Evolutionary dynamics of SARS-CoV-2
Trevor Bedford (@trvrb)
Associate Professor, Fred Hutchinson Cancer Research Center
26 Mar 2021
Pew Annual Meeting
Slides at: bedford.io/talks
Over 800k SARS-CoV-2 genomes shared to GISAID and evolution tracked in real-time at nextstrain.org

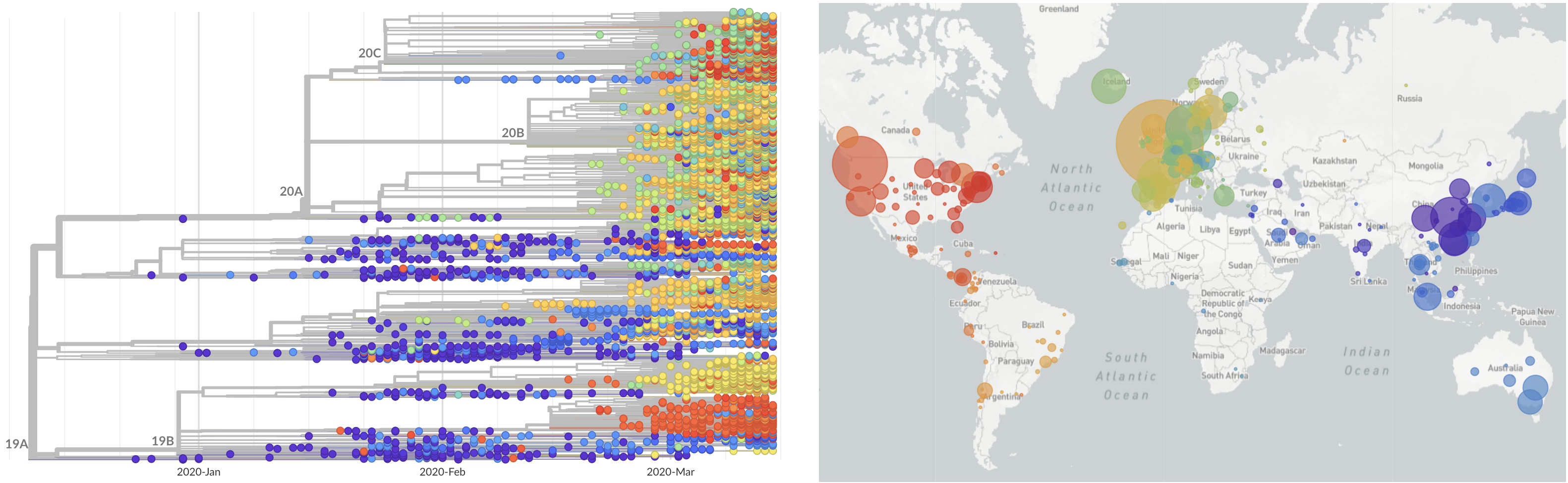
SARS-CoV-2 lineages establish globally in February and March
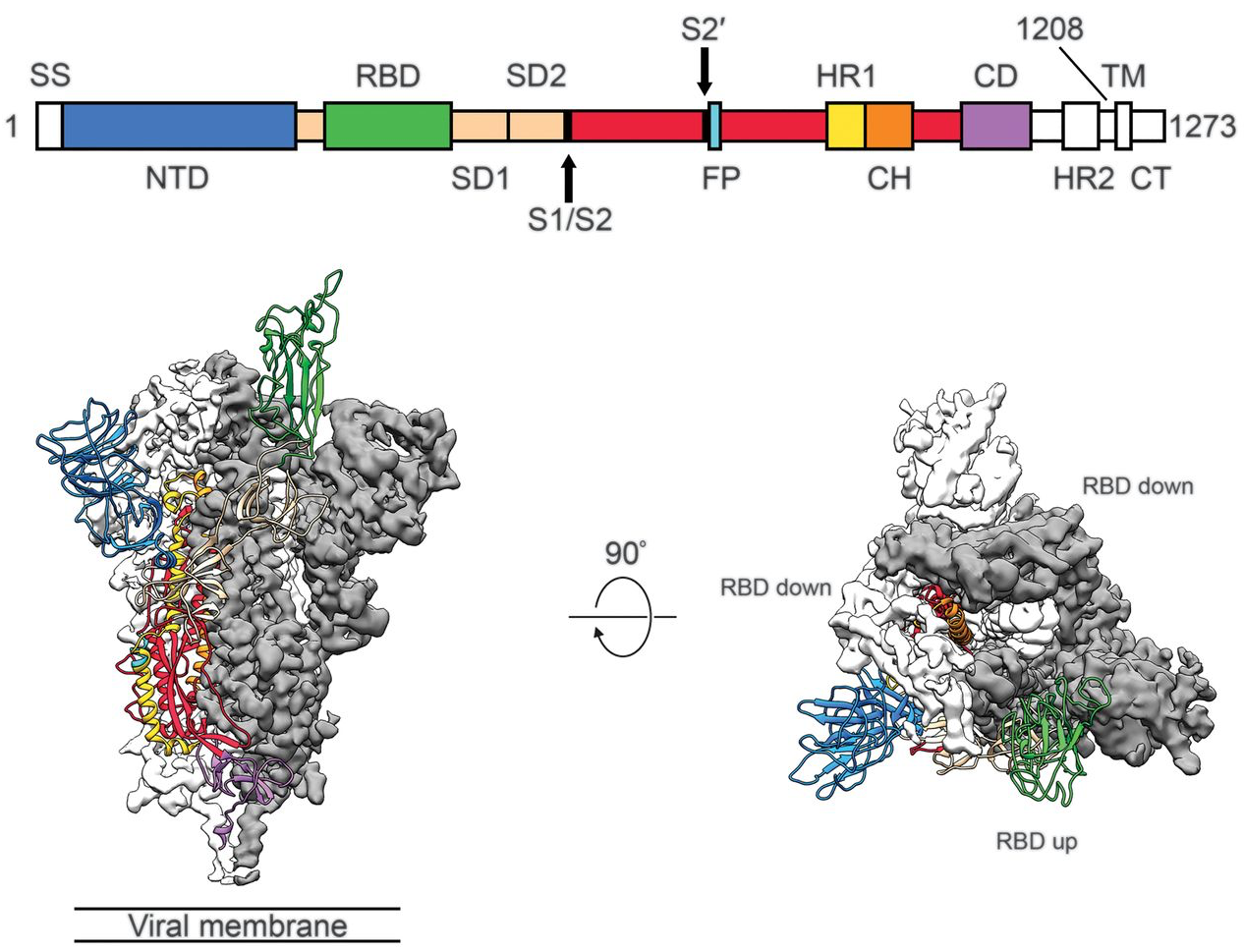
Spike protein is critical for cell invasion by the virus and is the primary target for human immune response
Spike mutations 484K and 501Y appearing repeatedly

