Potential for antigenic drift in SARS-CoV-2
Trevor Bedford (@trvrb)
11 Dec 2020
HB Friday Faculty Talk
Fred Hutch
Slides at: bedford.io/talks

How likely is it that antigenic variants of SARS-CoV-2 will emerge, circulate and thereby decrease vaccine efficacy?
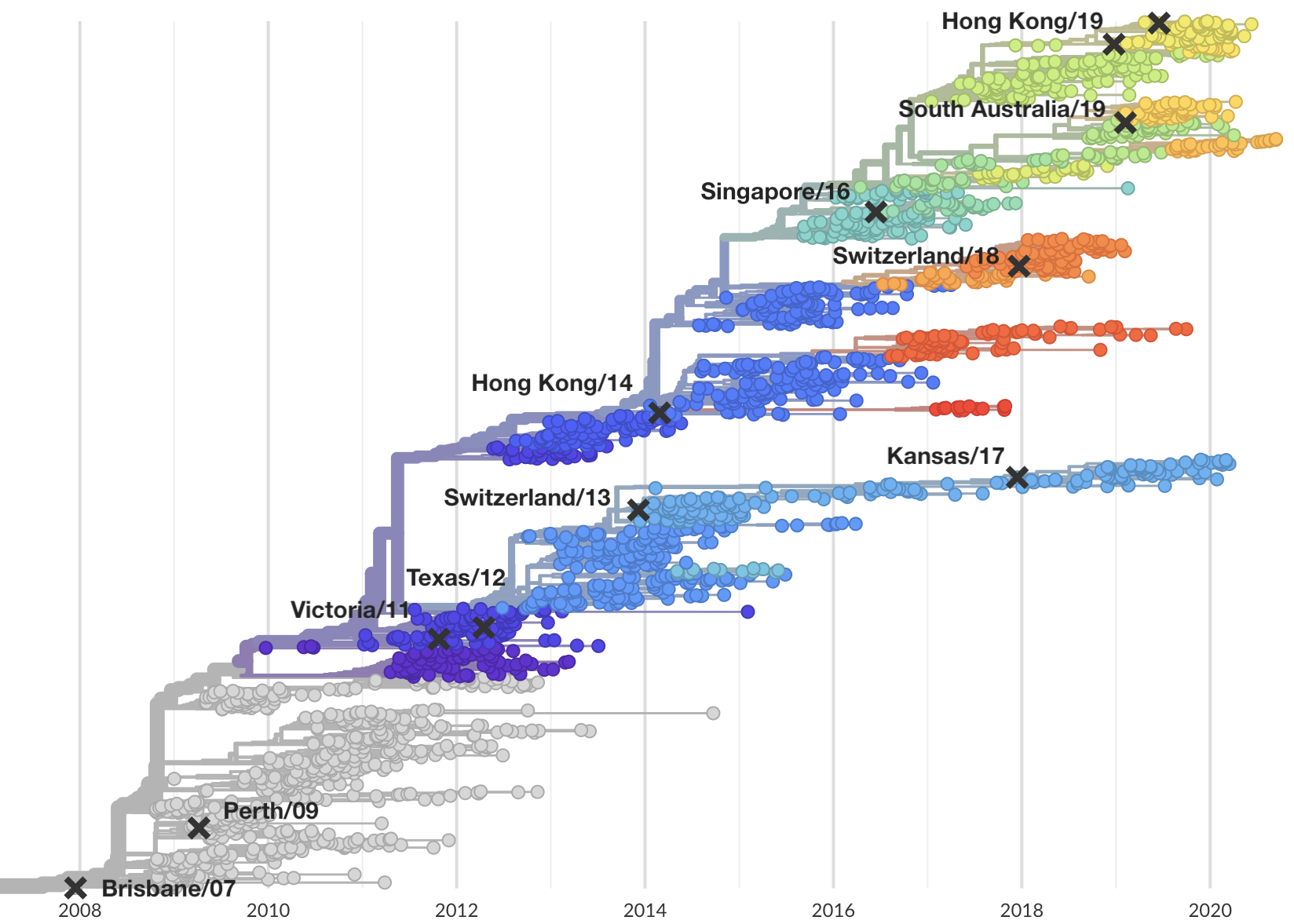
Integrating genotypes and phenotypes improves long-term forecasts of seasonal influenza A/H3N2 evolution
with ![]() John Huddleston, Richard Neher, Dave Wentworth, Becky Kondor, John McCauley, Hideki Hasegawa, Kanta Subbarao and others
John Huddleston, Richard Neher, Dave Wentworth, Becky Kondor, John McCauley, Hideki Hasegawa, Kanta Subbarao and others
Influenza viruses undergo antigenic drift
H3N2 vaccine updates occur every ~2 years

Vaccine strain selection by WHO
Select one strain from circulating diversity
Fitness models project strain frequencies
Future frequency $x_i(t+\Delta t)$ of strain $i$ derives from strain fitness $f_i$ and present day frequency $x_i(t)$, such that
$$\hat{x}_i(t+\Delta t) = x_i(t) \, \mathrm{exp}(f_i \, \Delta t)$$
Total strain frequencies at each timepoint are normalized. This captures clonal interference between competing lineages.
