Continuing evolution of SARS-CoV-2
Trevor Bedford
Fred Hutchinson Cancer Center / Howard Hughes Medical Institute
4 Dec 2023
Biozentrum Symposium on Adaptation and Immune escape of RNA viruses
University of Basel
Slides at: bedford.io/talks
Population immunity has driven a dramatic shift in death toll

But this immunity is now driving evolution of the virus

In which new variants emerge that escape from existing population immunity and spread rapidly

Novel variants sweep globally in months rather than years


We really should be at something past Psi at this point

1. SARS-CoV-2 genome evolution
2. Variant frequency dynamics
3. Evolution driving epidemics
4. Forecasting
5. Saltational evolution
SARS-CoV-2 genome evolution
Phylogeny of 2k viruses sampled globally in June and July 2022

Measure clade growth as a proxy for viral fitness

Across 2020 to 2023, clades with more S1 mutations grow faster

Rate of amino acid evolution and correlation between mutation and clade growth strongest in S1

Strength of adaptive evolution consistently high through time

Mutations at spike S1 propel escape from population immunity

These mutations are accumulating just as quickly post-Omicron

And are accruing much more rapidly than other endemic viruses

Variant frequency dynamics
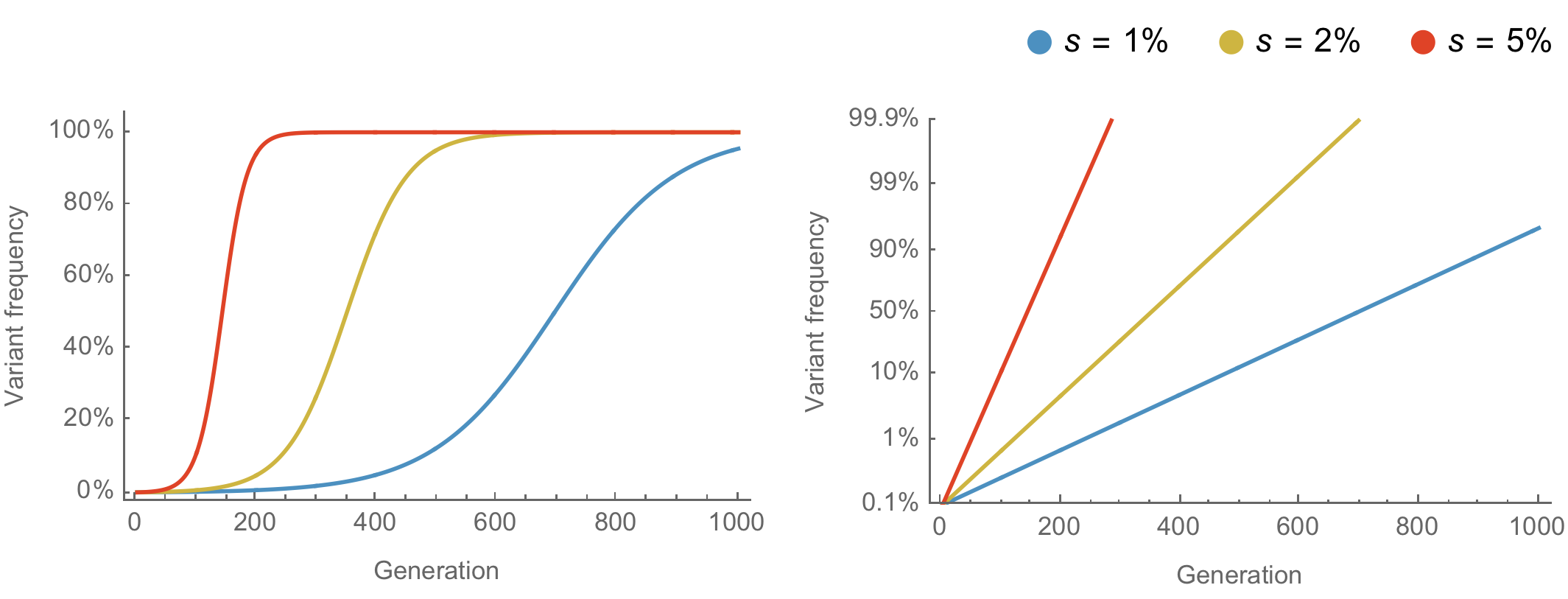
Population genetic expectation of variant frequency under selection
$x' = \frac{x \, (1+s)}{x \, (1+s) + (1-x)}$ for frequency $x$ over one generation with selective advantage $s$
