Selection and evolution in B cells
Most slides from Sarah Cobey ; some from Erick
B cells determine antibody diversity
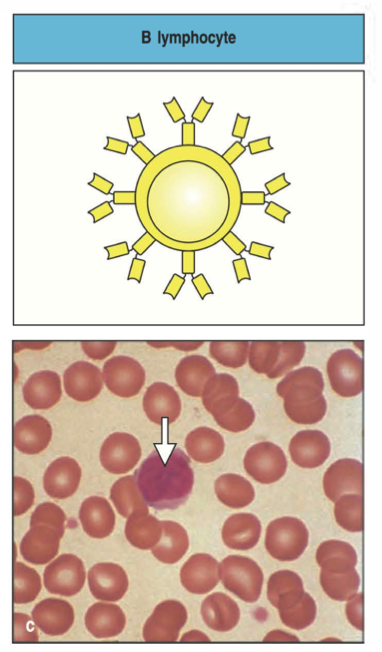
Structure of a B cell receptor
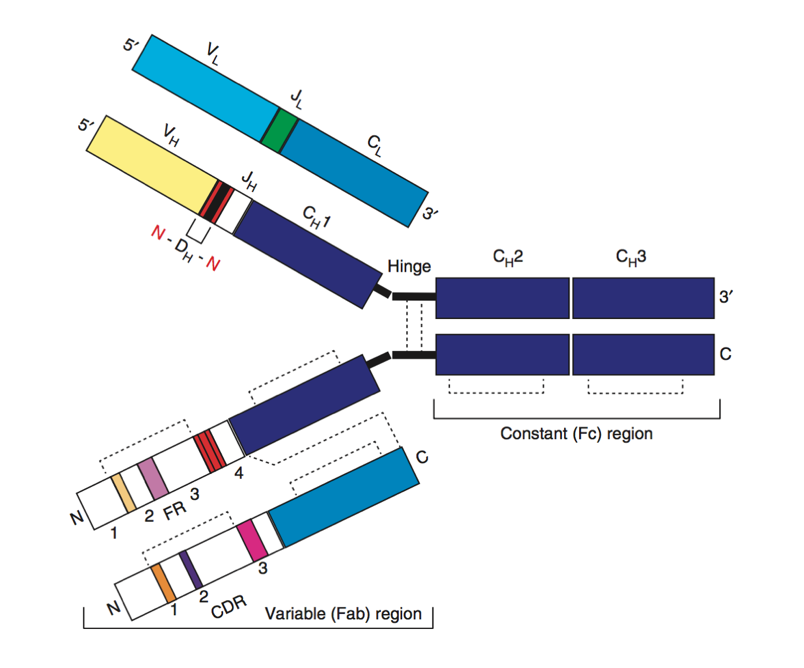
