Tracking and forecasting SARS-CoV-2 variant spread
Trevor Bedford (@trvrb)
Fred Hutchinson Cancer Center / Howard Hughes Medical Institute
30 Sep 2022
HB Friday Faculty Talk
Fred Hutch
1. SARS-CoV-2 evolution
2. Variant frequency dynamics
3. Emerging variants
4. Seasonality and near-term circulation
5. Continued work on forecasting
SARS-CoV-2 evolution
Rapid displacement of existing diversity by emerging variants

S1 evolved at a rate of 13 amino acid changes per year since pandemic start

Continued rapid accumulation from BA.2 onwards with 7 amino acid changes per year

S1 evolution remarkably fast relative to seasonal influenza
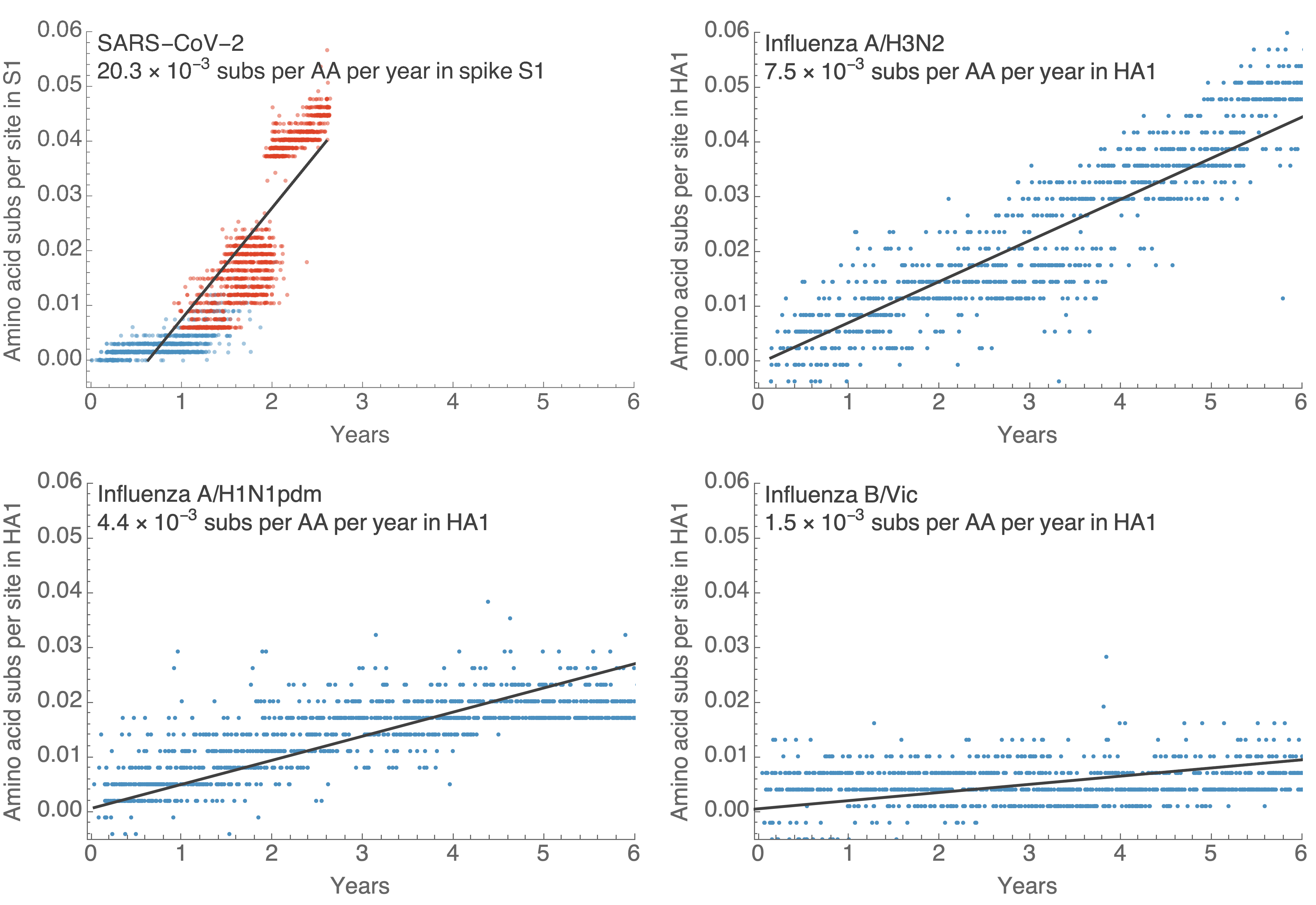
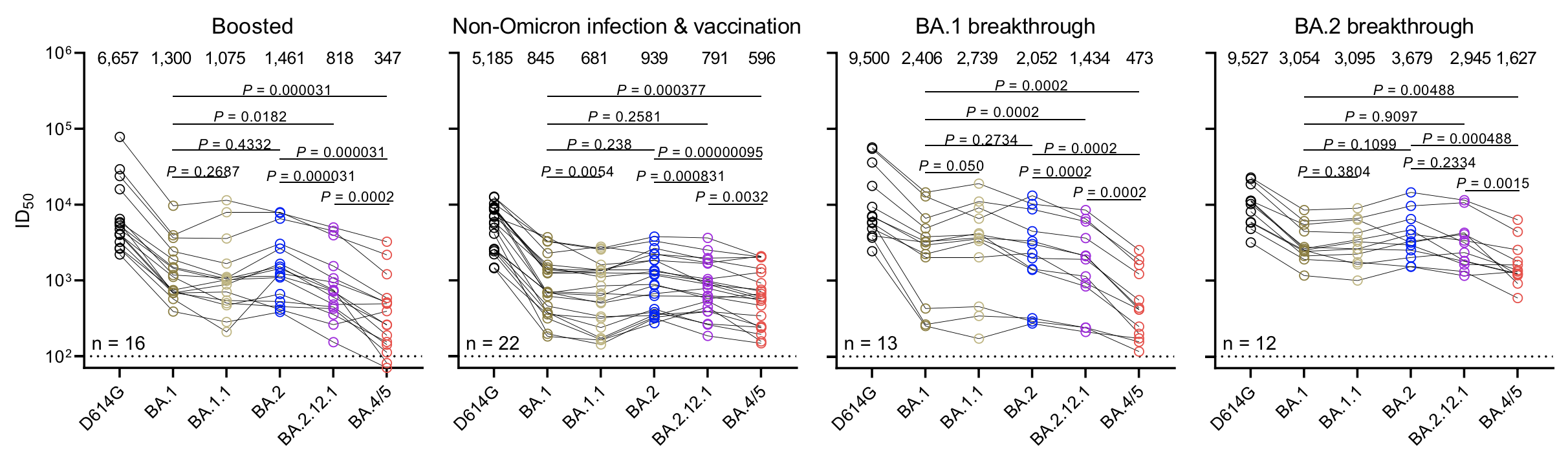
Continued escape from neutralization by existing population immunity
Variant frequency dynamics
Population genetic expectation of variant frequency under selection
$x' = \frac{x \, (1+s)}{x \, (1+s) + (1-x)}$ for frequency $x$ over one generation with selective advantage $s$
