SARS-CoV-2 genomics update
Trevor Bedford (@trvrb)
Fred Hutchinson Cancer Center / Howard Hughes Medical Institute
20 Sep 2022
Viruses and Vaccines Seminar Series
Brotman Baty Institute
Slides at: bedford.io/talks
Rapid displacement of existing diversity by emerging variants

S1 evolved at a rate of 13 amino acid changes per year since pandemic start

Continued rapid accumulation from BA.2 onwards with 7 amino acid changes per year

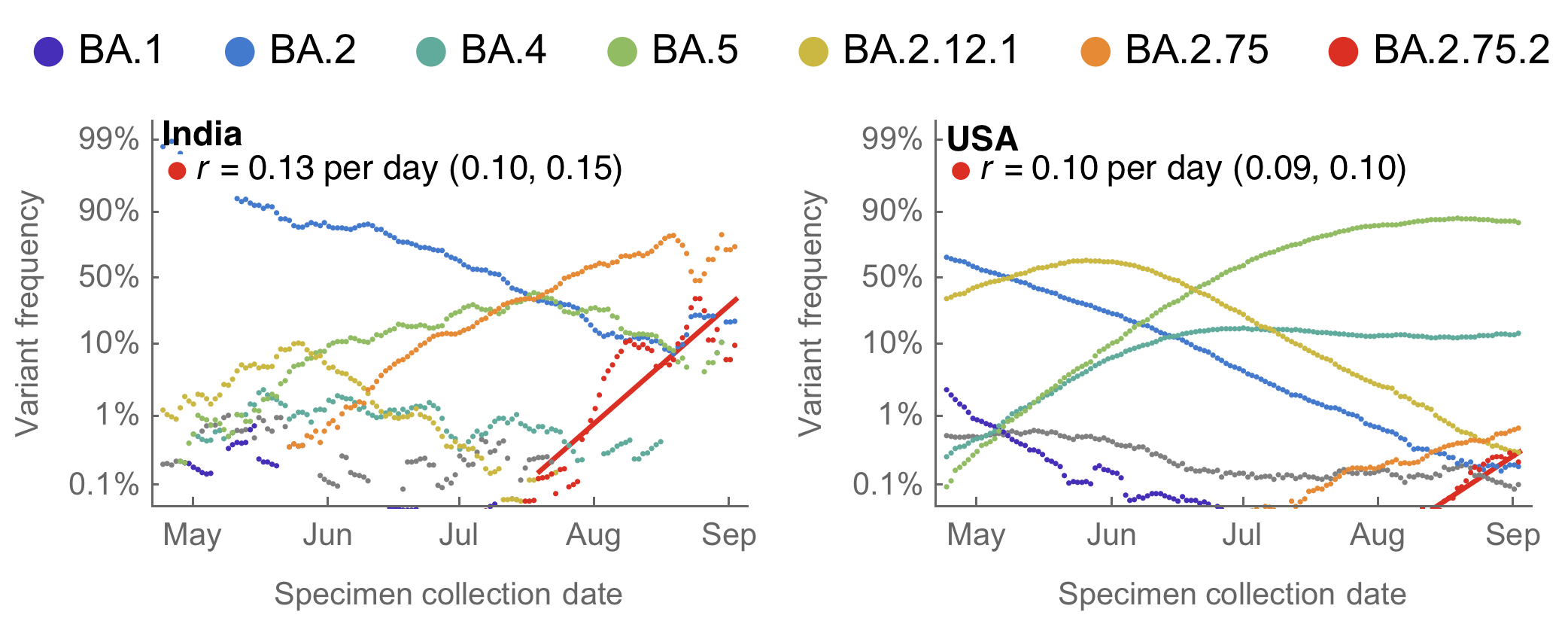
BA.2.75 and sublineage BA.2.75.2 emerging from India

BA.2.75(.2) shows consistent selective advantage in India and US
BA.2.75.2 has current Rt in the US of ~1.4
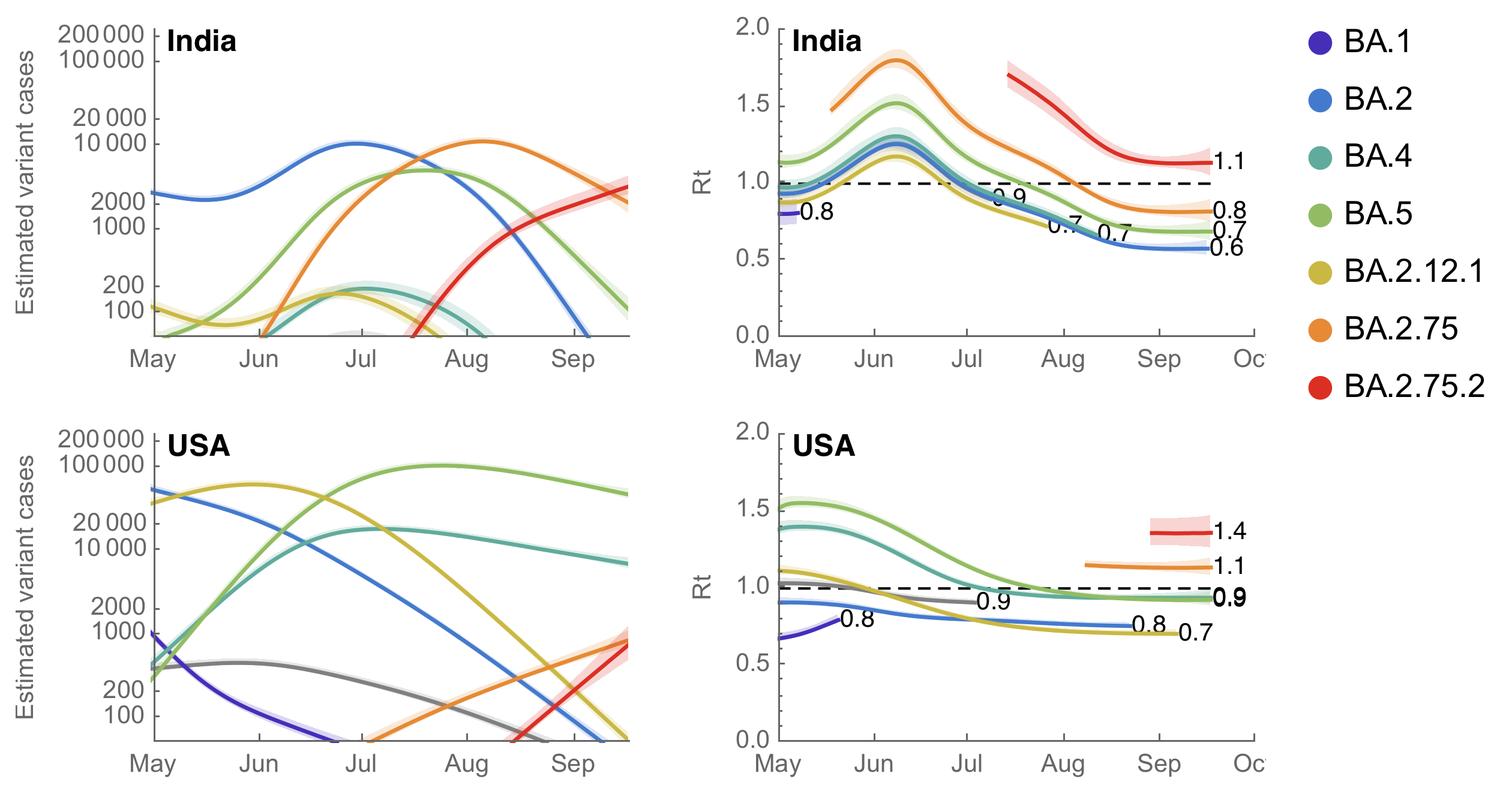
Model from Figgins and Bedford. 2022. medRxiv.
