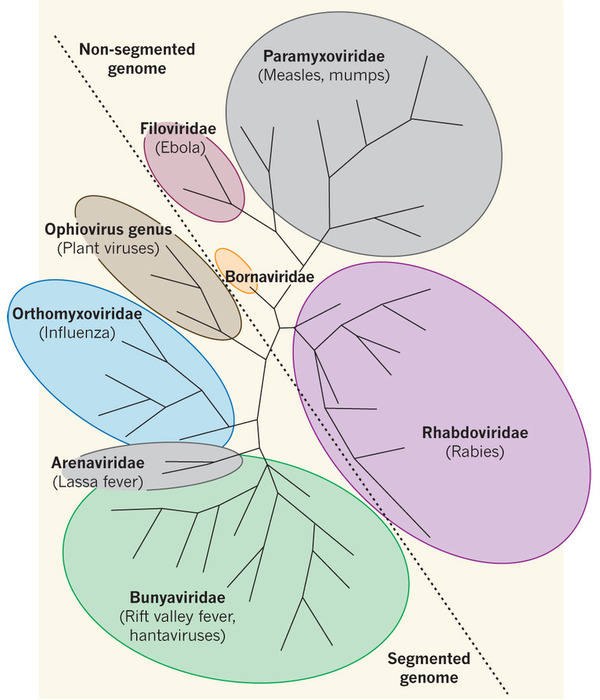
Negative-sense RNA viruses
- Influenza
- Measles
- Ebola
- Rabies
- Lassa
- Hanta
- Respiratory syncytial virus (RSV)
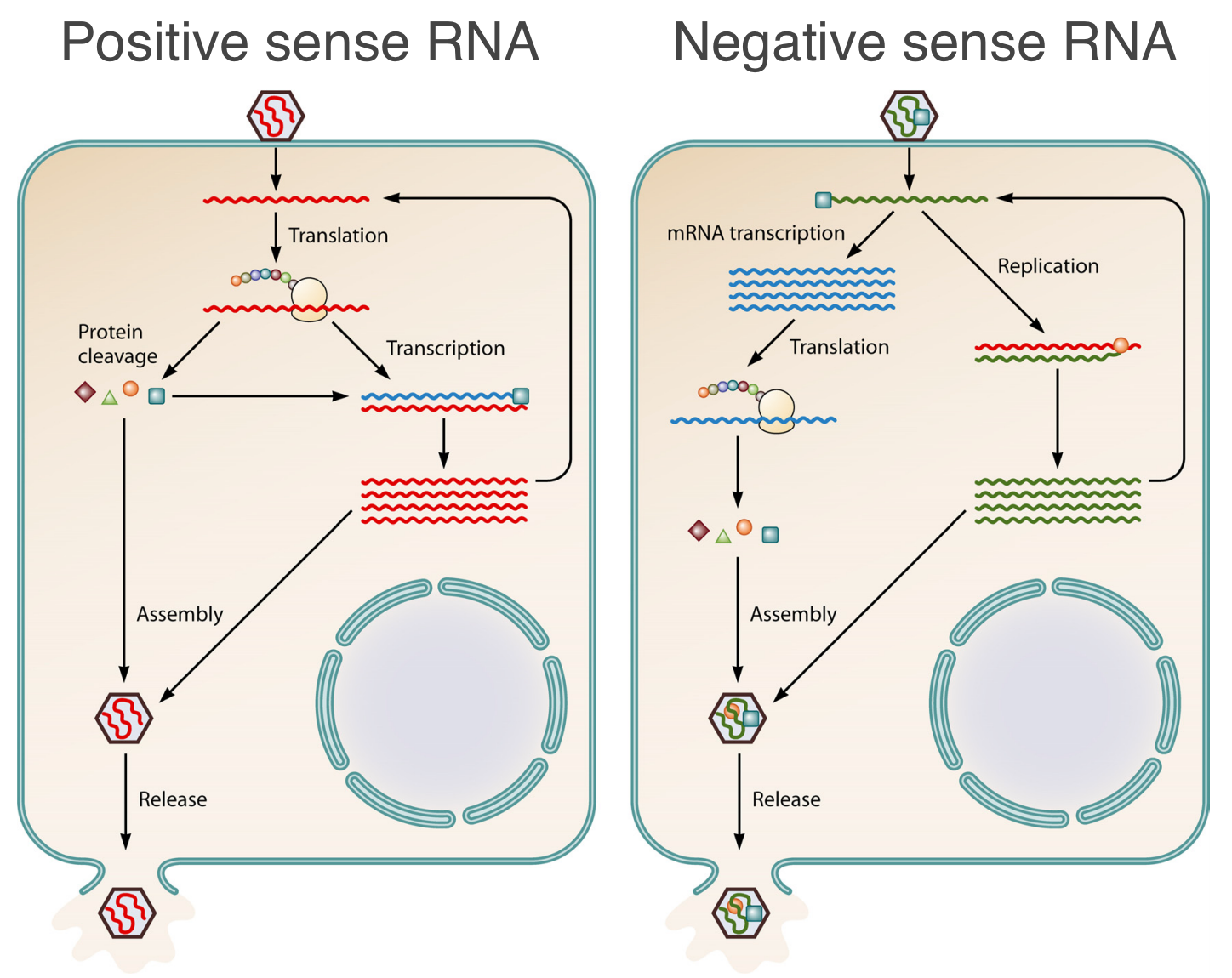
Positive-sense RNA viruses
- Flaviviruses: Yellow fever, West Nile, Dengue, Zika, HCV