$x(t) = \frac{x_0 \, (1+s)^t}{x_0 \, (1+s)^t + (1-x_0)}$ for initial frequency $x_0$ over $t$ generations
Trajectories are linear once logit transformed via $\mathrm{log}(\frac{x}{1 - x})$
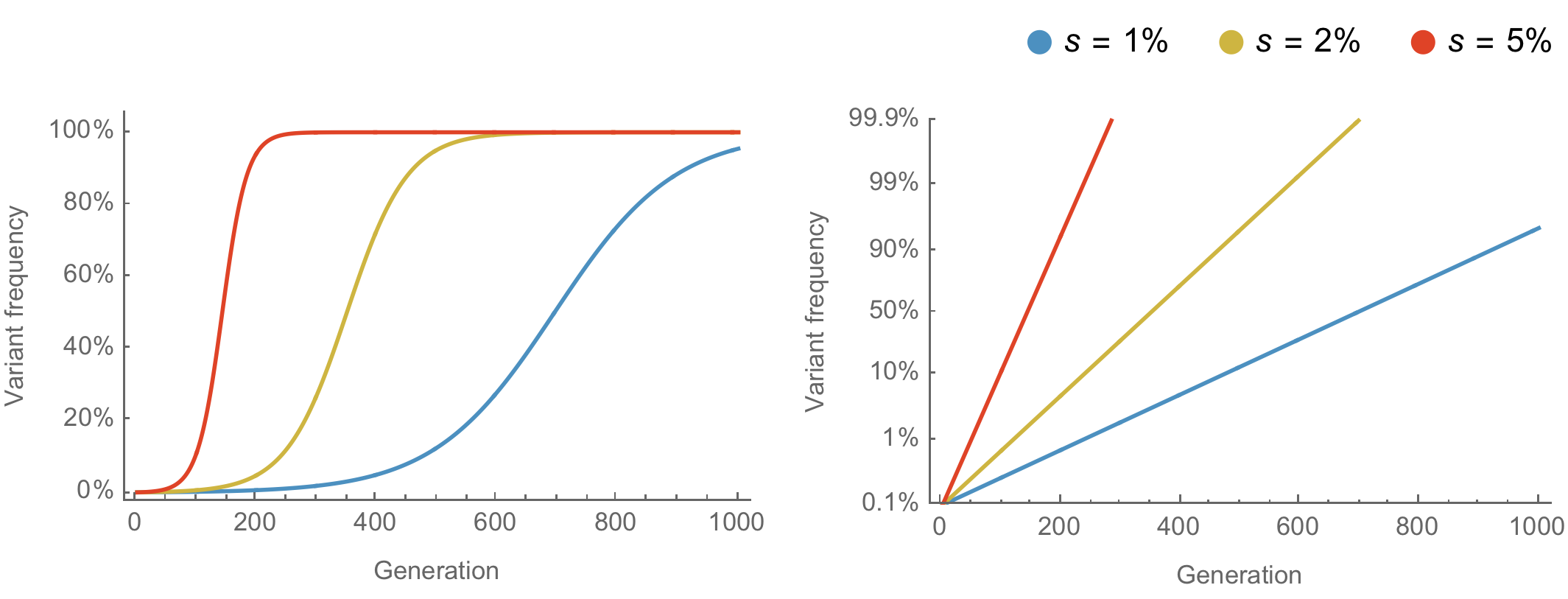
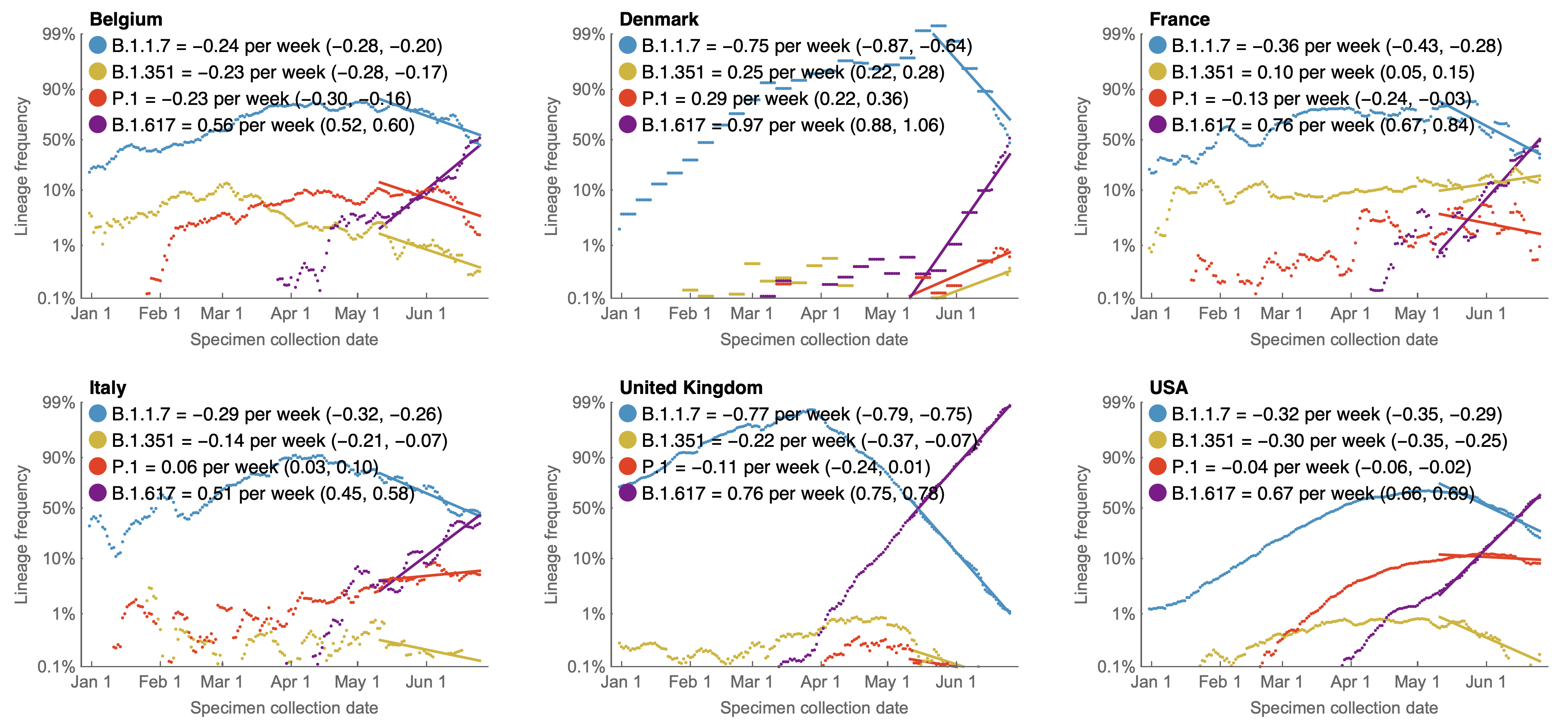
Variants show consistent frequency dynamics in logit space
Variants show consistent frequency dynamics in logit space
Multinomial logistic regression
Multinomial logistic regression models the probability of a virus sampled at time $t$ belonging to variant $i$ as
$$\mathrm{Pr}(X = i) = x_i(t) = \frac{p_i \, \mathrm{exp}(f_i \, t)}{\sum_{1 \le j \le n} p_j \, \mathrm{exp}(f_j \, t) }$$
where the model has $2n$ parameters consisting of $p_i$ the frequency of variant $i$ at initial timepoint and $f_i$ the growth rate or fitness of variant $i$ for $n$ variants.
The model is fit to minimize "log loss" of predicted variant vs observed variant across observations in dataset.
