SARS-CoV-2 variants and
evolutionary dynamics
Trevor Bedford (@trvrb)
Associate Professor
Fred Hutchinson Cancer Research Center
18 Oct 2021
1. Emergence and spread of variants of concern
2. Assessing adaptive evolution
3. Variant transmission dynamics
Emergence and spread of variants of concern
Over 3.4M SARS-CoV-2 genomes shared to GISAID and evolution tracked in real-time at nextstrain.org
![]() Richard Neher,
Richard Neher,
![]() Emma Hodcroft,
Emma Hodcroft,
![]() James Hadfield,
James Hadfield,
![]() Thomas Sibley,
Thomas Sibley,
![]() John Huddleston,
John Huddleston,
![]() Ivan Aksamentov,
Ivan Aksamentov,
![]() Moira Zuber,
Moira Zuber,
![]() Eli Harkins,
Eli Harkins,
![]() Jover Lee,
Jover Lee,
![]() Cassia Wagner,
Cassia Wagner,
![]() Louise Moncla,
Louise Moncla,
![]() Misja Ilcisin,
Misja Ilcisin,
![]() Kairsten Fay,
Kairsten Fay,
![]() Allison Black,
Allison Black,
![]() Miguel Paredes,
Miguel Paredes,
![]() Sidney Bell,
Sidney Bell,
![]() Denisse Sequeira
Denisse Sequeira
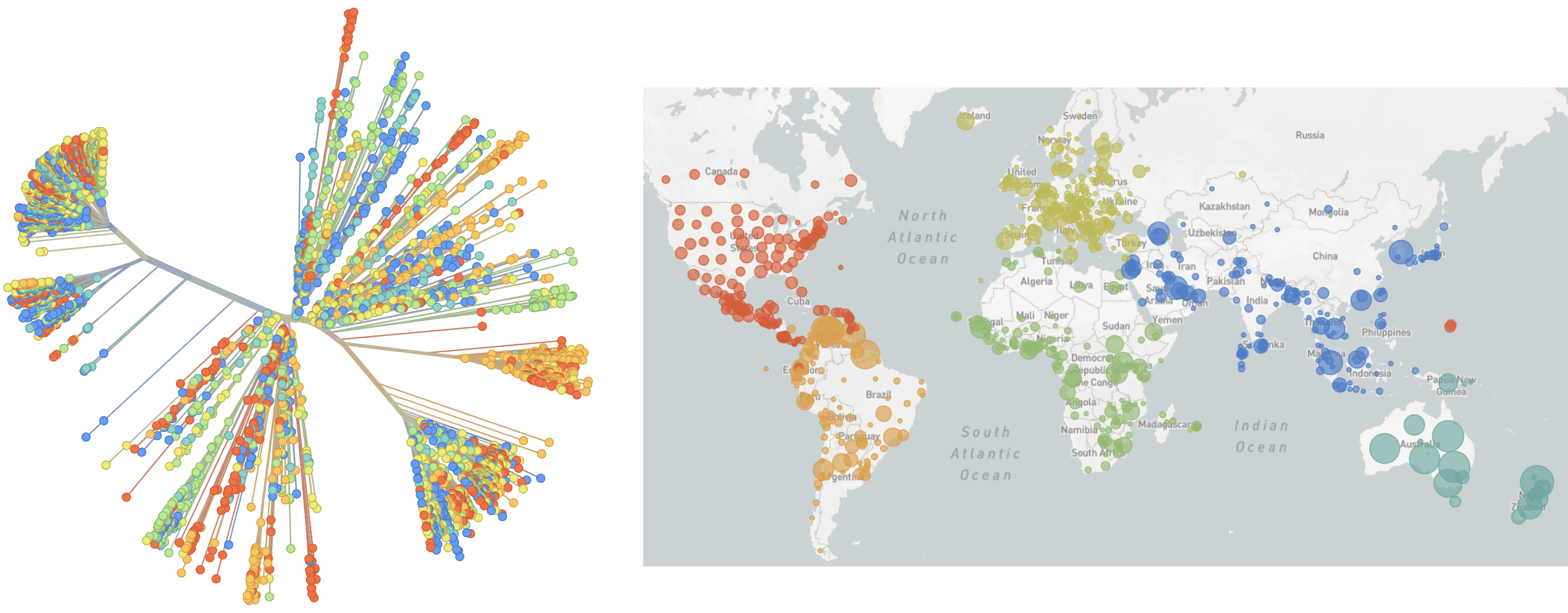
SARS-CoV-2 lineages establish globally in Feb and Mar 2020
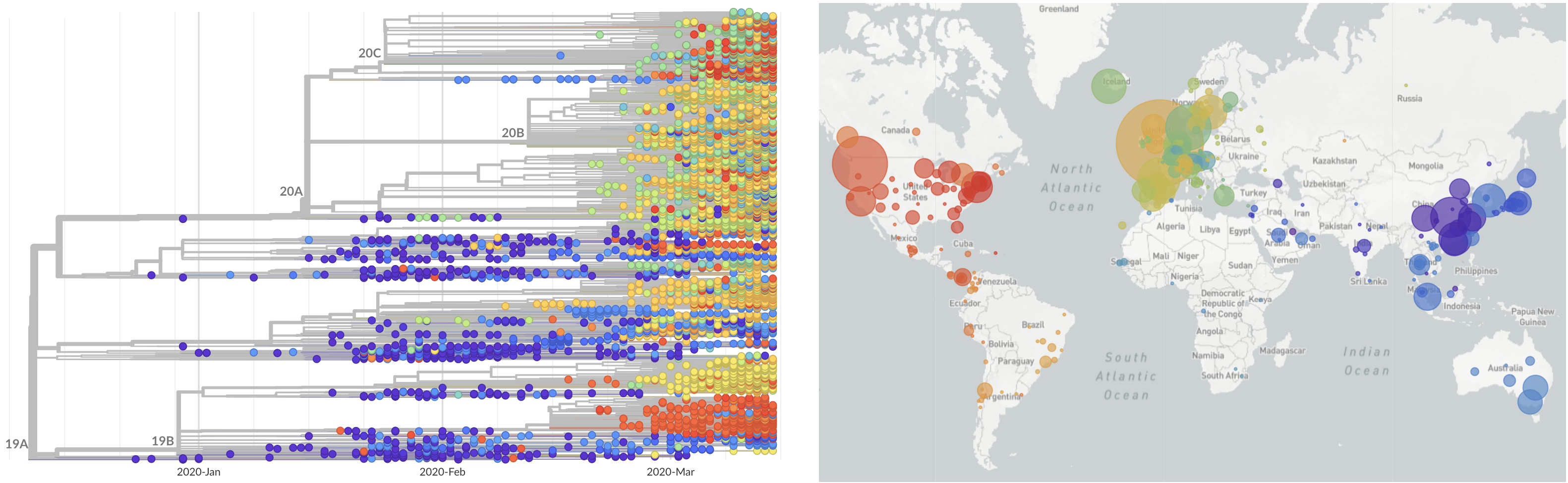
Limited early mutations like spike D614G spread globally during initial wave
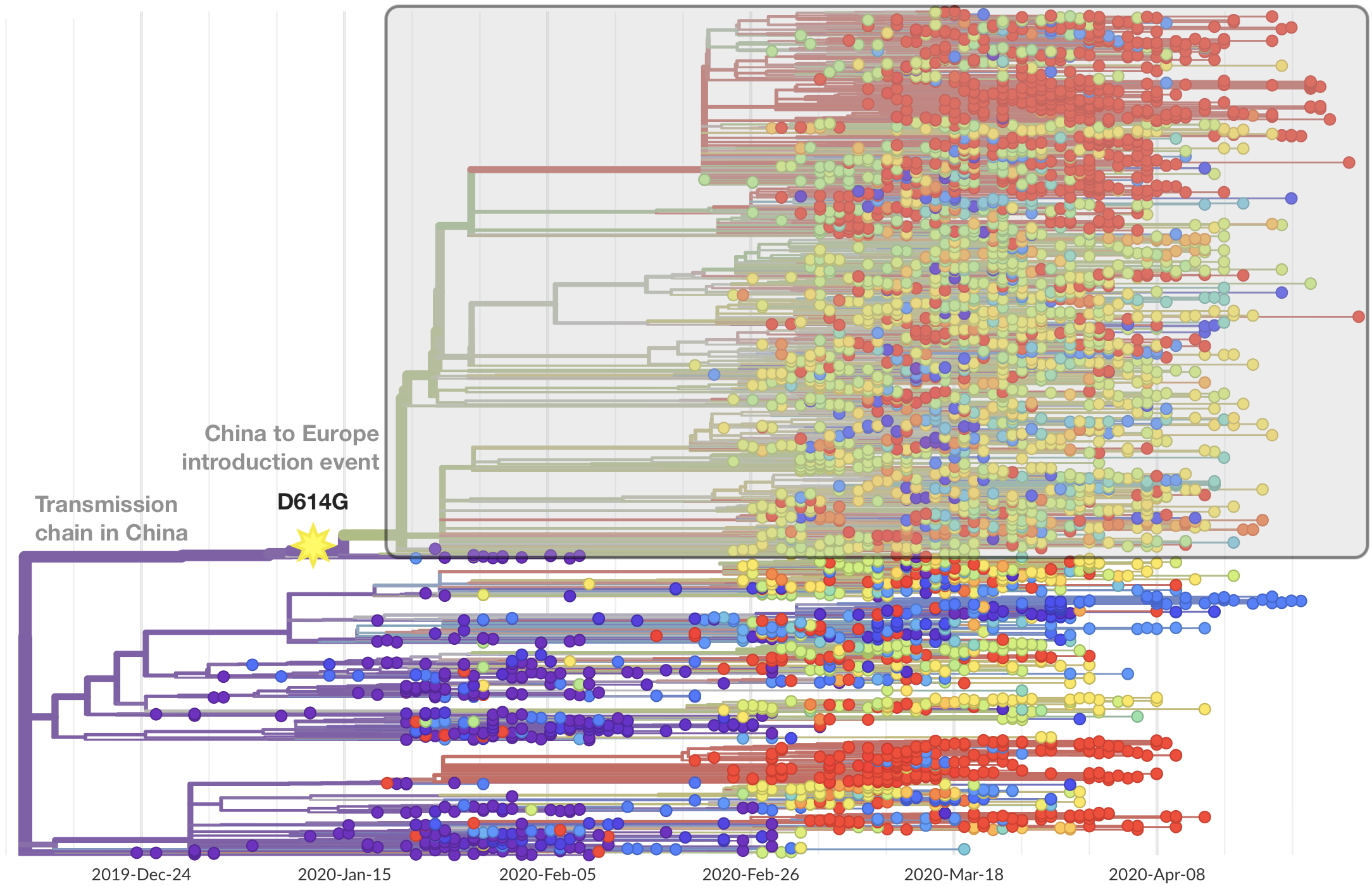
After initial wave, with mitigation
efforts and decreased travel,