$x(t) = \frac{x_0 \, (1+s)^t}{x_0 \, (1+s)^t + (1-x_0)}$ for initial frequency $x_0$ over $t$ generations
Trajectories are linear once logit transformed via $\mathrm{log}(\frac{x}{1 - x})$
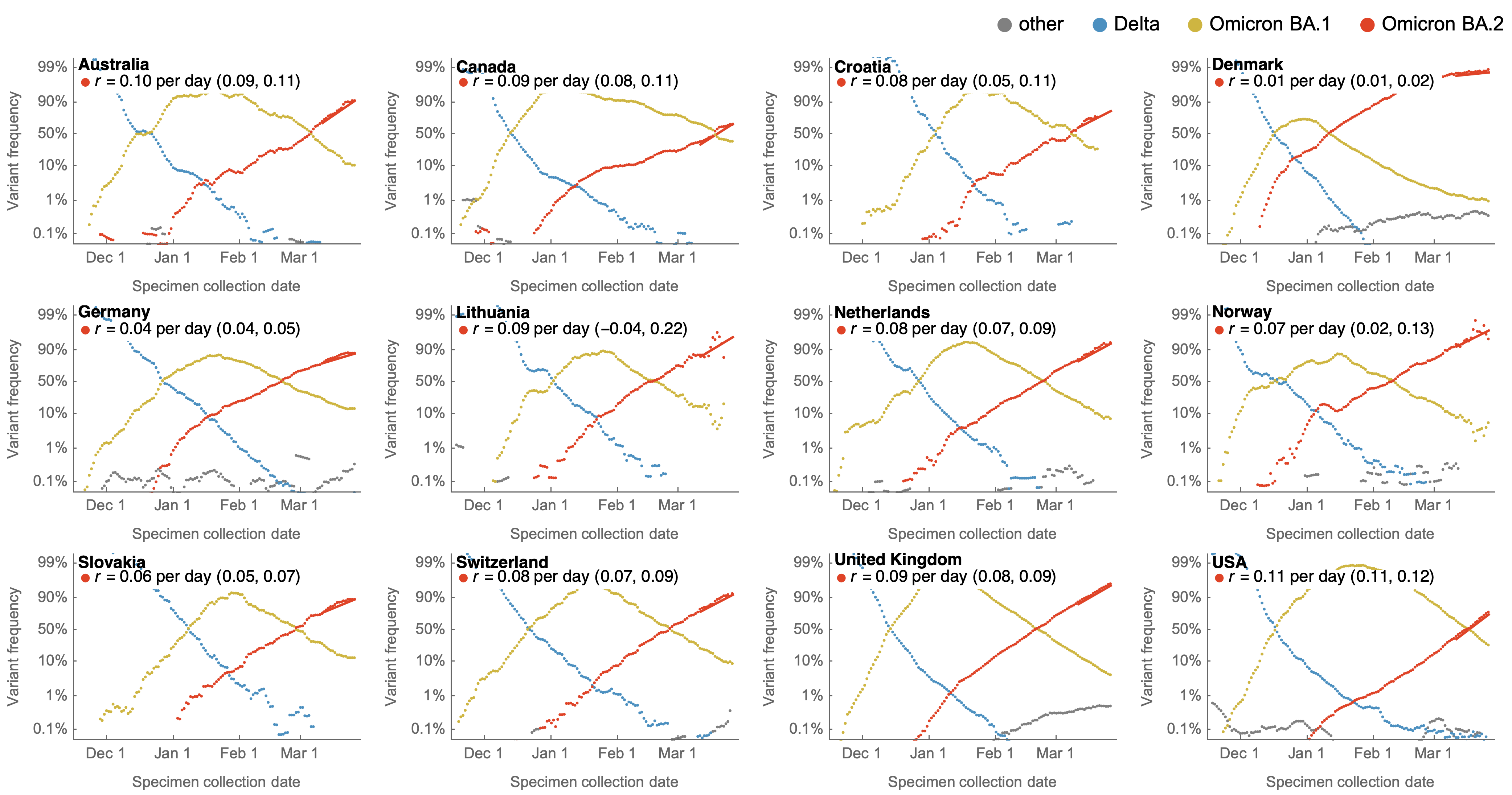
Consistent frequency dynamics in logit space (BA.2 Mar 2022)
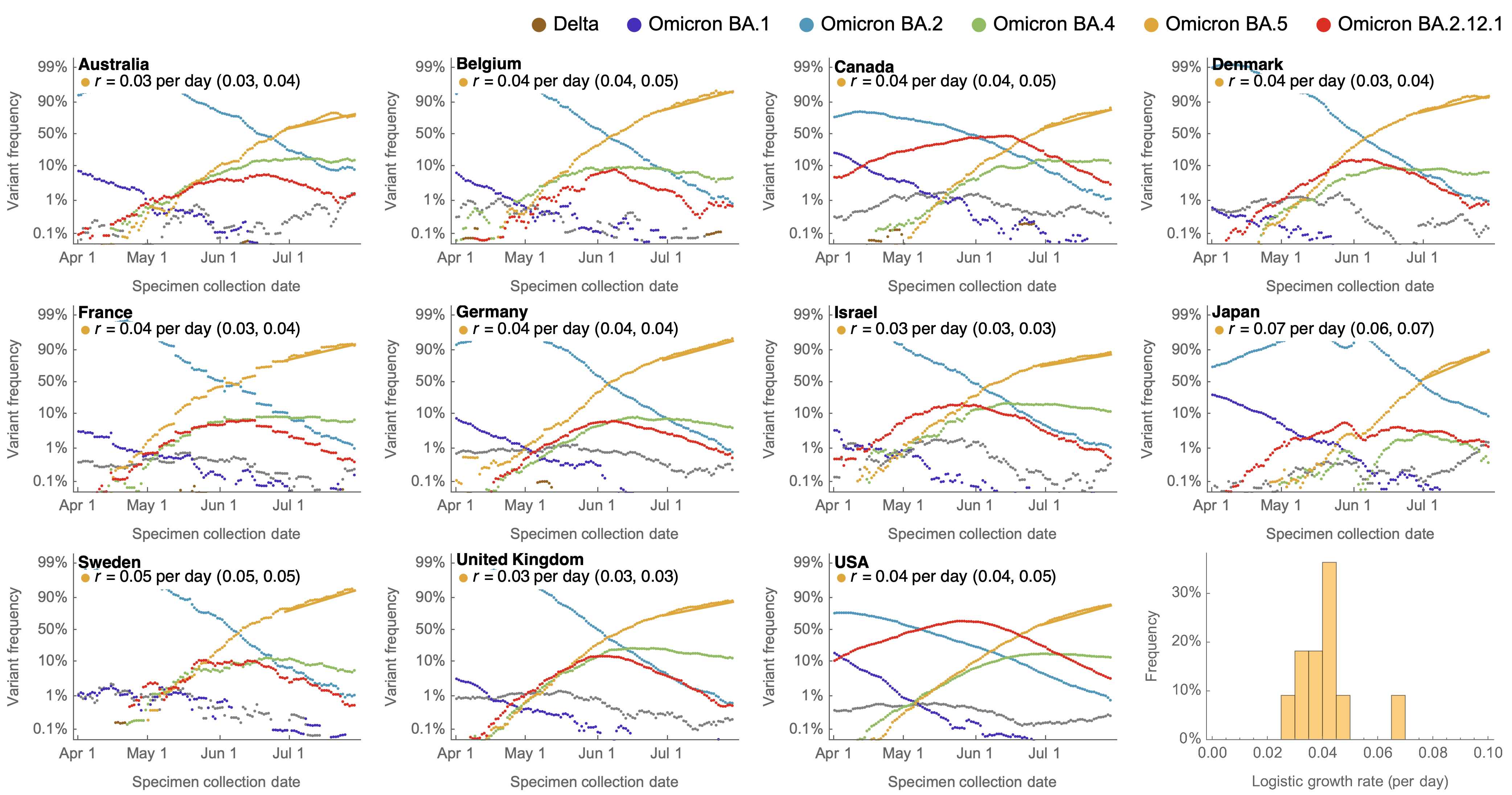
Consistent frequency dynamics in logit space (BA.5 Jul 2022)
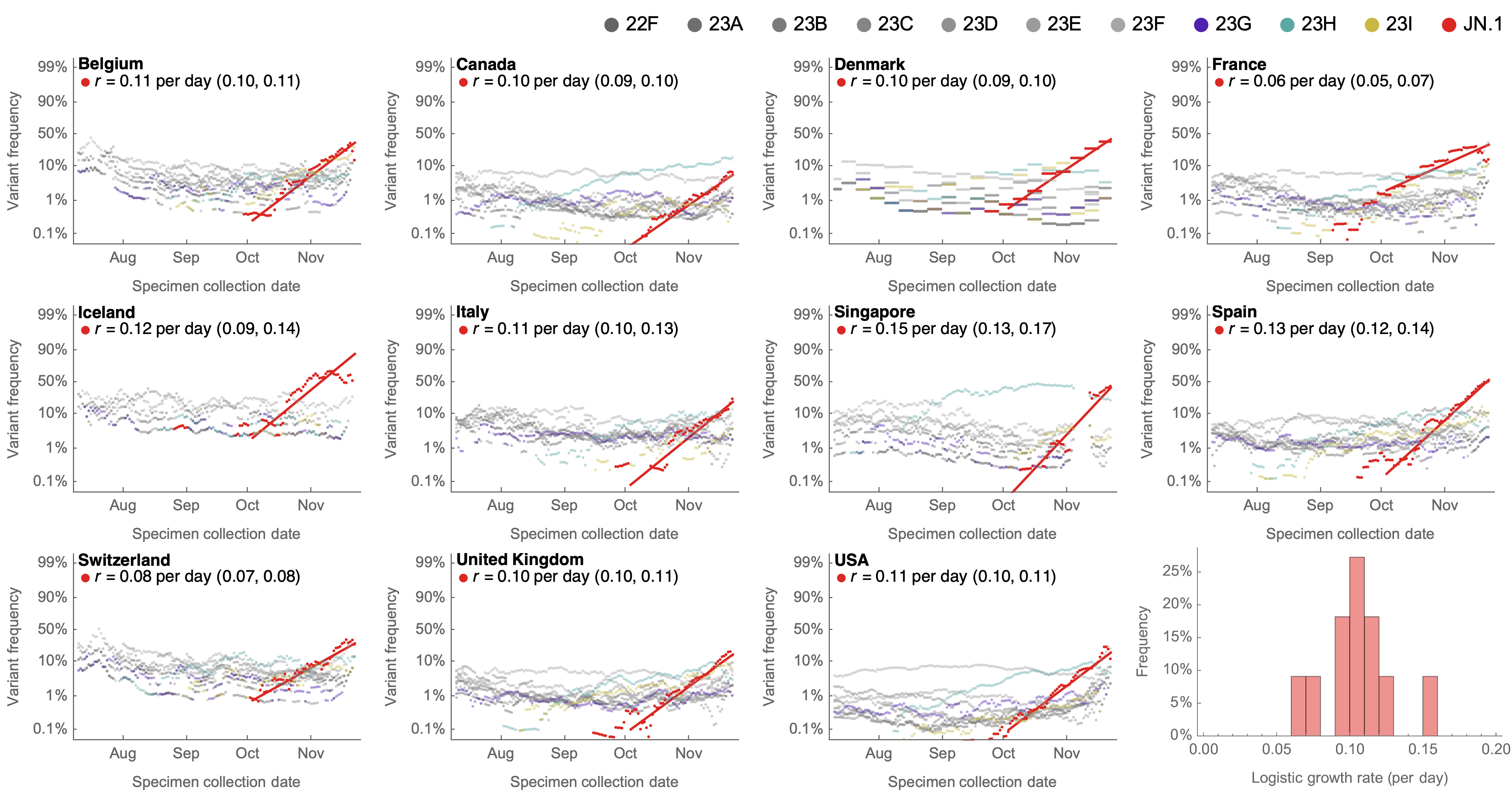
Consistent frequency dynamics in logit space (JN.1 Nov 2023)
Multinomial logistic regression
Multinomial logistic regression across $n$ variants models the probability of a virus sampled at time $t$ belonging to variant $i$ as
$$\mathrm{Pr}(X = i) = x_i(t) = \frac{p_i \, \mathrm{exp}(f_i \, t)}{\sum_{1 \le j \le n} p_j \, \mathrm{exp}(f_j \, t) }$$