- Coronaviruses, including SARS-CoV-2
- Rhinovirus
- Polio
- Norovirus
- Chikungunya
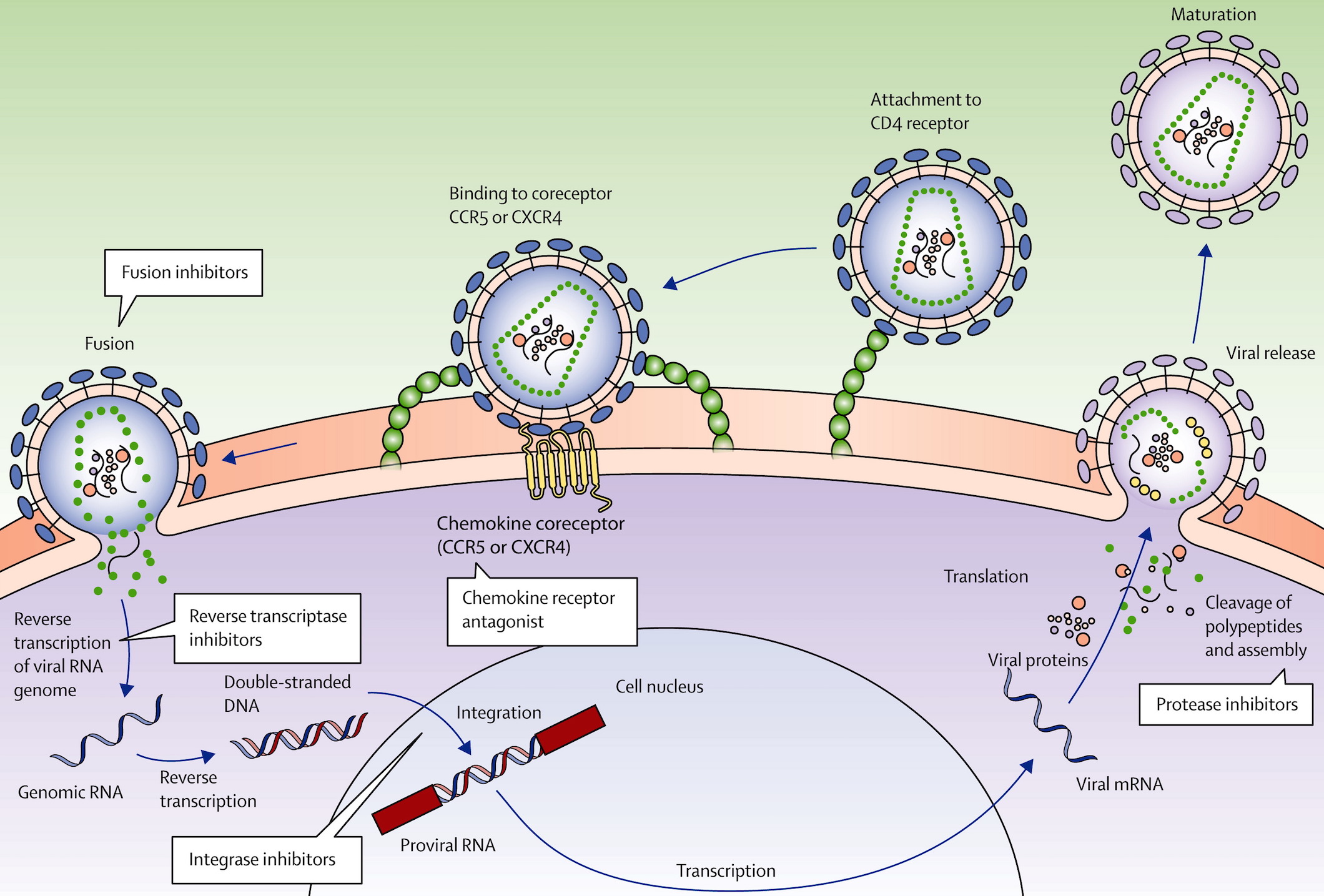
Retrovirus lifecycle
DNA virus genomes
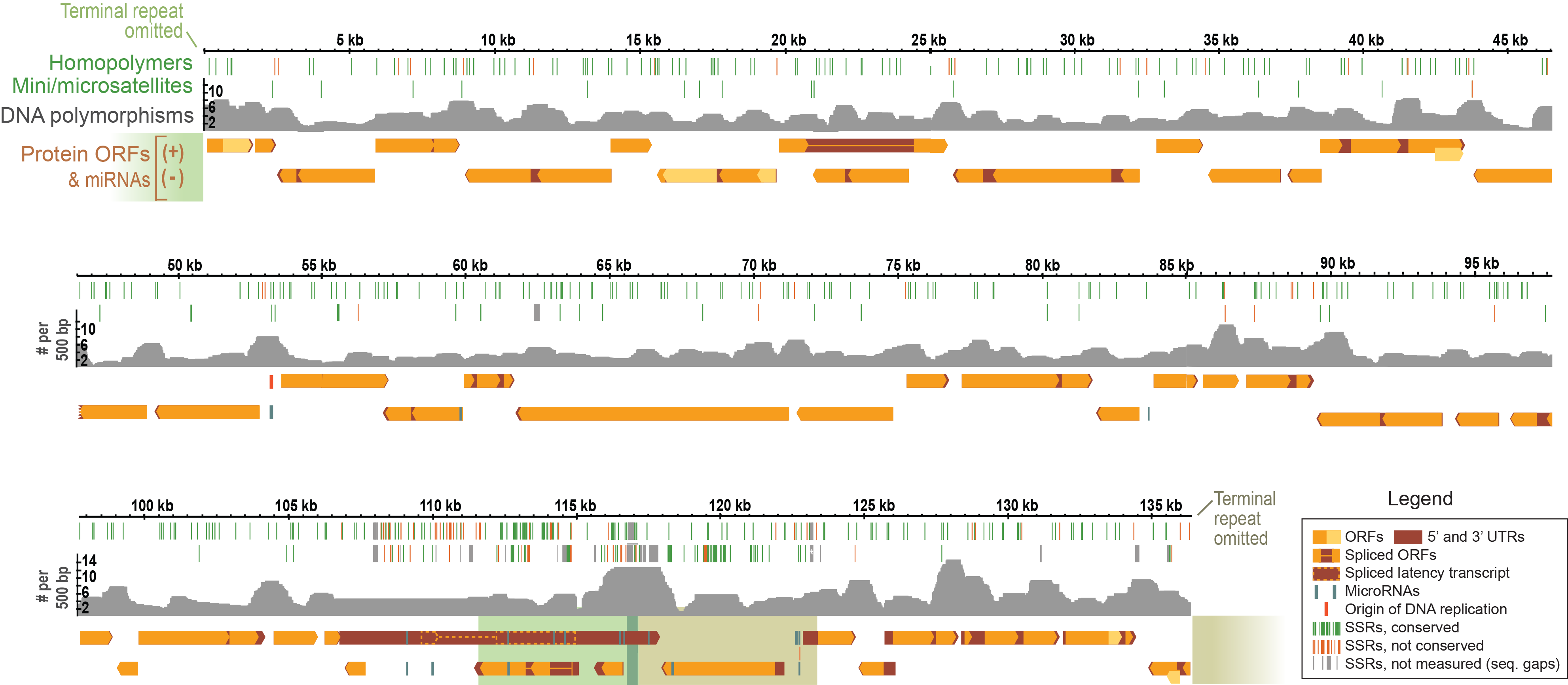
Bigger genome allows for more functions
DNA viruses
- Herpesviruses: HSV, Epstein–Barr virus (EBV), Varicella, Cytomegalovirus (CMV), Kaposi's sarcoma-associated herpesvirus (KSHV)
- Papillomavirus (HPV)
- Smallpox, monkeypox
Bacterial cell
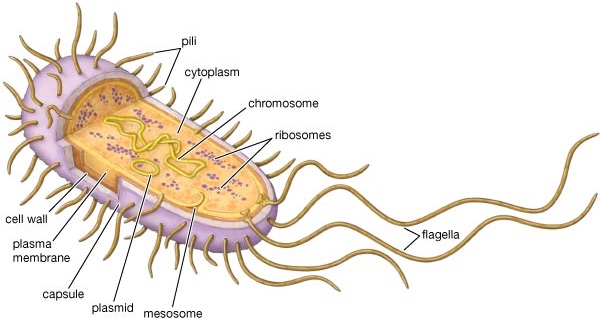
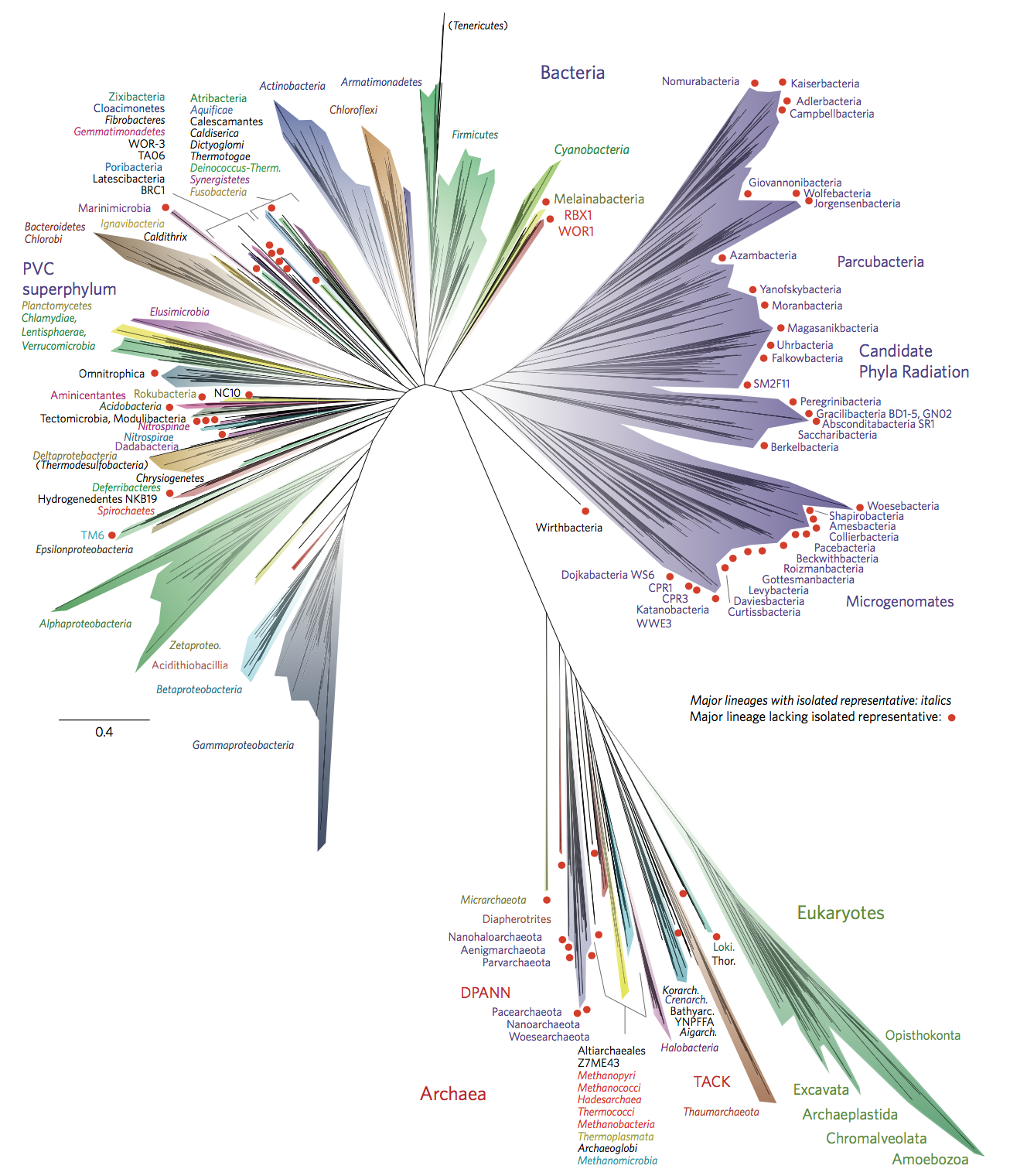
Bacterial diversity
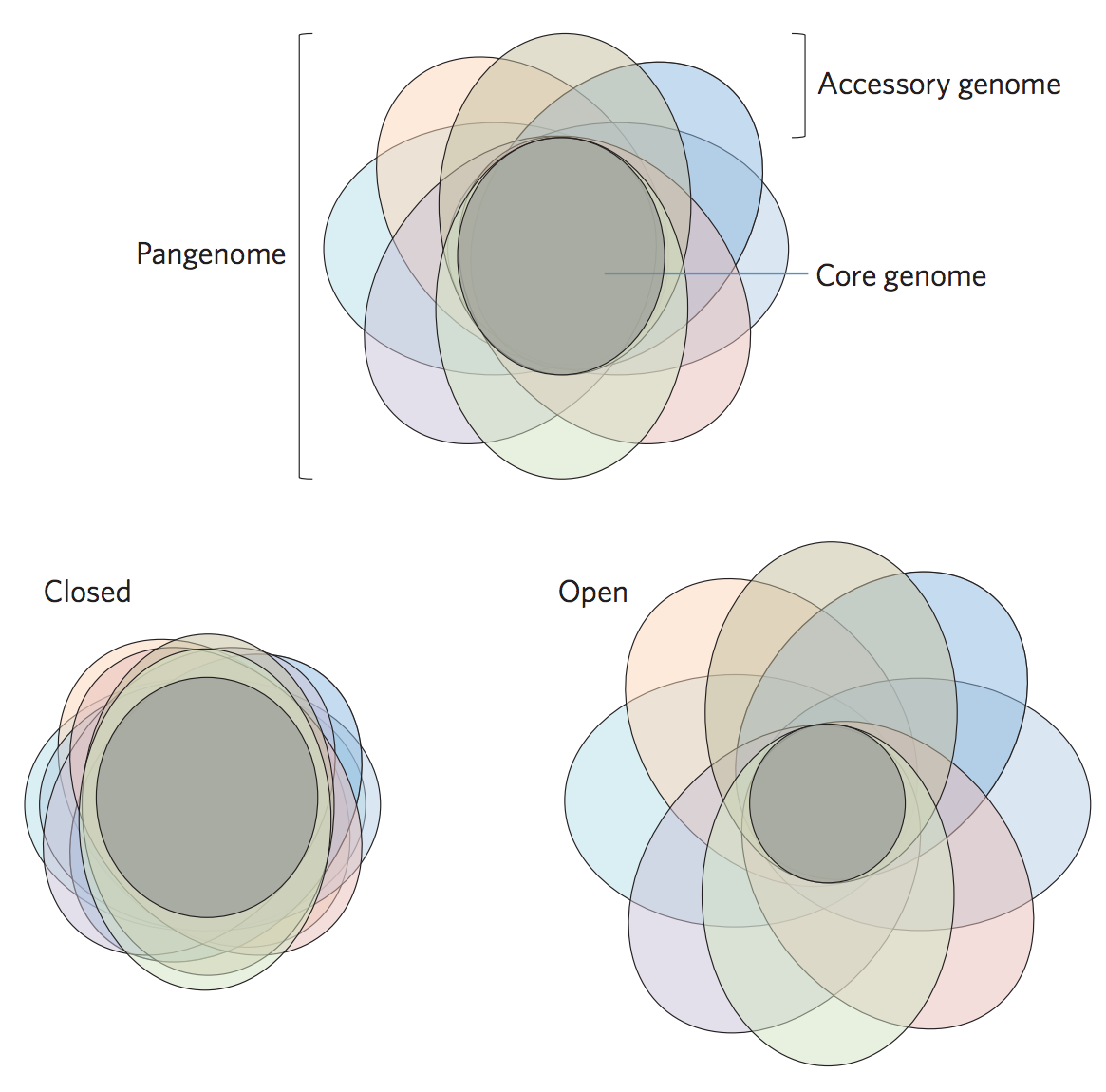
Core vs pangenome