Origins of B cell receptor diversity
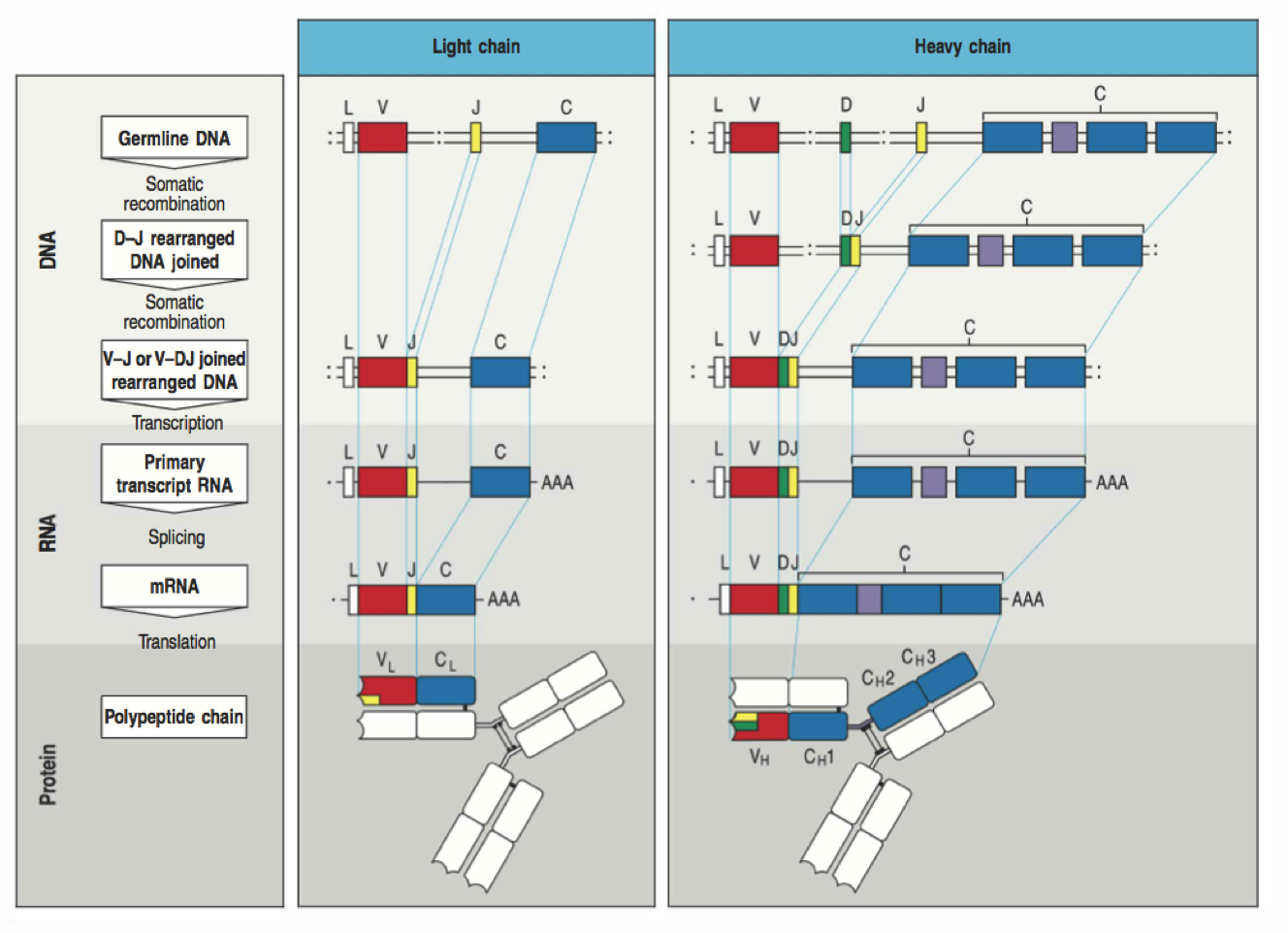
VDJ recombination
Affinity maturation
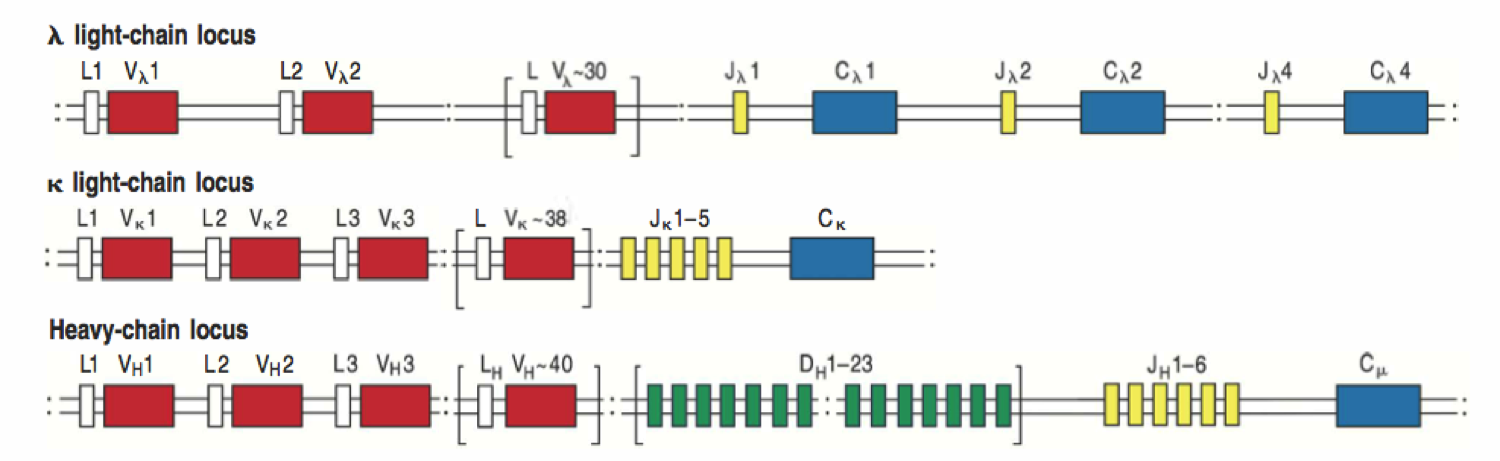
VDJ loci
VDJ recombination
Selection on naive repertoire
Stability
Autoreactivity
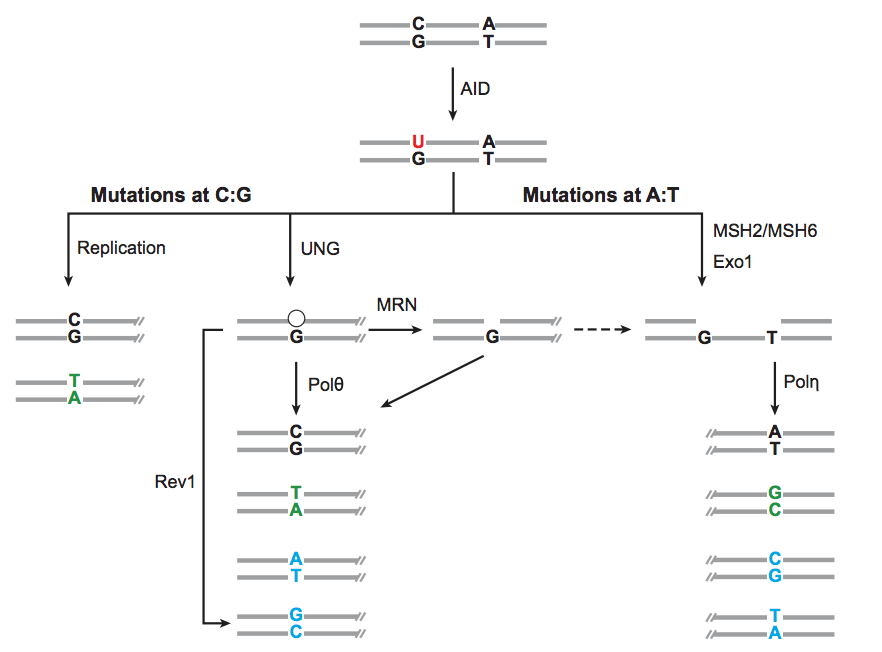