regional clades emerge
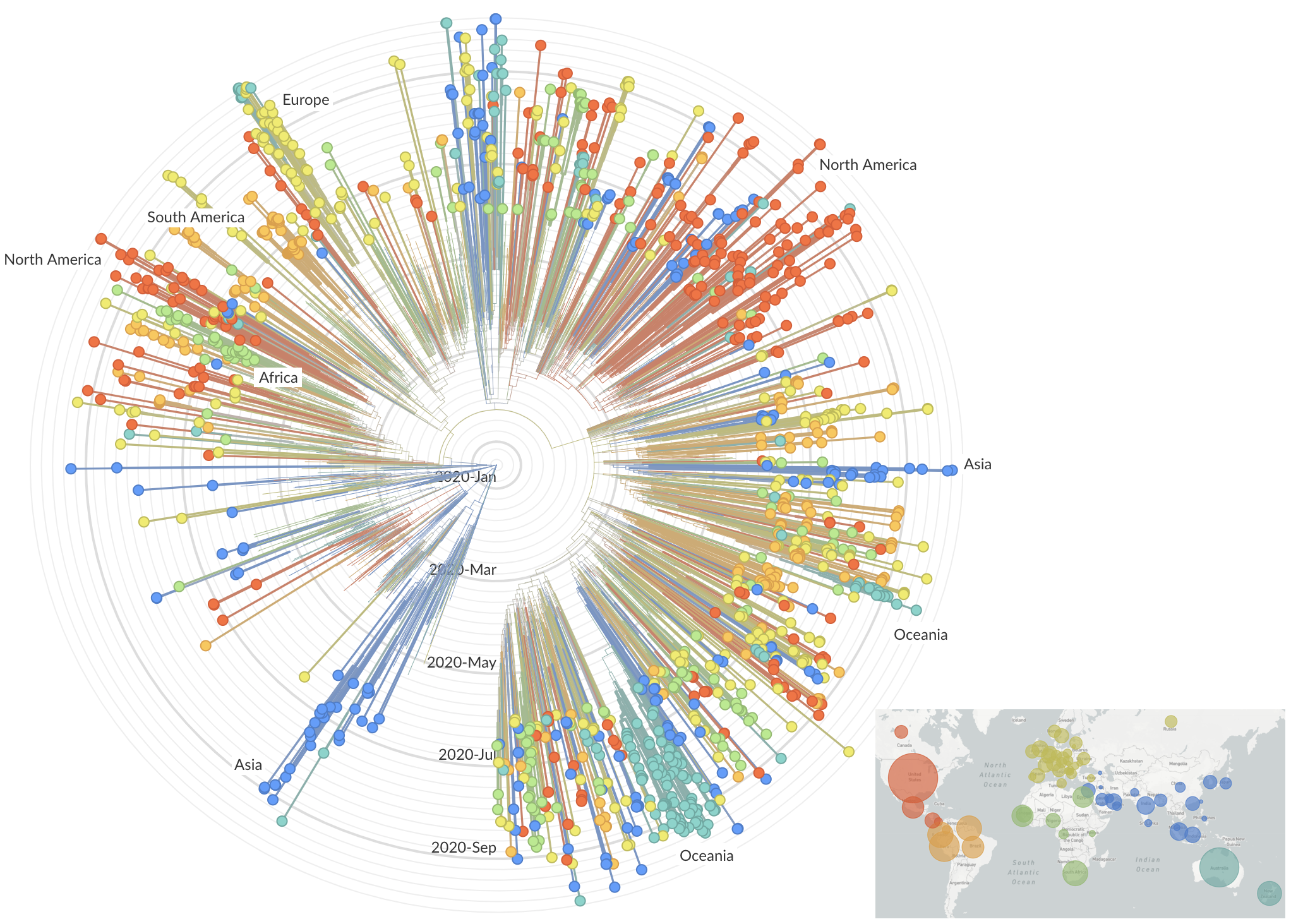
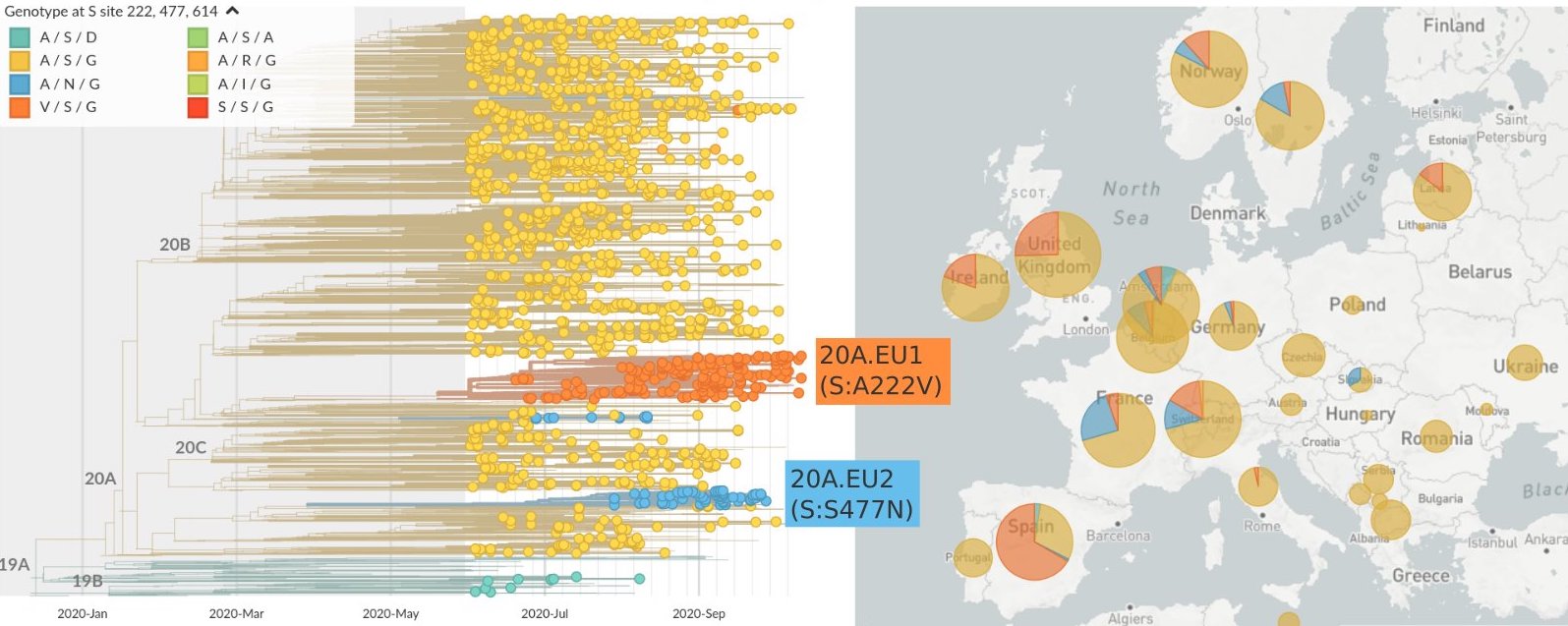
Mutations in summer and fall 2020 were confined to regional dominance
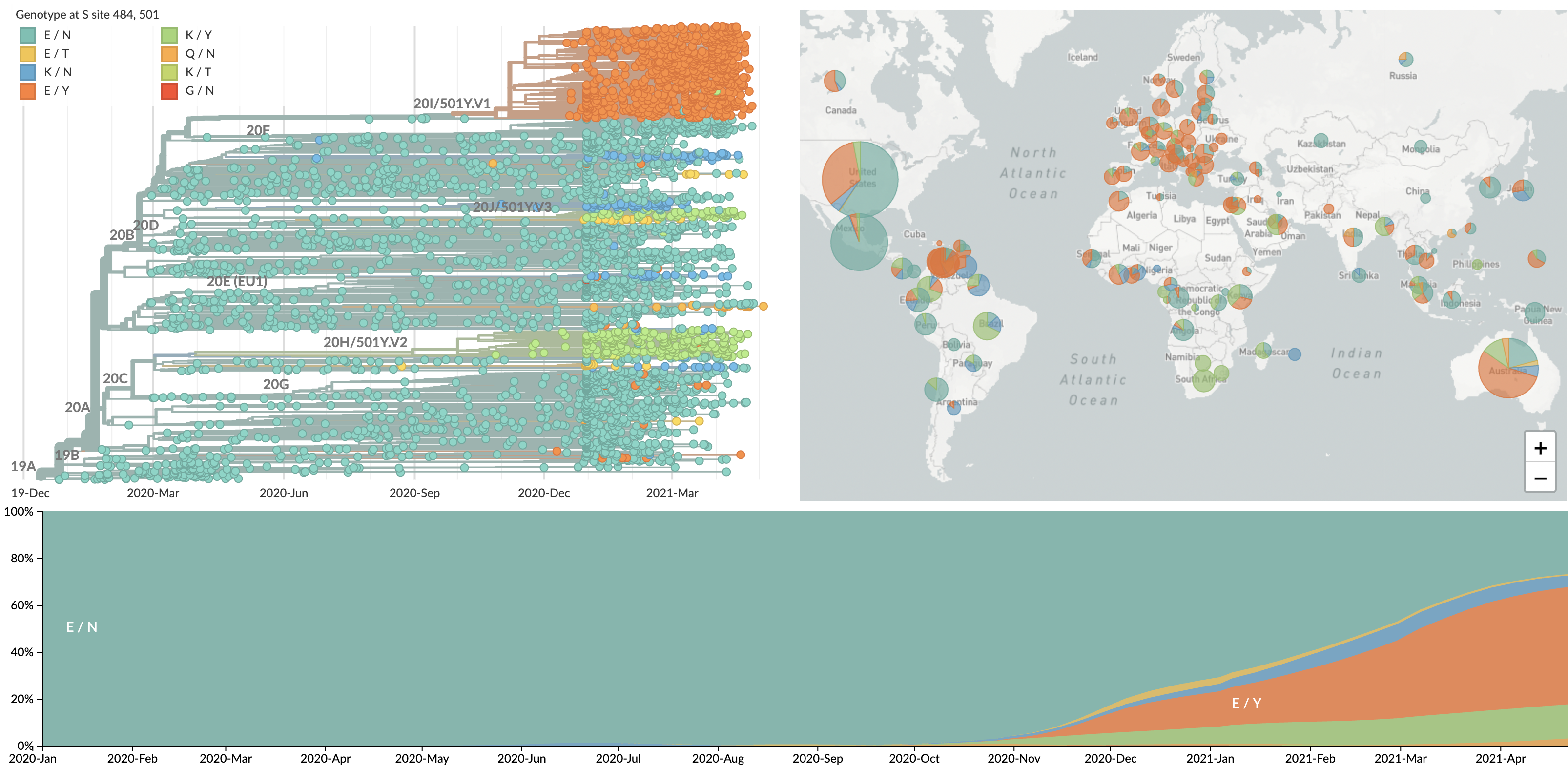
Repeated emergence of 484K and 501Y across the world
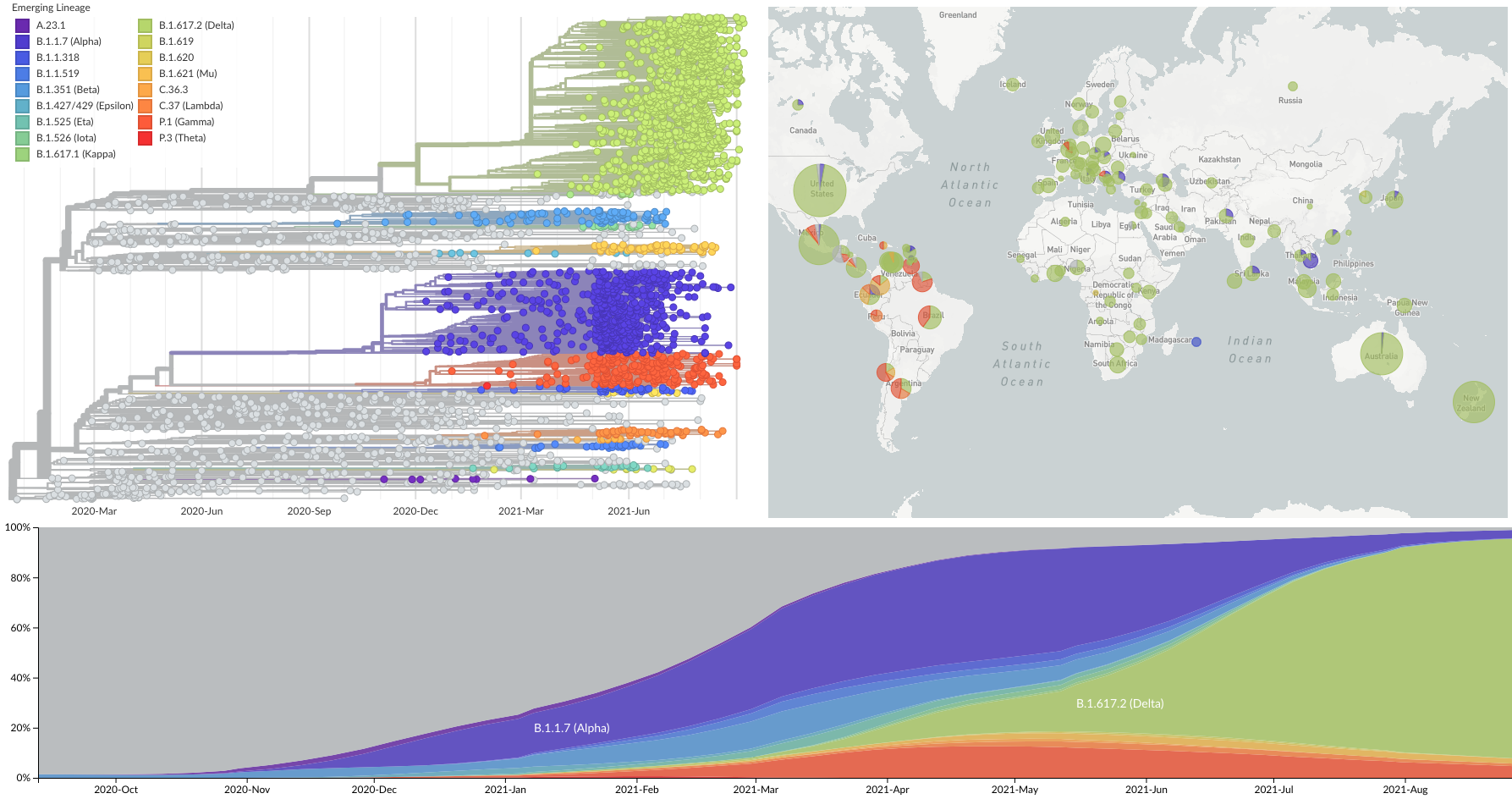
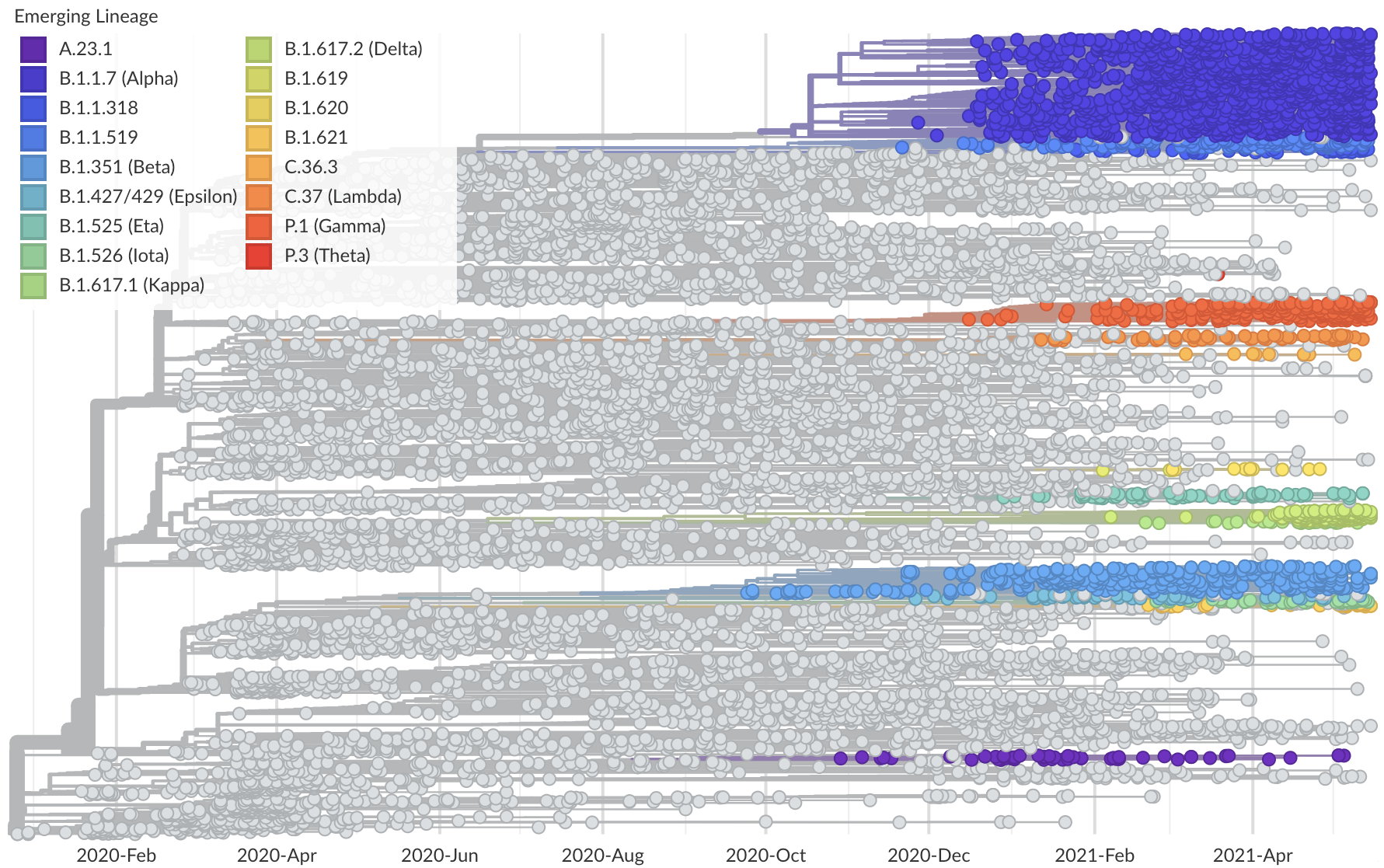
Spread of VOC / VOI lineages across the world
Delta is outcompeting other variants and is on track to sweep
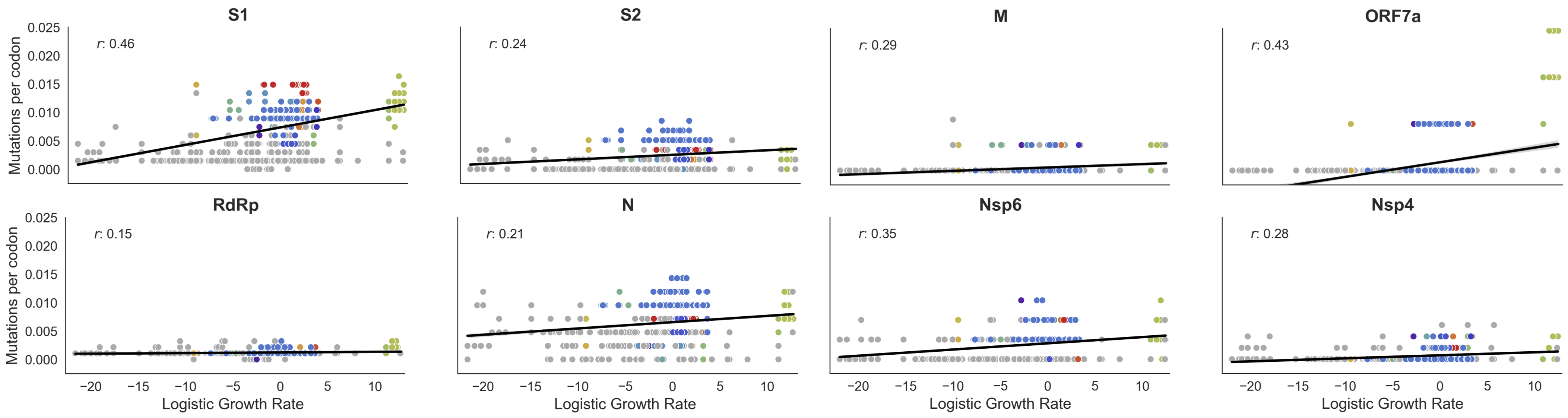
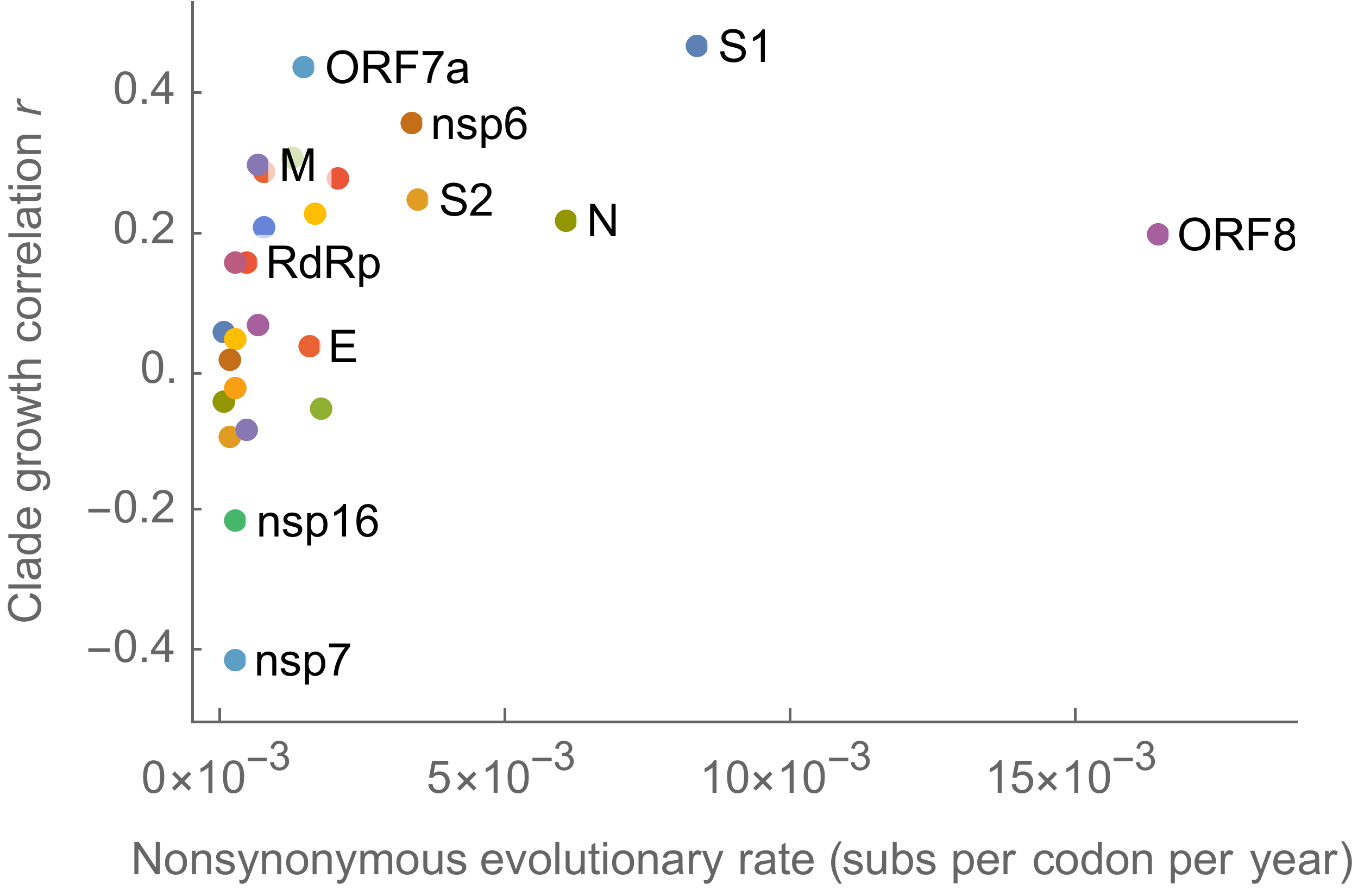
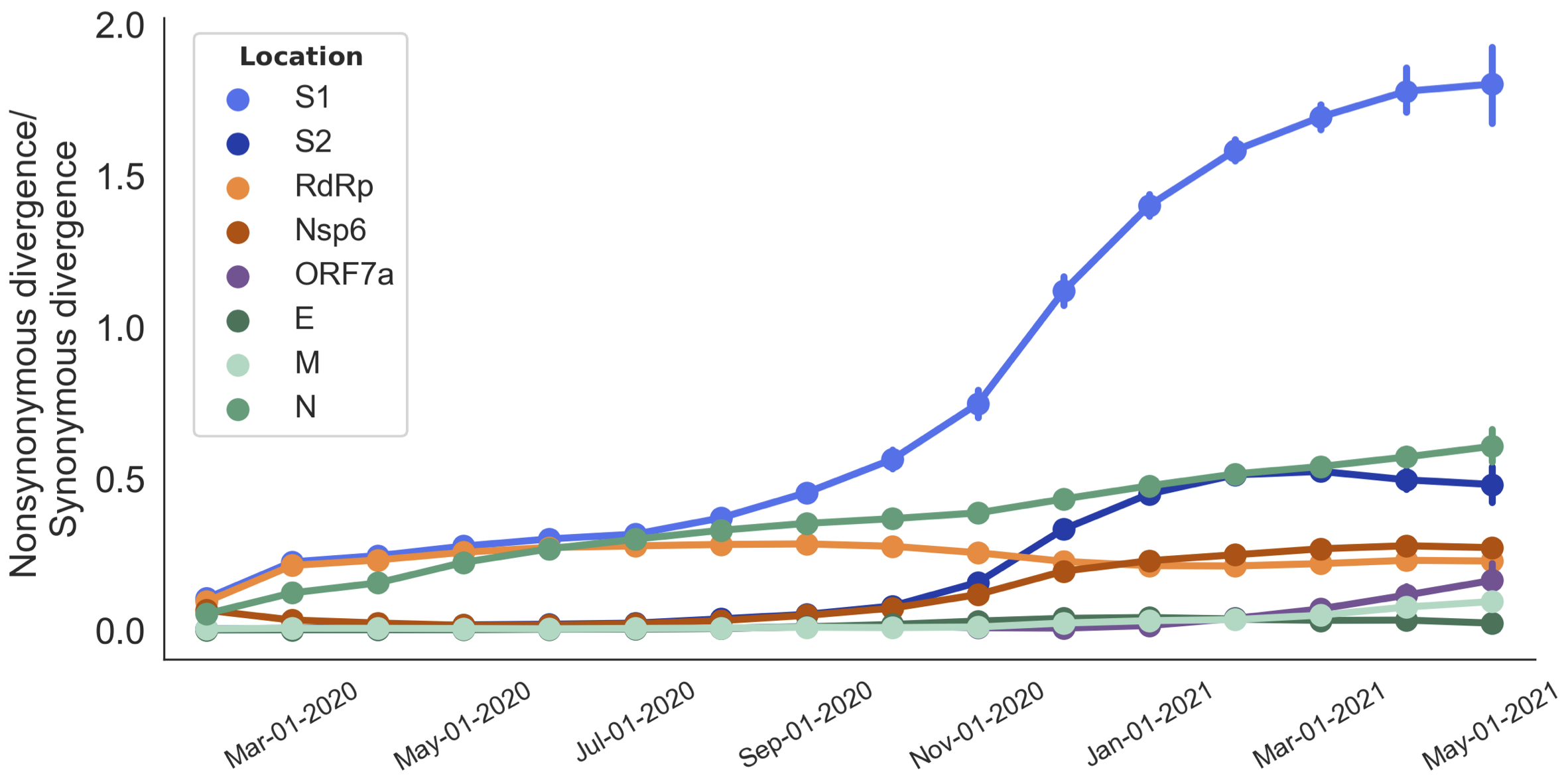
Variants show excess mutations across the genome
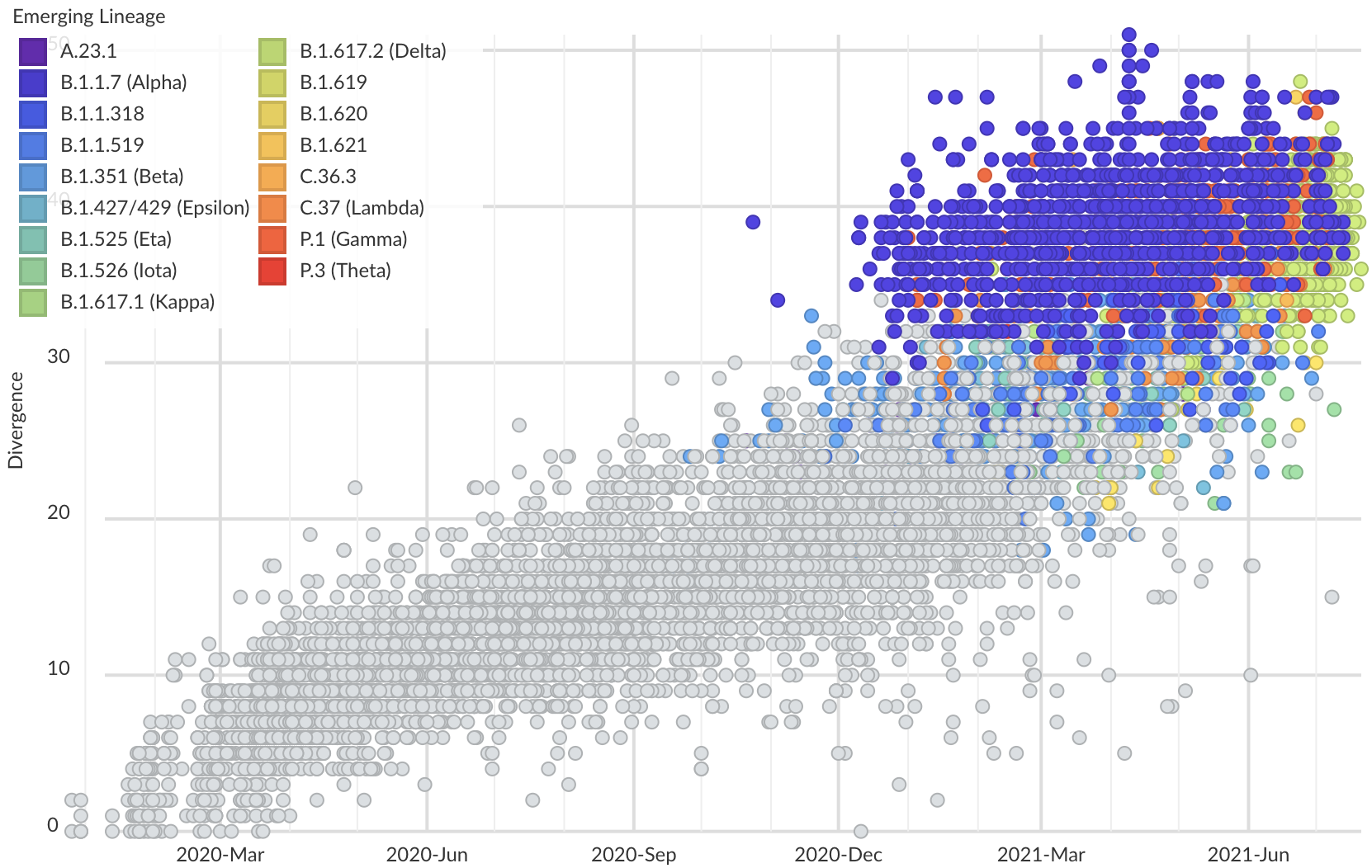
