Evolutionary dynamics of SARS-CoV-2
Trevor Bedford (@trvrb)
Associate Professor, Fred Hutchinson Cancer Research Center
22 Mar 2021
Comp Bio Faculty Lunch
Fred Hutch
Slides at: bedford.io/talks
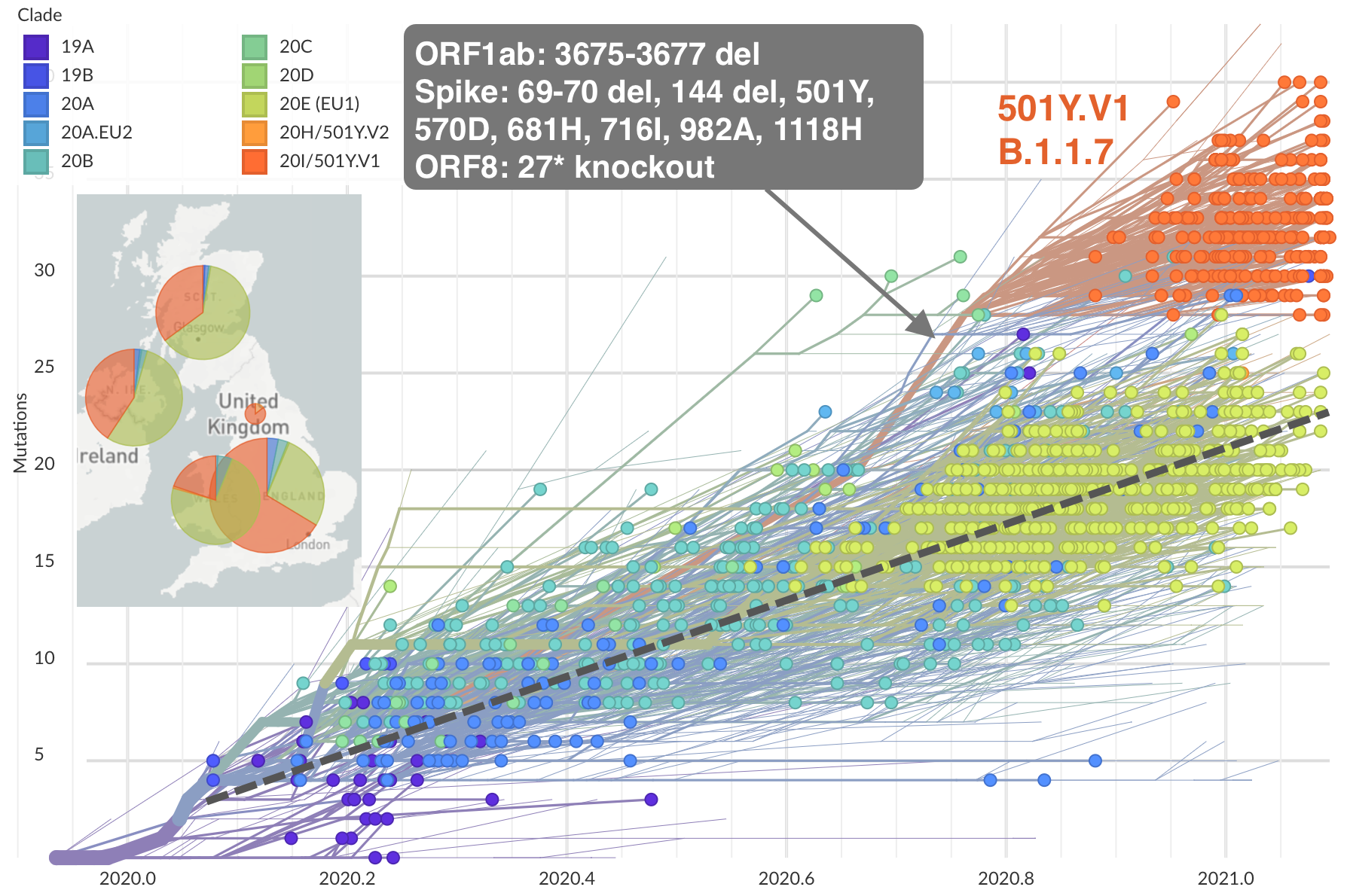
Rapid replacement of existing diversity by lineage B.1.1.7