Somatic hypermutation is AID + repair
Mutation rate varies over BCR
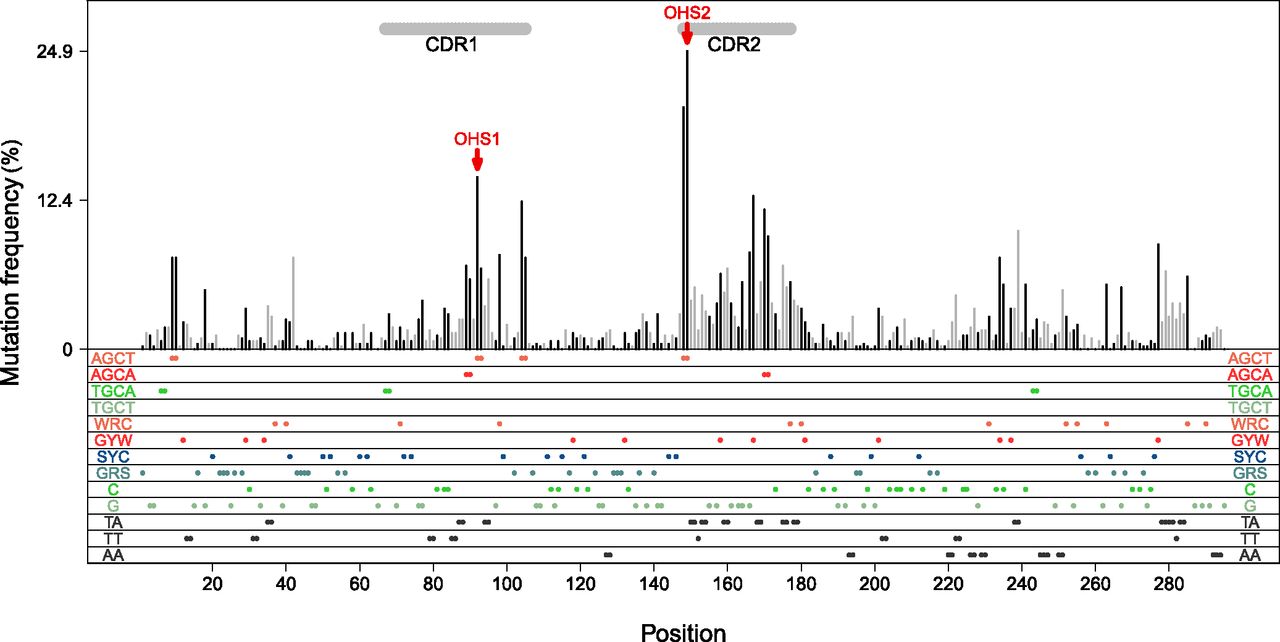
The mutations have peculiar biases
The mutations have peculiar biases
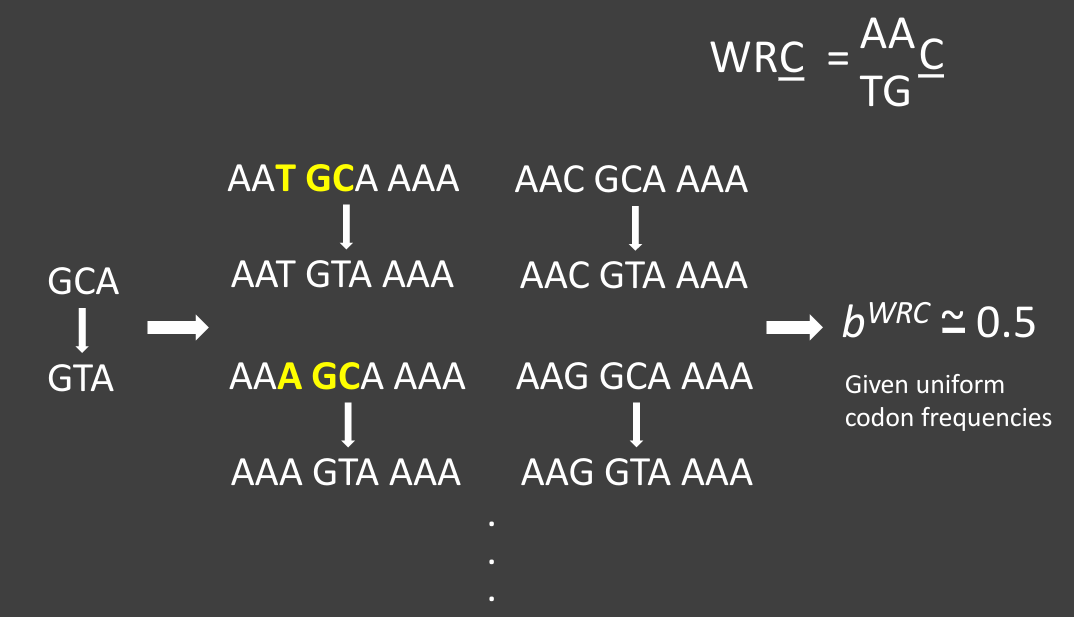