But show most substantial excess in the S1 domain of spike
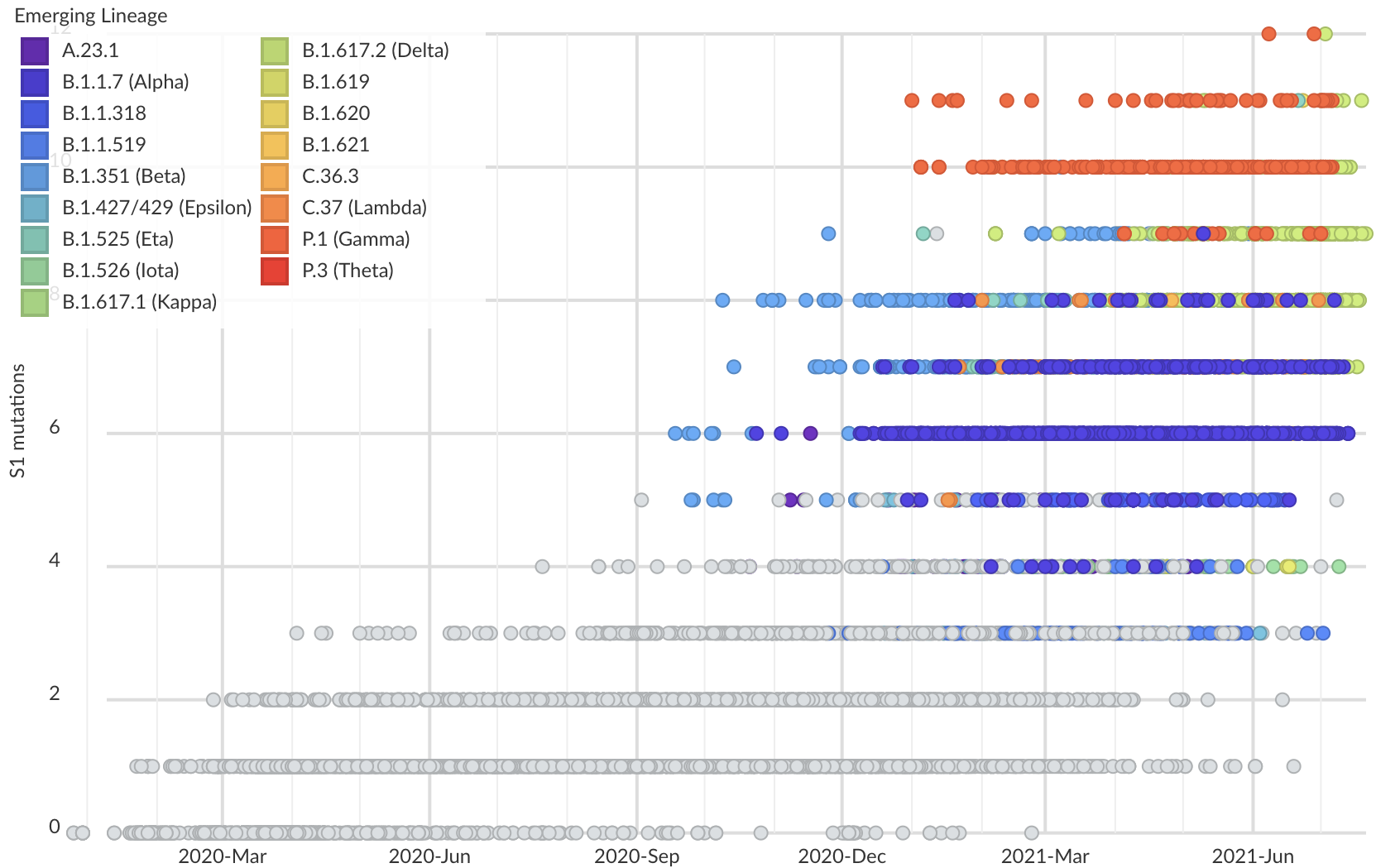
Assessing adaptive evolution
Rapid and parallel adaptive mutations in spike S1 drive clade success in SARS-CoV-2
 Katie Kistler,
Katie Kistler,
 John Huddleston
John Huddleston
Phylogeny of 10k genomes equitably sampled in space and time
Measure clade growth as a proxy for viral fitness
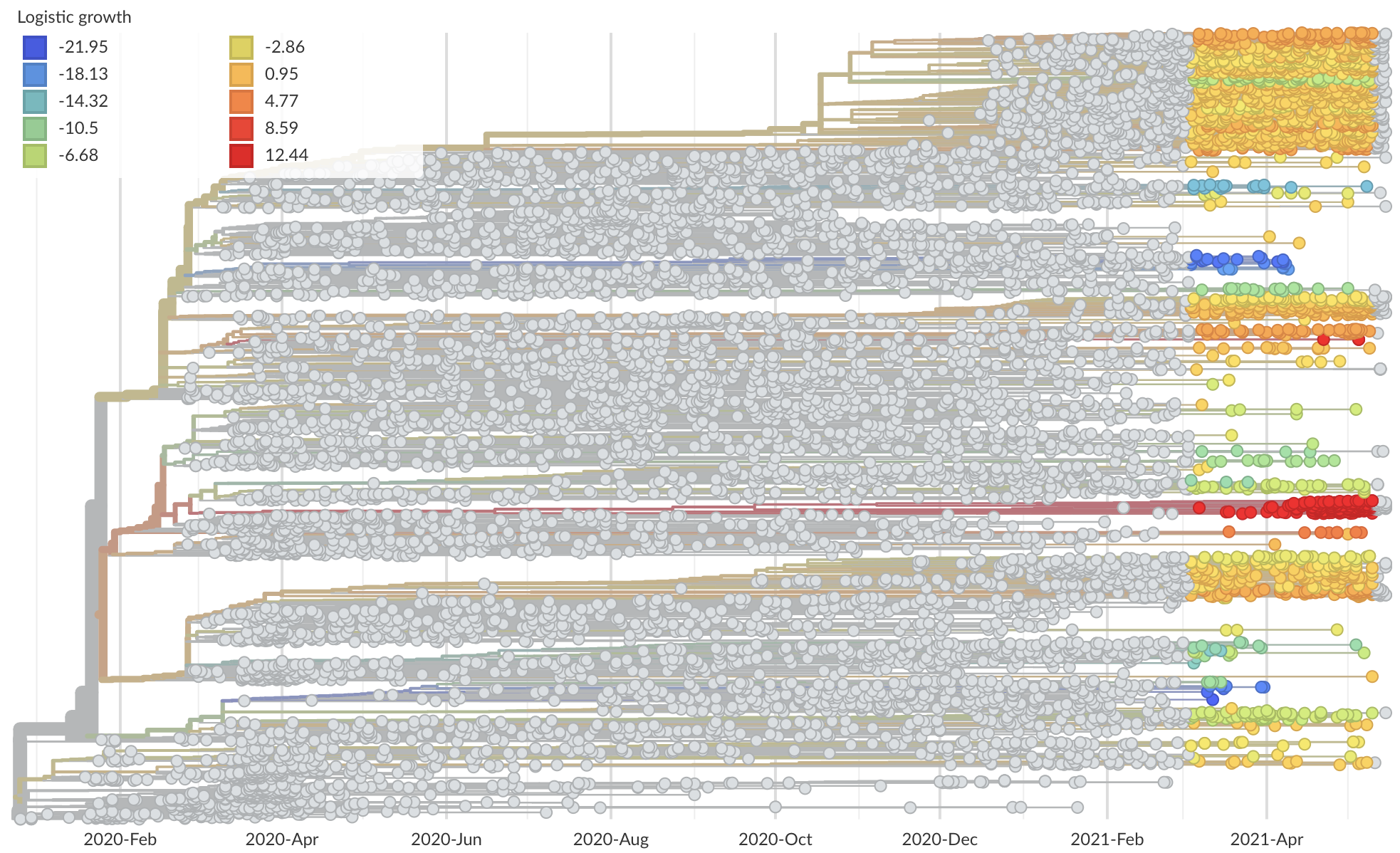
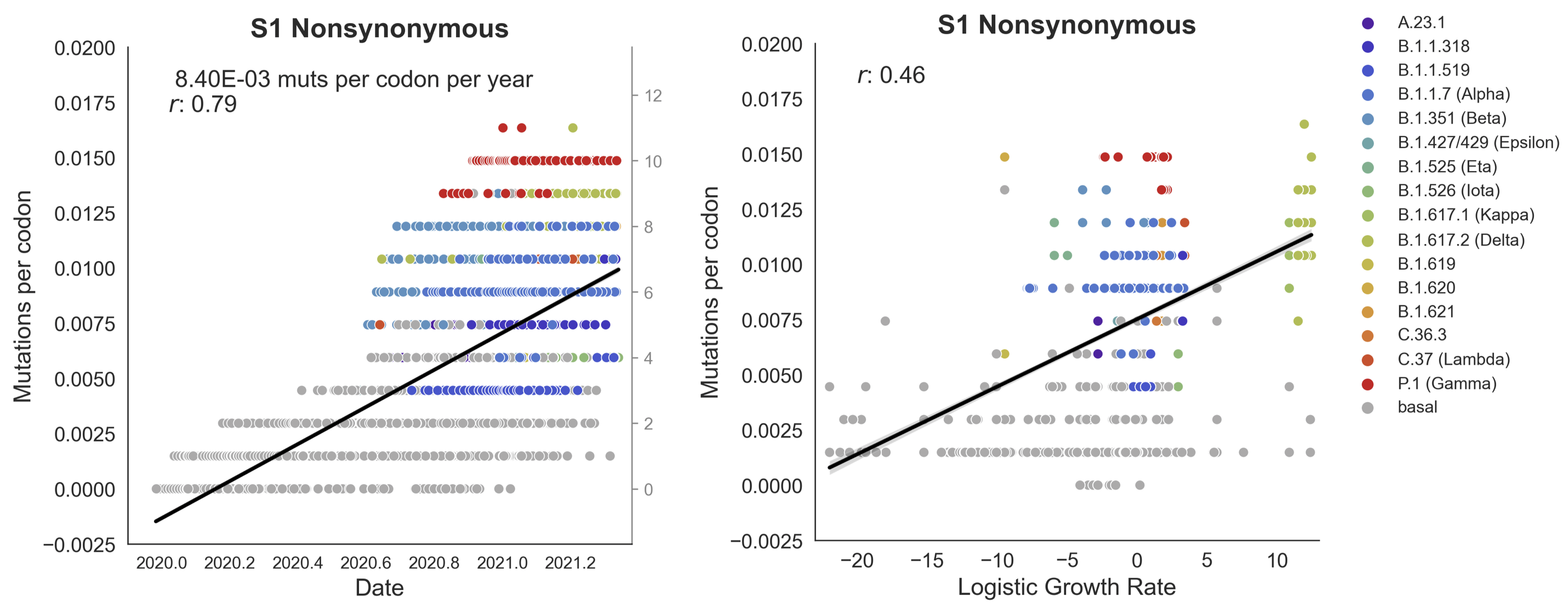
Clades with more S1 nonsynonymous mutations grow faster
This correlation is absent in control regions
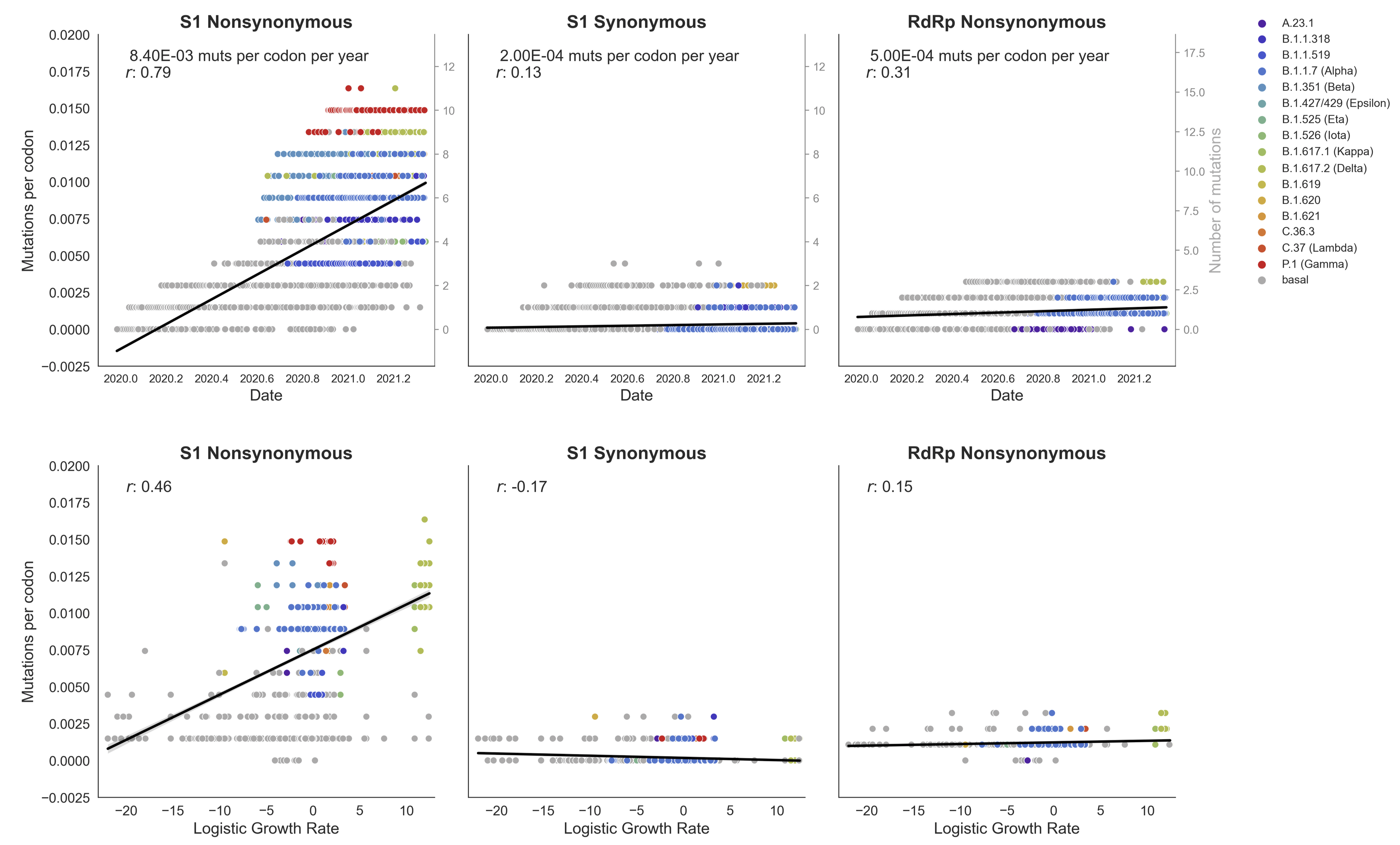
This correlation is stronger in S1 than other genes