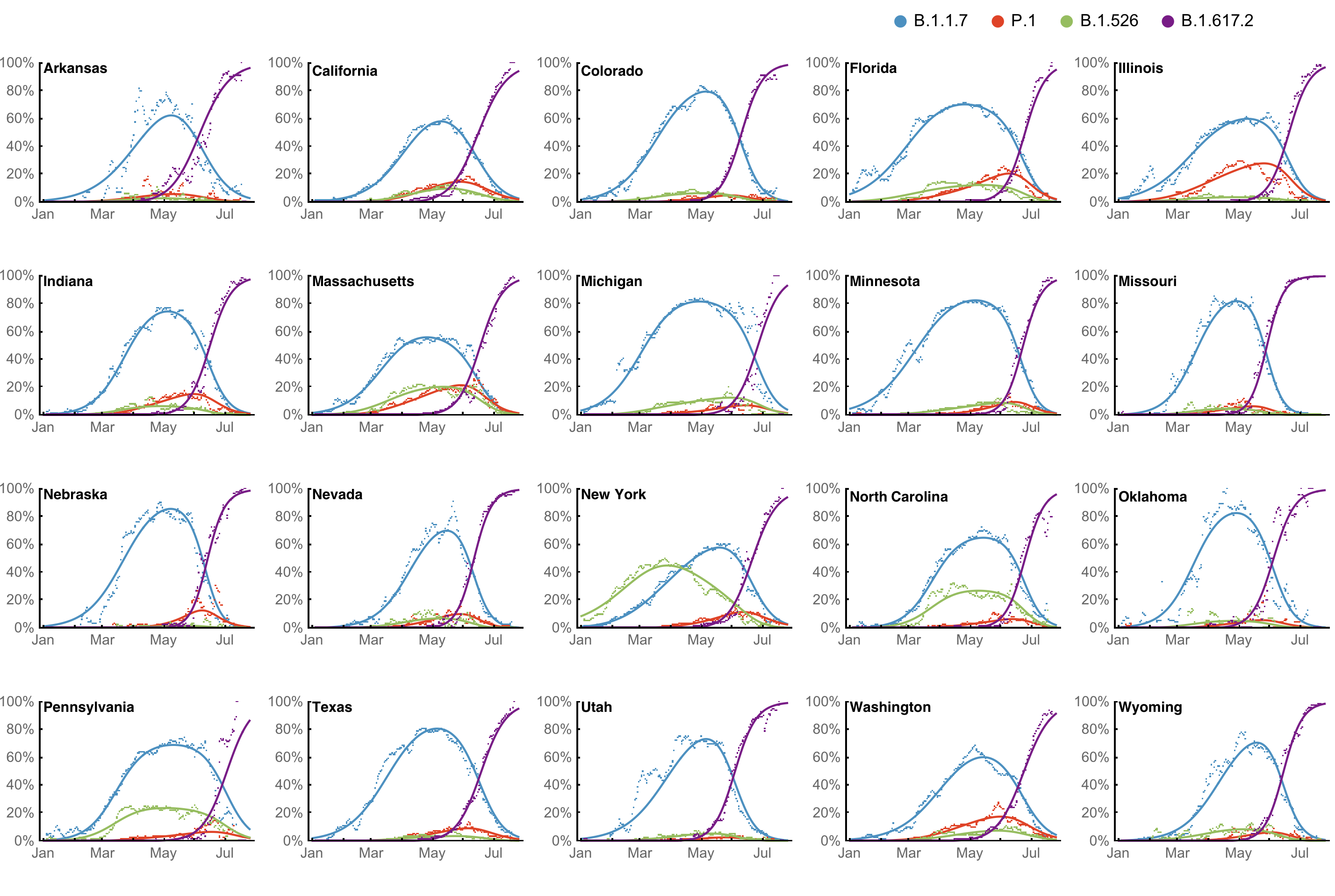
Multinomial logistic regression fits variant frequencies well
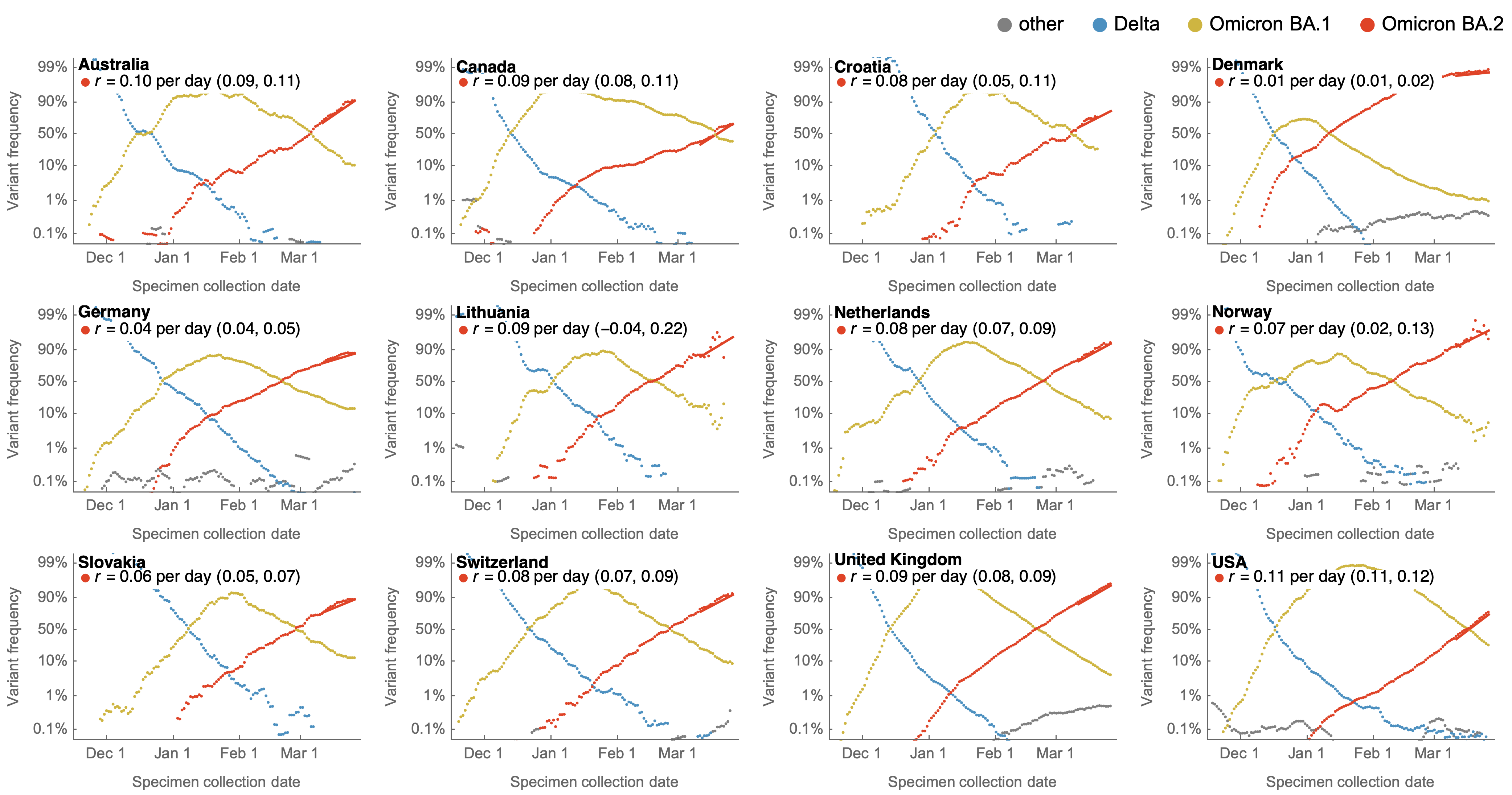
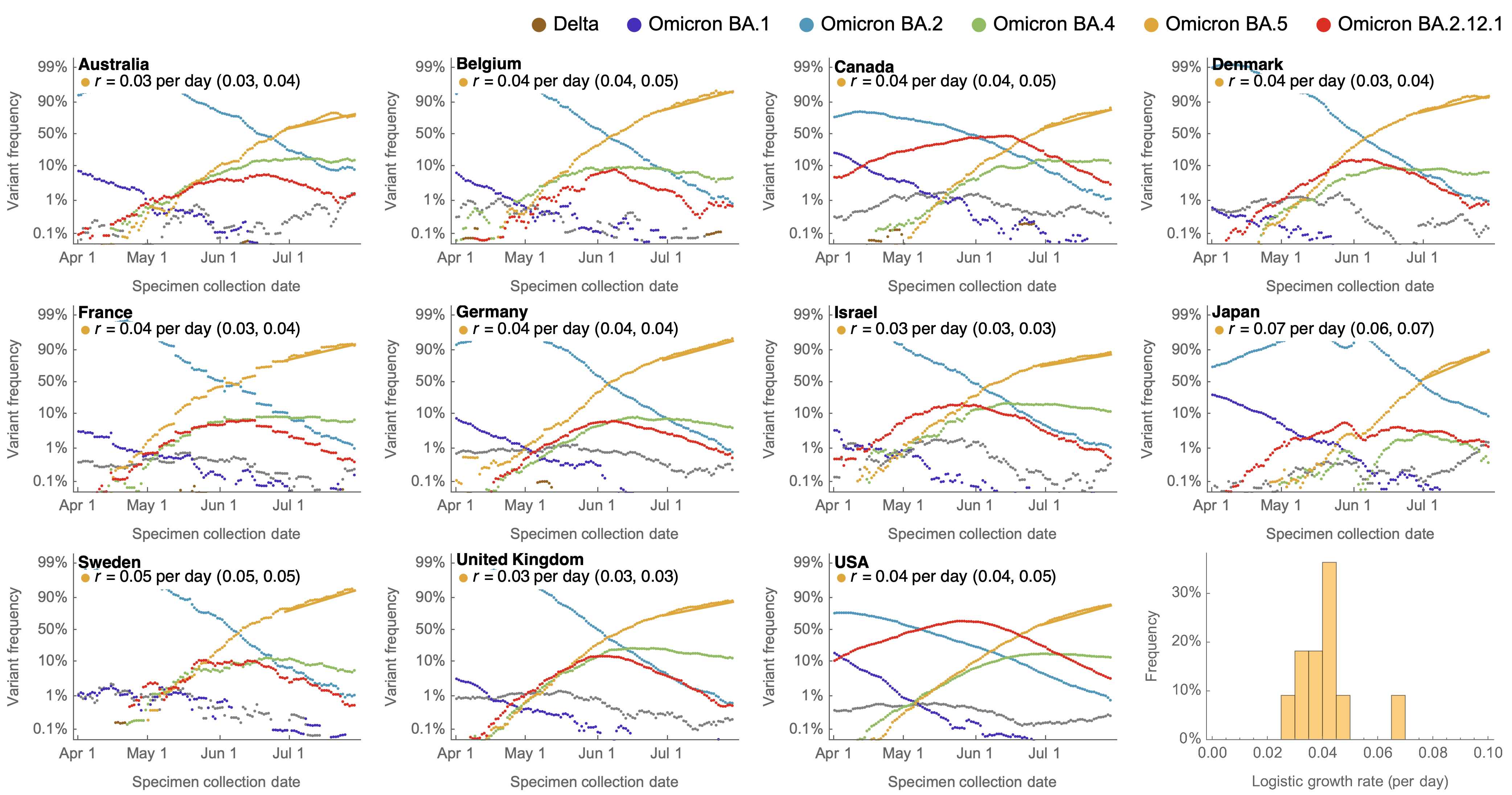
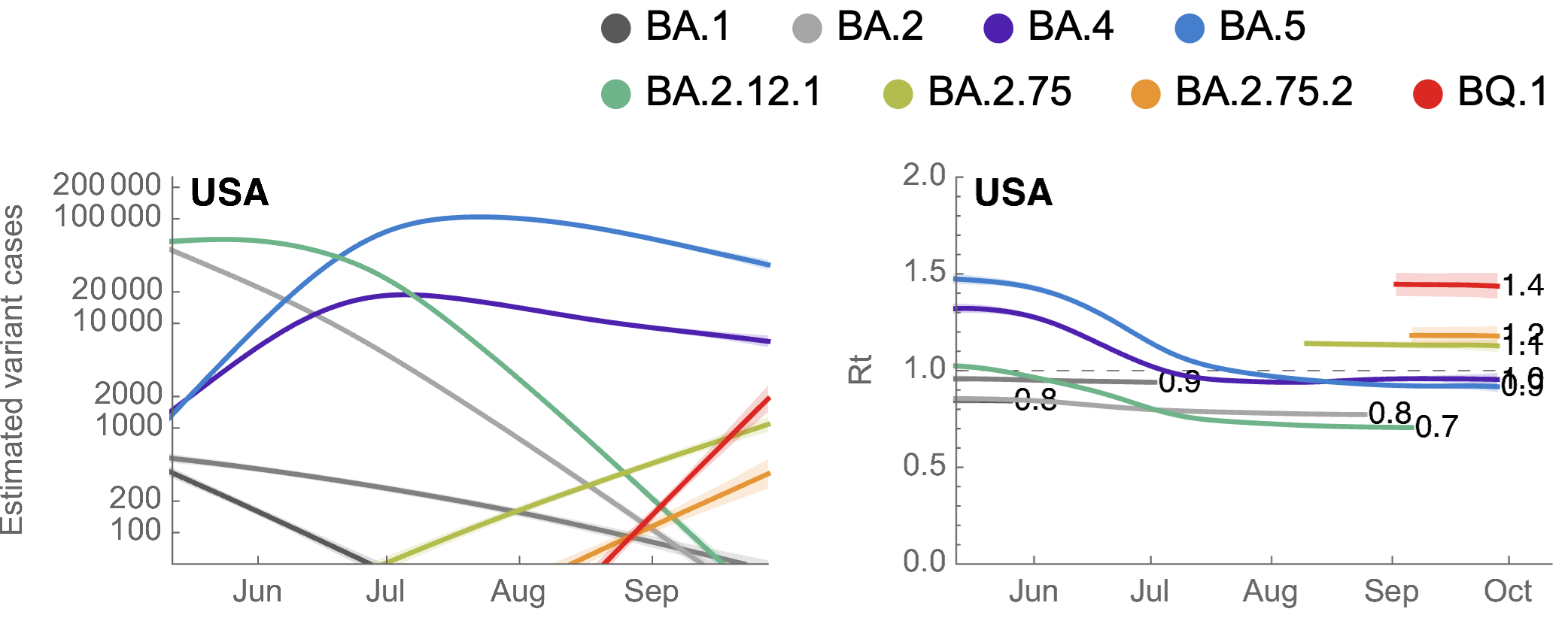
Consistent fitness advantage of BA.5 across countries
Despite similar rates of displacement, BA.5 epidemics vary
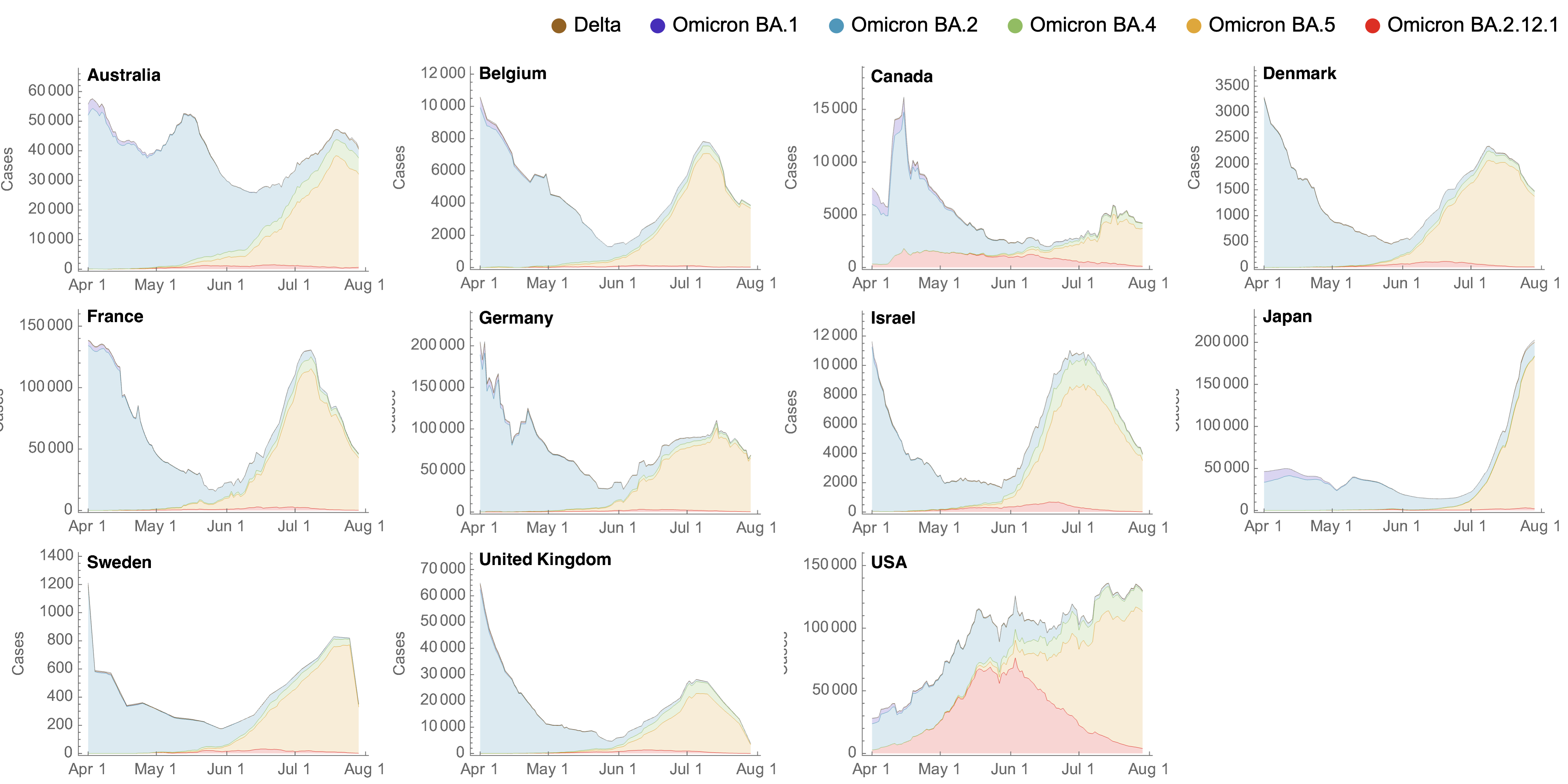
Despite similar rates of displacement, BA.5 epidemics vary
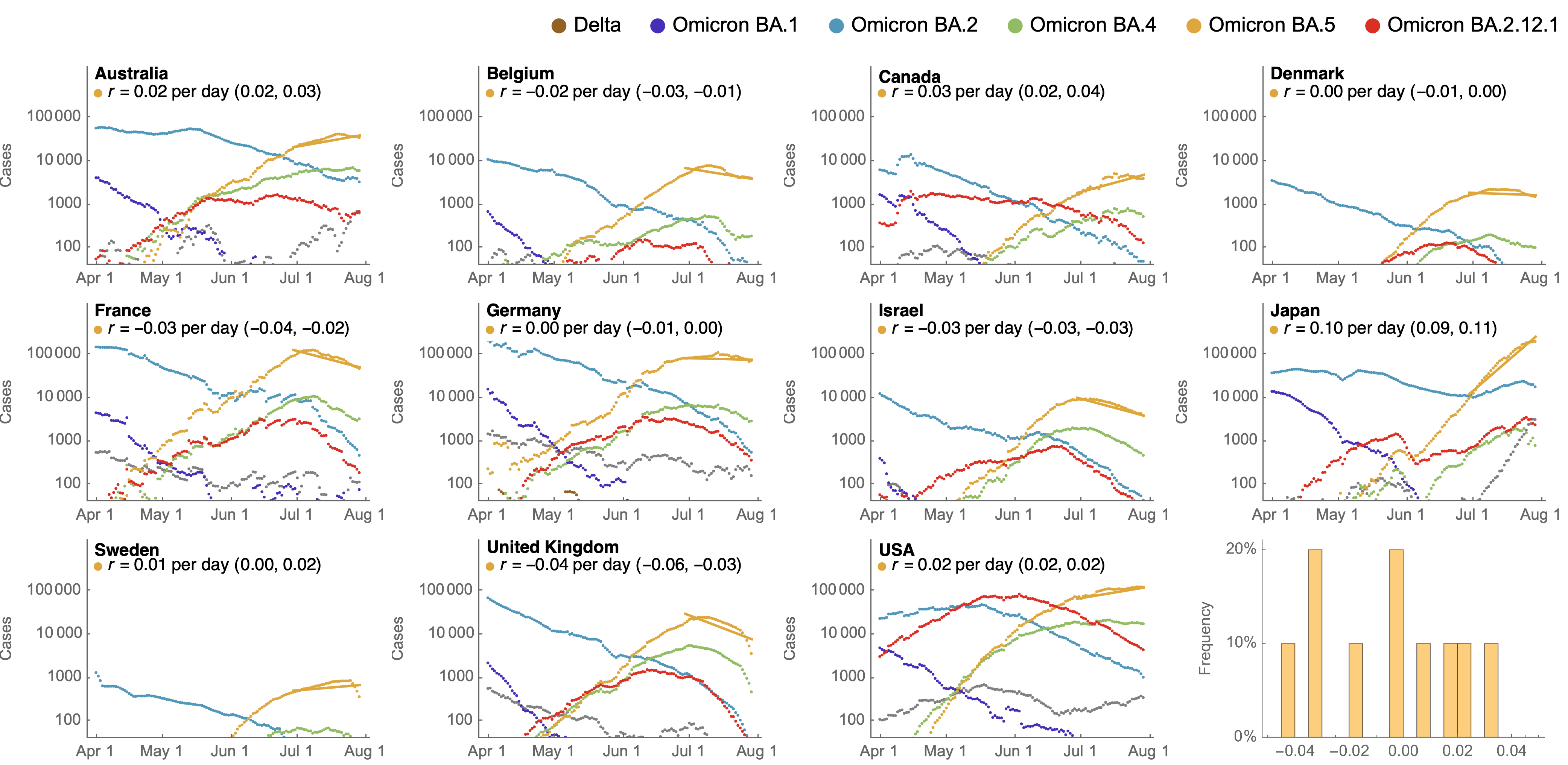
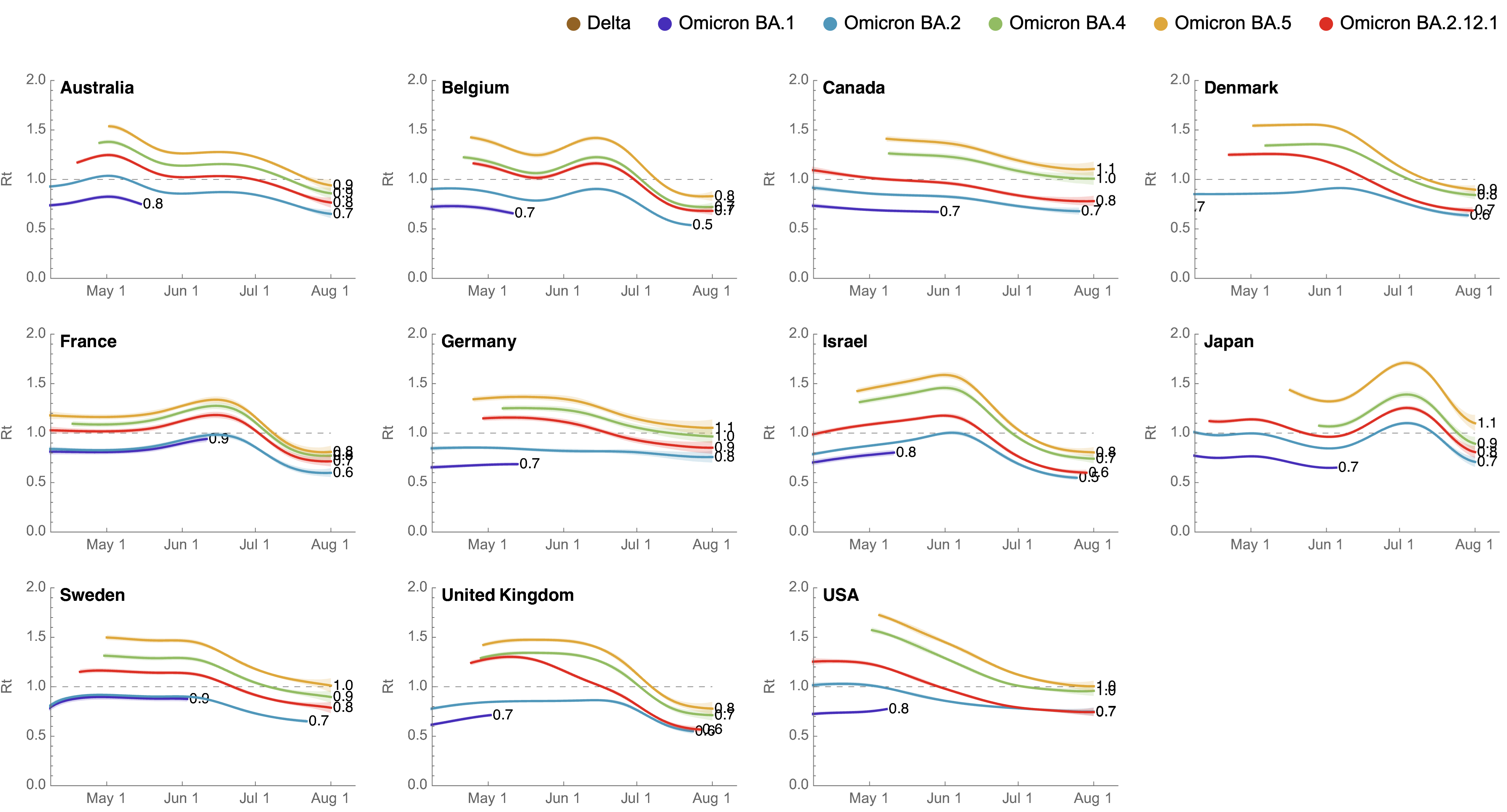