One would like to quantify these biases
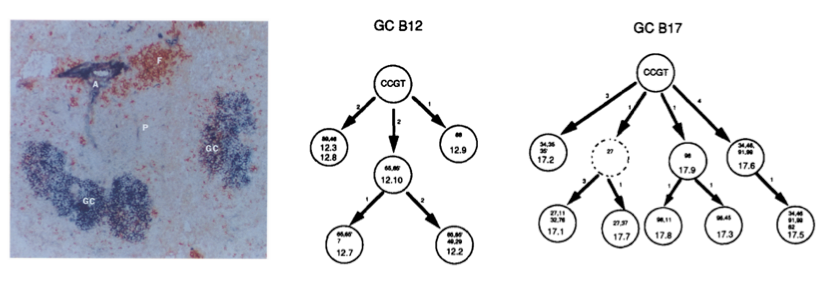
Phylogenetics is not straightforward
GC B cells evolve
Permissive selection vs complex antigens
Memory cells can affinity mature
Antibody isotypes
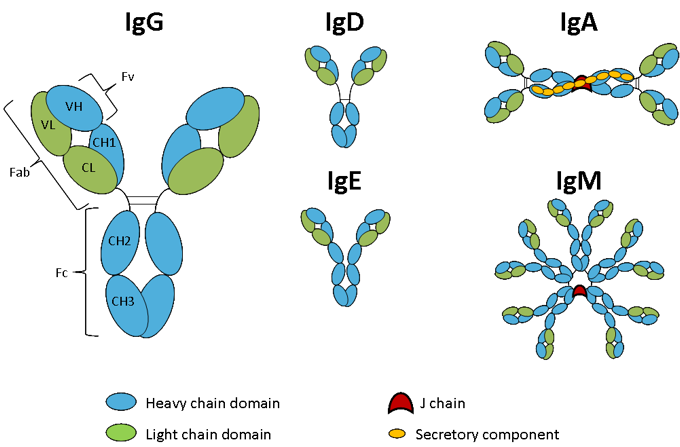