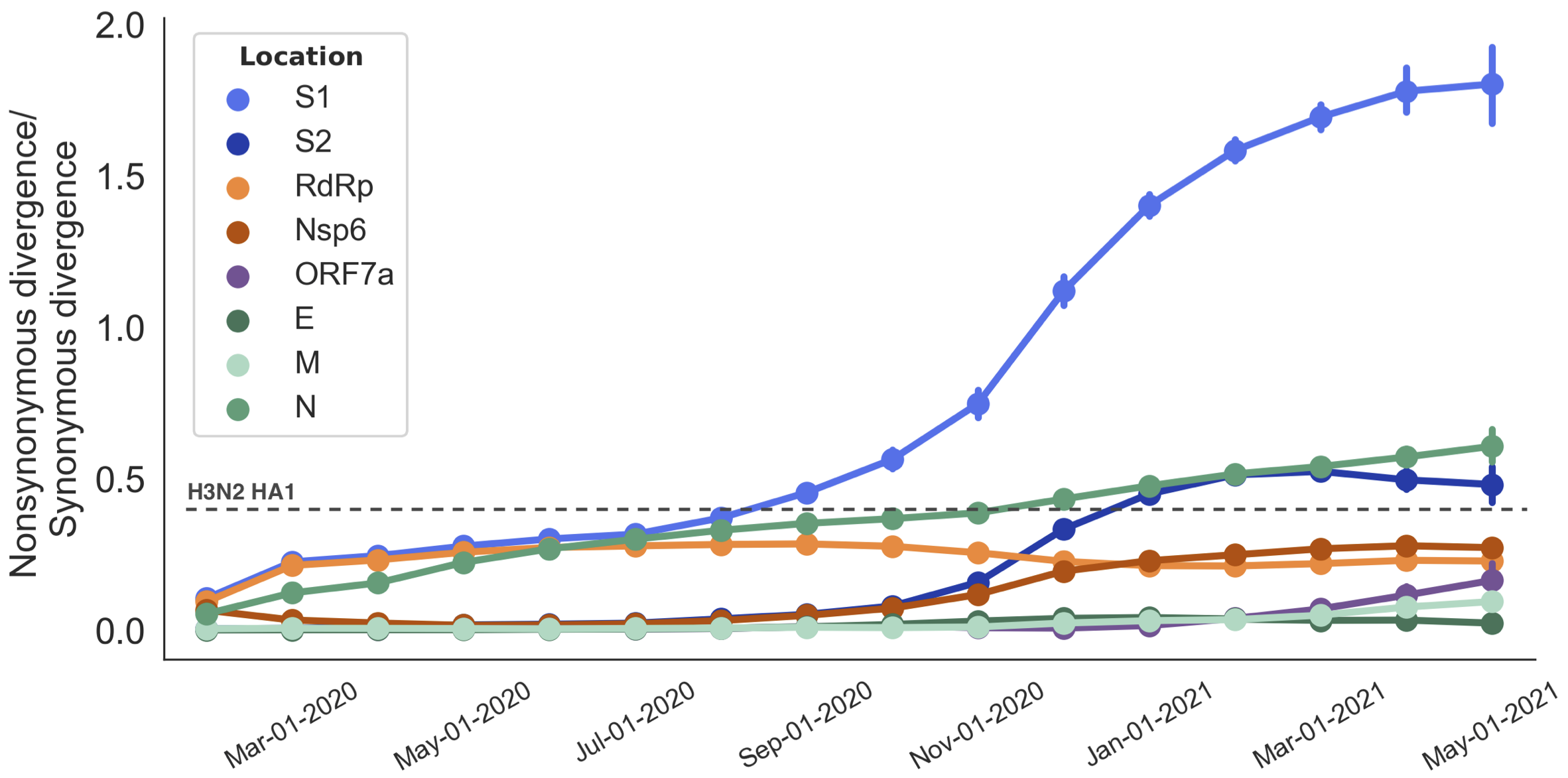
S1 is quickly evolving and highly correlated with clade growth
dN/dS through time further highlights adaptive evolution
Rapid pace of adaptive evolution relative to H3N2 influenza
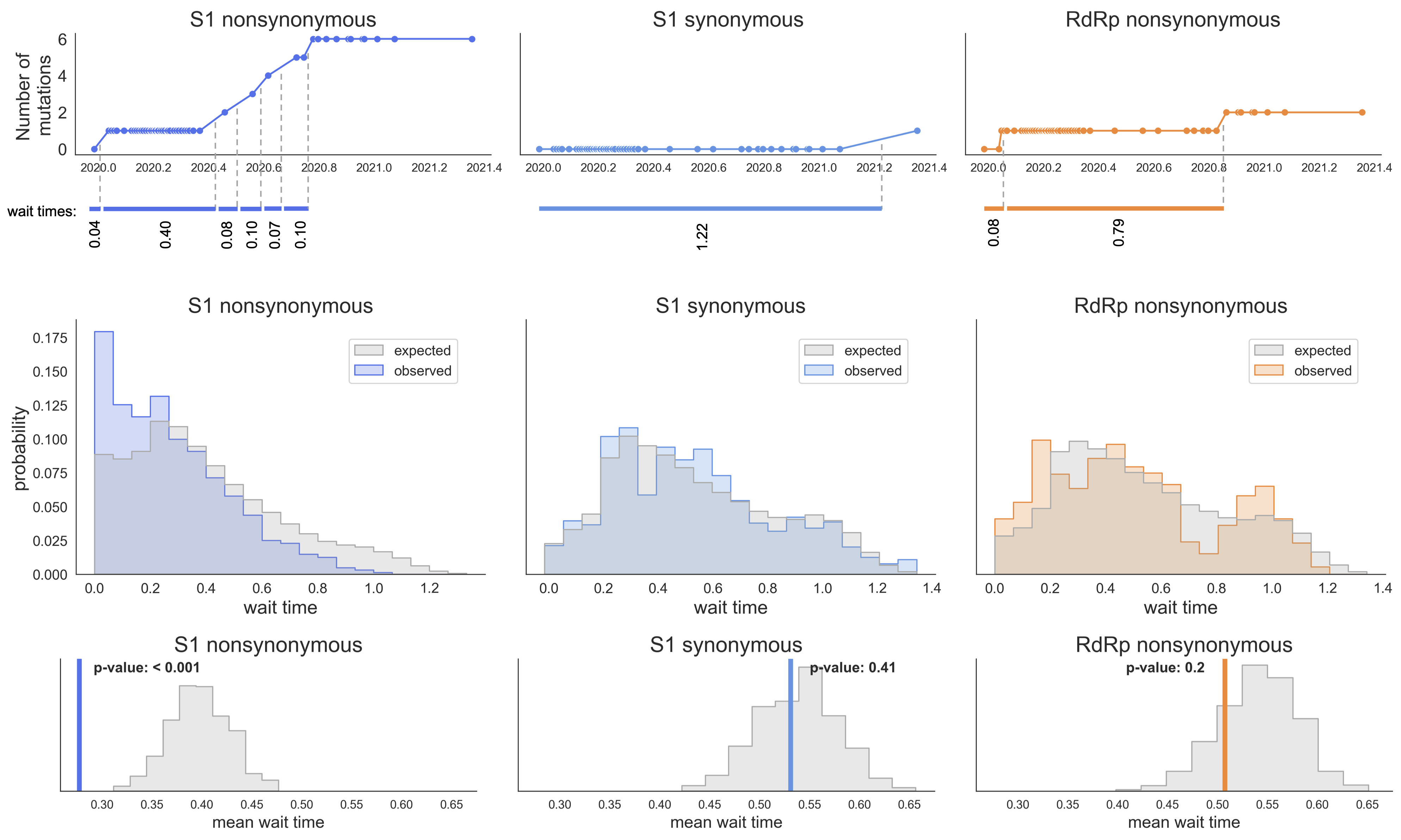
S1 mutations cluster phylogenetically, suggesting bursts of evolution
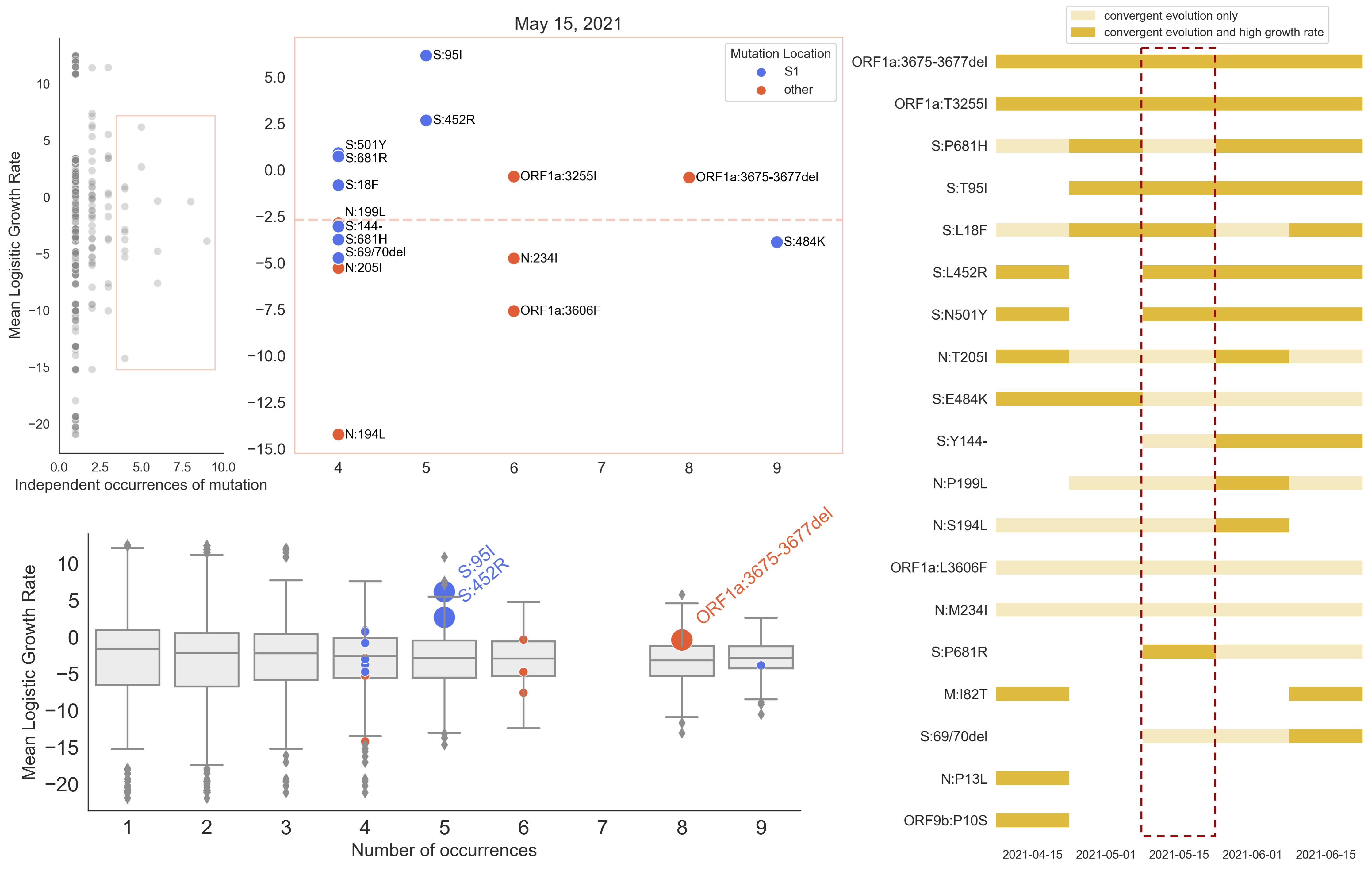
Many S1 mutations show strong convergent evolution
Mutations in S1 arising via within-host pressures result increase viral fitness and are enriched in the viral population by natural selection
Although selection has not been primarily for antigenic drift, observed level of adaptability suggests its potential