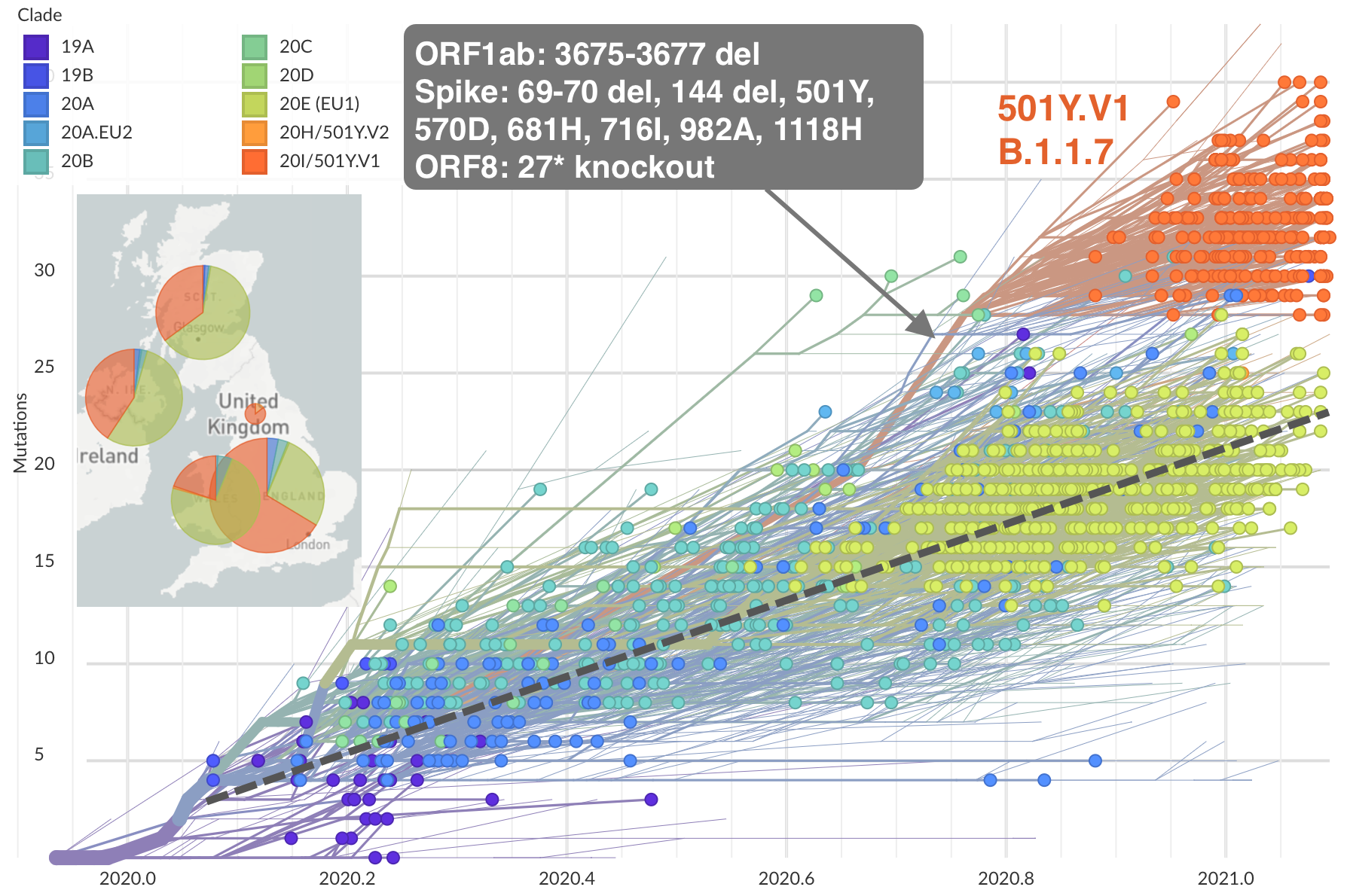
Emergence of 501Y.V1 (B.1.1.7) in the UK
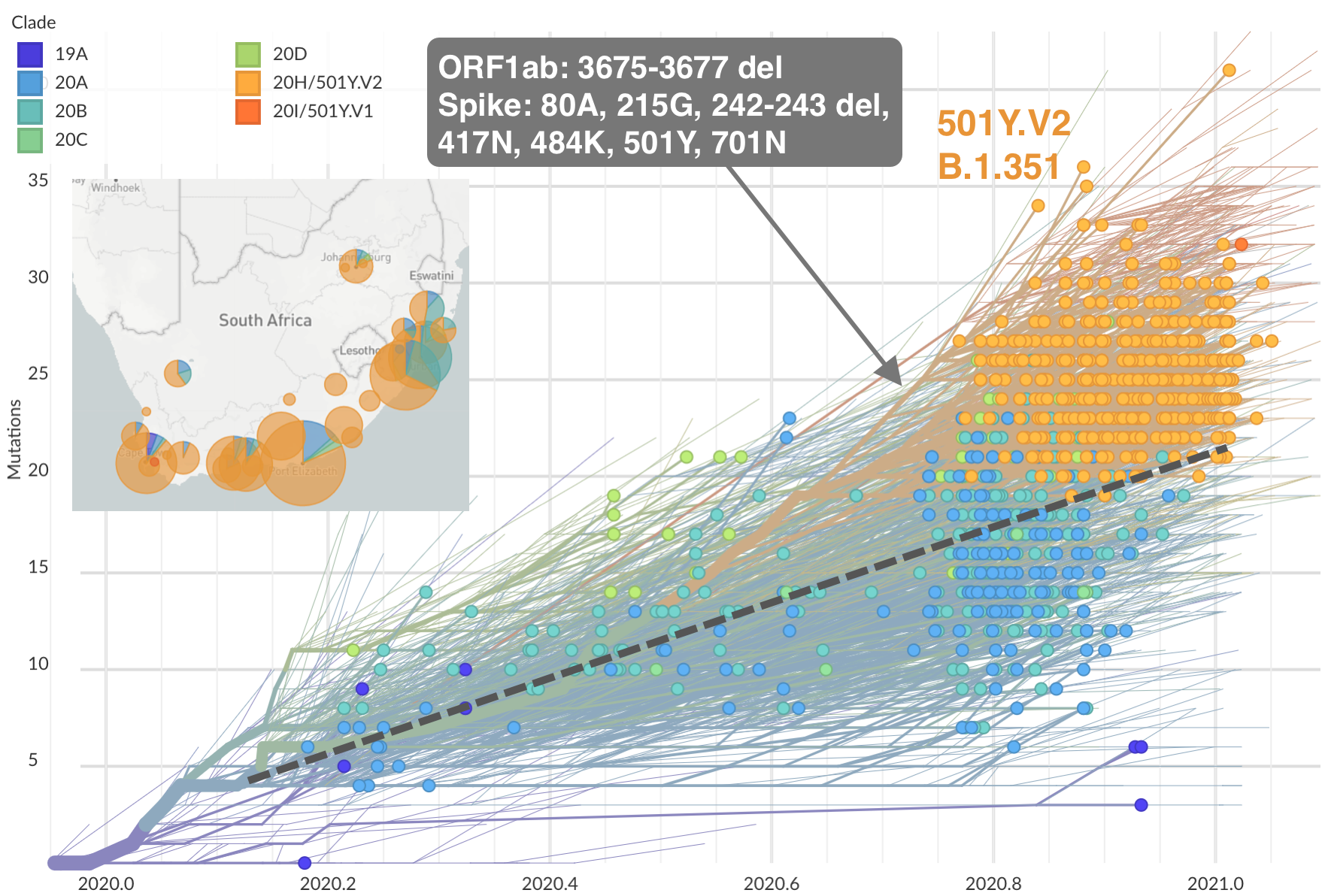
Emergence of 501Y.V2 (B.1.351) in the South Africa
Emergence of 501Y.V3 (P.1) in the Brazil