Isotype switching

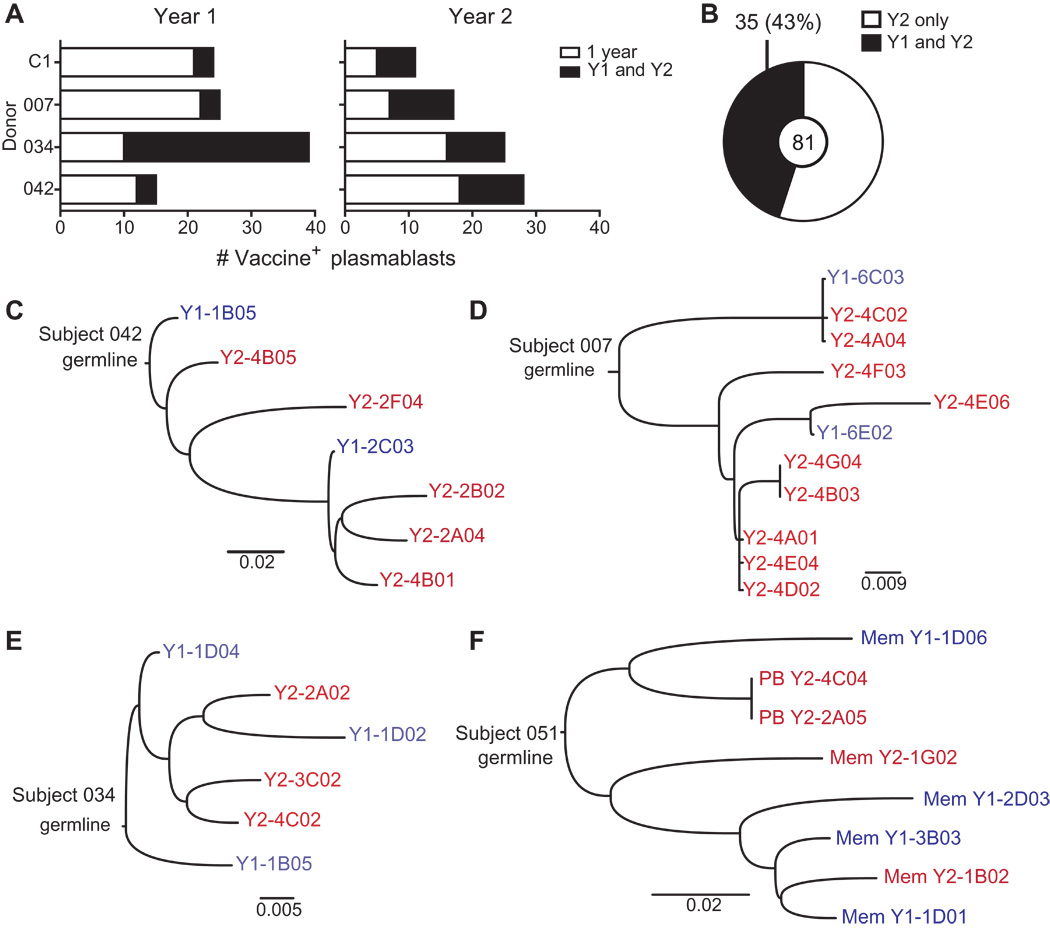
Clones can be long-lived
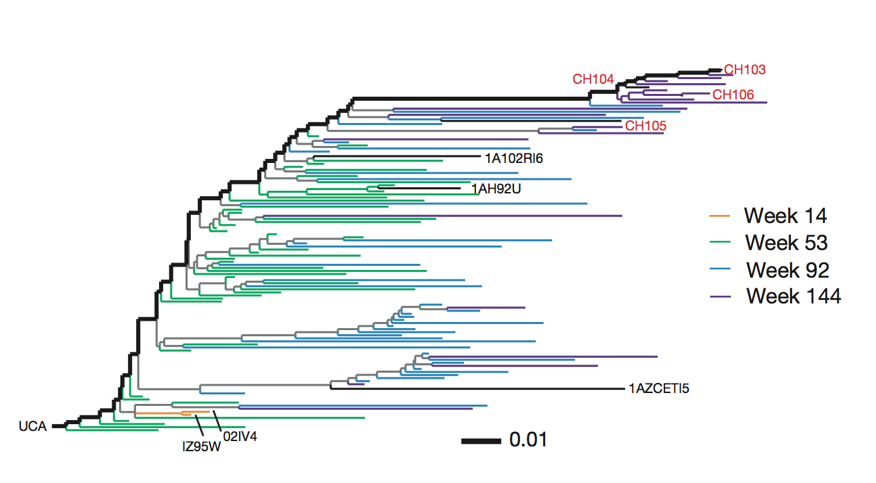
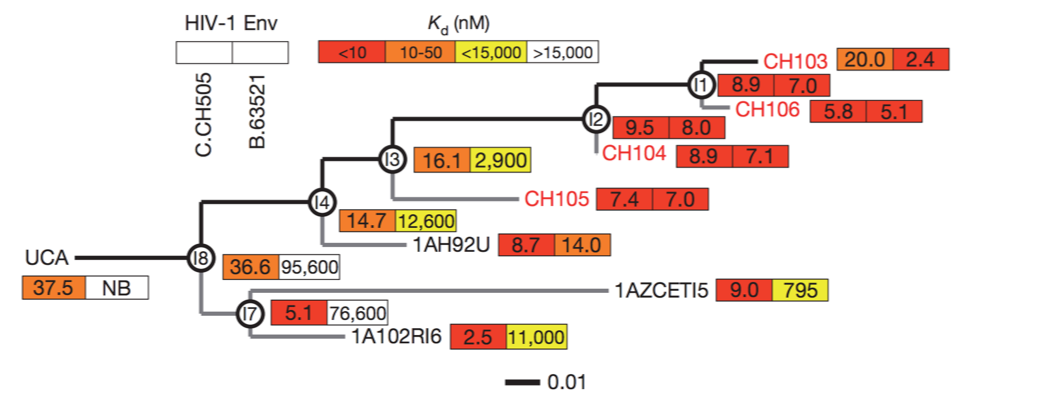
Broadly neutralizing antibodies are often highly diverged
Broadly neutralizing antibodies to flu elusive