1. Real-time tracking of SARS-CoV-2 evolution
2. Emergence of variants of concern
3. Current circulation patterns
4. Expectations for antigenic evolution
5. Scale of genomic surveillance required
Real-time tracking of SARS-CoV-2 evolution
Spike protein is critical for cell invasion by the virus and is the primary target for human immune response
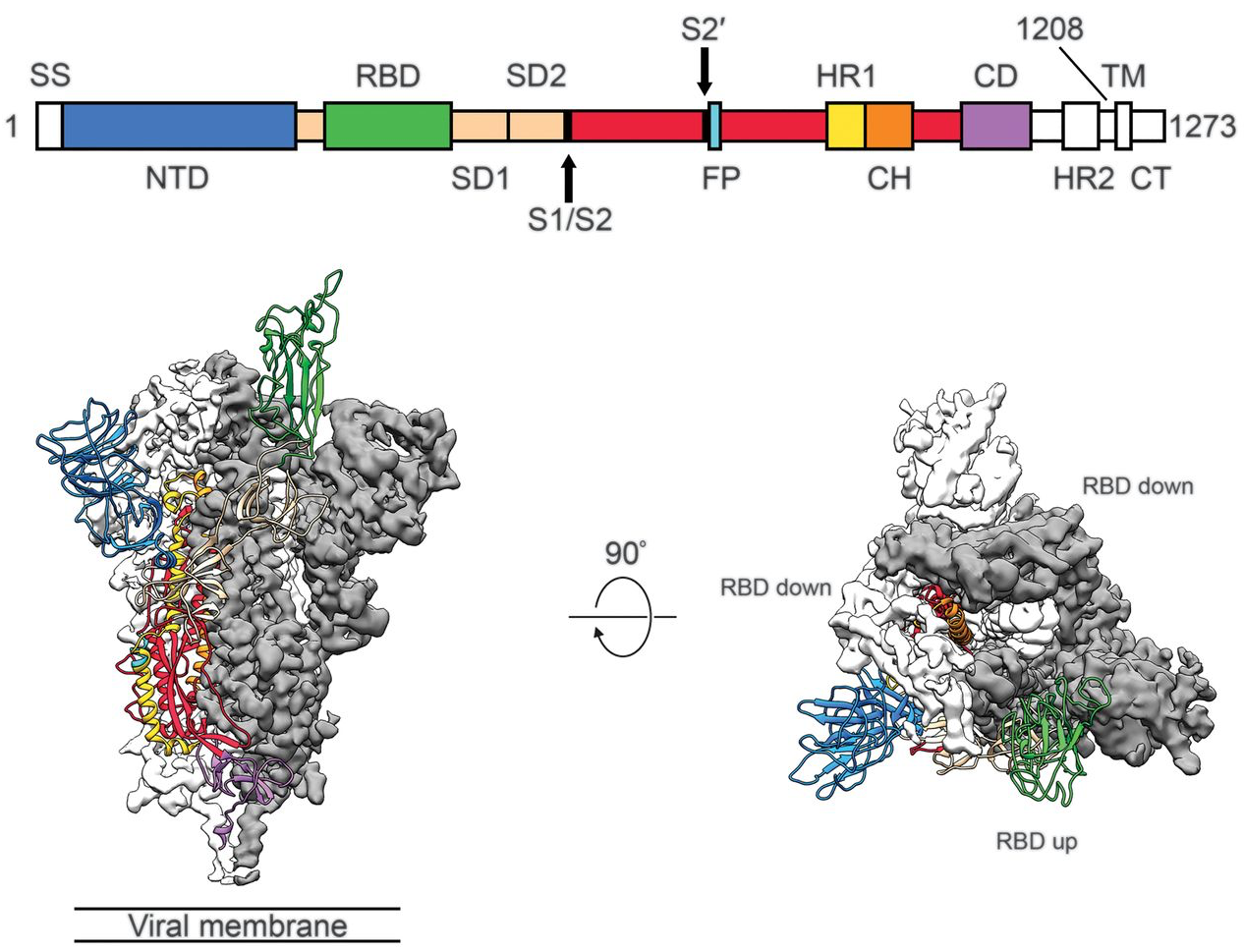
Over 800k SARS-CoV-2 genomes shared to GISAID and evolution tracked in real-time at nextstrain.org