These differences can be explained by consistent growth advantage of BA.5, but different baseline Rt across countries
May 17: fitness advantage of BA.4/BA.5 clear even though rare
Multinomial logistic regression should work well for SARS-CoV-2 prediction, except emergence of new variants limits prediction horizon

Emerging variants
Over 350 Pango lineages designated in 2022
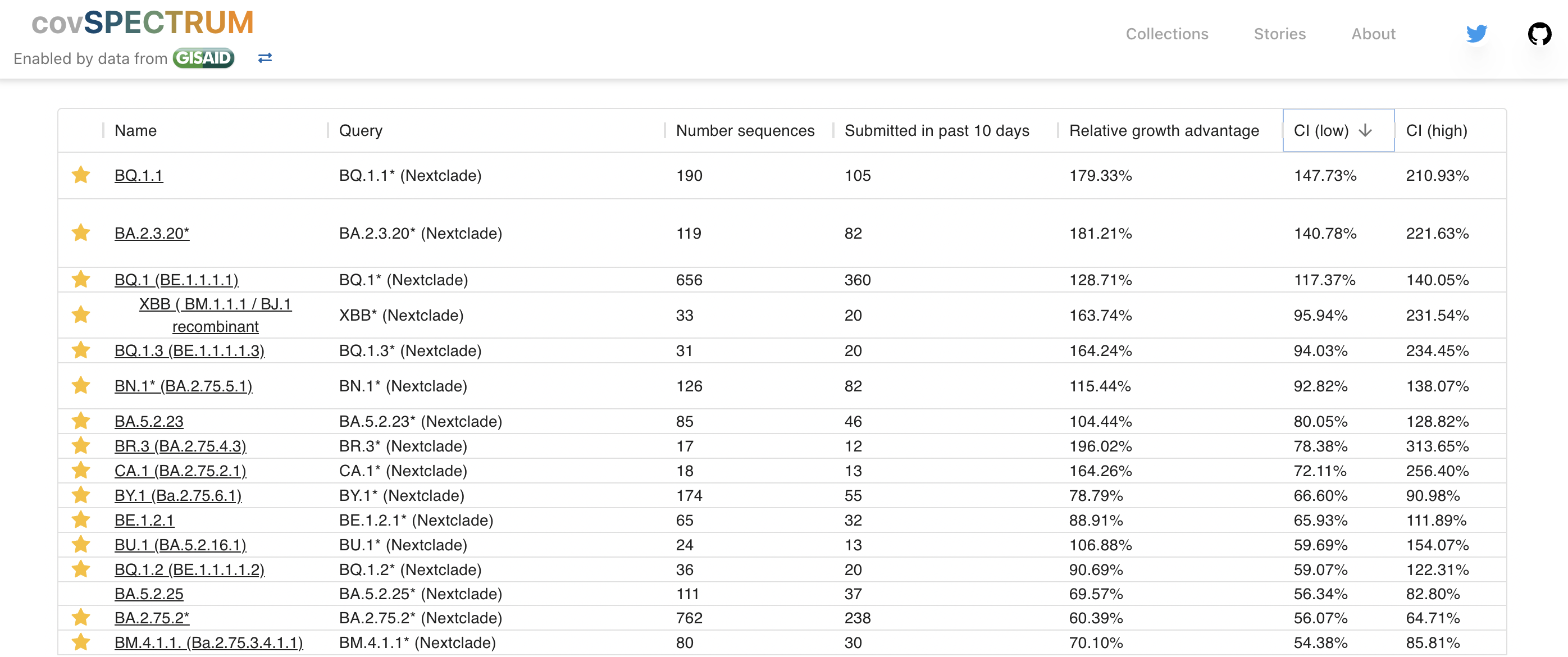
Emerging sublineages within BA.2.75 focusing on BA.2.75.2 and emerging lineages within BA.5 focusing on BA.5.3.1.1.1.1.1 ie BQ.1

BA.2.75 and sublineage BA.2.75.2 emerging from India