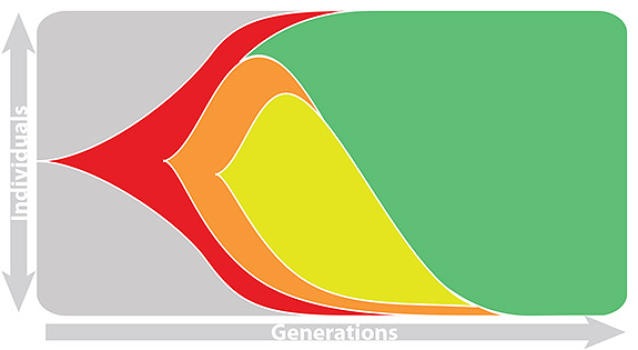
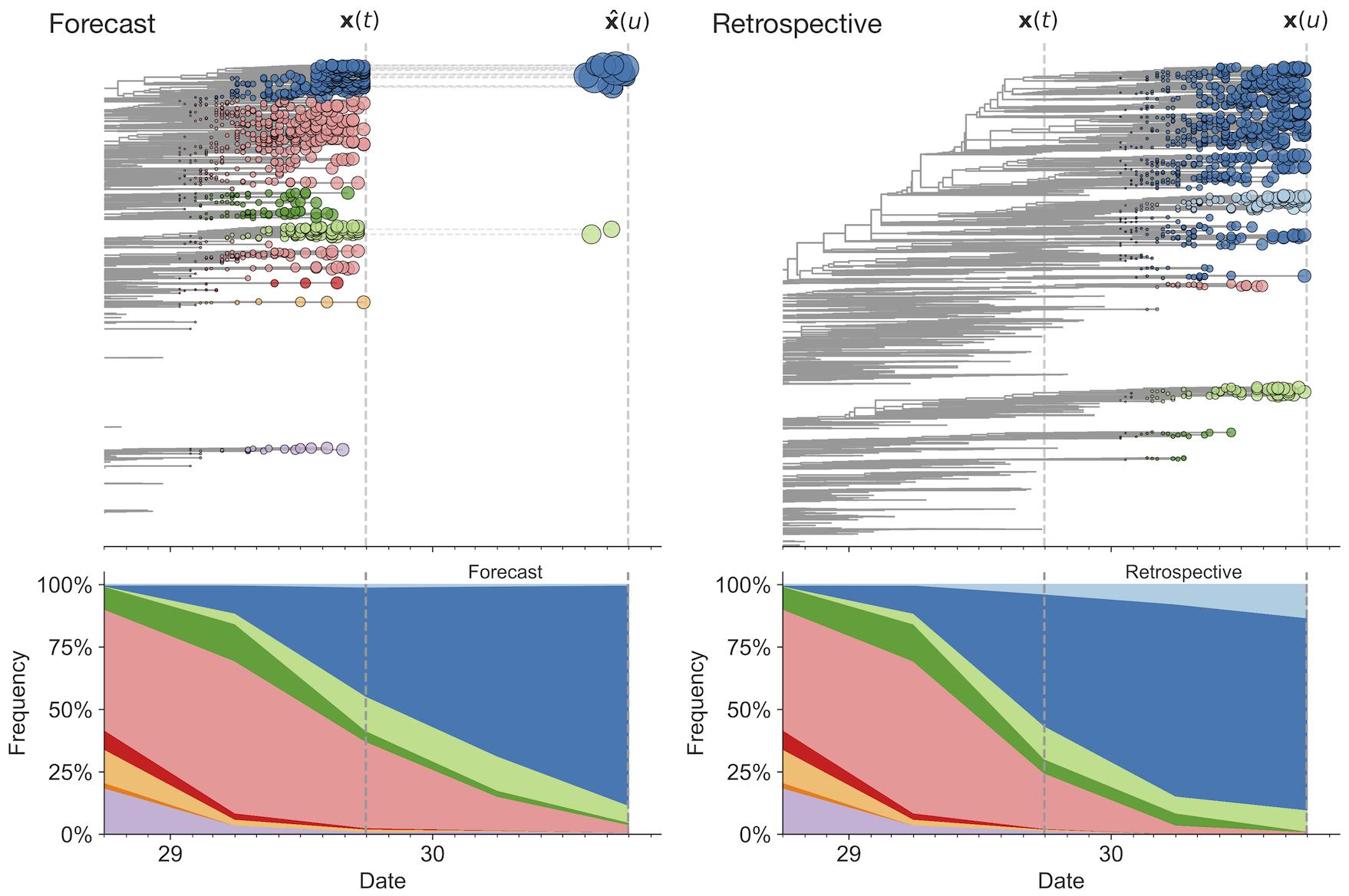
Match strain forecast to retrospective circulation
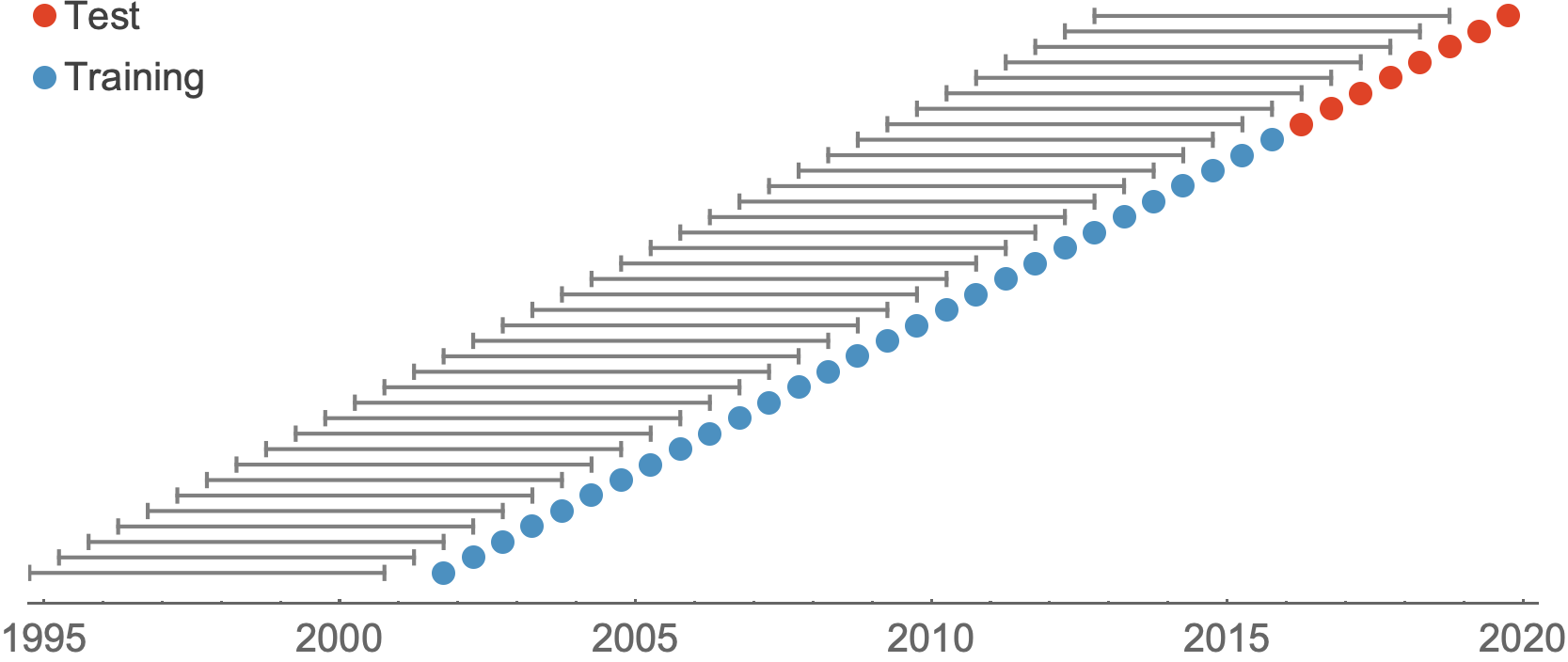
Train in 6-year sliding windows from 1995 to 2015 with most recent years held out as test
Composite models favor combinations of HI drift, local branching index and non-epitope fitness