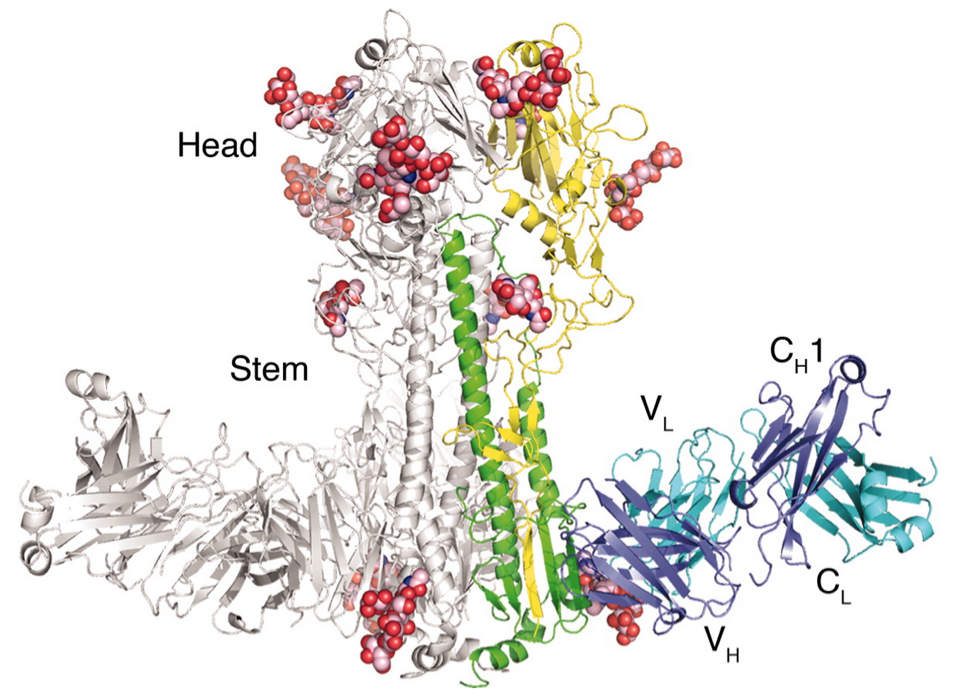
Why do some people develop broadly neutralizing antibodies?
Can we induce them in everyone?
Will they dominate?
"Restricted" responses to epitopes
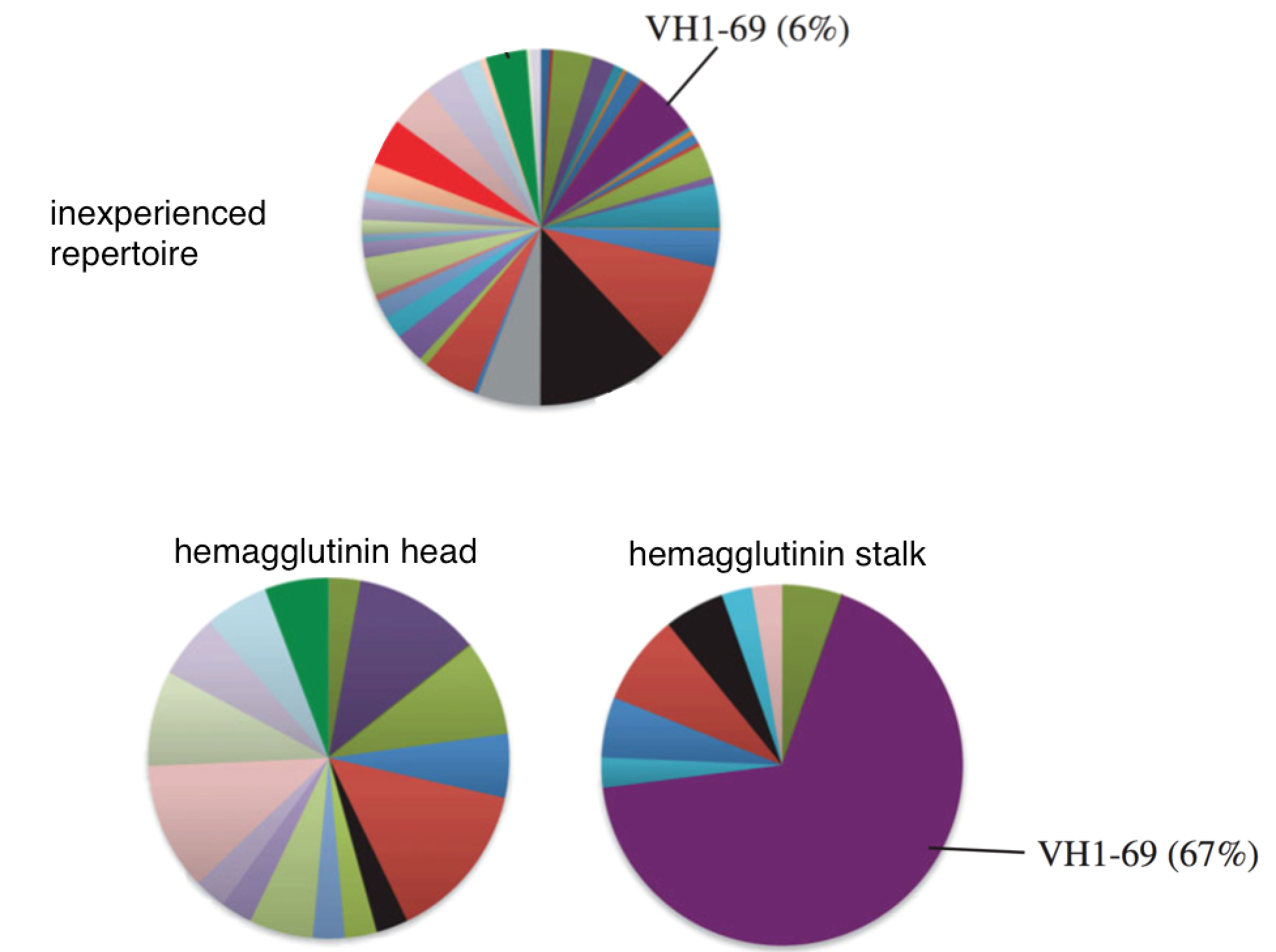