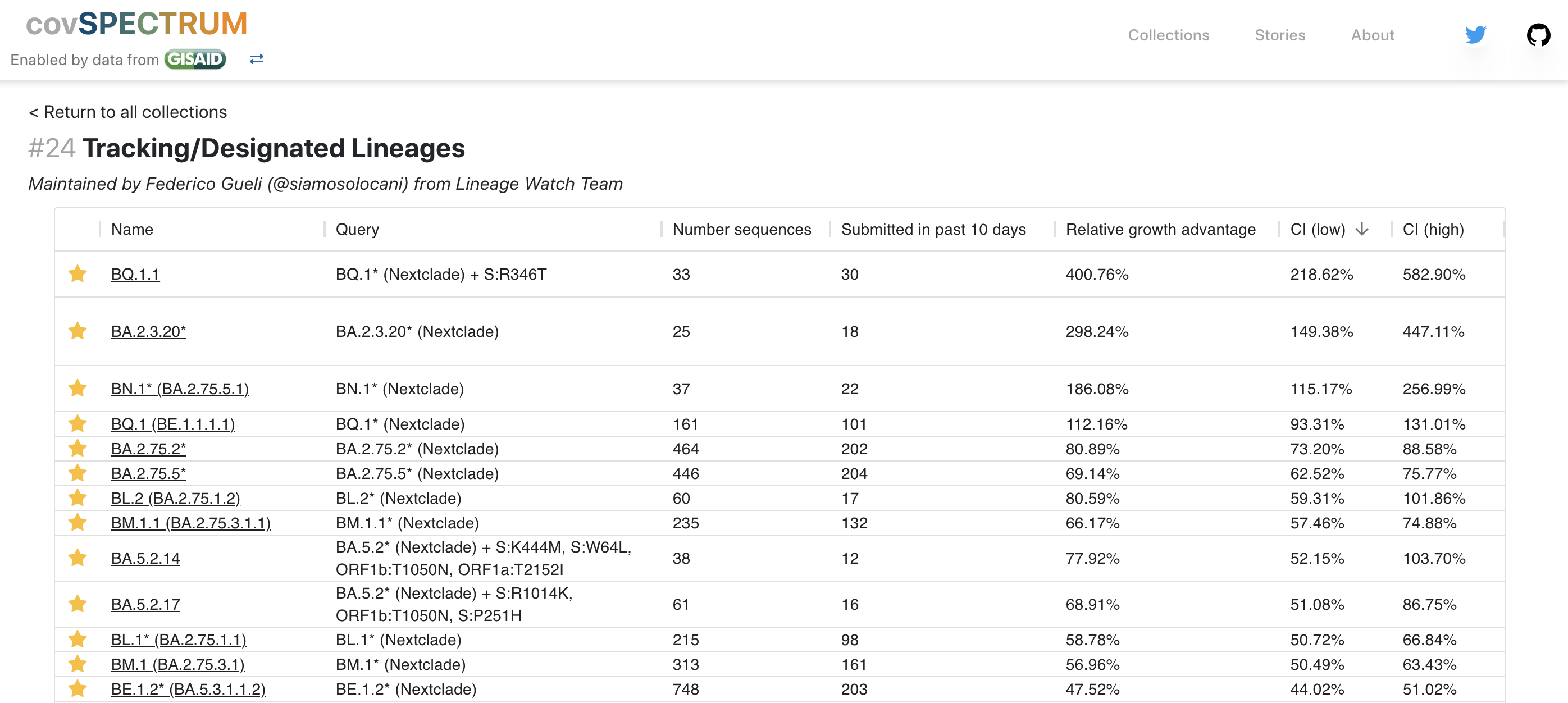
However, multiple low frequency Pango lineages that may compete with BA.2.75.2
Acknowledgements
SARS-CoV-2 genomic epi: Data producers from all over the world, GISAID and the Nextstrain team
Bedford Lab:
![]() John Huddleston,
John Huddleston,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Maya Lewinsohn,
Maya Lewinsohn,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Cassia Wagner,
Cassia Wagner,
![]() Miguel Paredes,
Miguel Paredes,
![]() Nicola Müller,
Nicola Müller,
![]() Marlin Figgins,
Marlin Figgins,
![]() Denisse Sequeira,
Denisse Sequeira,
![]() Victor Lin,
Victor Lin,
![]() Jennifer Chang,
Jennifer Chang,
![]() Jennifer Chang,
Jennifer Chang,
![]() Donna Modrell
Donna Modrell