Variant transmission dynamics
Multiple approaches to modeling fitness differences between circulating variants
Multinomial logistic regression models work well on frequencies
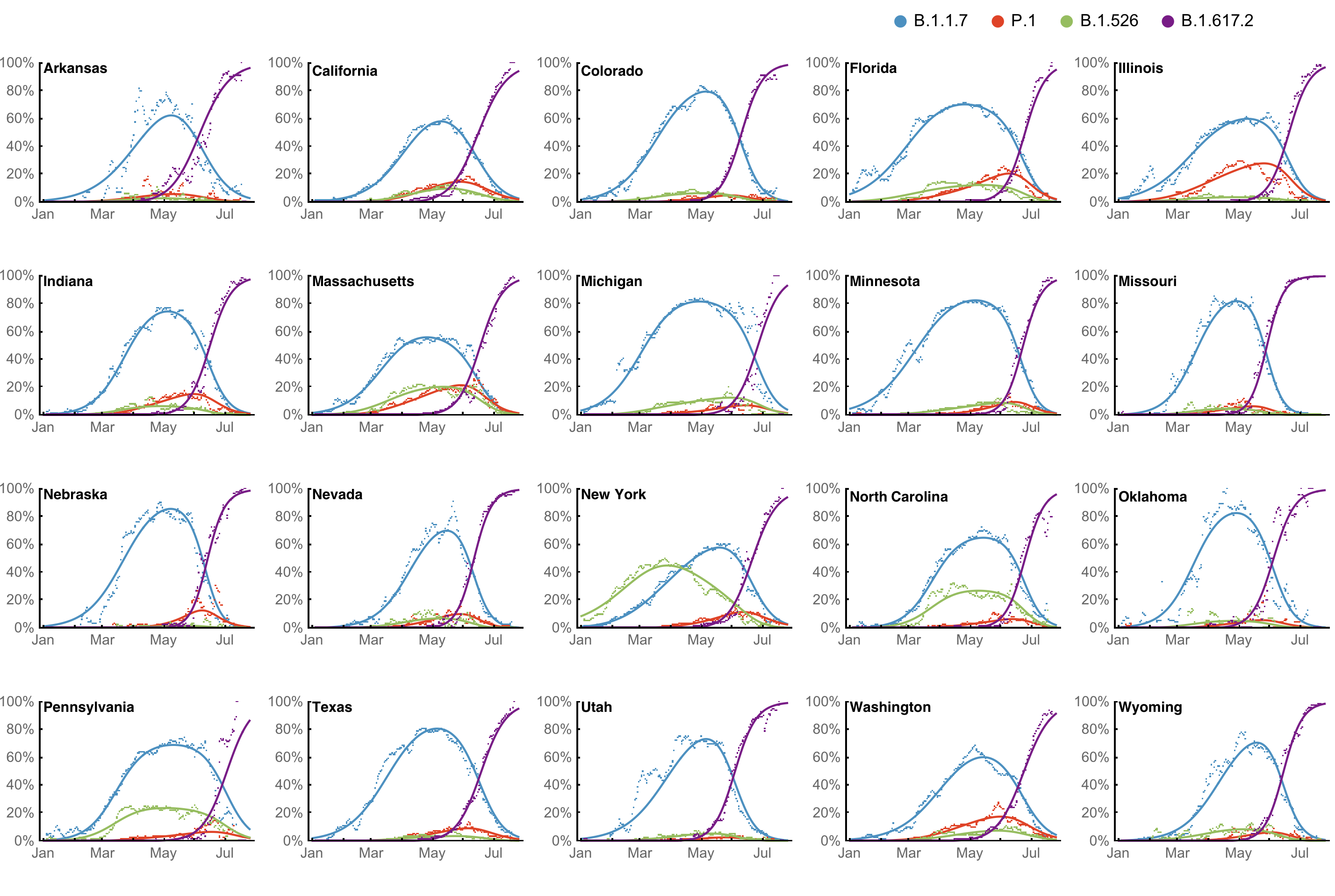
However, frequency of a variant may rise while cases fall
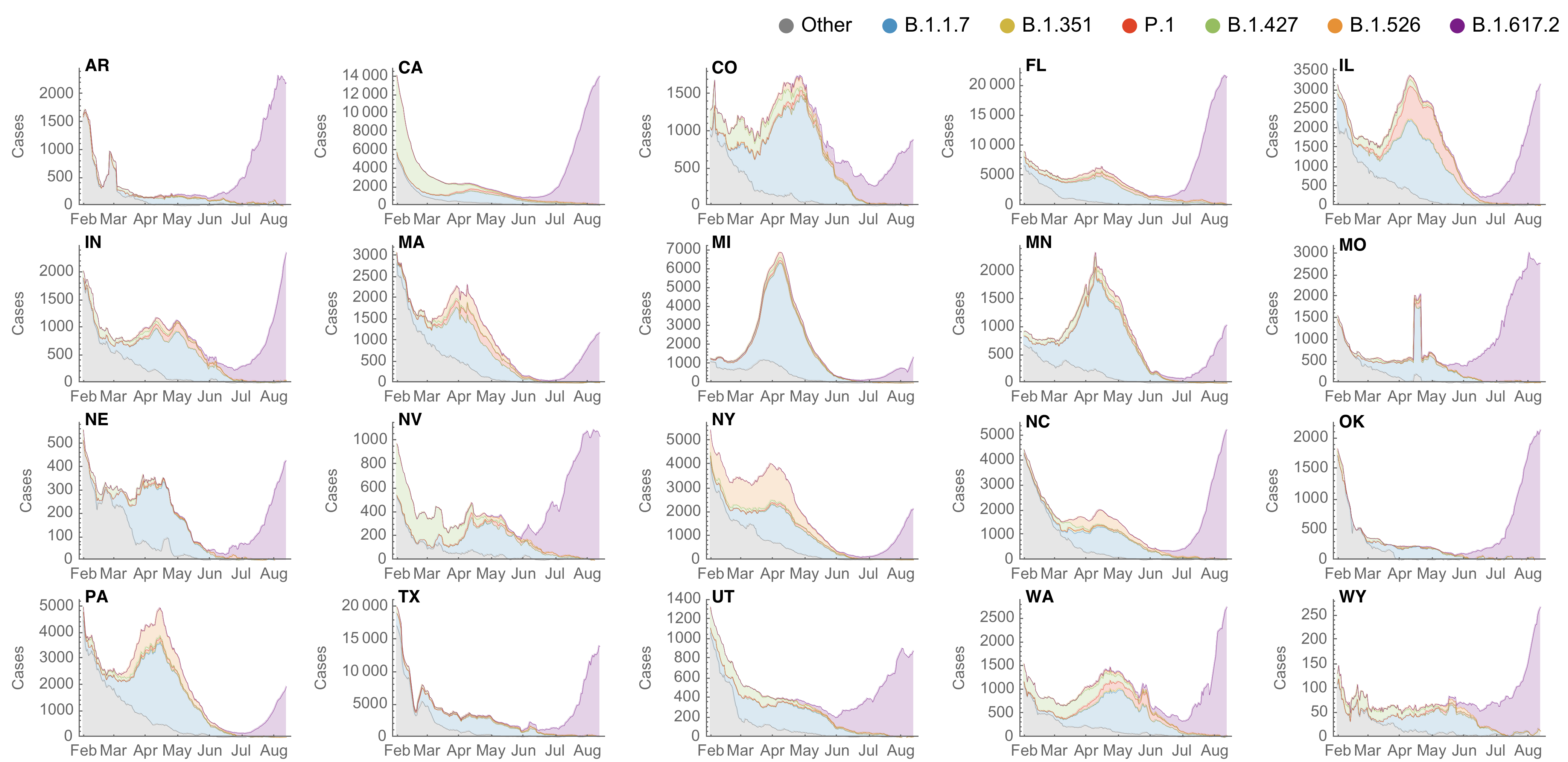
SARS-CoV-2 variant dynamics across US states show consistent differences in transmission rates
 Marlin Figgins
Marlin Figgins
Estimation of variant-specific Rt through time using state-level data
State-level case counts are partitioned based on frequencies of sequenced cases