BA.2.75.2 and BQ.1 show selective advantage in UK and US
BA.2.75.2 has R346T and F486S on top of BA.2.75 and BQ.1 has K444T, N460K on top of BA.5
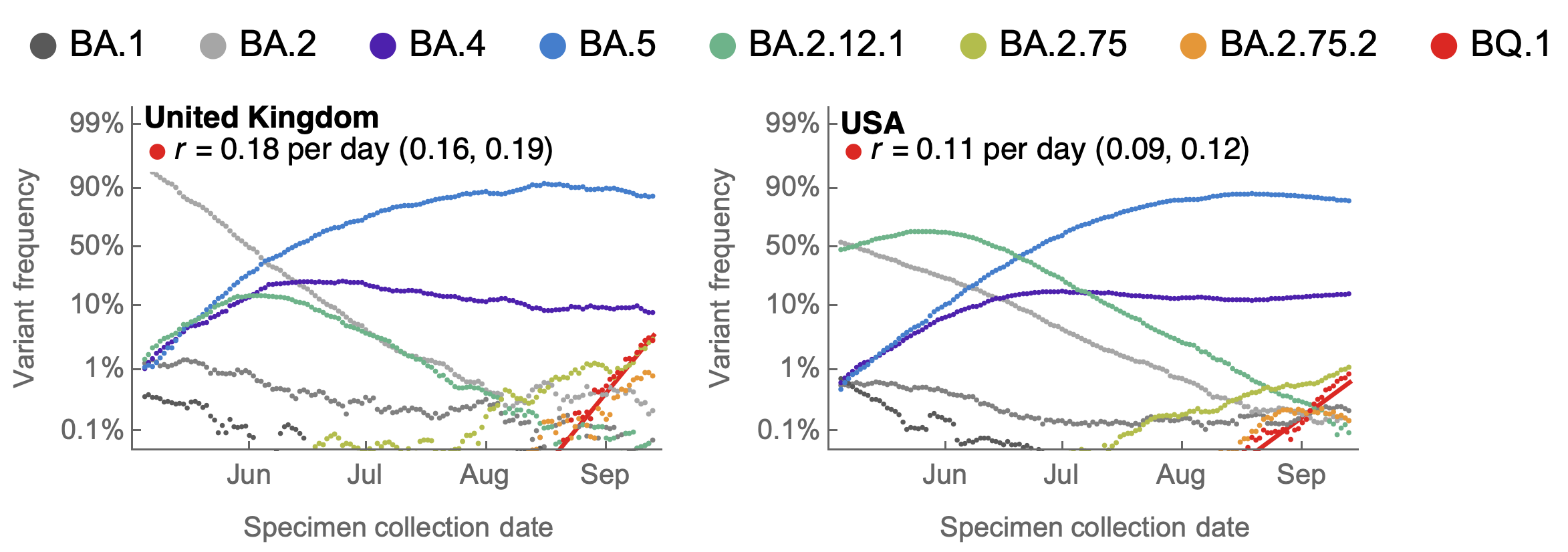
BQ.1 has Rt in the US of ~1.4, approaching that of BA.5 in May
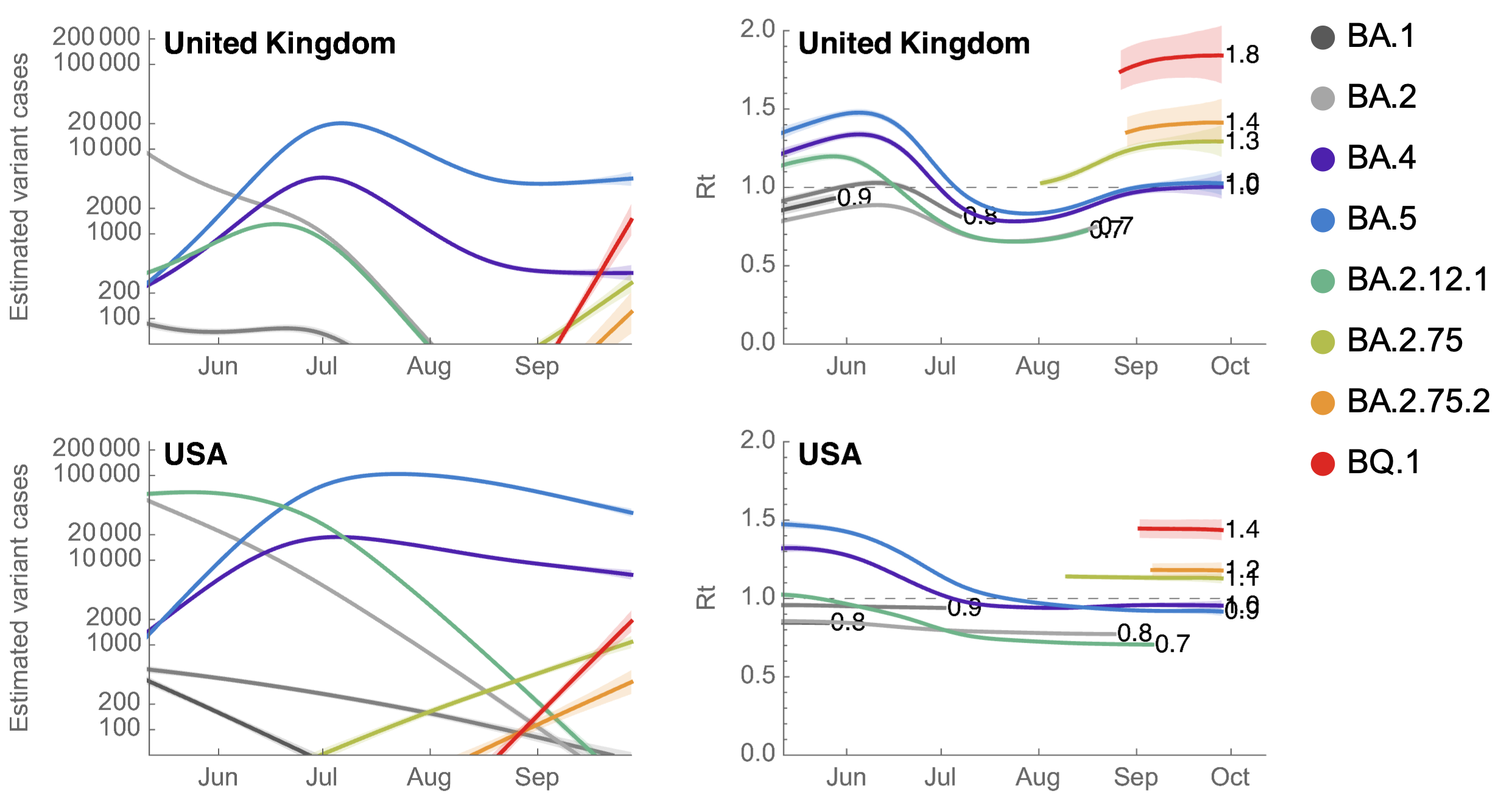
However, multiple low frequency Pango lineages that may compete with BA.2.75.2 and BQ.1
Seasonality and near-term circulation
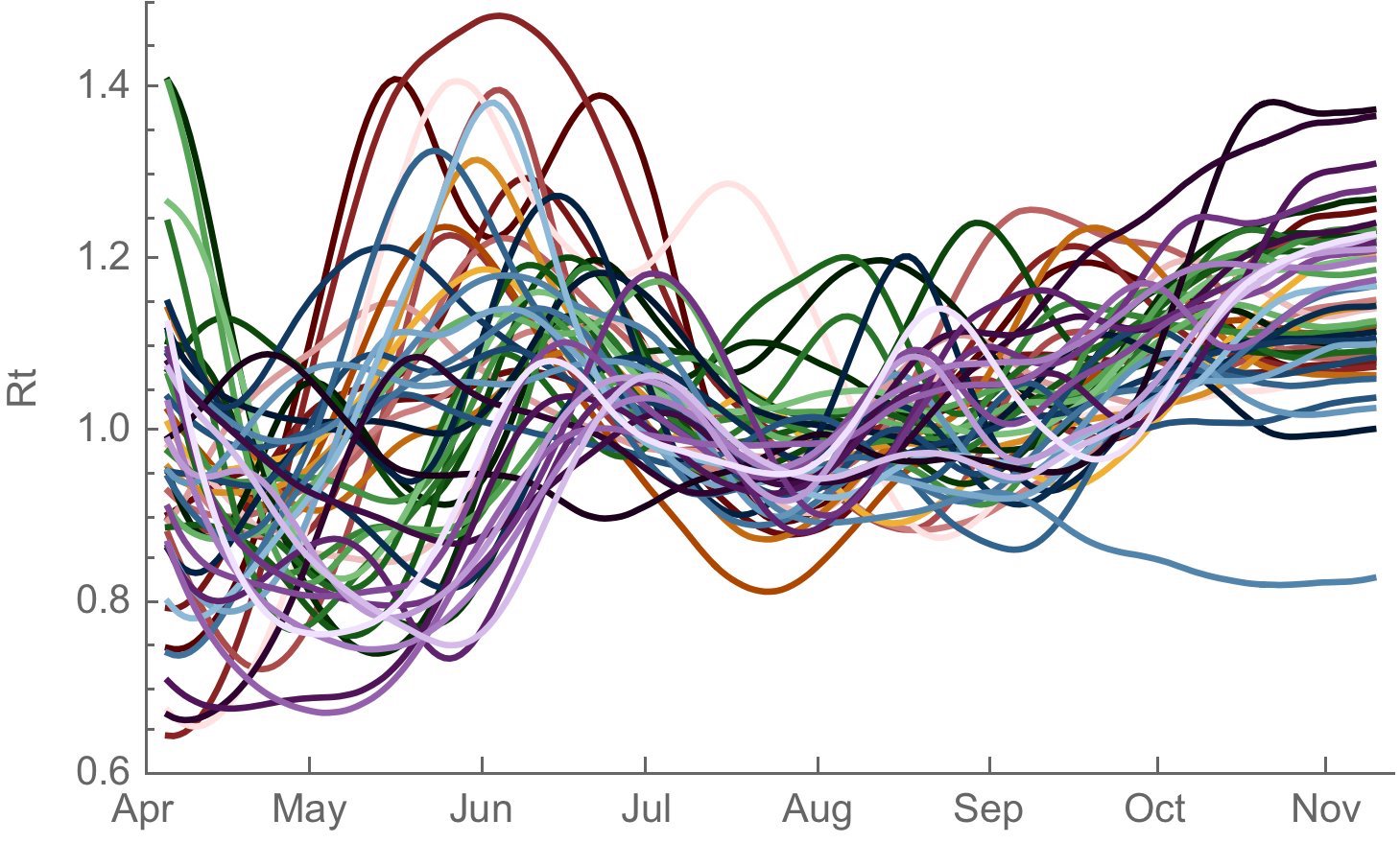
Seasonality clearly evidence in 2020-2021 winter epidemic