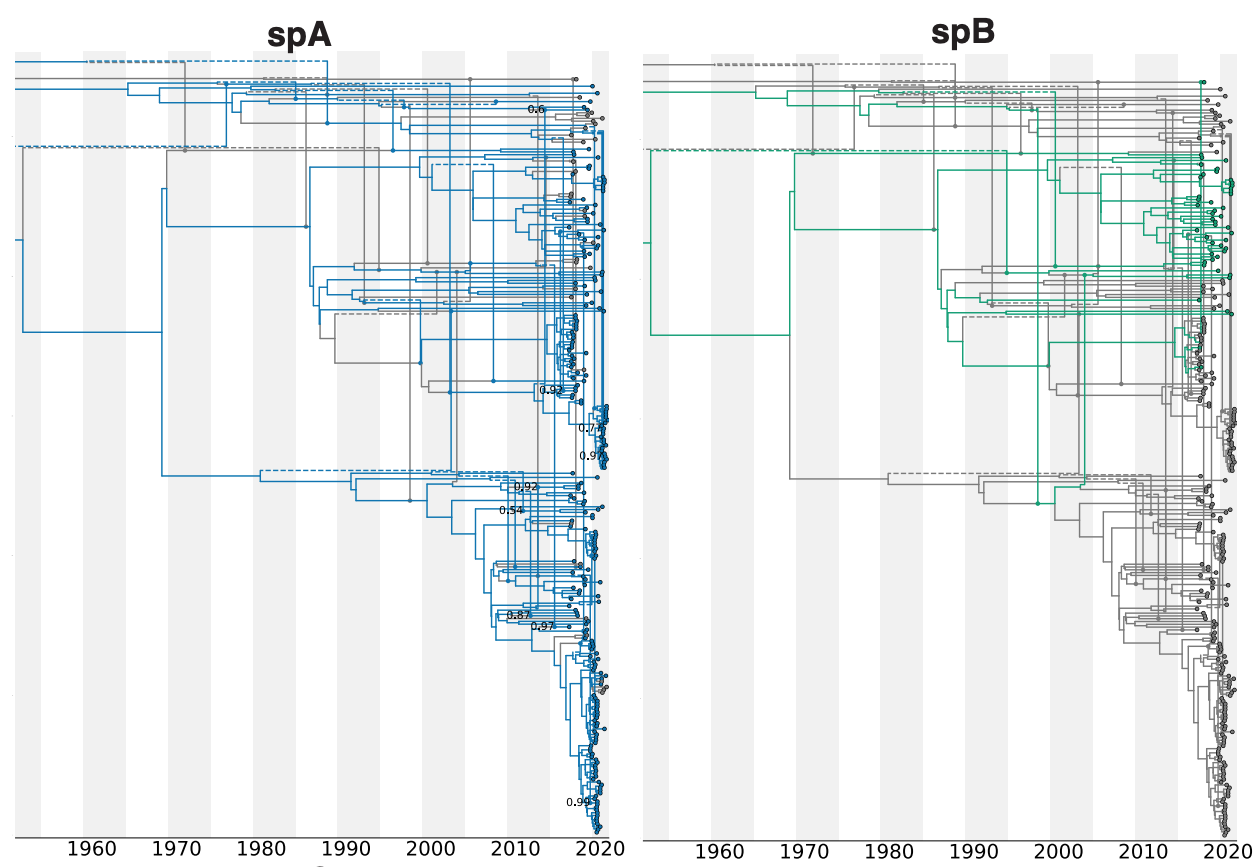
Divergent evolutionary histories between core chromosome and plasmids
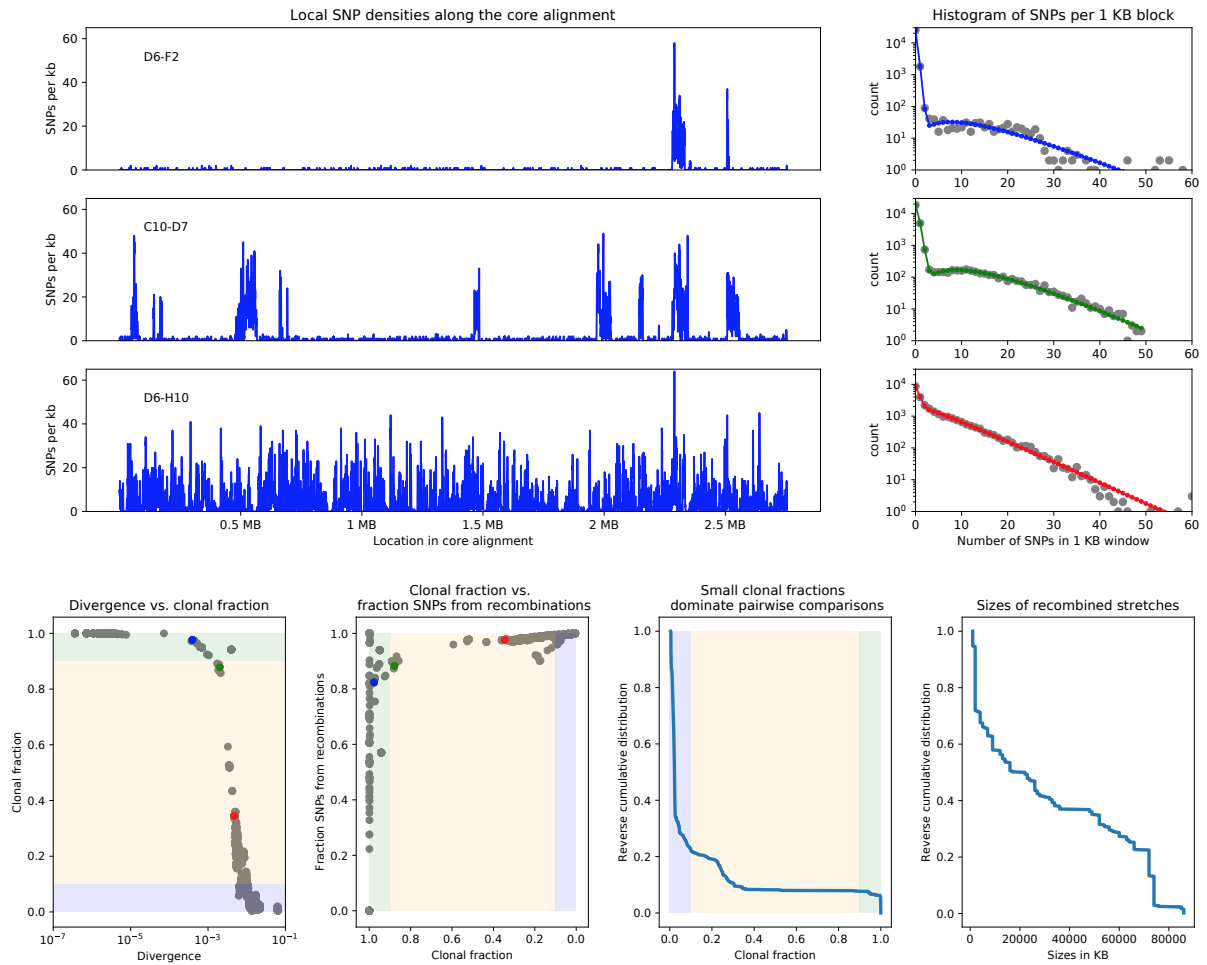
Recombination is often rampant
Bacterial pathogens
- C. difficile
- Cholera
- Klebsiella
- Neisseria gonorrhoeae
- S. aureus (MRSA)
- Shigella
- Streptococcus pneumoniae