Genotype affects response to H5N1 vaccination
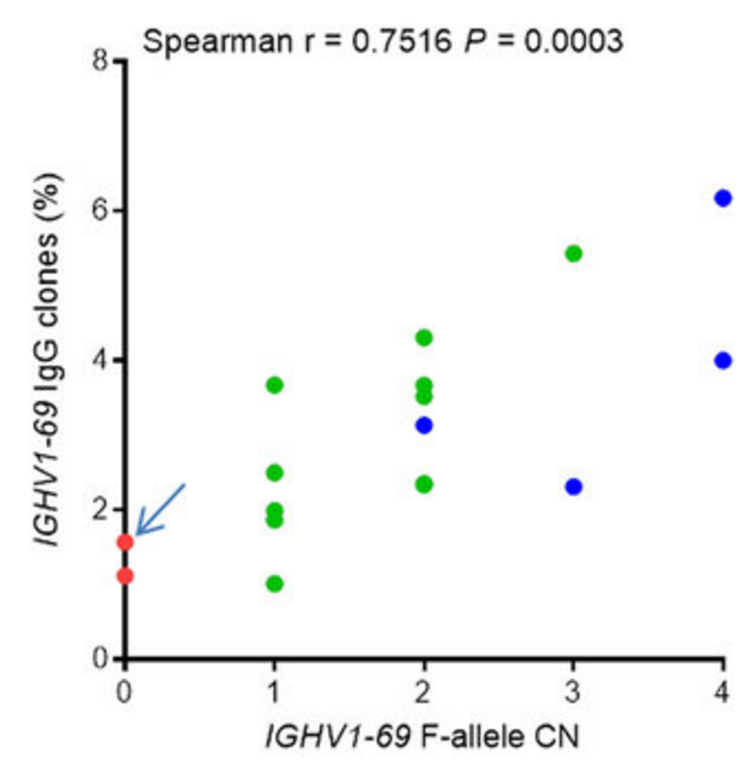
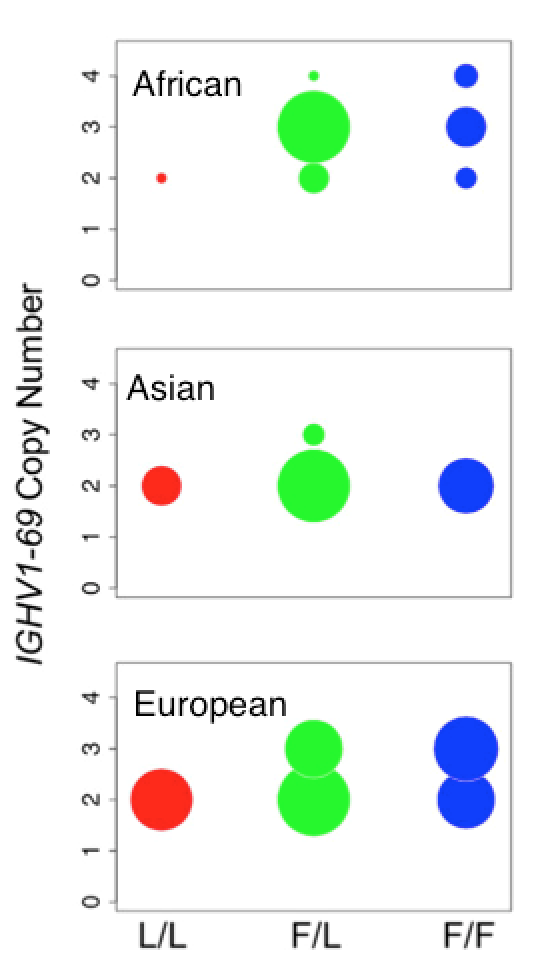
Allele copy number affects clone frequency
Ethnic differences in copy number
How can we learn using probabilistic models?