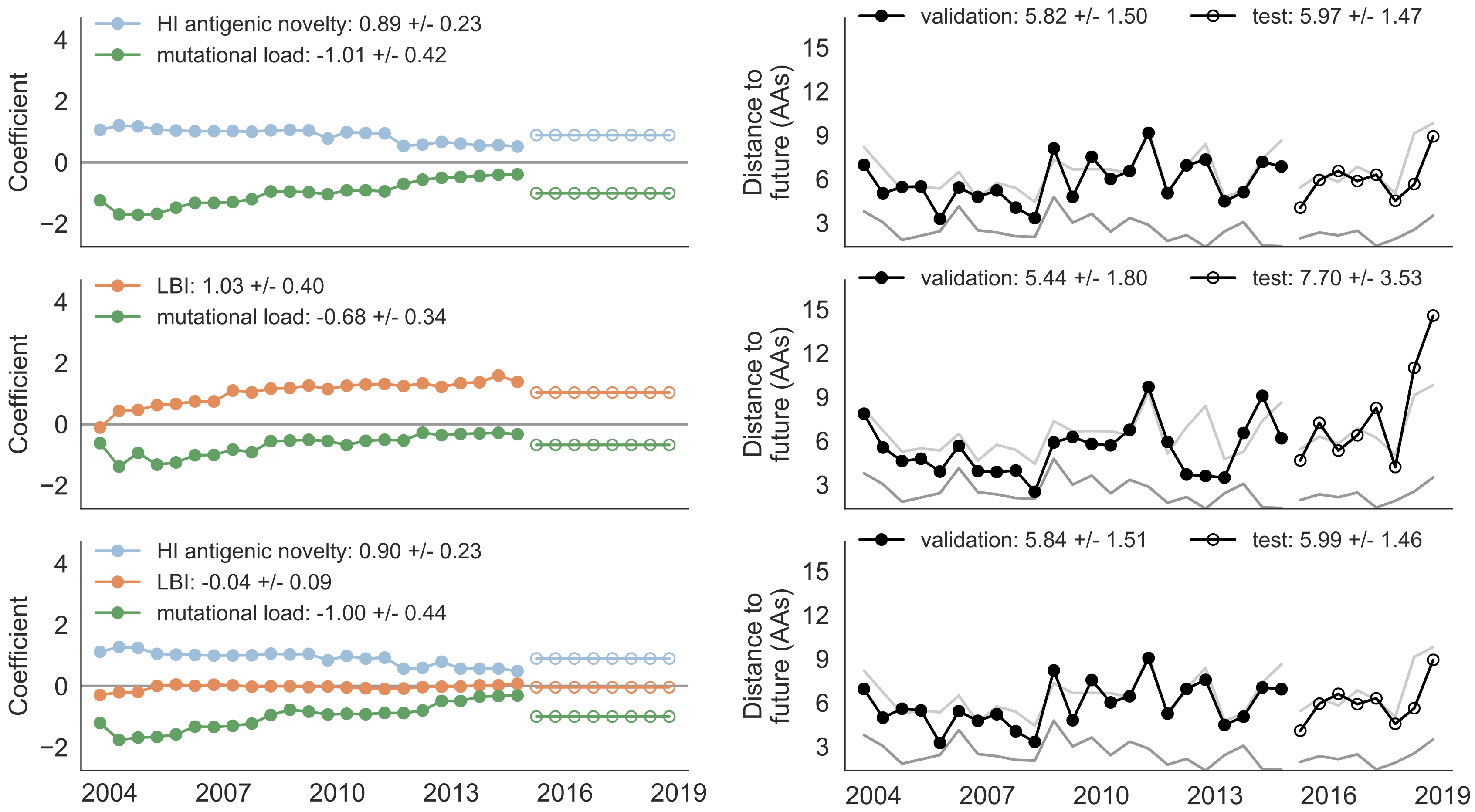
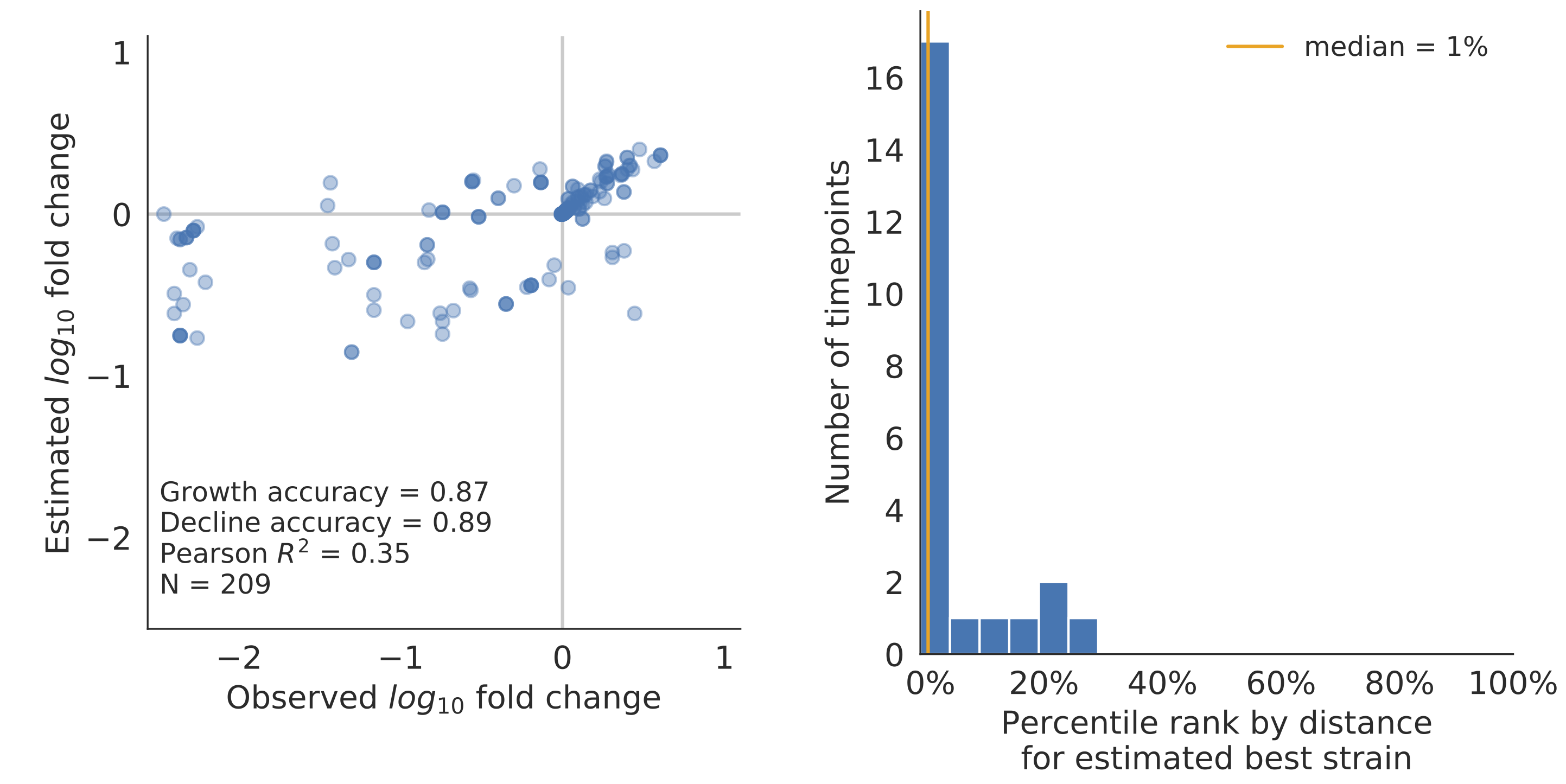
Model successfully predicts clade growth and best pick from model is generally close to best possible retrospective pick
Forecasts are live at nextstrain.org/flu, but are currently fraught due to lack of recent isolates