- Tuberculosis
- Typhoid
- Y. pestis
Parasites
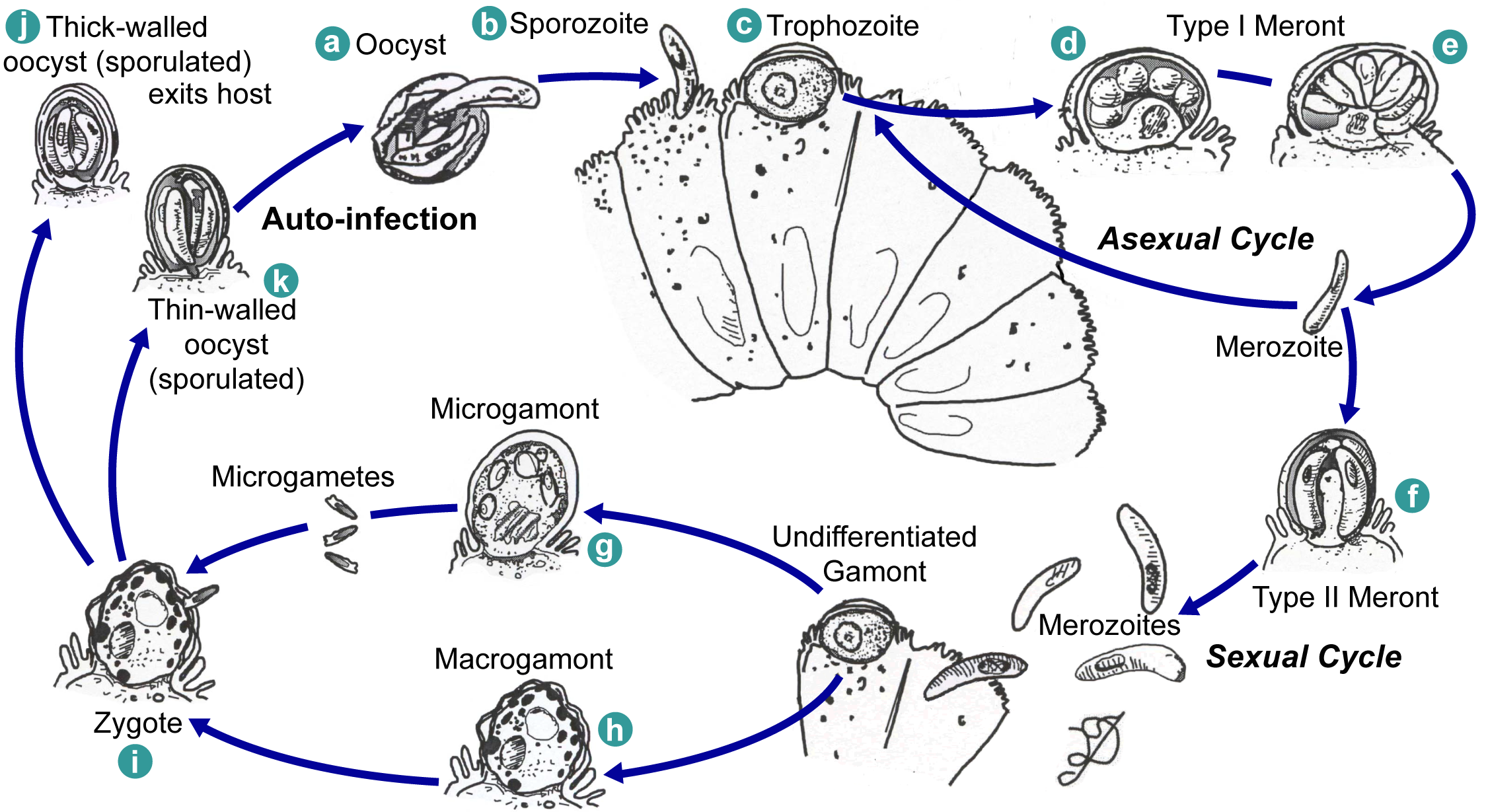
Parasite lifecycle
- Malaria
- Cryptosporidium
- Trypanosomes
- Giardia
- Leishmaniasis
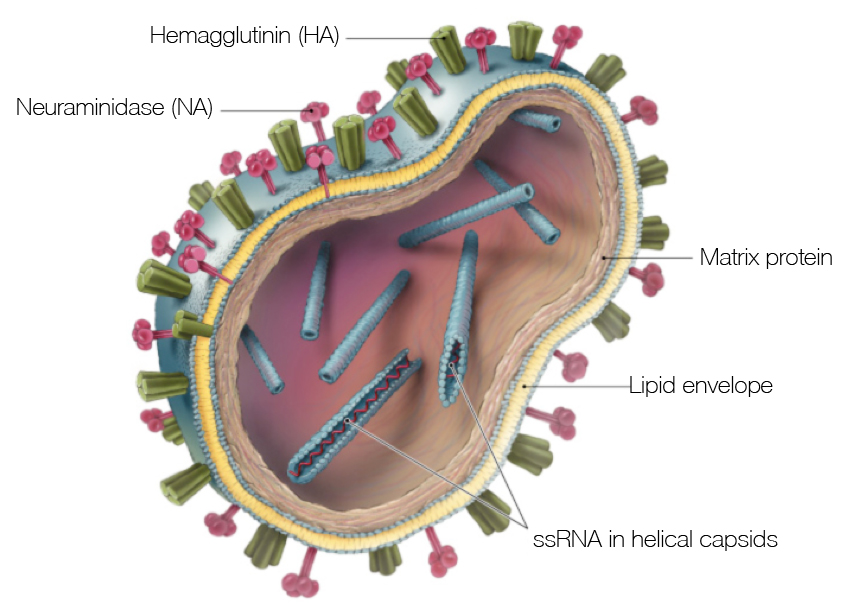
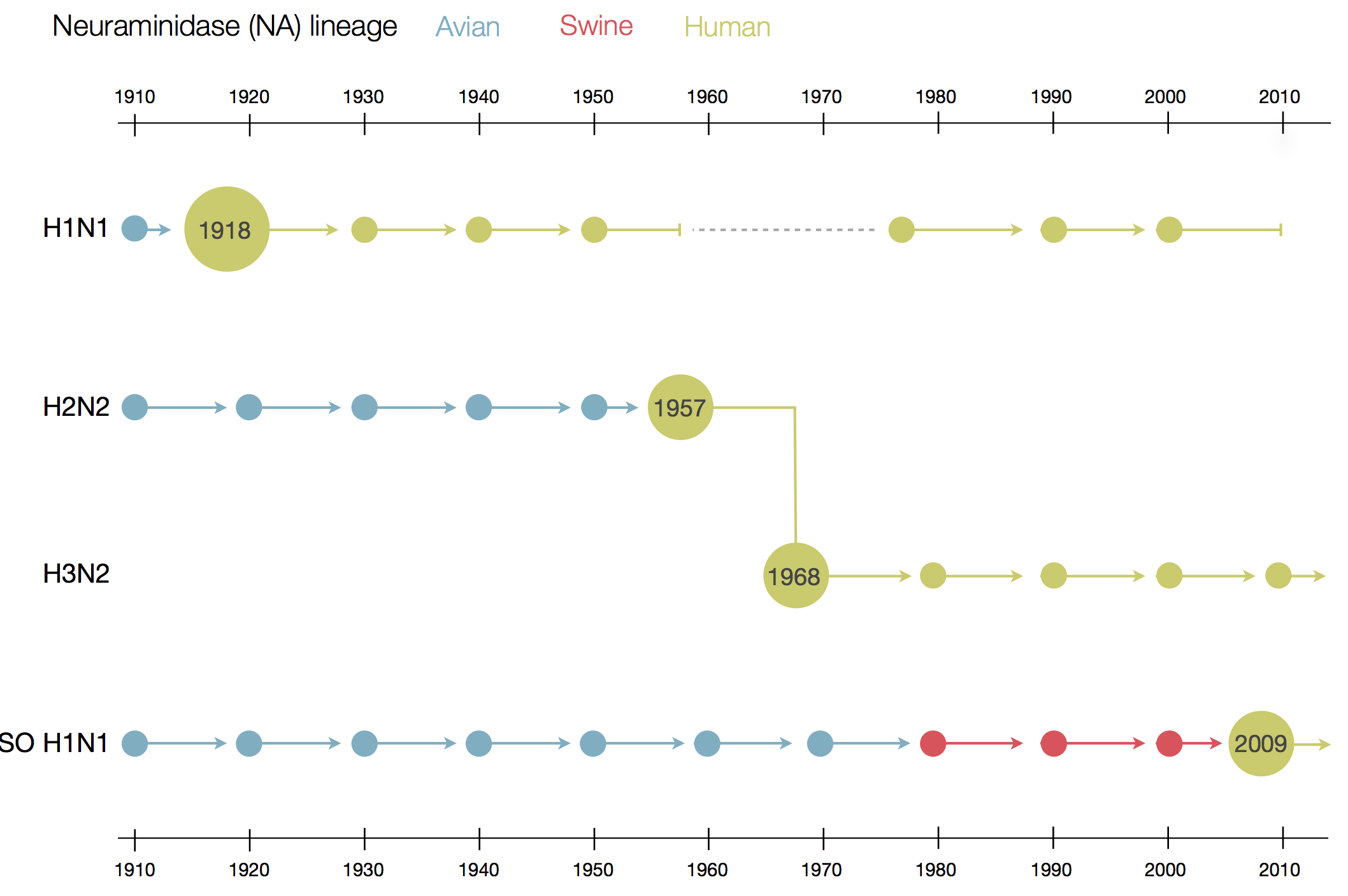
Influenza
Prototypical antigenically evolving pathogen