Repeated convergent evolution across sites in spike

Up to December, my expectation was evolution as seen in seasonal coronaviruses with flu B-like rates of evolution

Rapid spike S1 amino acid evolution in SARS-CoV-2

SARS-COV-2 spike S1 evolution similar to evolution of flu H3N2

501Y.V2 shows an ~8-fold drop in neutralization titer, equivalent to ~3 years of H3N2 evolution

Generally, I suspect we're looking at a data-rich version of the influenza strain selection process
- mRNA vaccines allow faster turnarounds and may push for more regional strategies
- Work directly from human serological data rather than naive animals
- Multiple valencies possible, clinical trials should be conducted now comparing regimens
Acknowledgements
SARS-CoV-2 genomic epi: Data producers from all over the world, GISAID and the Nextstrain team
Bedford Lab:
![]() Alli Black,
Alli Black,
![]() John Huddleston,
John Huddleston,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Louise Moncla,
Louise Moncla,
![]() Maya Lewinsohn,
Maya Lewinsohn,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Kairsten Fay,
Kairsten Fay,
![]() Misja Ilcisin,
Misja Ilcisin,
![]() Cassia Wagner,
Cassia Wagner,
![]() Miguel Paredes,
Miguel Paredes,
![]() Nicola Müller,
Nicola Müller,
![]() Marlin Figgins,
Marlin Figgins,
![]() Eli Harkins
Eli Harkins