Evidence for adaptive evolution in the receptor-binding domain of seasonal coronaviruses
with ![]() Katie Kistler
Katie Kistler
Four species of seasonal coronavirus (OC43, 229E, NL63, HKU1) cause ~30% of common colds

Focus on spike protein as target for immune response and antigenic escape
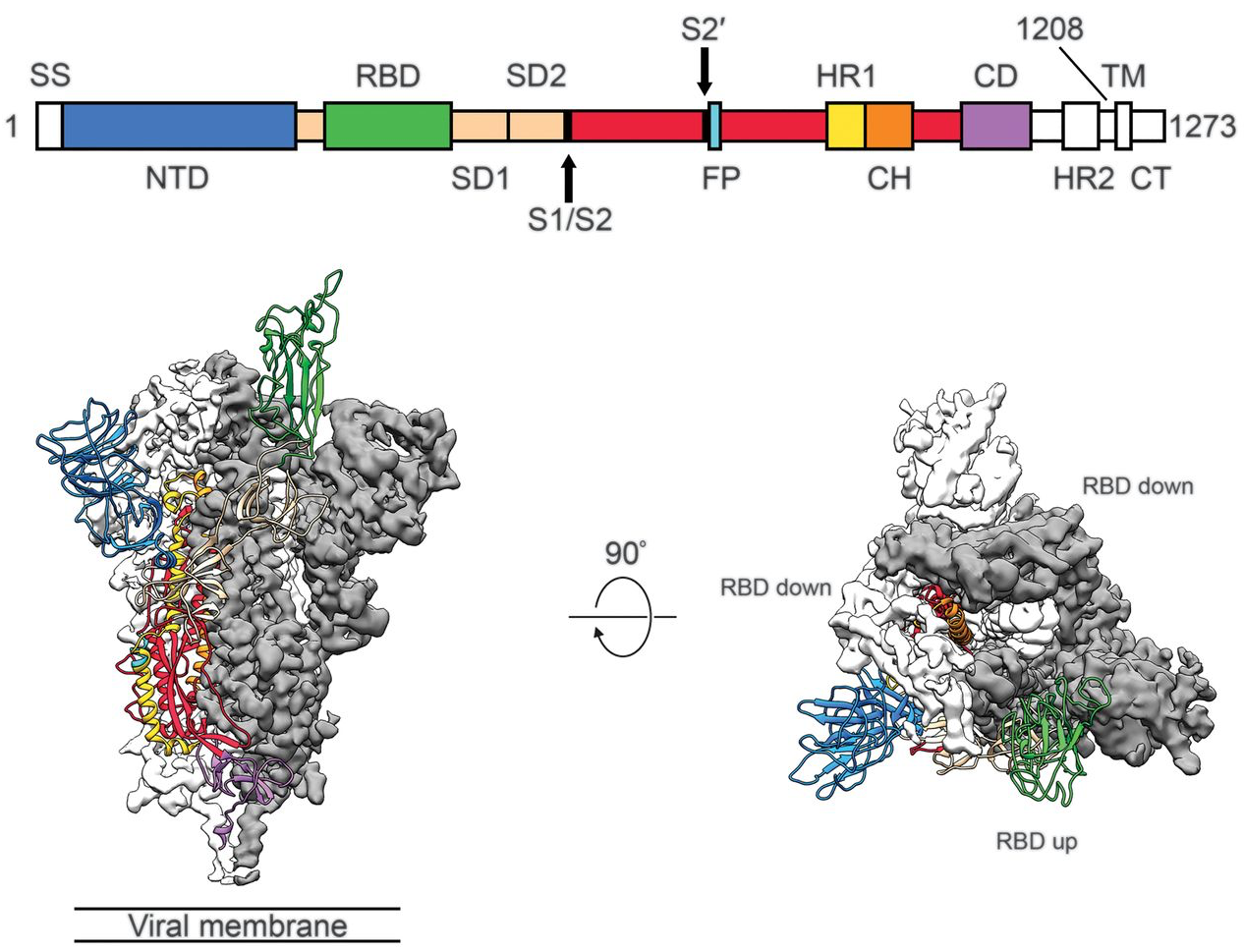
Spike gene phylogenies for OC43 and 229E

Substitutions occur more often in S1 subunit than S2

Nonsynonymous substitutions accumulate rapidly in exposed portions of spike

Diversity and divergence used to estimate accumulation of adaptive substitutions

This analysis can be summarized as rate of adaptive substitutions per year (about ~0.5 adaptive subs per year for OC43)

And compared to rates of adaptive substitutions in other viruses

Adaptive evolution in spike is clearly present in OC43 and 229E. It's likely this results in antigenic evolution and contributes to reinfection observed to occur every ~3 years.