SARS-CoV-2 lineages establish globally in February and March
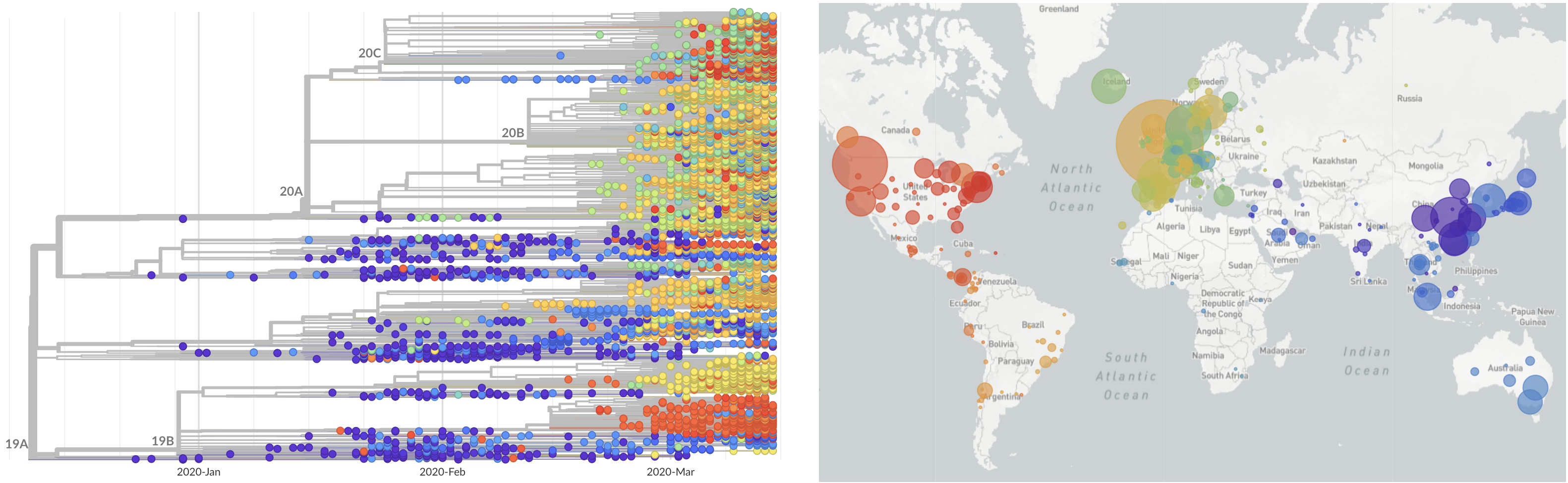
Limited early mutations like D614G spread globally during initial wave