Partitioned case counts used to estimate variant-specific Rt through time
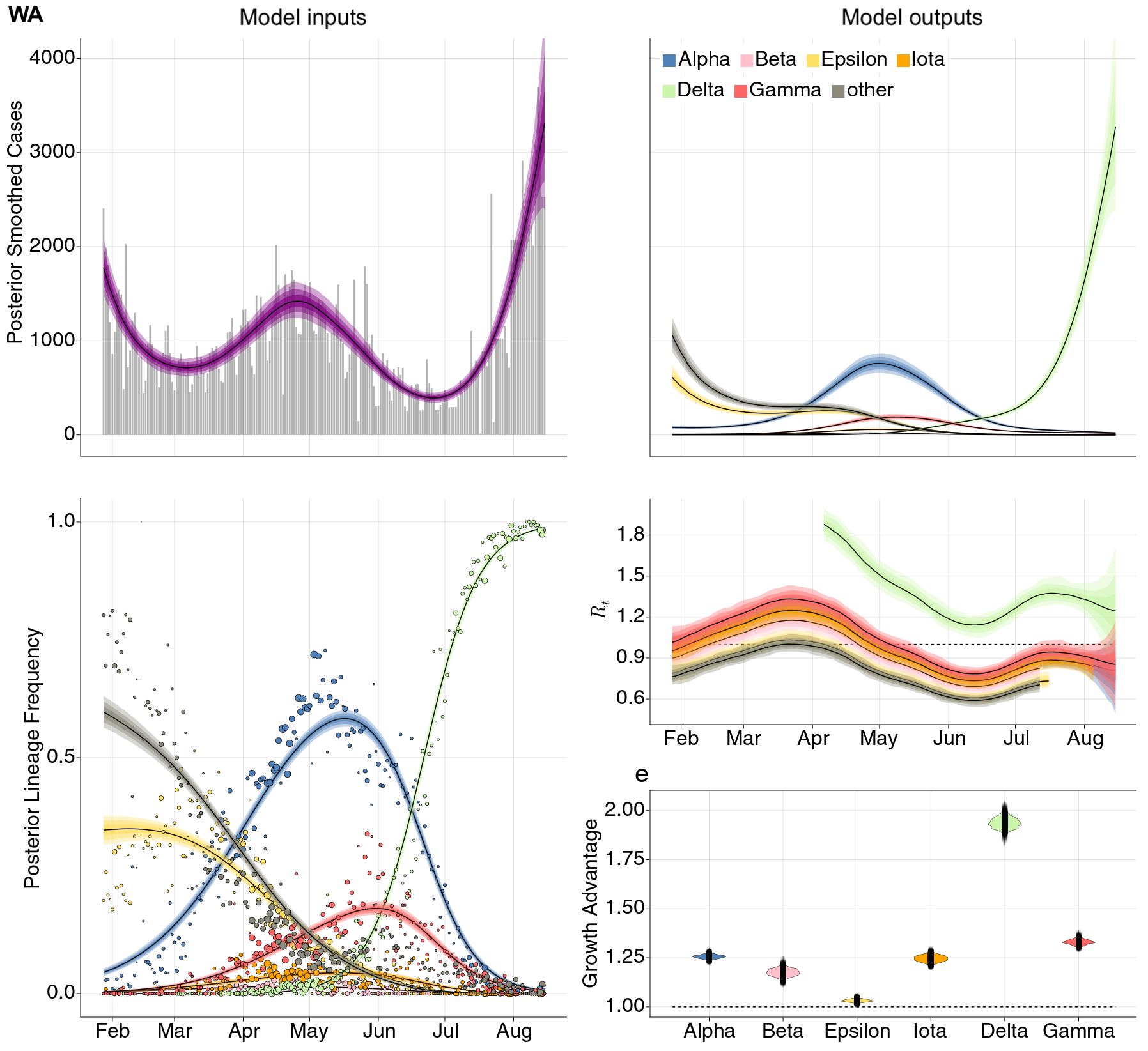
Differences in intrinsic Rt across variants, but all trending downwards
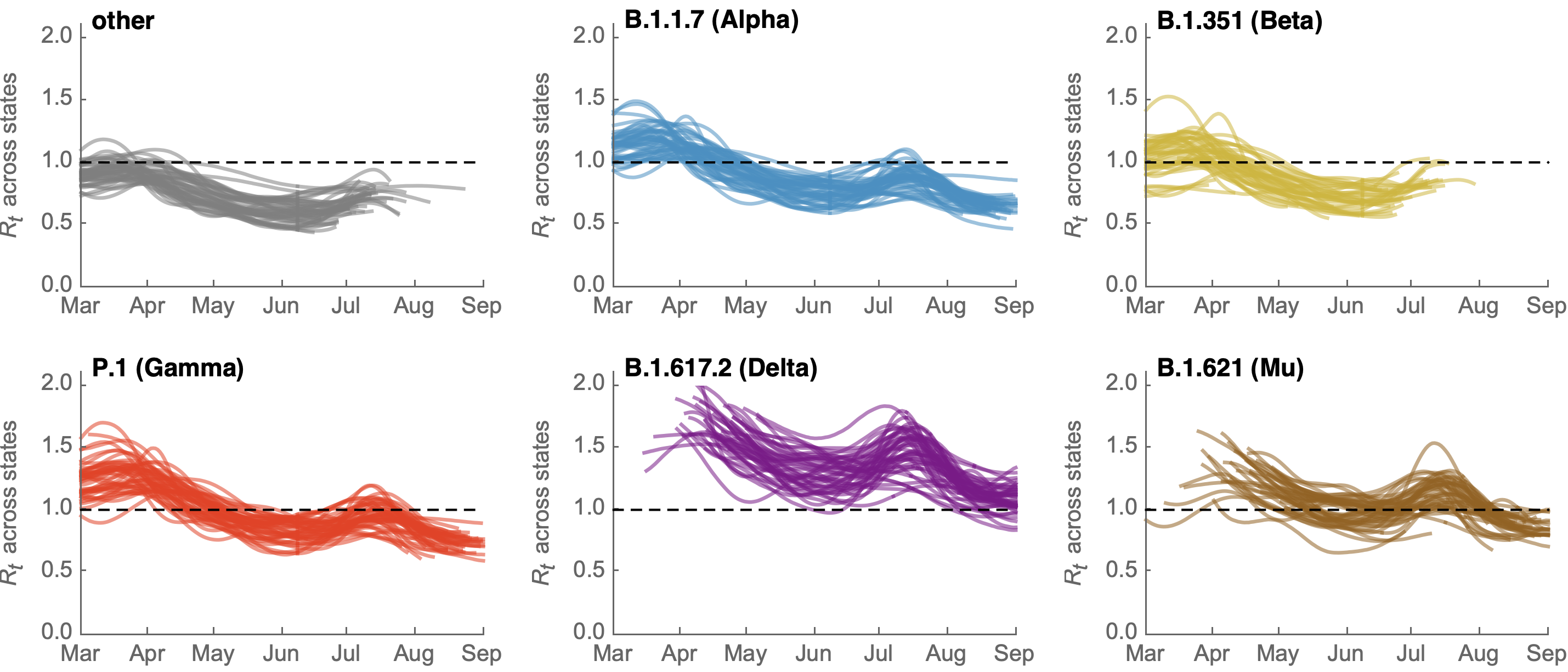
Consistent differences in variant-specific transmission rate across states
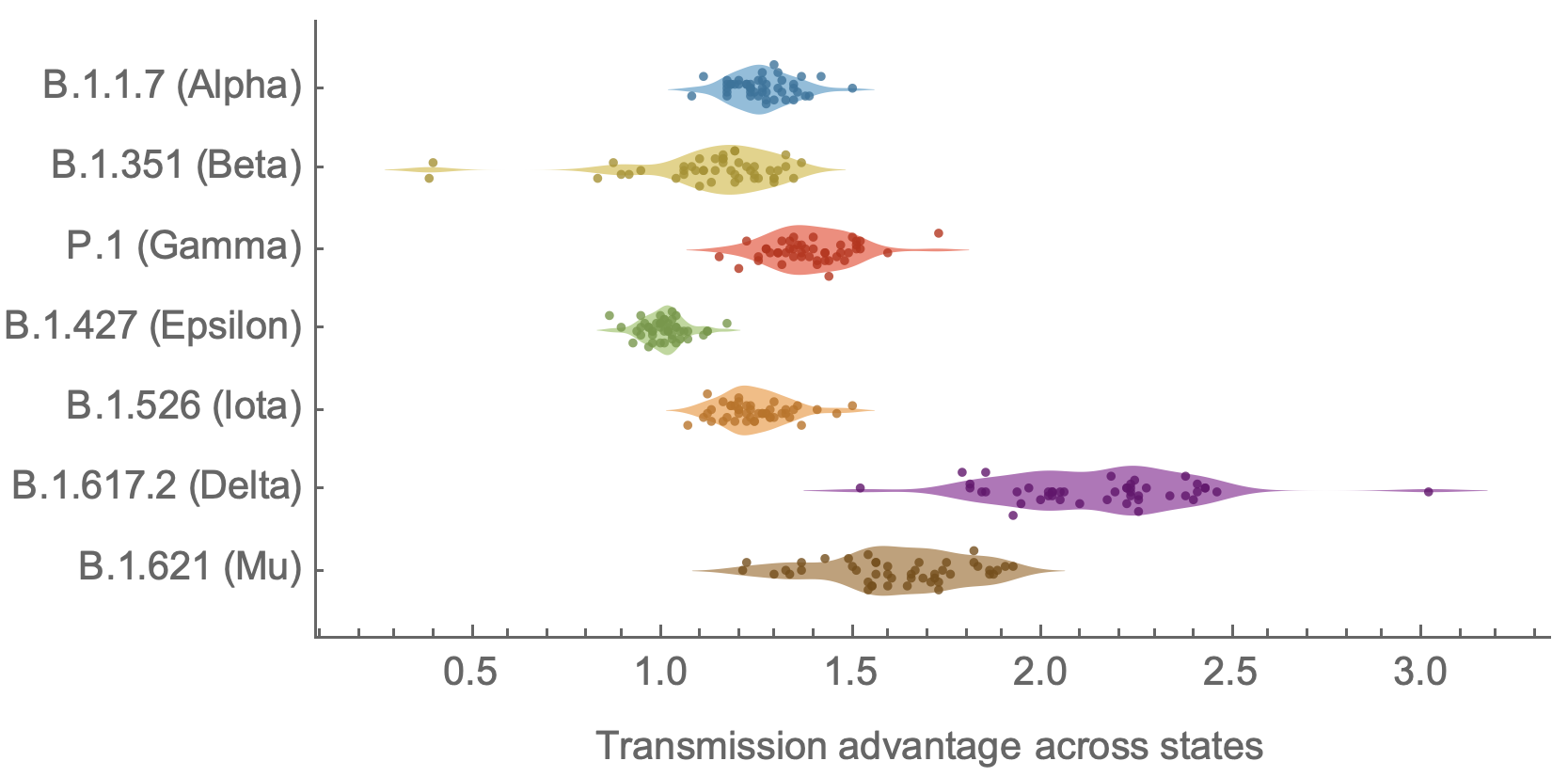
Consistent reductions in variant-specific Rt from vaccination
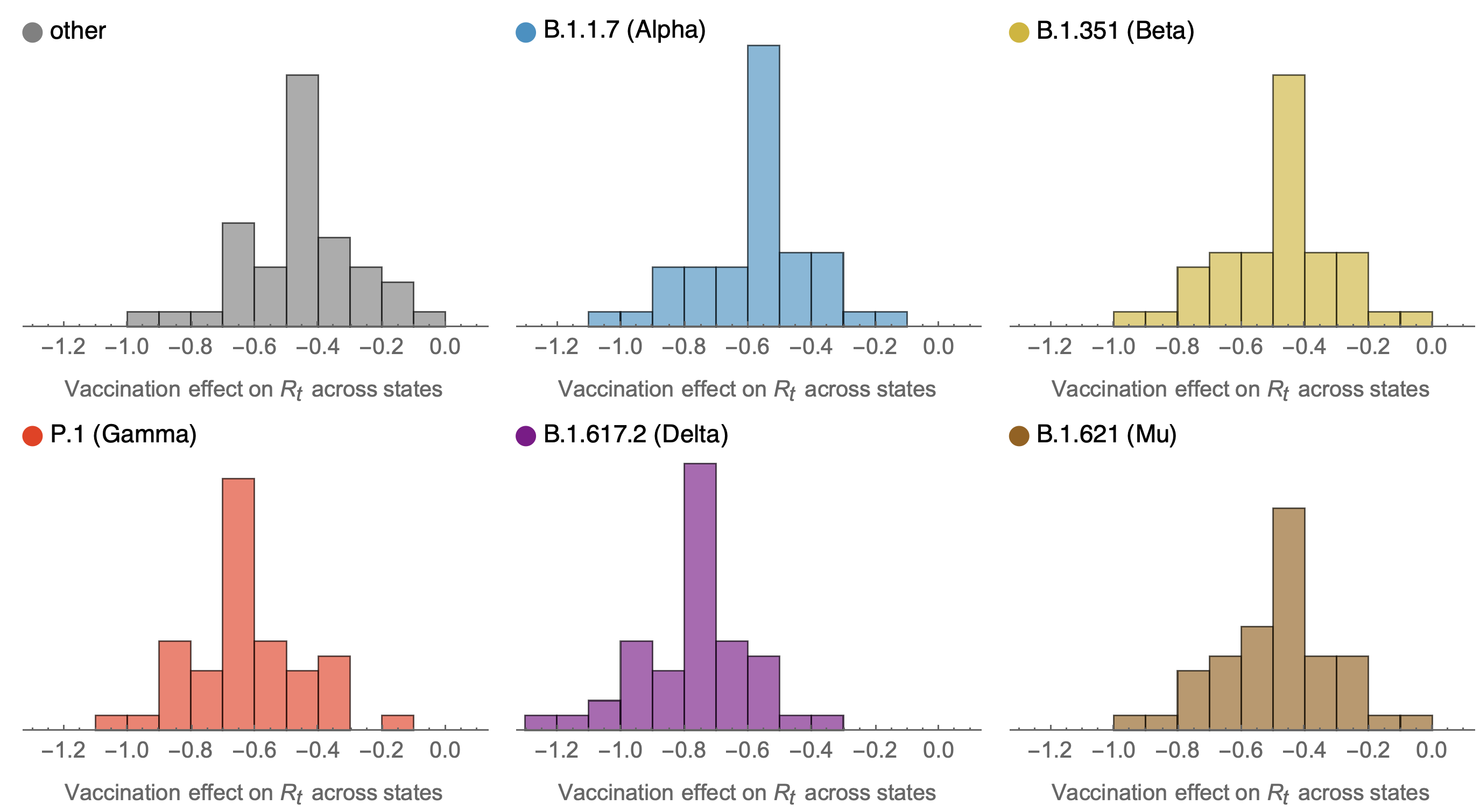
Future work would ideally tie together granular empirical estimates of viral fitness from frequency data together with mutational and phenotypic predictors to learn what is driving variant success
Generally expect a transition from evolution being driven by selection for increased transmission to sustained selection for antigenic drift
Acknowledgements
SARS-CoV-2 genomic epi: Data producers from all over the world, GISAID and the Nextstrain team
Bedford Lab:
![]() John Huddleston,
John Huddleston,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Louise Moncla,
Louise Moncla,
![]() Maya Lewinsohn,
Maya Lewinsohn,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Cassia Wagner,
Cassia Wagner,
![]() Miguel Paredes,
Miguel Paredes,
![]() Nicola Müller,
Nicola Müller,
![]() Marlin Figgins,
Marlin Figgins,
![]() Eli Harkins,
Eli Harkins,
![]() Denisse Sequeira
Denisse Sequeira