Influenza virion
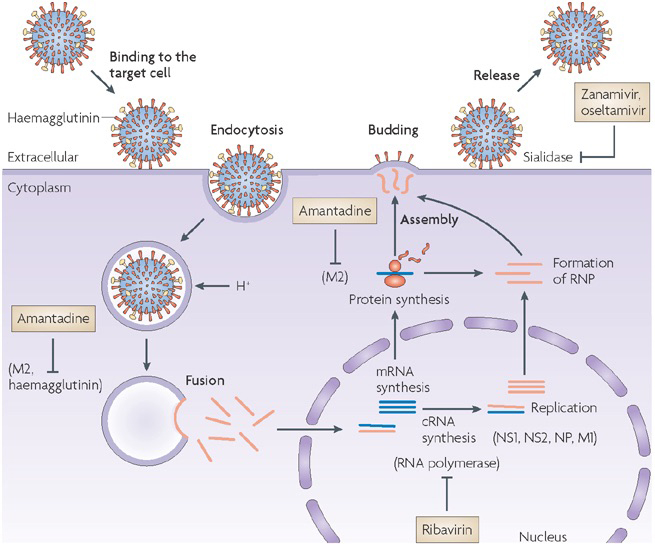
Influenza life cycle
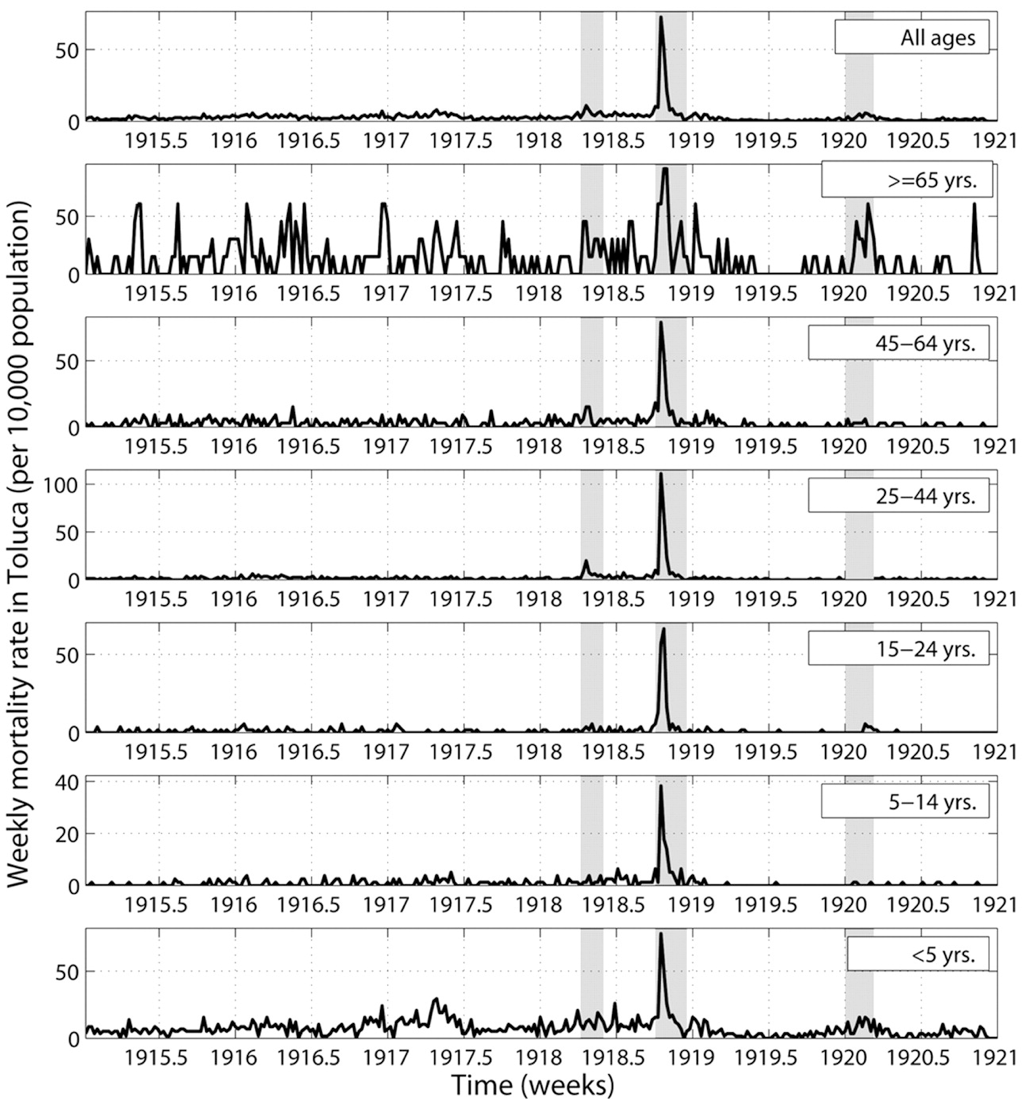
1918 "Spanish flu"
Flu pandemics caused by host switch events
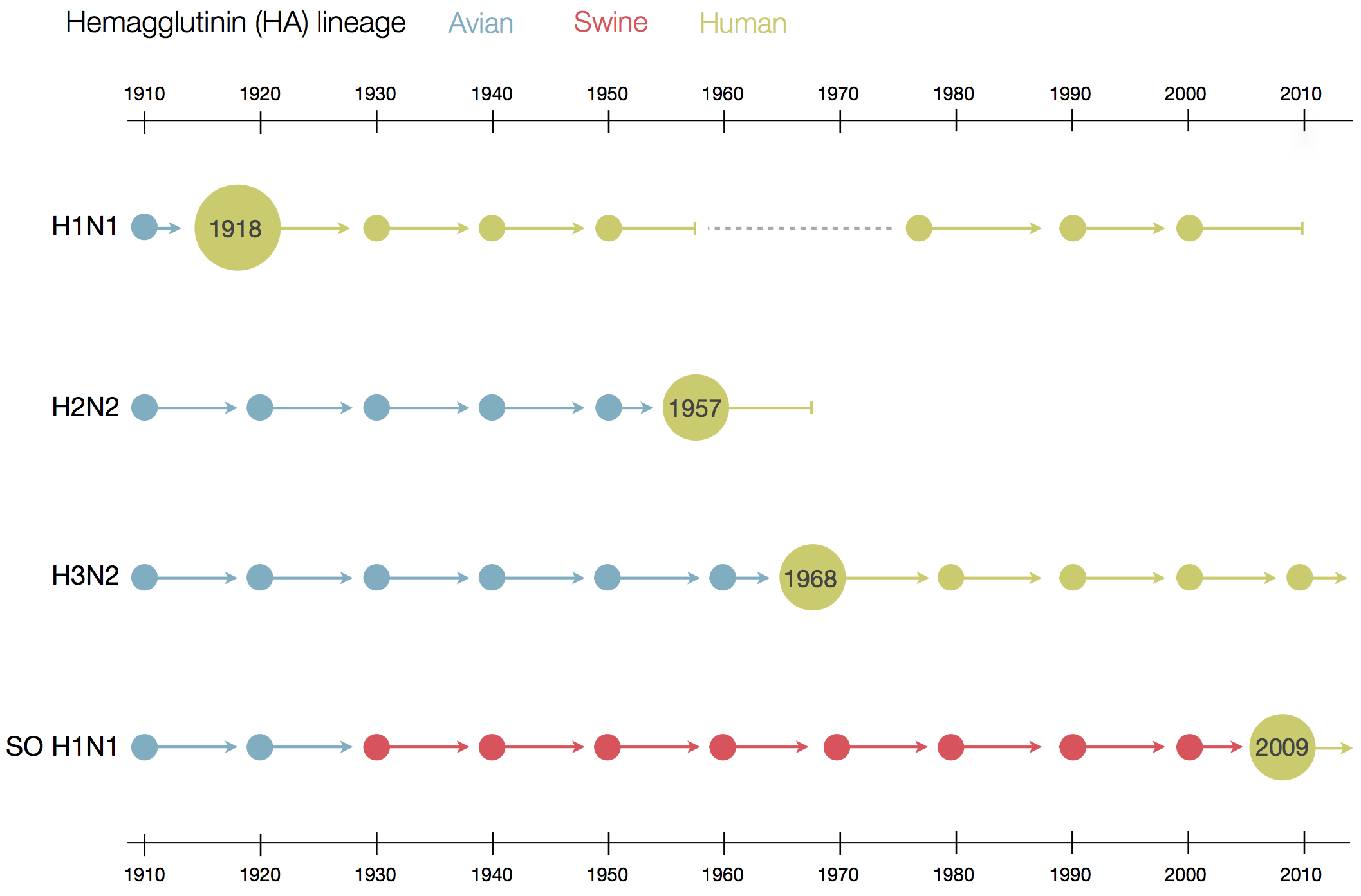
Host switch events often occur through reassortment