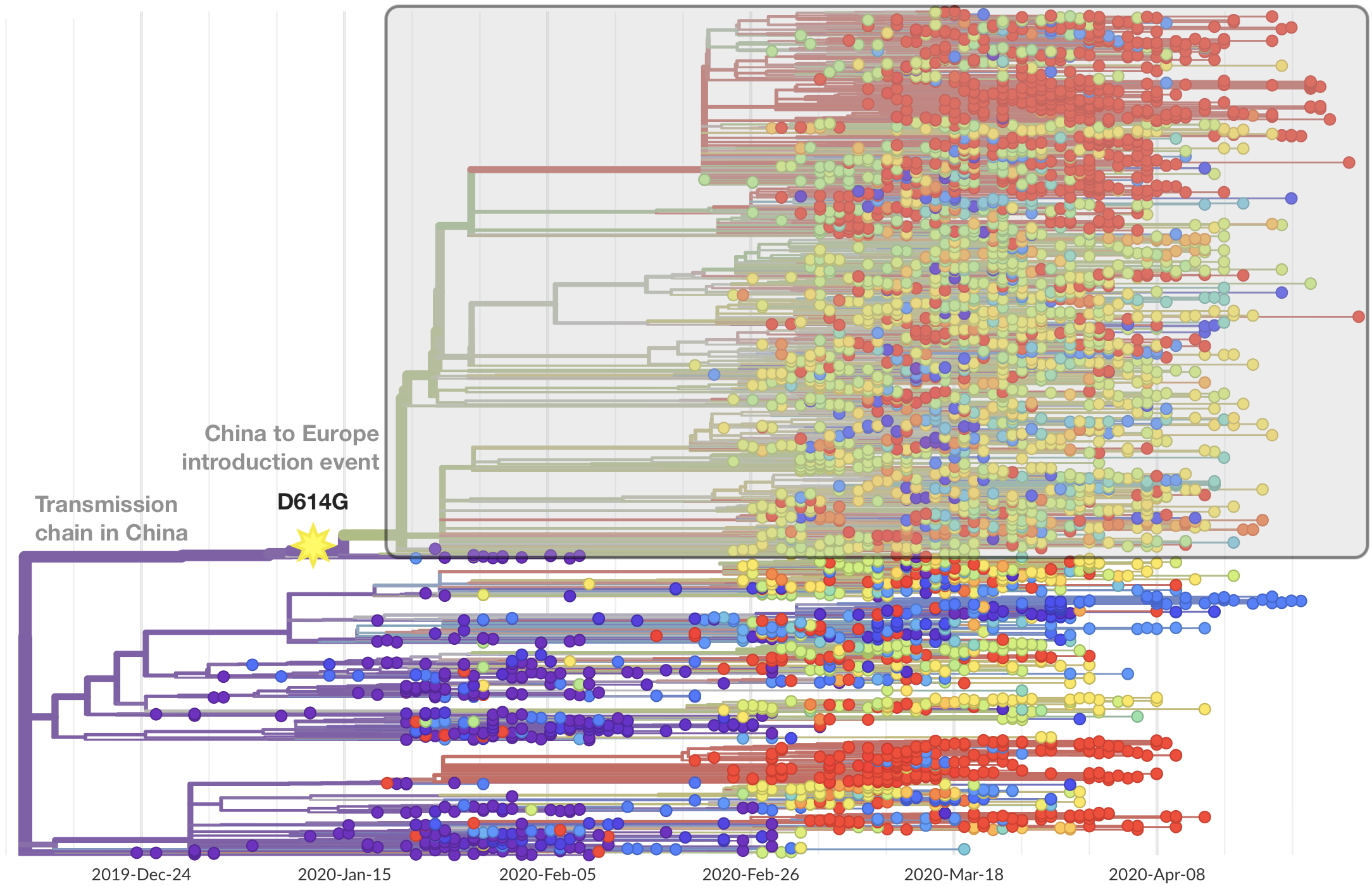
After initial wave, with mitigation
efforts and decreased travel,
regional clades emerge
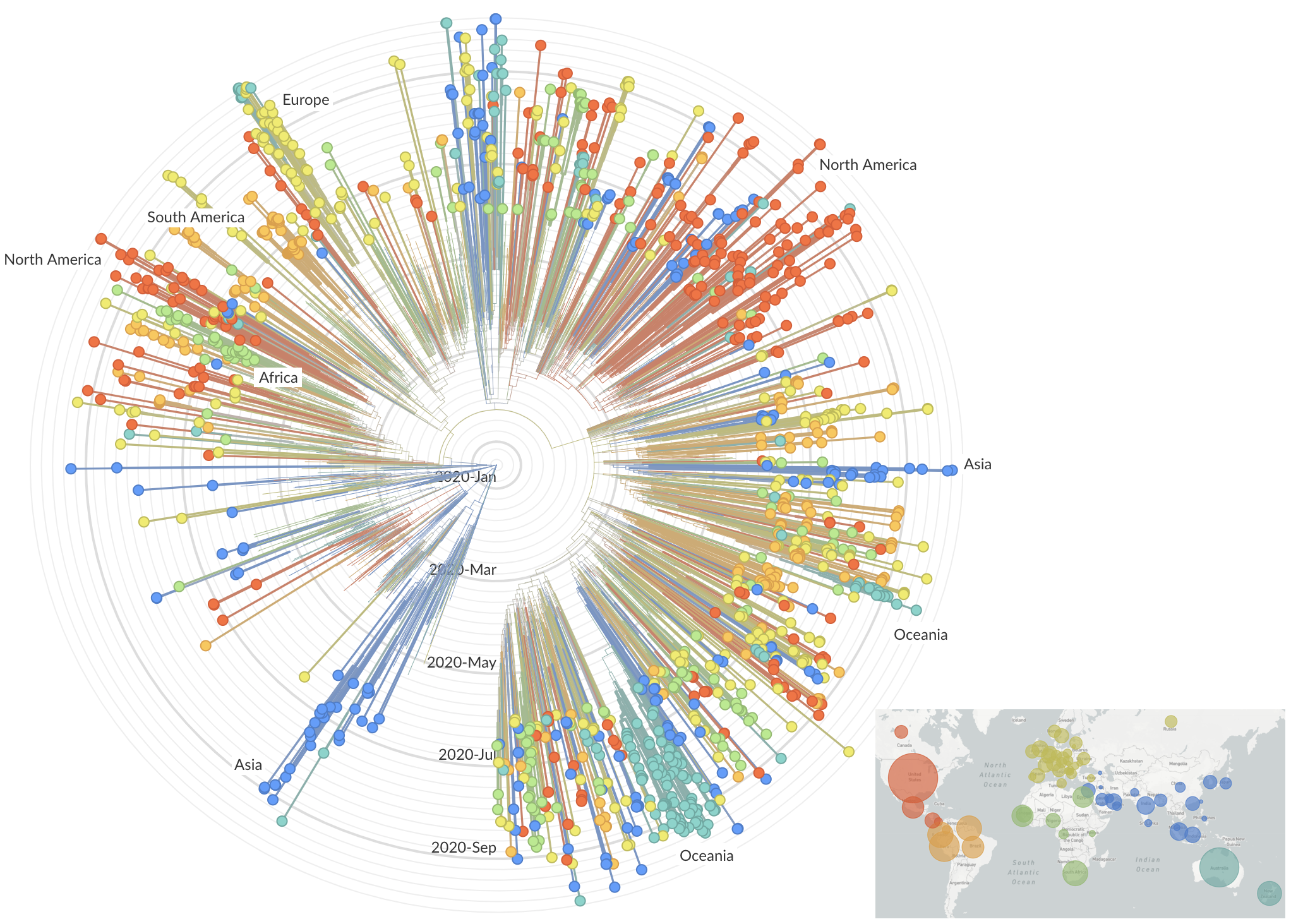