with $2n$ parameters consisting of $p_i$ the frequency of variant $i$ at initial timepoint and $f_i$ the growth rate or fitness of variant $i$.
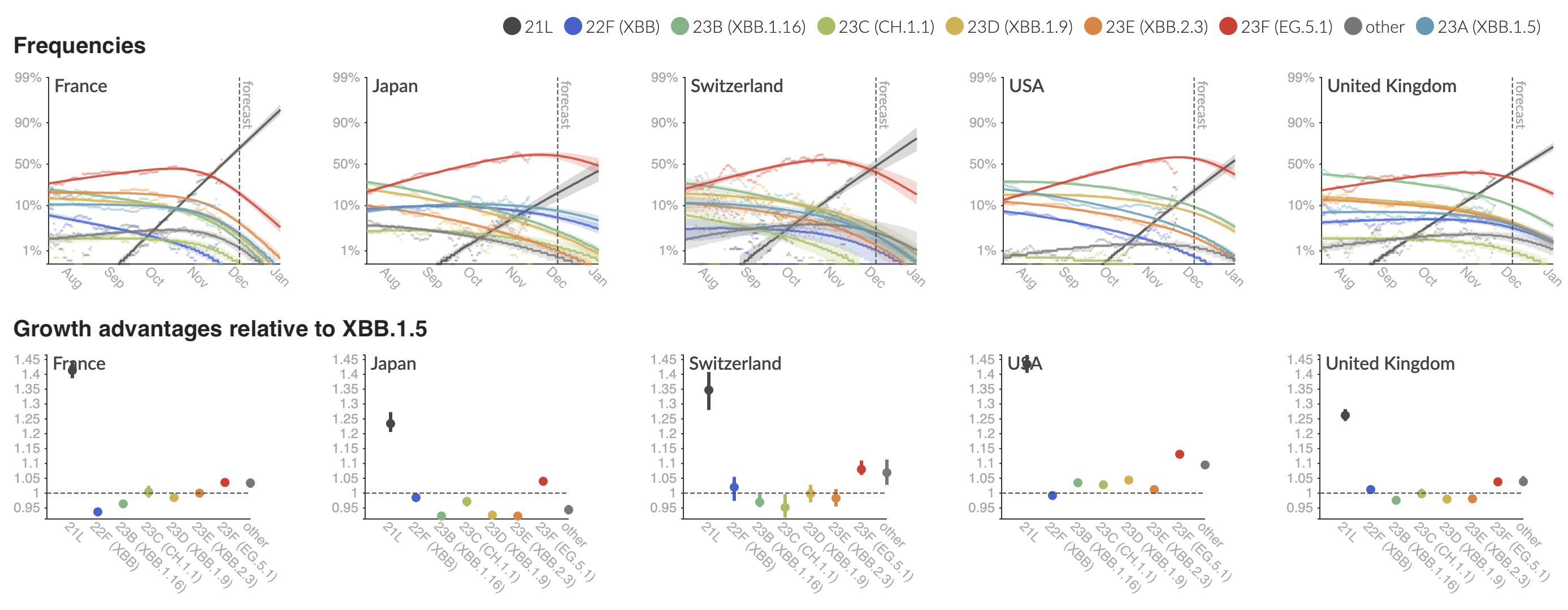
Variant frequencies across countries from Feb 2022 to present
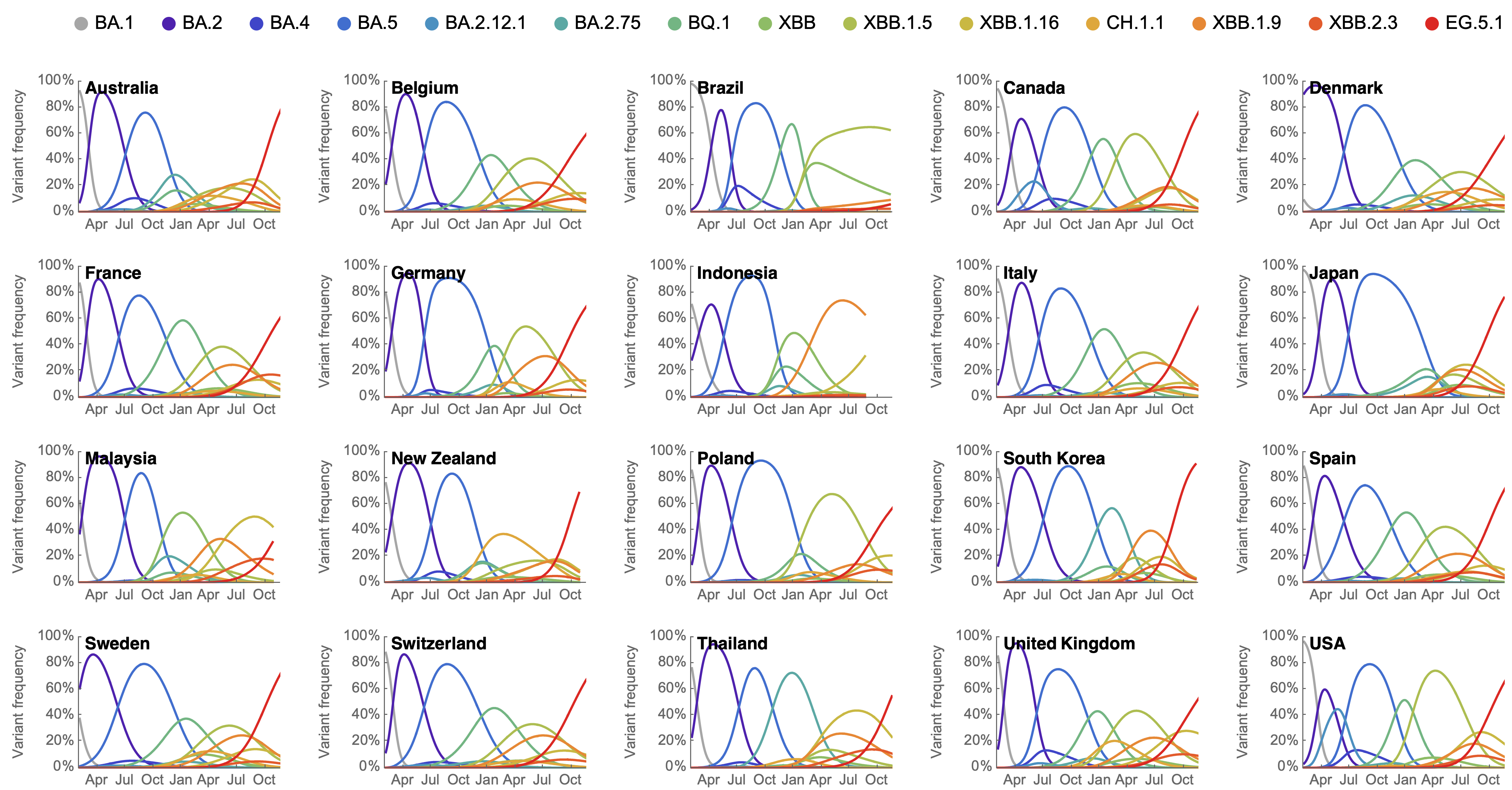
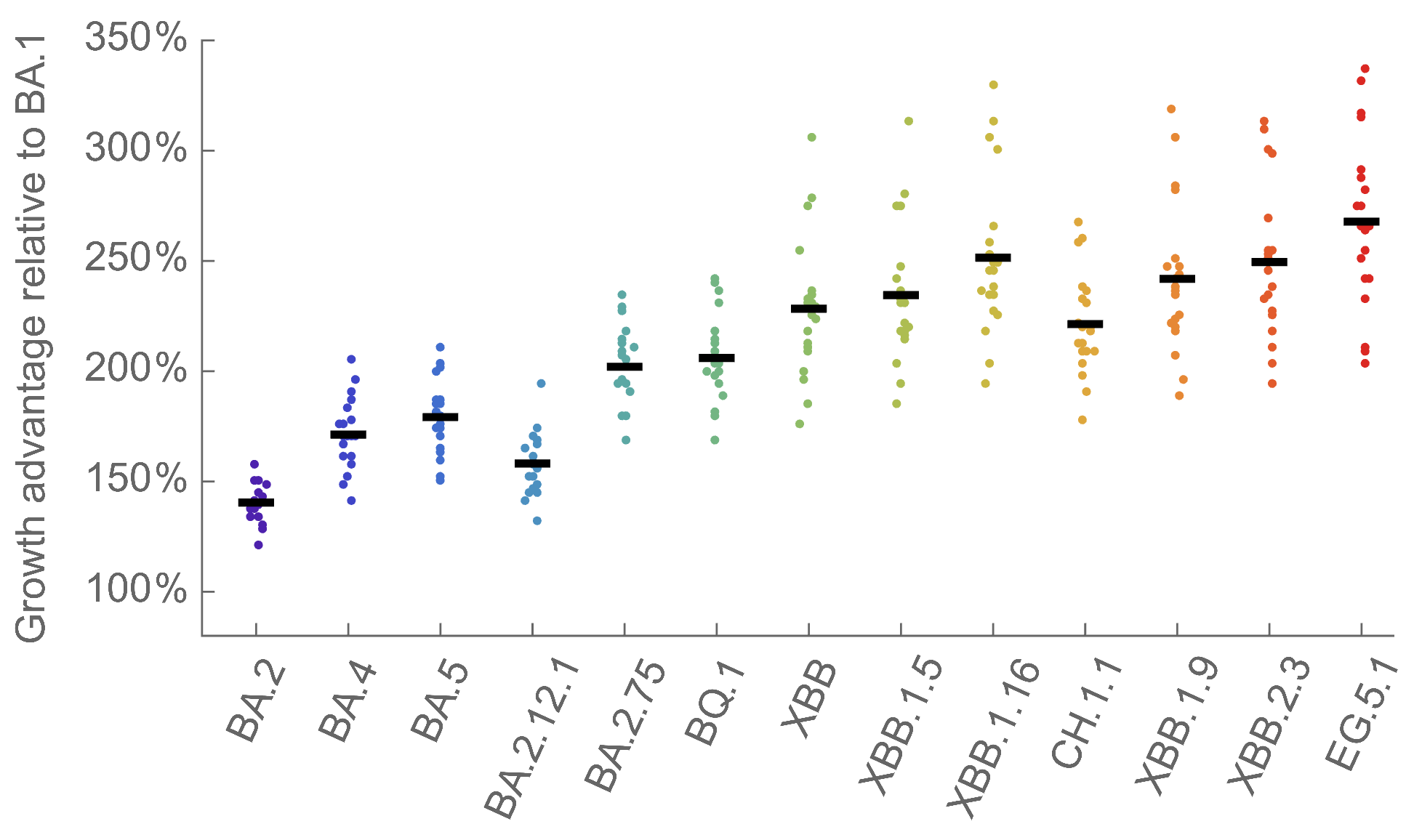
We find that recent variants like EG.5.1 are ~250% fitter than original Omicron BA.1
Evolution driving epidemics
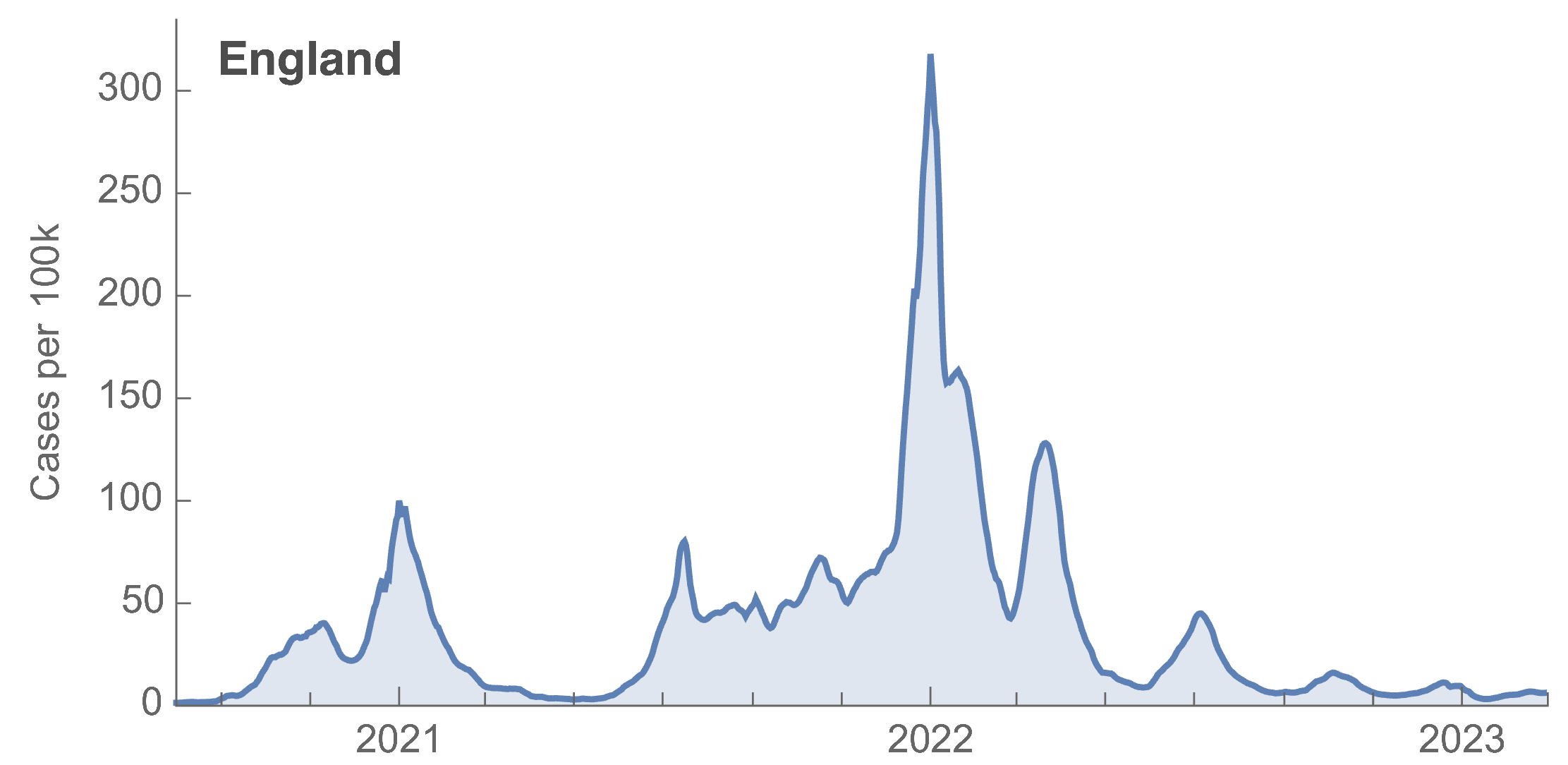
Many fewer reported cases in England post-Omicron
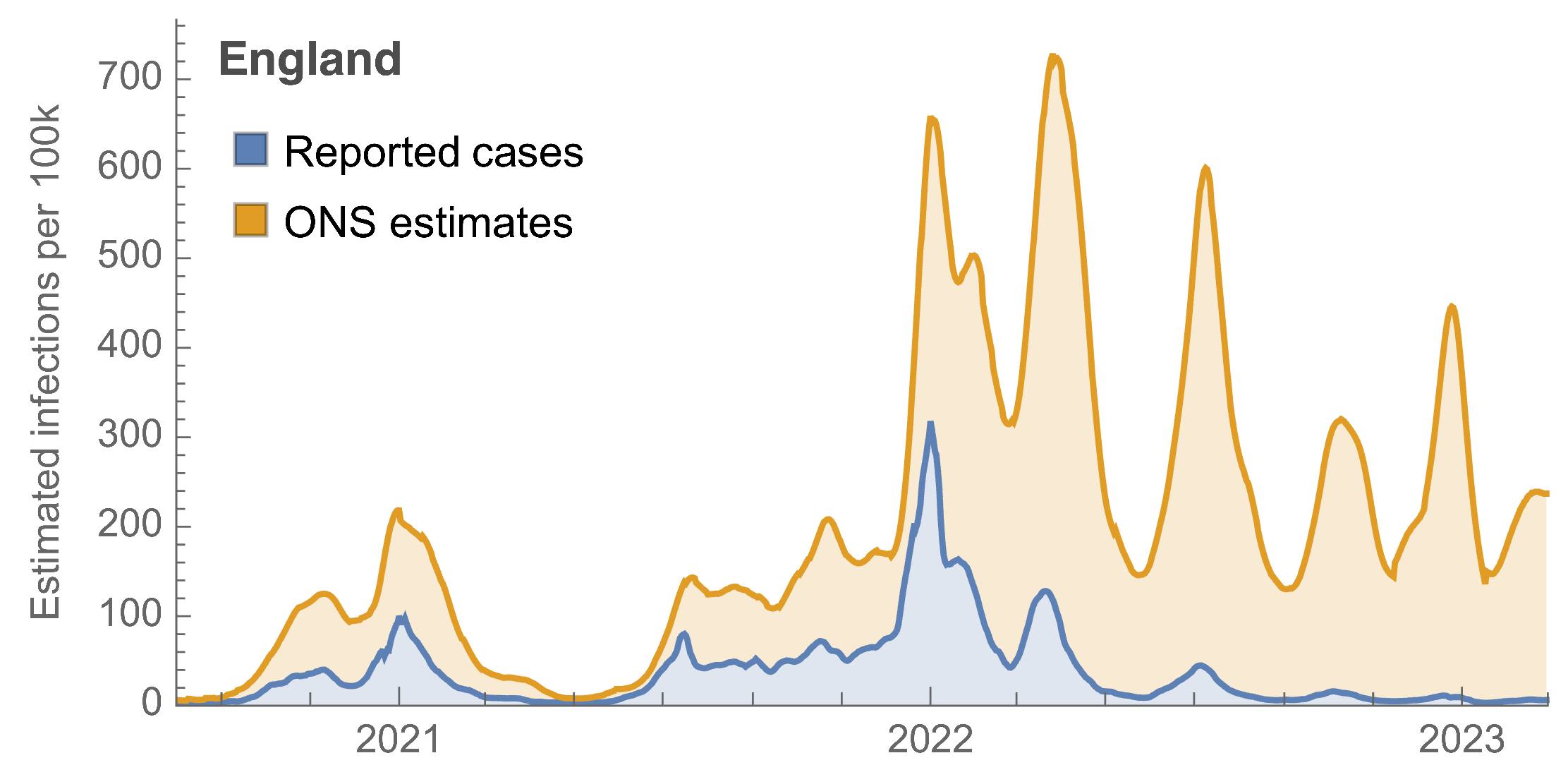
ONS Infection Survey provides unique source of ground truth
Roughly 1 in 3 infections detected in 2021, while 1 in 40 in Mar 2023 at end of study period
Partitioning ONS incidence based on sequencing data shows variant-driven epidemics