Rt analysis suggests a ~30% transmission advantage going from Aug 2020 to Nov 2020
Seasonality alone unlikely to drive BA.5 epidemic, but seasonality + novel variants will likely drive epidemics
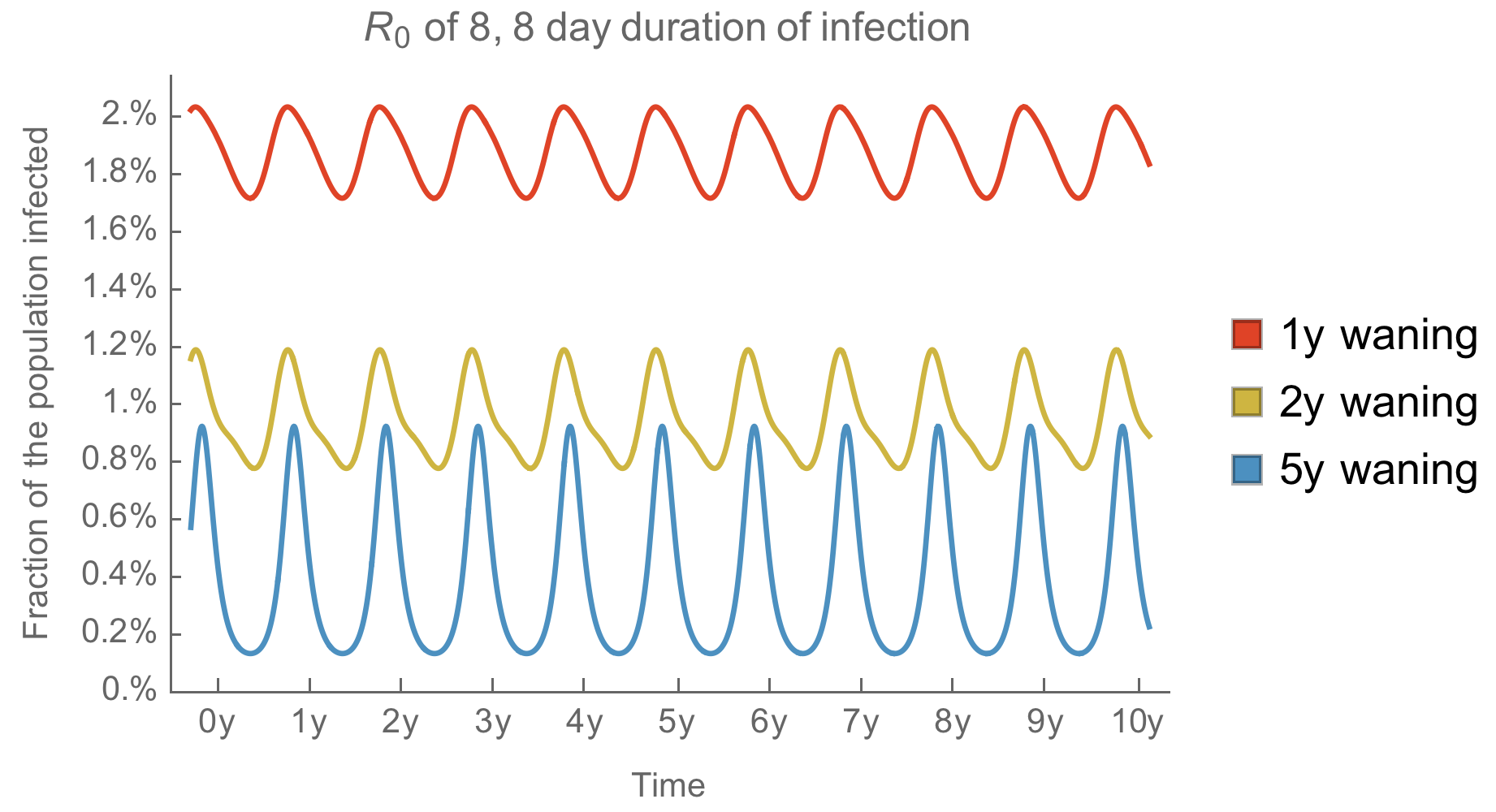
Generally, we expect faster antigenic drift to smooth summer vs winter circulation
Heading into fall, we very roughly have:
- 40% of the population with BA.2, BA.4, BA.5 in Mar 1 to Sep 15 (16.3M confirmed cases and 8x under-reporting)
- 40% of the population with BA.1 infection in Jan/Feb (24.1M confirmed cases and 5x under-reporting)