Summer and fall mutations were confined to regional dominance
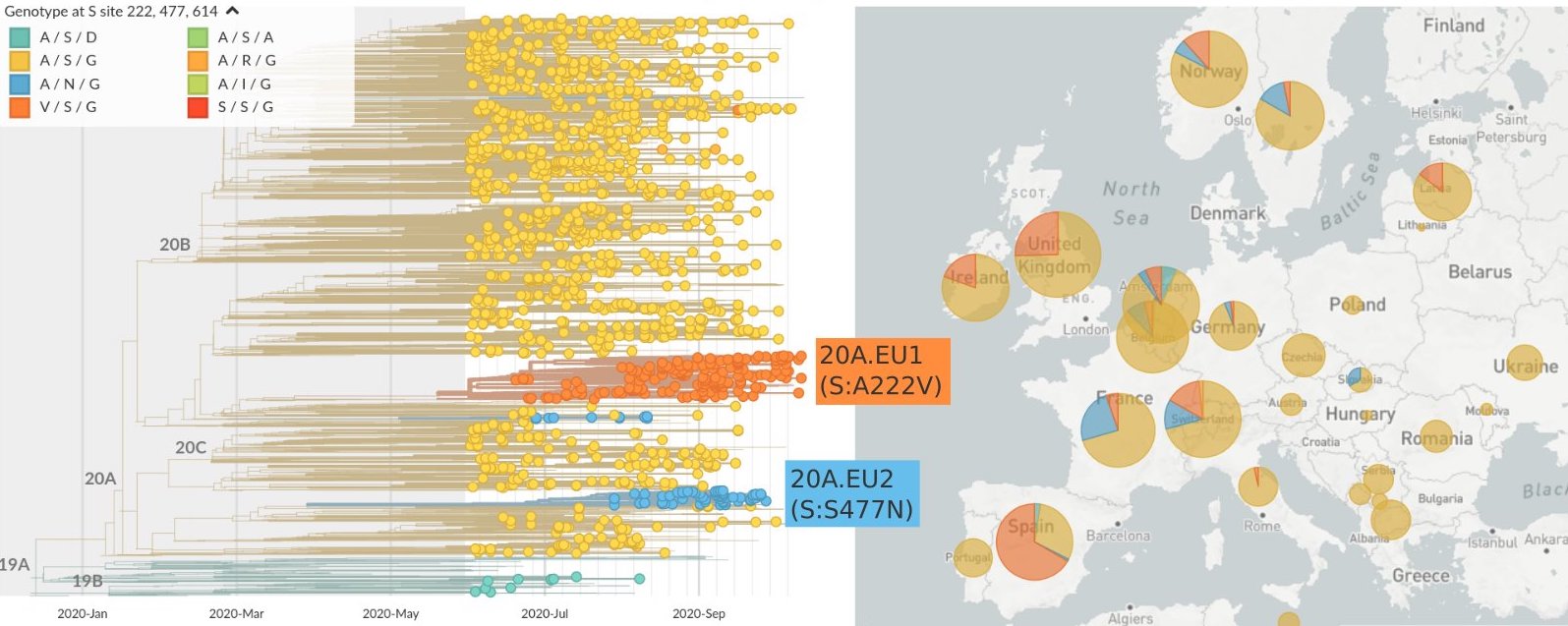
Emergence of variants of concern
484K and 501Y repeatedly emerging across the world