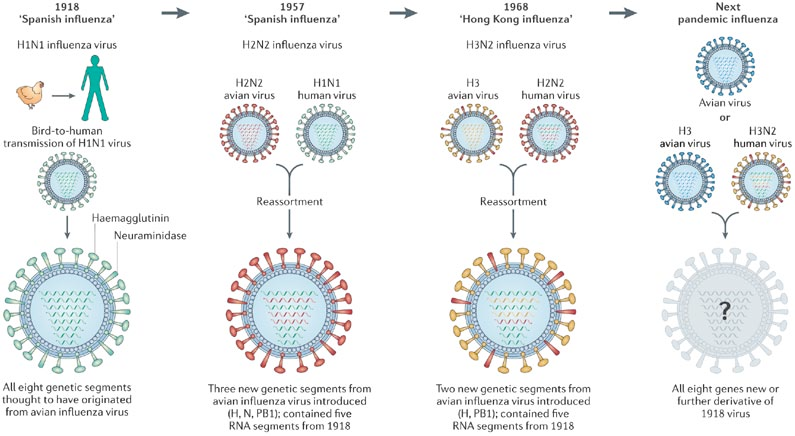
Host switch events often occur through reassortment
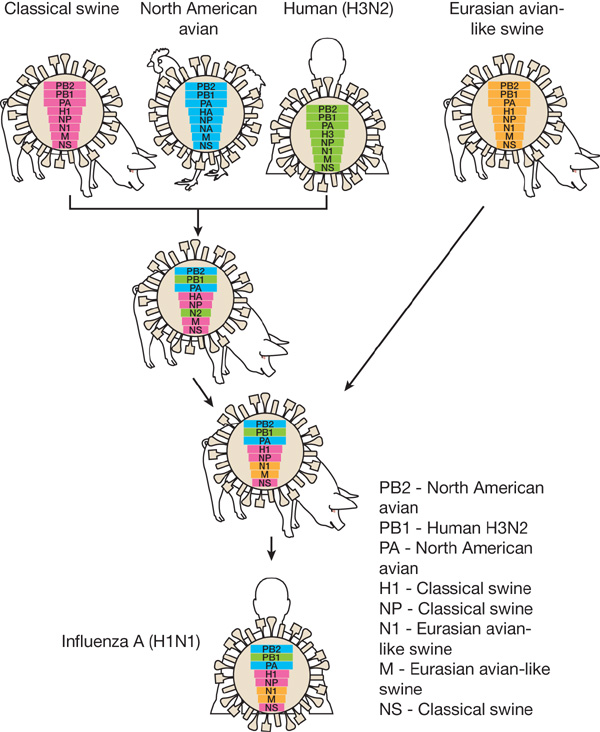
Reassortment creates different histories
Influenza B does not have pandemic potential
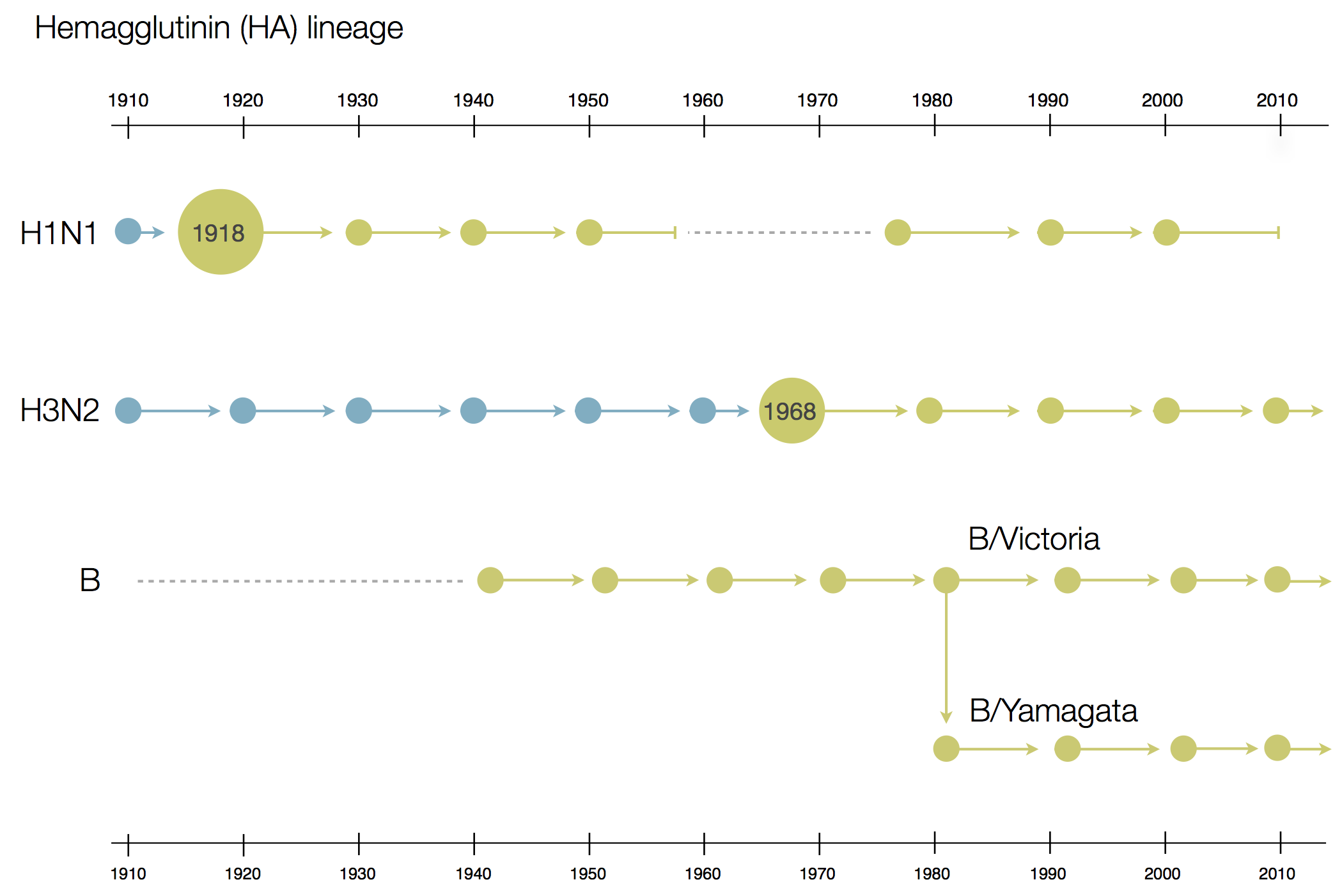
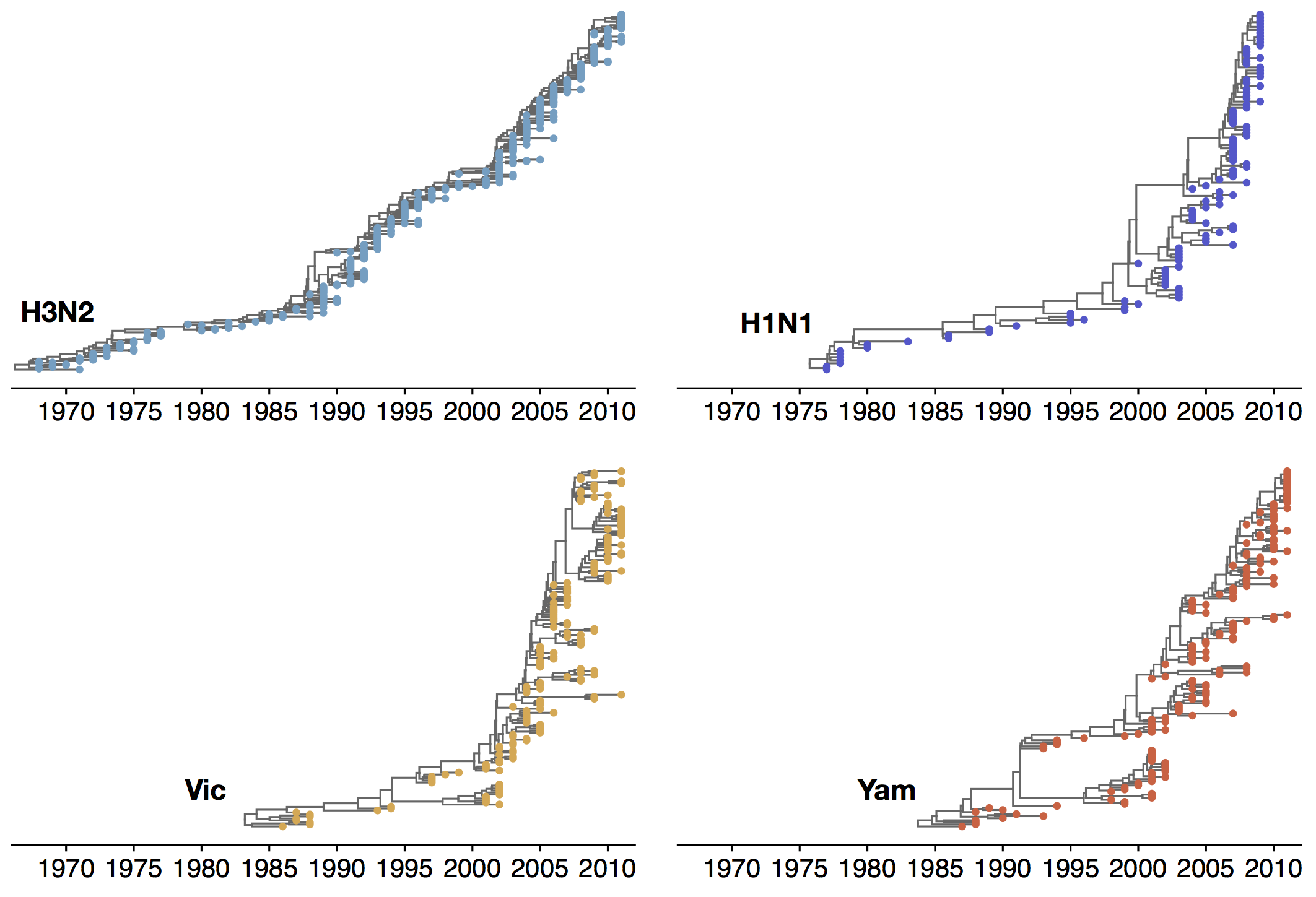