we only need models and one principle: likelihood.
We can sequence BCRs in high throughput
How do we learn from BCR sequences?
- Develop probabilistic model of biological process and how data is generated.
- Find parameter choices that maximize the likelihood of generating the observed data.
When a field of bioinformatics is mature, it becomes statistics.
E.g. Maximum-likelihood phylogenetics, HMMER, DESeq2, etc.
VDJ rearrangement
HMM intro: dishonest casino
HMM intro: dishonest casino
HMM intro: dishonest casino
HMM intro: dishonest casino
- What is the likelihood of the data under this path (write equation)
- If \(p\) is close to 0.5, what is an alternate path that will have a higher likelihood? What is this likelihood?
Biased die roll \(\leftrightarrow\) read base (under mutation)
Switching dice \(\leftrightarrow\) changing between V, D, J genes
VDJ annotation problem: from where did each nucleotide come?
A: Take maximum-likelihood HMM path.
What are probabilities?
Distributions are reproducibly weird!
Distributions are reproducibly weird!
Distributions are reproducibly weird!
Find clonal families
Find clonal families
Did two sequences come from the same VDJ recombination?
Say we are given two sequences
Double roll \(\leftrightarrow\) Pair HMM
Double roll \(\leftrightarrow\) Pair HMM
Pick double roll hypothesis if it has higher likelihood of generating data.
What about BCRs?
Do two sequences come from a single rearrangement event?
Probability of generating observed sequence \(x\) from HMM:
\[ \mathbb P(x) = \sum_{\text{paths}\ \sigma} \mathbb P(x;\sigma), \]
Probability of generating two sequences \(x\) and \(y\) from the same path through the HMM (i.e. from the same rearrangement event):
\[ \mathbb P(x,y) = \sum_{\text{paths}\ \sigma} \mathbb P(x,y;\sigma), \]
\[ \text{Calculate: } \frac{\mathbb P(x, y)}{\mathbb P(x) \mathbb P(y)} = \frac{\mathbb P(\text{single rearrangement})}{\mathbb P(\text{independent rearrangements})} \]
Do sets of sequences come from a
single rearrangement event?
\[ \frac{\mathbb P(A \cup B)}{\mathbb P(A) \mathbb P(B)} = \frac{\mathbb P(A \cup B \ | \ \text{single rearrangement})}{\mathbb P(A,B \ | \ \text{independent rearrangements})} \]
Use this for agglomerative clustering; stop when the ratio < 1.
- Munshaw, S., & Kepler, T. B. (2010). SoDA2: a Hidden Markov Model approach for identification of immunoglobulin rearrangements. Bioinformatics.
- Murugan, Mora, Walczak, & Callan (2012). Statistical inference of the generation probability of T-cell receptors from sequence repertoires. PNAS.
- Ralph & M. (2016). Consistency of VDJ Rearrangement and Substitution Parameters Enables Accurate B Cell Receptor Sequence Annotation. PLOS Computational Biology.
- Ralph & M. (2016). Likelihood-based inference of B-cell clonal families.PLOS Computational Biology.
- Elhanati, Sethna, Marcou, Callan, Mora, \& Walczak (2015). Inferring processes underlying B-cell repertoire diversity. Philosophical Transactions of the Royal Society of London.
- Elhanati, Marcou, Mora, & Walczak (2016). repgenHMM: a dynamic programming tool to infer the rules of immune receptor generation from sequence data. Bioinformatics.