Emergence of 501Y.V1 (B.1.1.7) in the UK
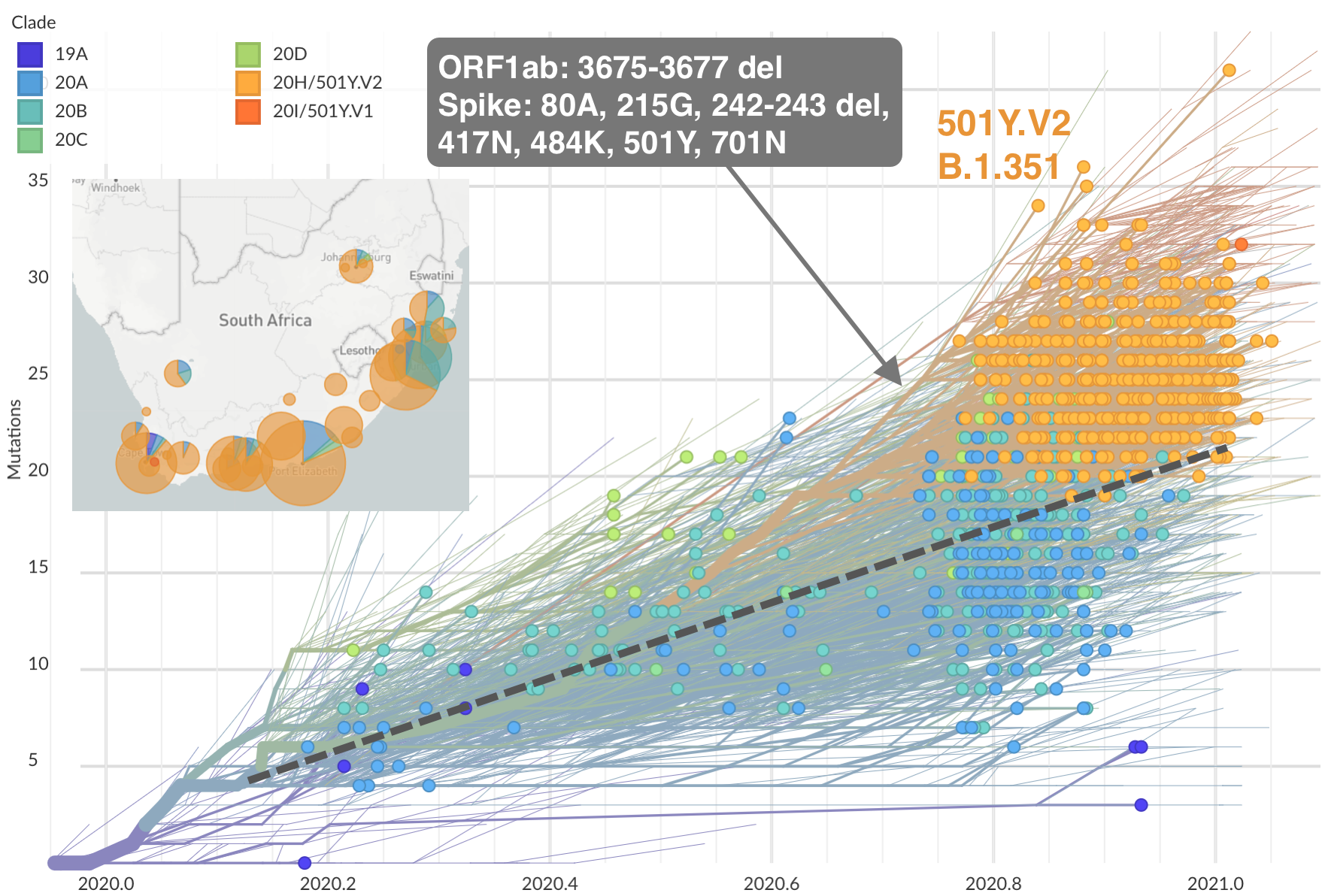
Emergence of 501Y.V2 (B.1.351) in the South Africa
Emergence of 501Y.V3 (P.1) in the Brazil