SARS-CoV-2
Sequencing and data sharing in almost real-time, now with over 250k genomes
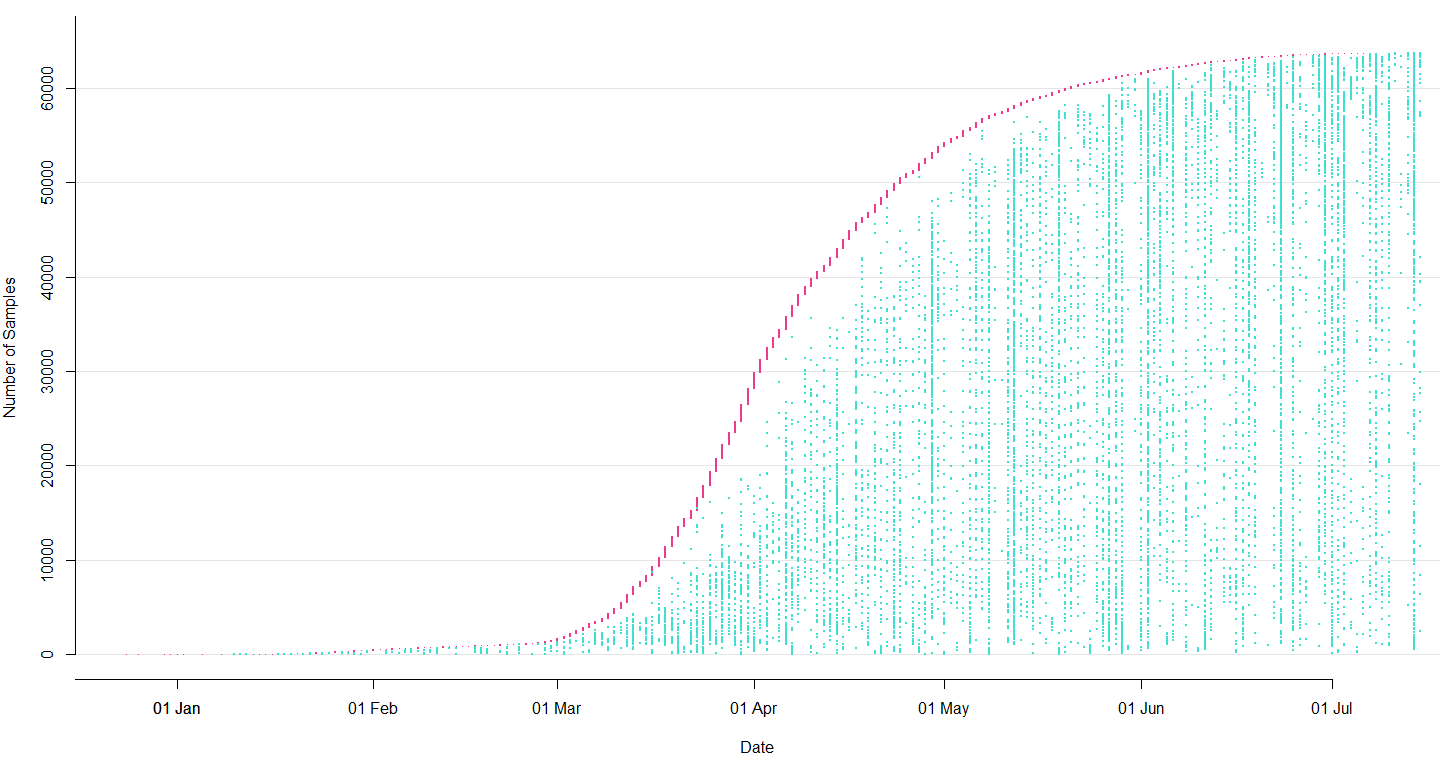
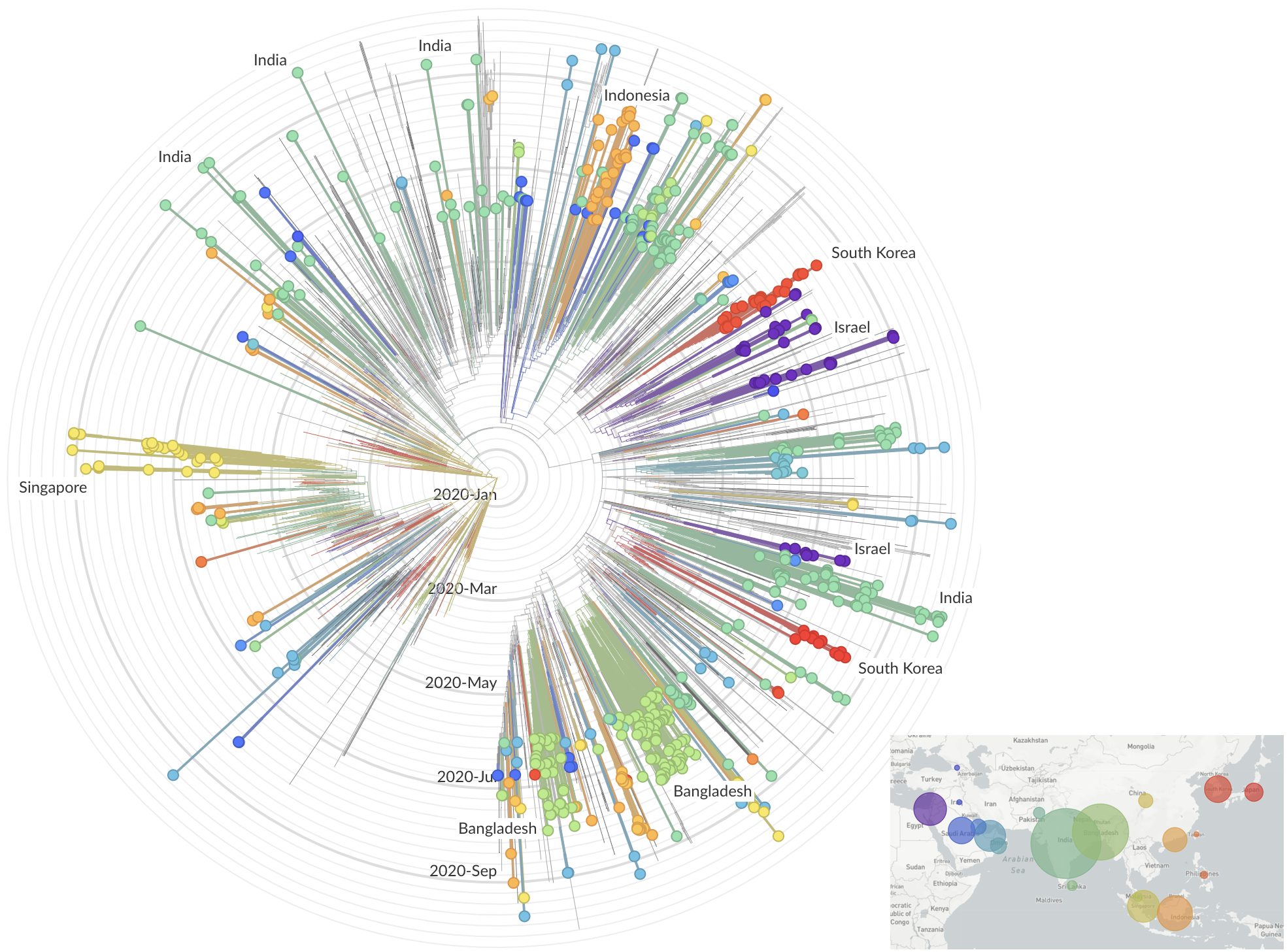