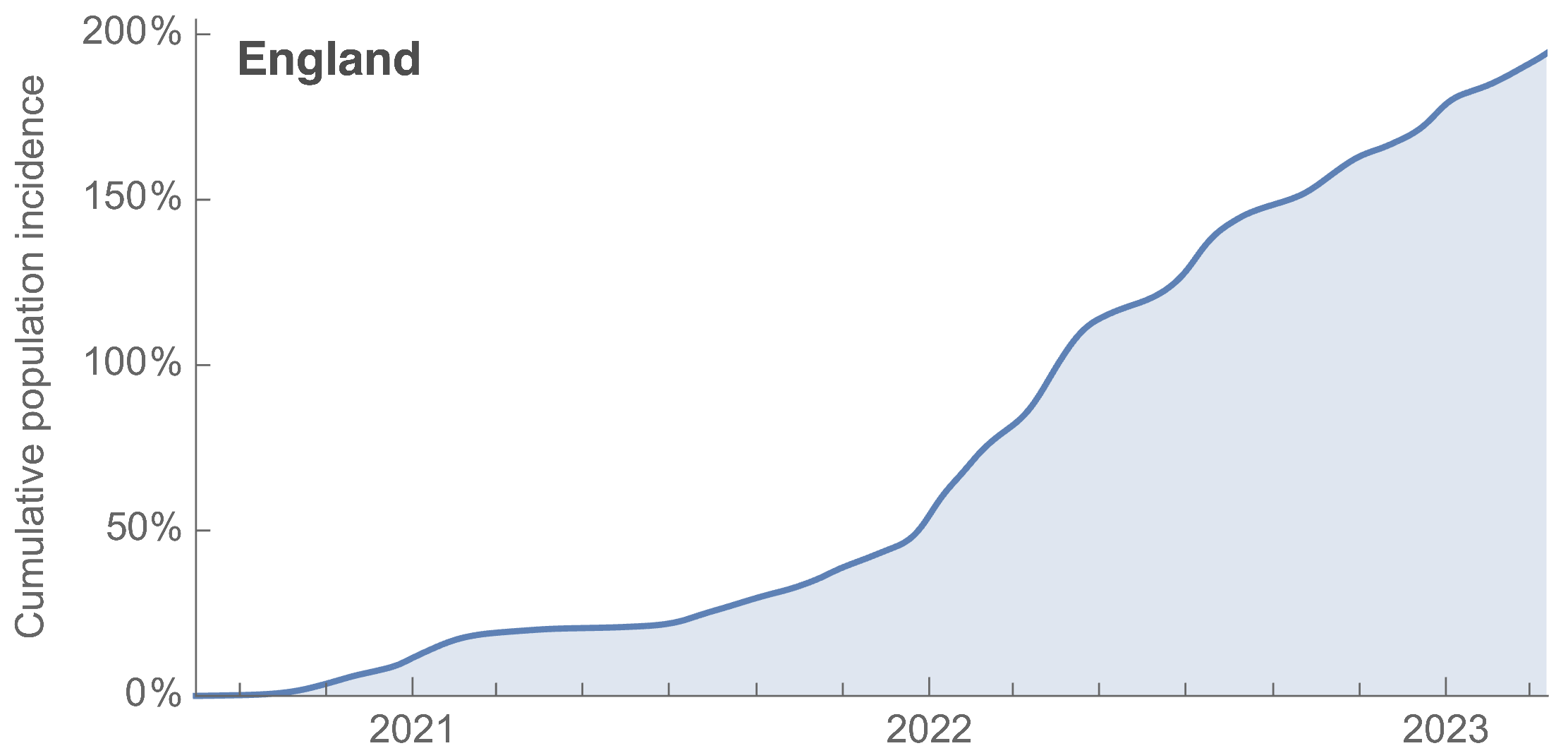
~110% population attack rate from March 2022 to March 2023
~27k deaths in ~61M infections yields IFR of 0.04%
Forecasting
Assessing MLR models for short-term frequency forecasting
Retrospective projections twice monthly during 2022
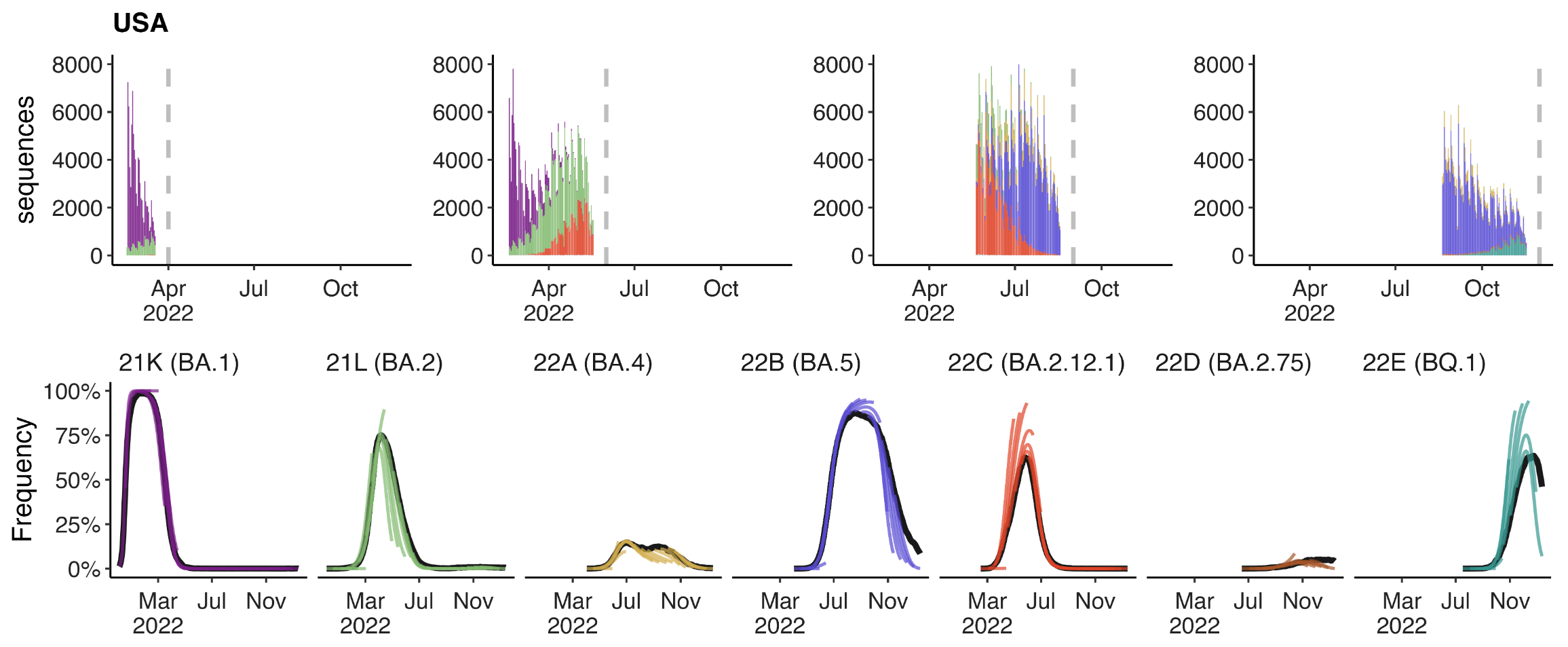
+30 day short-term forecasts across different countries
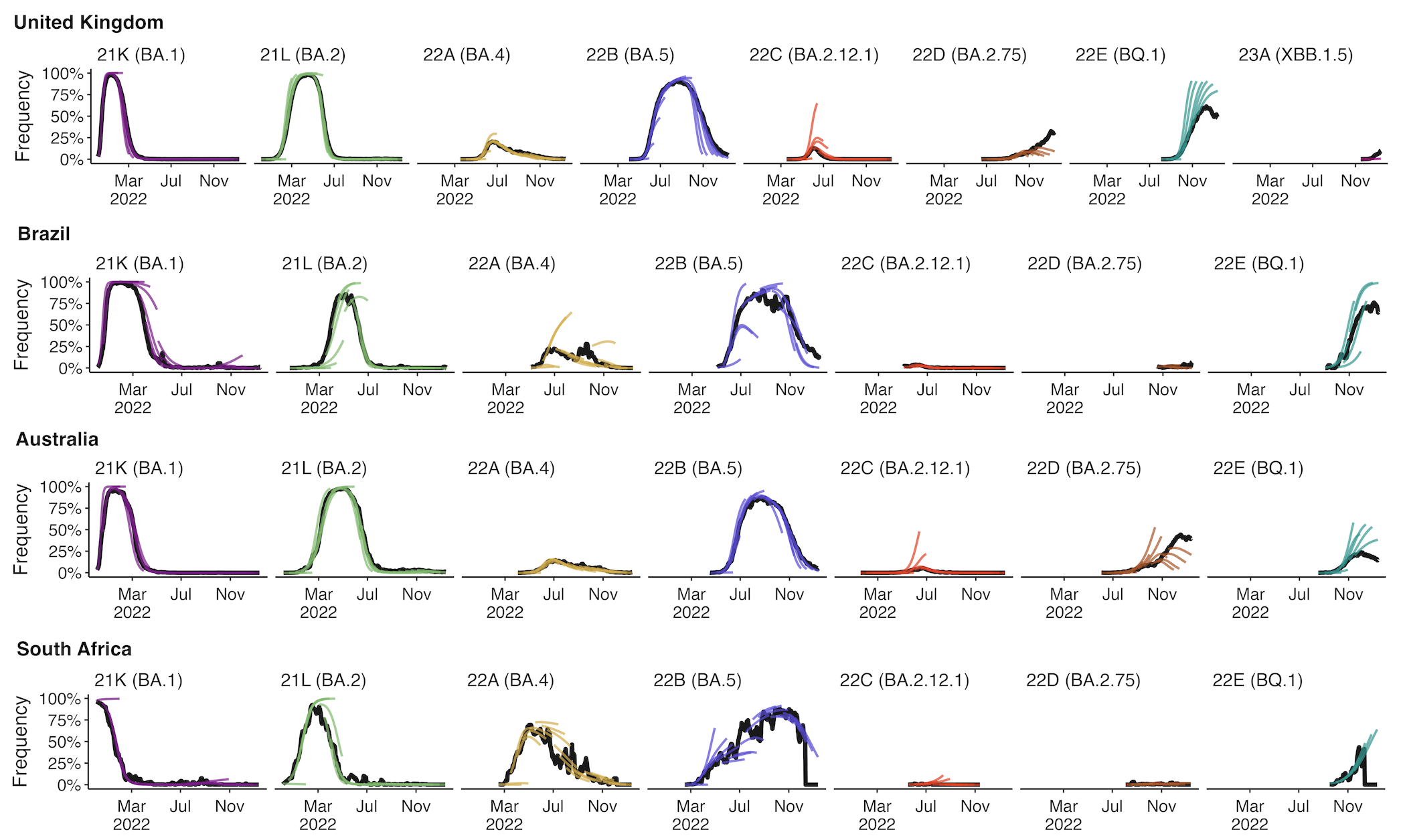
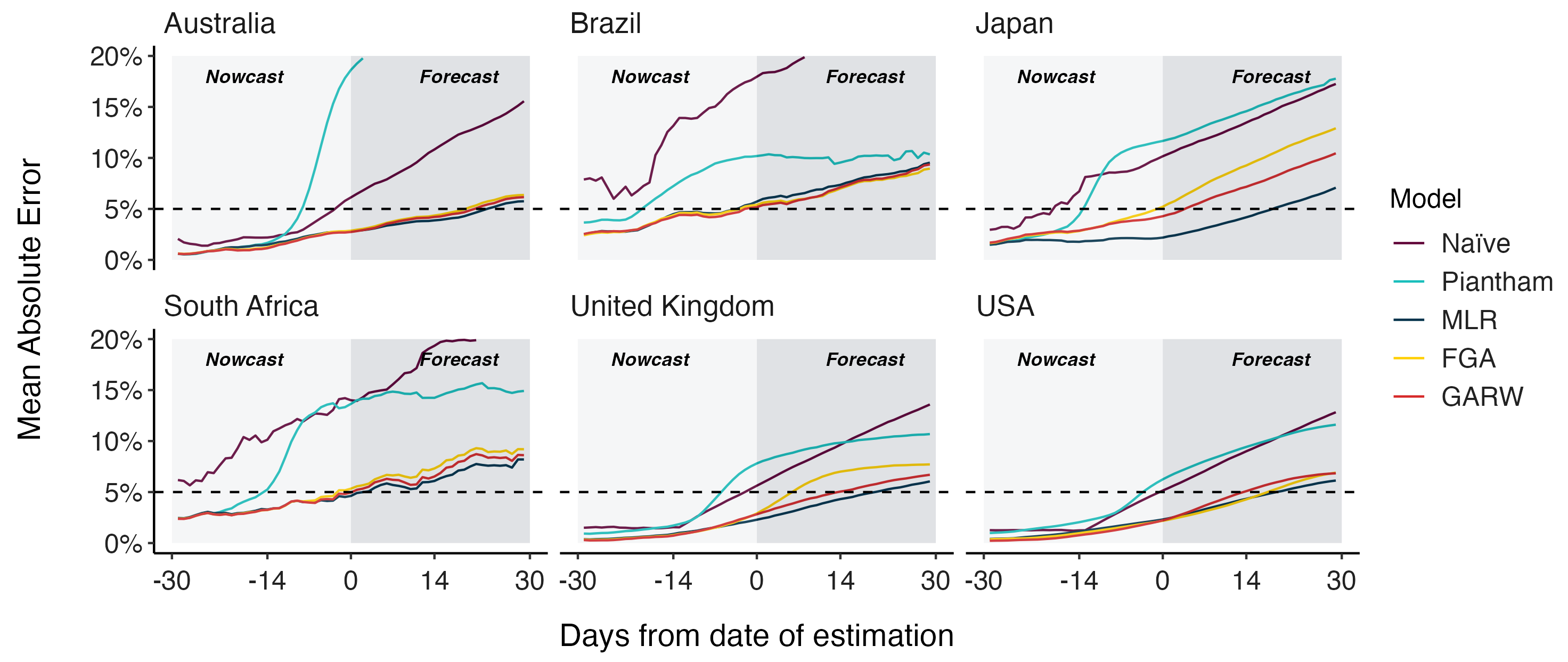
MLR models generate accurate short-term forecasts
30 days out, countries range from 6 to 10% mean absolute error