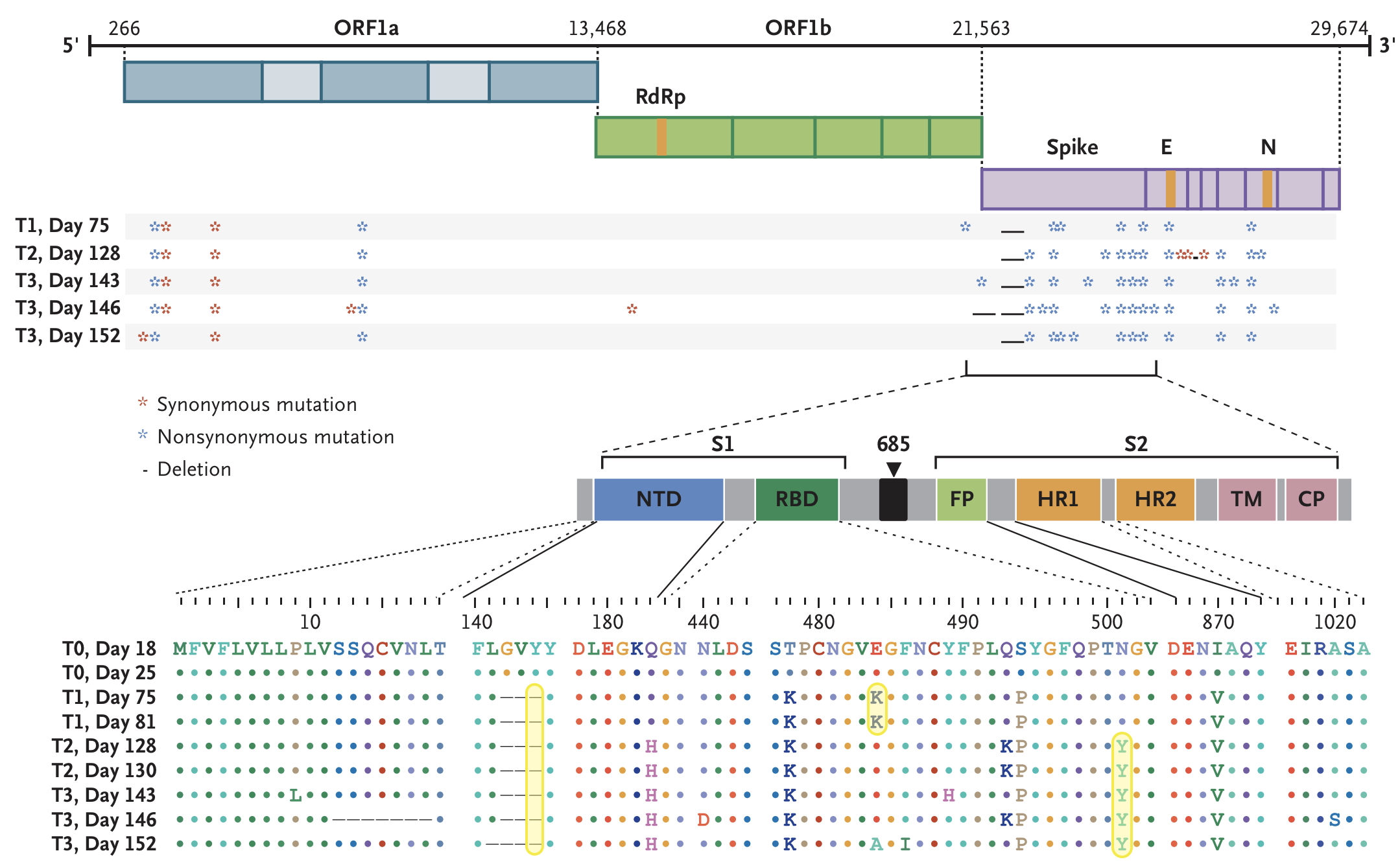
Working hypothesis of within-host evolution occurring during prolonged infection, driven by natural selection for immune escape.
Rapid within-host evolution during persistent infection
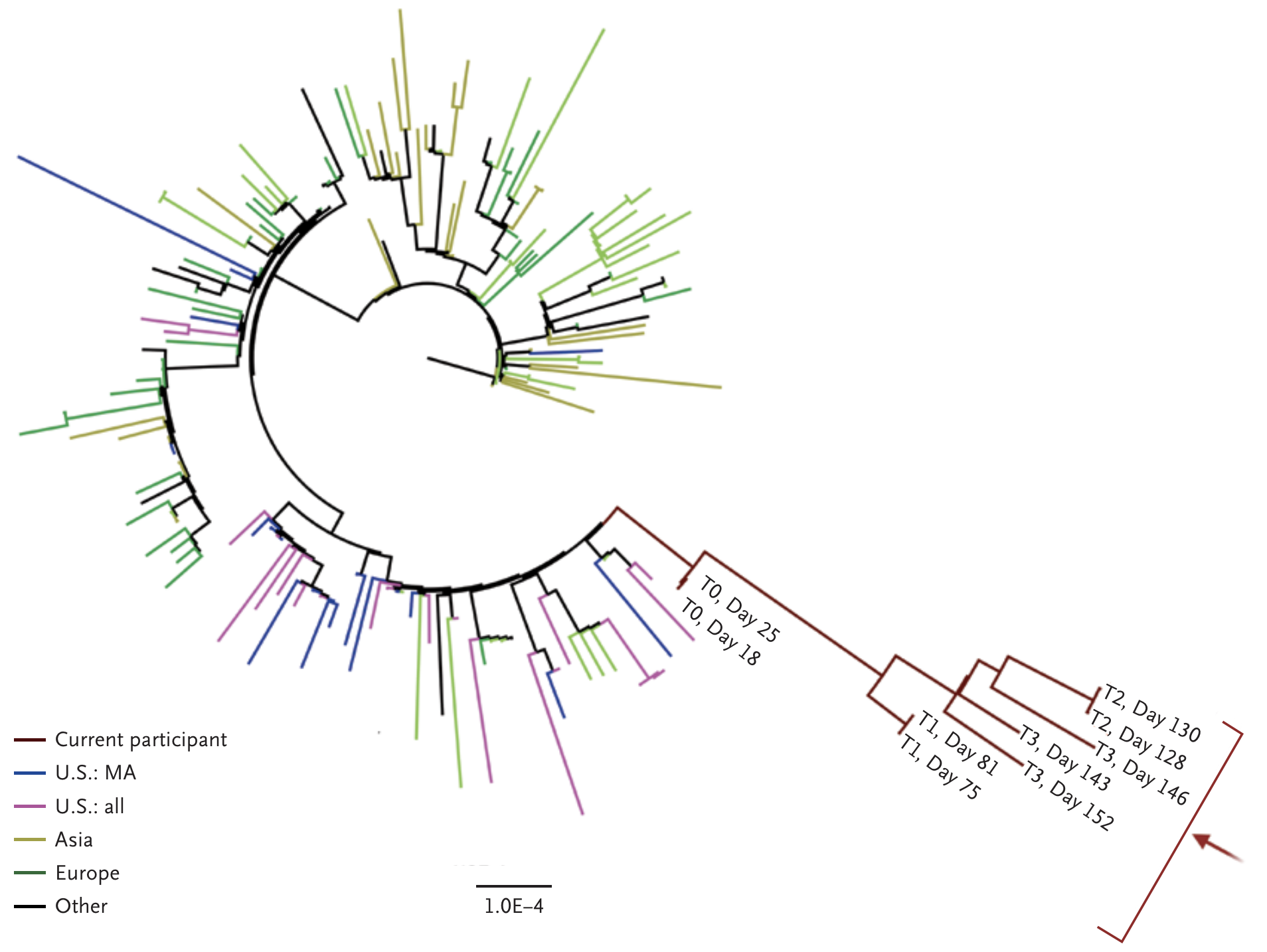
484K and 501Y observed during this evolution
Current circulation patterns
VOCs are growing in frequency with B.1.1.7 leading the curve