- 20% of the population vaccinated, boosted or with pre-Omicron infection
Continued work on forecasting
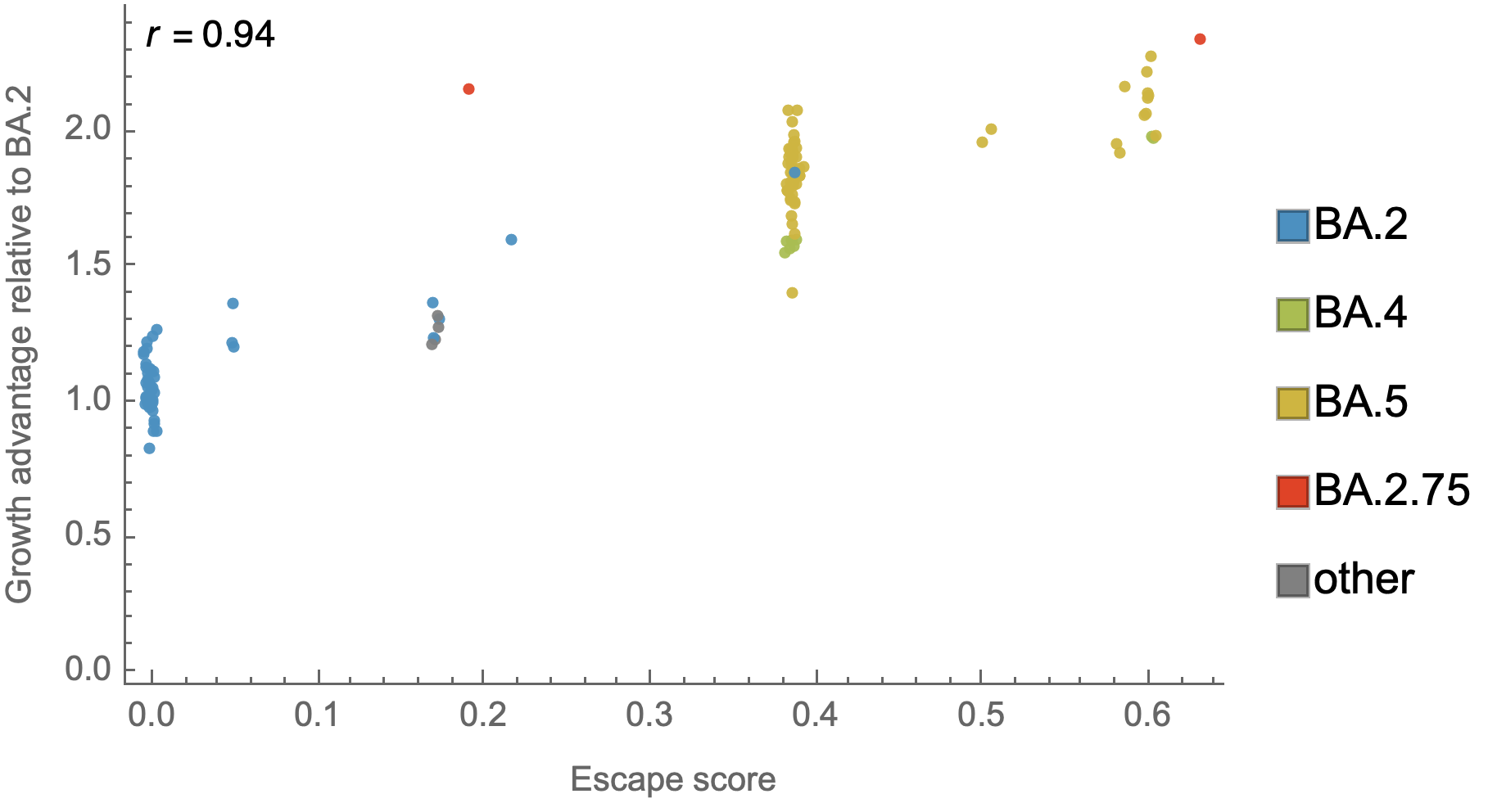
Could we predict the spread of new mutations using DMS data?
Perhaps this worked for 486V in BA.4/BA.5


Now working from Jesse's calculator using panel of monoclonals known to neutralize BA.2


Proof-of-principle that we can use immune escape to predict lineage fitness
Acknowledgements
SARS-CoV-2 genomic epi: Data producers from all over the world, GISAID and the Nextstrain team
Bedford Lab:
![]() John Huddleston,
John Huddleston,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Maya Lewinsohn,
Maya Lewinsohn,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Cassia Wagner,
Cassia Wagner,
![]() Miguel Paredes,
Miguel Paredes,
![]() Nicola Müller,
Nicola Müller,
![]() Marlin Figgins,
Marlin Figgins,
![]() Denisse Sequeira,
Denisse Sequeira,
![]() Victor Lin,
Victor Lin,
![]() Jennifer Chang,
Jennifer Chang,
![]() Allison Li,
Allison Li,
![]() Eslam Abousamra,
Eslam Abousamra,
![]() Donna Modrell,
Donna Modrell,
![]() Nashwa Ahmed
Nashwa Ahmed