Clade and lineage forecasts continuously updated at nextstrain.org
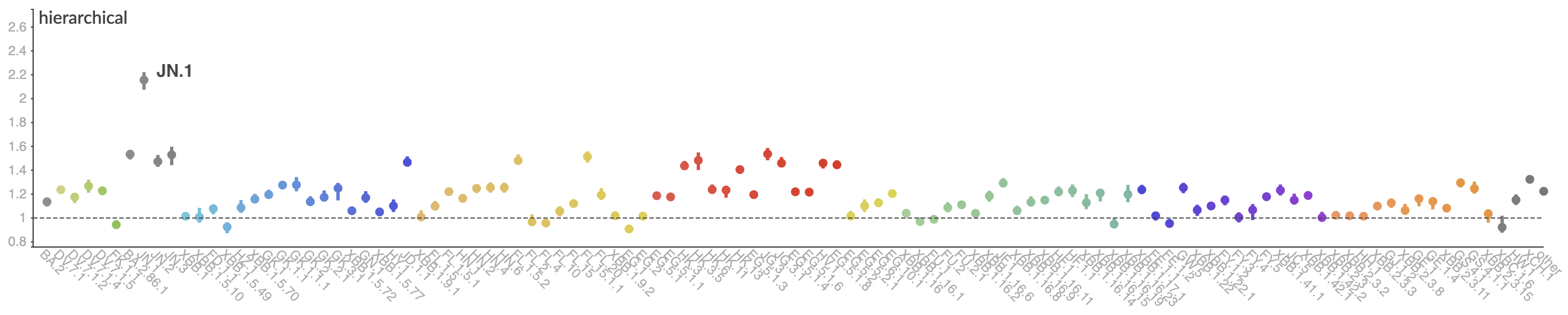
Pango-level growth advantages place JN.1 far ahead of the curve
Multinomial logistic regression should work well for SARS-CoV-2 prediction, except new variants have been emerging fast enough that the prediction horizon is really quite short
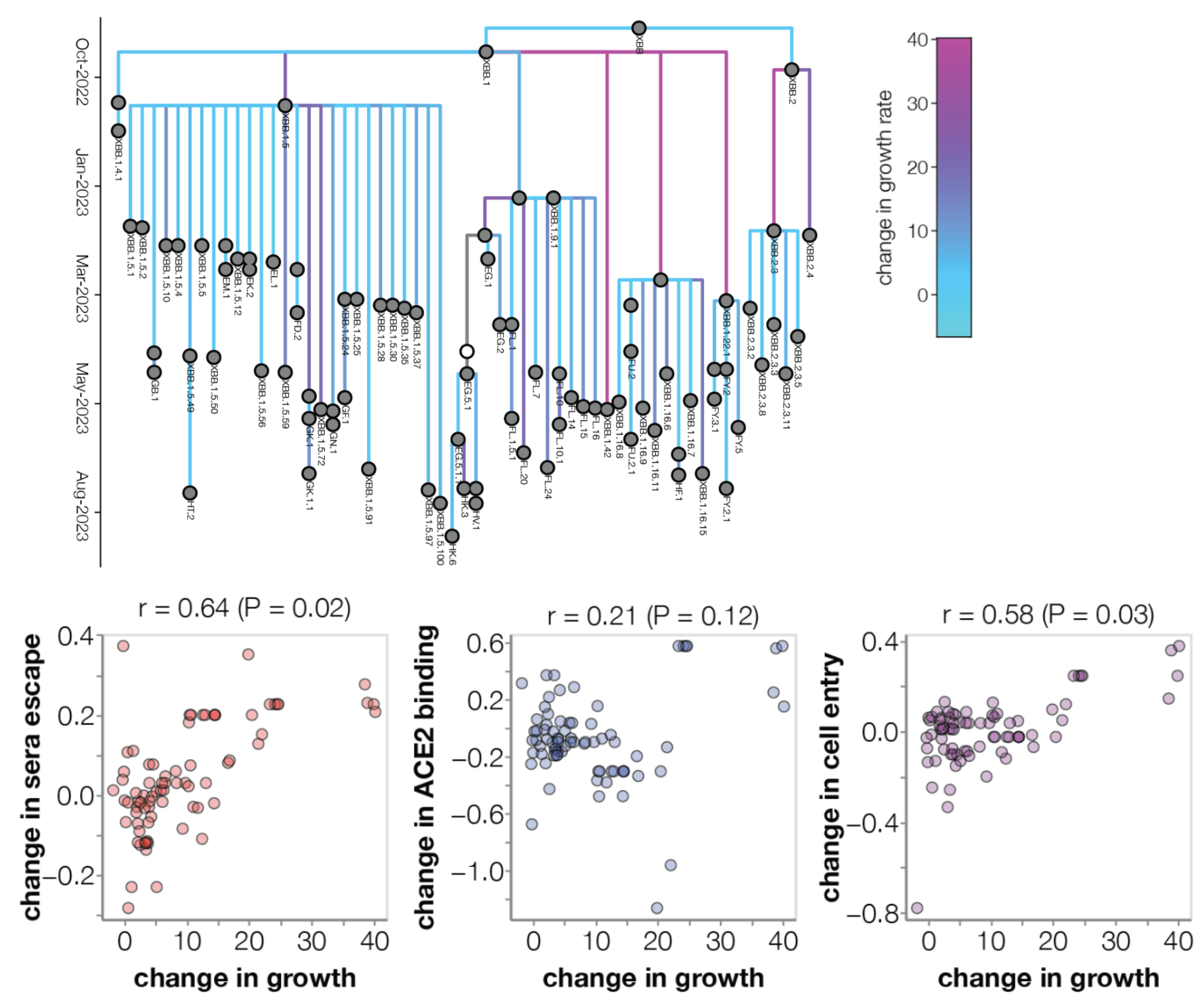
Could we predict the spread of new mutations using DMS data?
Escape from antibodies that potently neutralize BA.2
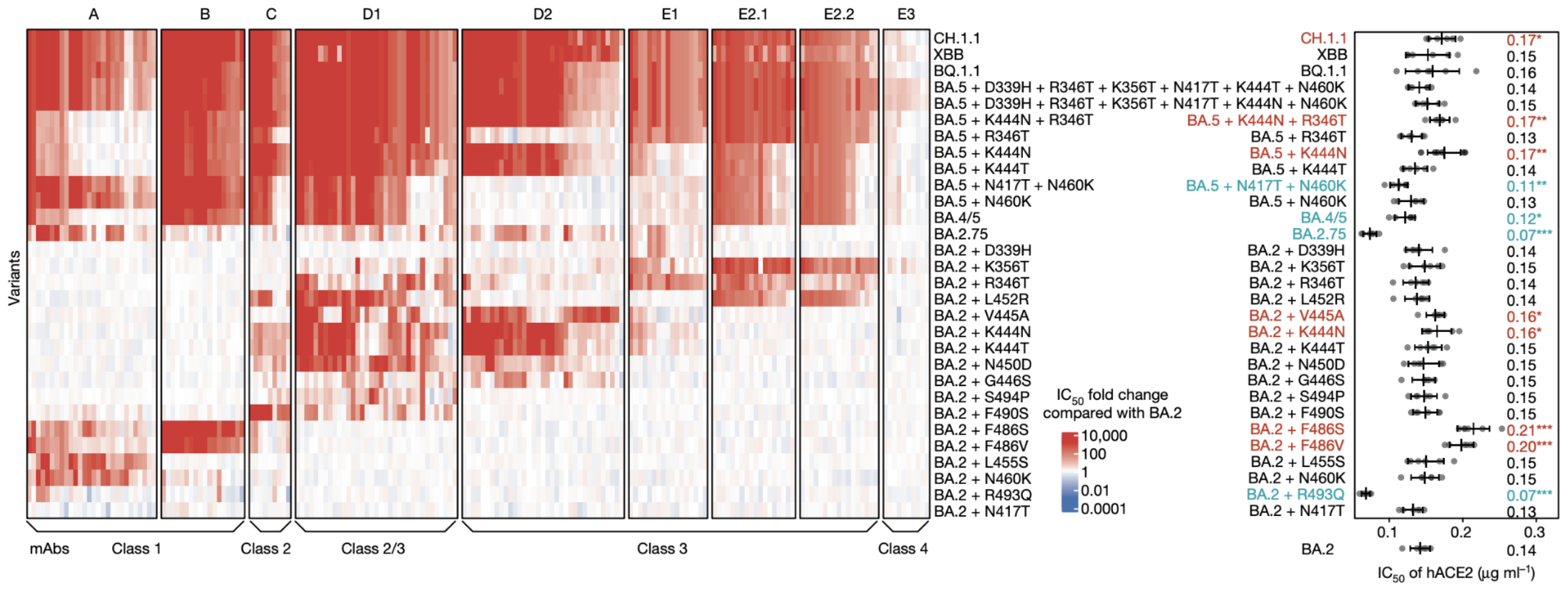
Can calculate escape of arbitrary RBD against antibodies known to neutralize BA.2