Phylogenetic trees of different influenza lineages
SARS-CoV-2
Genetic relationships of globally sampled SARS-CoV-2 to present
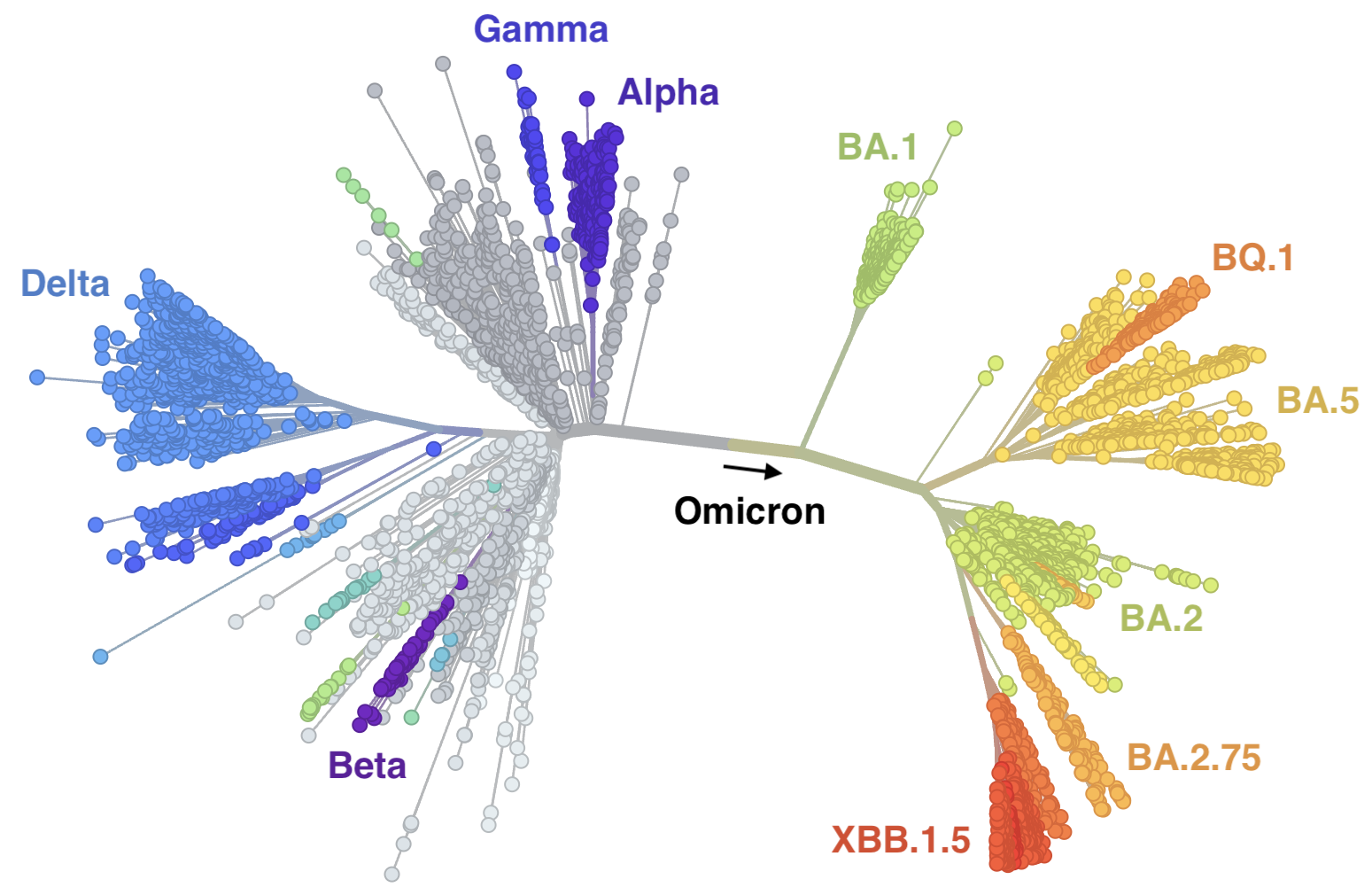
New variants rapidly replace existing diversity
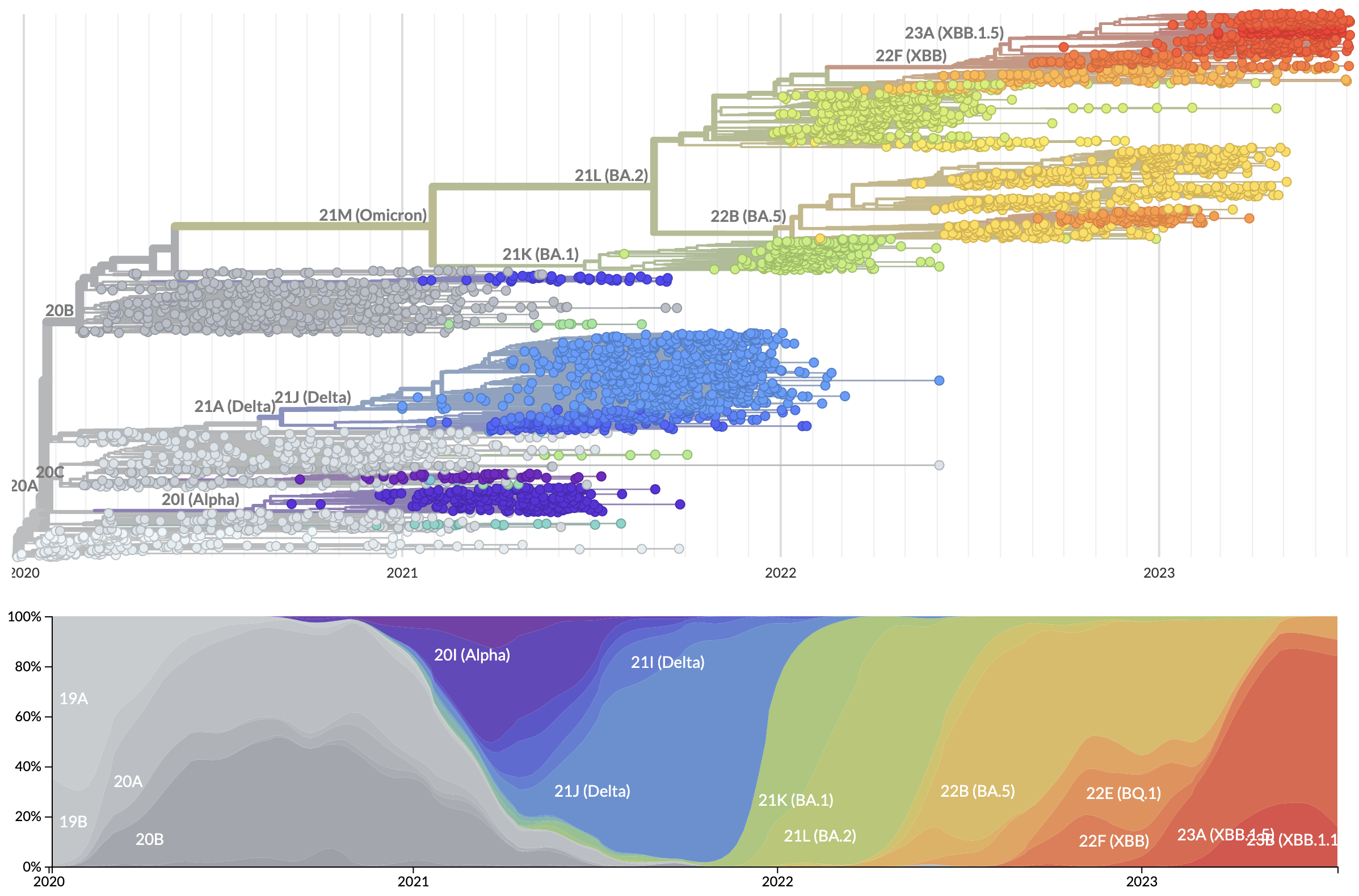
S1 evolution remarkably fast relative to seasonal influenza