Repeated convergent evolution across sites in spike (452R)

Repeated convergent evolution across sites in spike (681H)

Repeated convergent evolution across sites in spike (69del)

Repeated convergent evolution across sites in spike (combined)

Certain viruses with substantial S1 divergence

Particular mutations increasing globally

Interested in how constellations behave

Expectations for antigenic evolution
Up to December, my expectation was evolution as seen in seasonal coronaviruses

OC43 and 229E show flu B-like rates of adaptive evolution in S1

~23 mutations per year across SARS-CoV-2 genome

Substantial increase in spike S1 amino acid substitutions in VOCs

Accumulation of substitutions faster in NTD than RBD

Spike S1 rate of 2.9 subs per year similar to rate in HA1 of flu A

Influenza H3N2 drifts at ~1 two-fold HI dilution per year
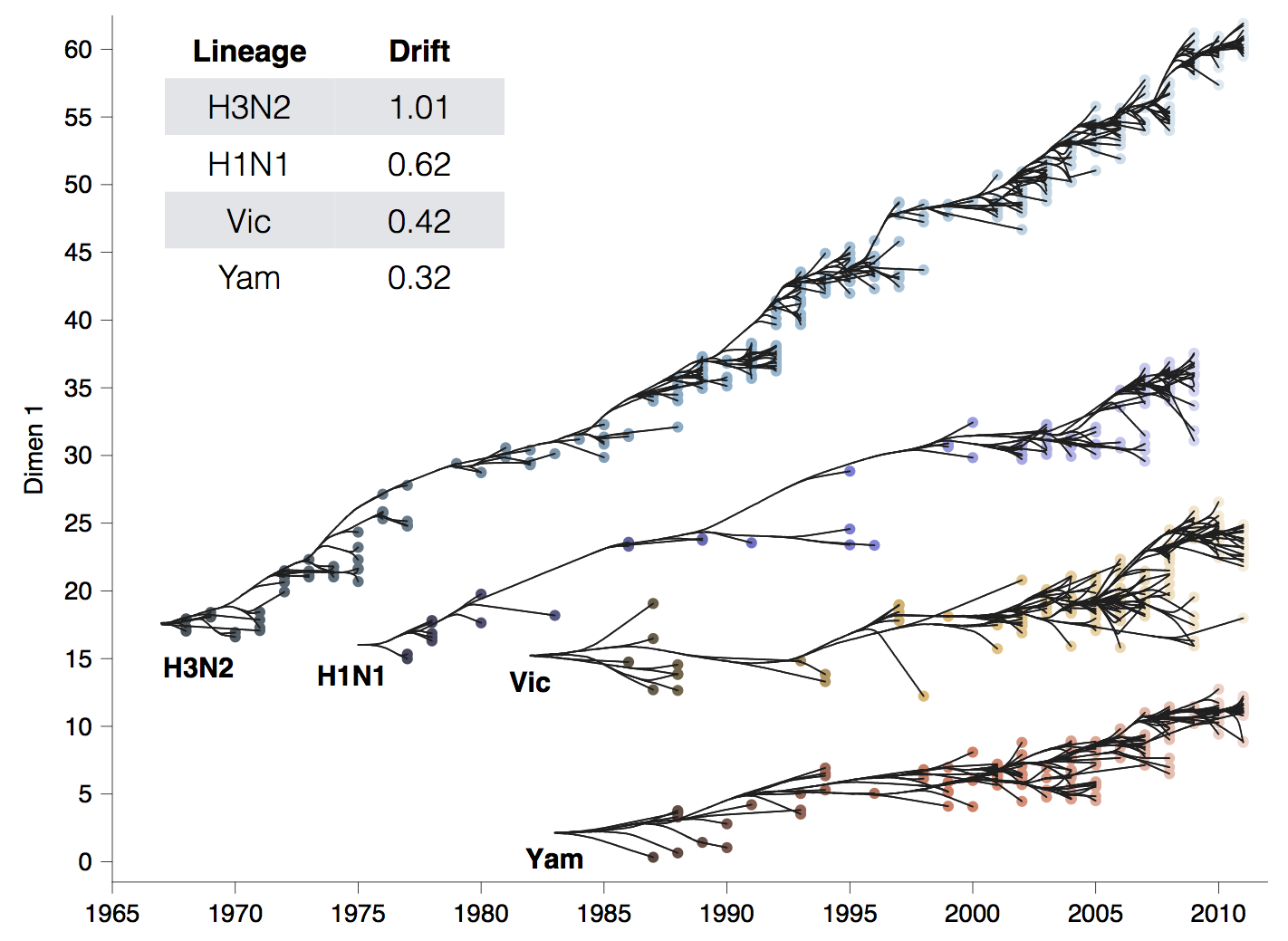
501Y.V2 shows an ~8-fold drop in neutralization titer, equivalent to ~3 years of H3N2 evolution

Genomic surveillance
Sequences generated and shared at an unprecedented pace with 185k genomes shared in Feb 2021 alone

Data from gisaid.org
My favorite metric is number of sequences available from samples collected in the past 30 days

Data from gisaid.org
Front-runners of the UK, Denmark and the US have dramatically increased throughput, but other regions lag

Data from gisaid.org
If the goal is early detection of emerging variants to lay groundwork for vaccine updates, another 1000 genomes per month would be far more valuable from South America or Africa than 1000 more genomes from the US or the UK
Aim for rapid turnaround of recent samples rather than volume
Important variant bearing 141-143del, 484K, 501Y and 681H detected in the Philippines

Generally, I suspect we're looking at a data-rich version of the influenza strain selection process
- mRNA vaccines allow faster turnarounds and may push for more regional strategies
- Work directly from human serological data rather than naive animals
- Multiple valencies possible, clinical trials should be conducted now comparing regimens
Acknowledgements
SARS-CoV-2 genomic epi: Data producers from all over the world, GISAID and the Nextstrain team
Bedford Lab:
![]() Alli Black,
Alli Black,
![]() John Huddleston,
John Huddleston,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Louise Moncla,
Louise Moncla,
![]() Maya Lewinsohn,
Maya Lewinsohn,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Kairsten Fay,
Kairsten Fay,
![]() Misja Ilcisin,
Misja Ilcisin,
![]() Cassia Wagner,
Cassia Wagner,
![]() Miguel Paredes,
Miguel Paredes,
![]() Nicola Müller,
Nicola Müller,
![]() Marlin Figgins,
Marlin Figgins,
![]() Eli Harkins
Eli Harkins