Strong correlation between DMS immune escape and lineage-level MLR growth advantage
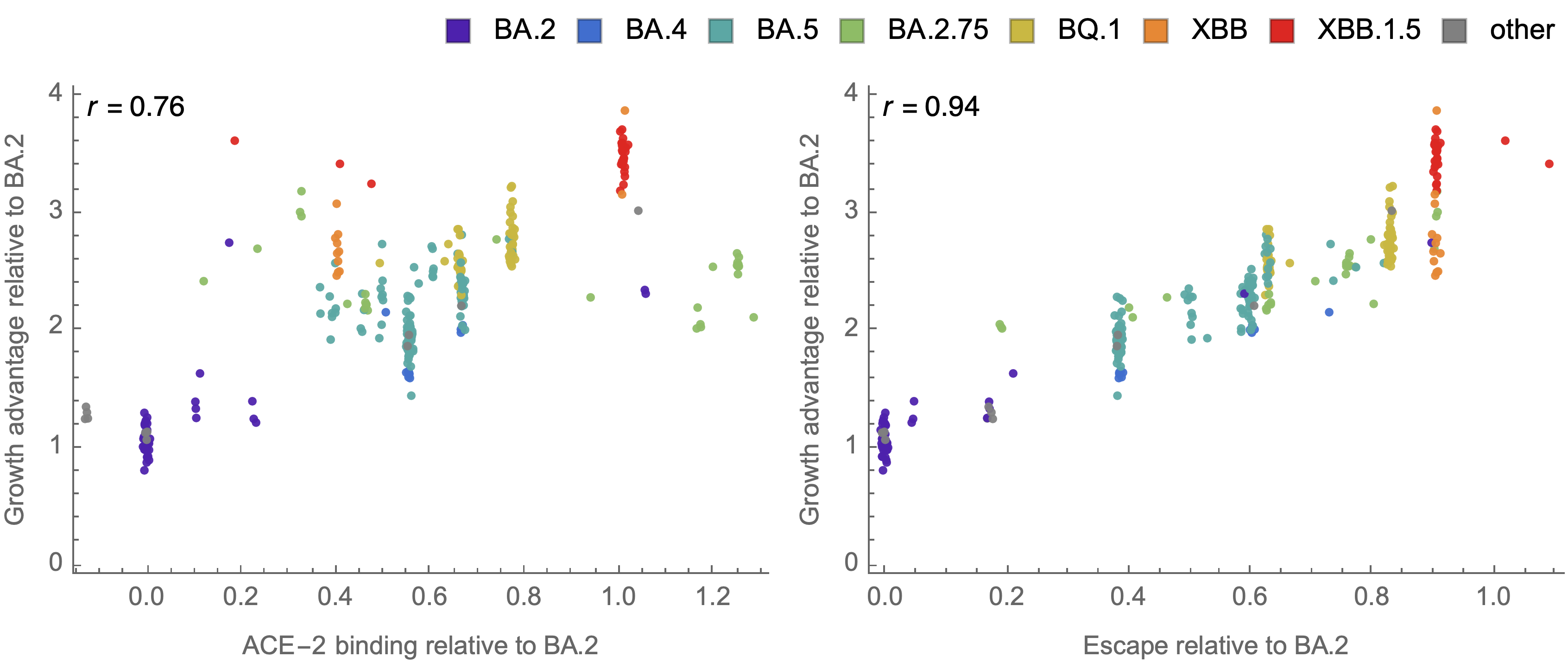
Similar results for new DMS platform measuring cell entry vs ACE2 binding vs escape from serum panel
Saltational evolution
How likely are further Omicron-like saltational events?

Highly derived "cryptic lineages" commonly being found in wastewater

Occasional chronic infection sequences uploaded that are highly distinct

XBB is derived from putative within-host lineage BJ.1


BA.2.86 emerged as "saltational" variant with ~30 spike mutations and was first detected in August

BA.2.86 with JN.1 is rapidly headed for global dominance

Even without further "saltational" events, ongoing evolution is rapid and so far shows little evidence of slowing

Research priorities to deal with this rapid evolution
- Continued genomic surveillance and evolutionary forecasting
- Development of new antivirals
-
Vaccination strategies to combat antigenic evolution and OAS
- Universal vaccines?
- Cocktail vaccines?
- Masking epitope sites to reduce imprinted response?
Acknowledgements
SARS-CoV-2 genomic epi: Data producers from all over the world and GISAID
Nextstrain: Richard Neher, Ivan Aksamentov, Jennifer Chang, James Hadfield, Emma Hodcroft, John Huddleston, Jover Lee, Victor Lin, Cornelius Roemer, Thomas Sibley
Bedford Lab:
![]() John Huddleston,
John Huddleston,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Cassia Wagner,
Cassia Wagner,
![]() Miguel Paredes,
Miguel Paredes,
![]() Nicola Müller,
Nicola Müller,
![]() Marlin Figgins,
Marlin Figgins,
![]() Victor Lin,
Victor Lin,
![]() Jennifer Chang,
Jennifer Chang,
![]() Allison Li,
Allison Li,
![]() Eslam Abousamra,
Eslam Abousamra,
![]() Donna Modrell,
Donna Modrell,
![]() Nashwa Ahmed,
Nashwa Ahmed,
![]() Cécile Tran Kiem
Cécile Tran Kiem