SARS-CoV-2 lineages establish globally in February and March
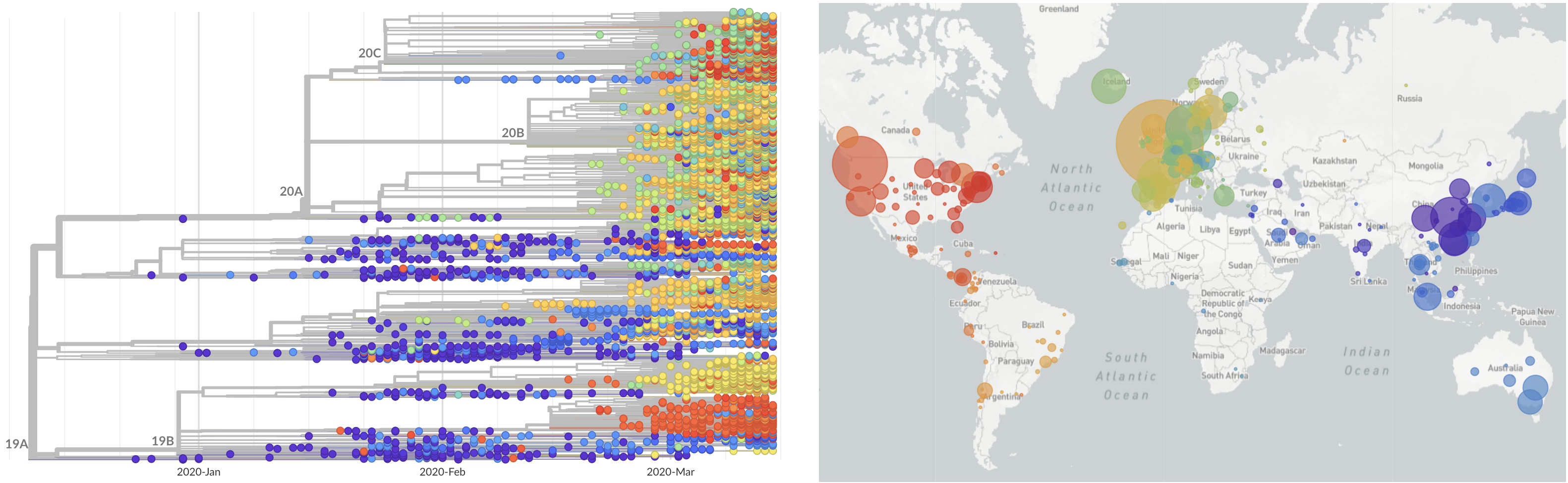
decreased international travel,
regional clades have emerged
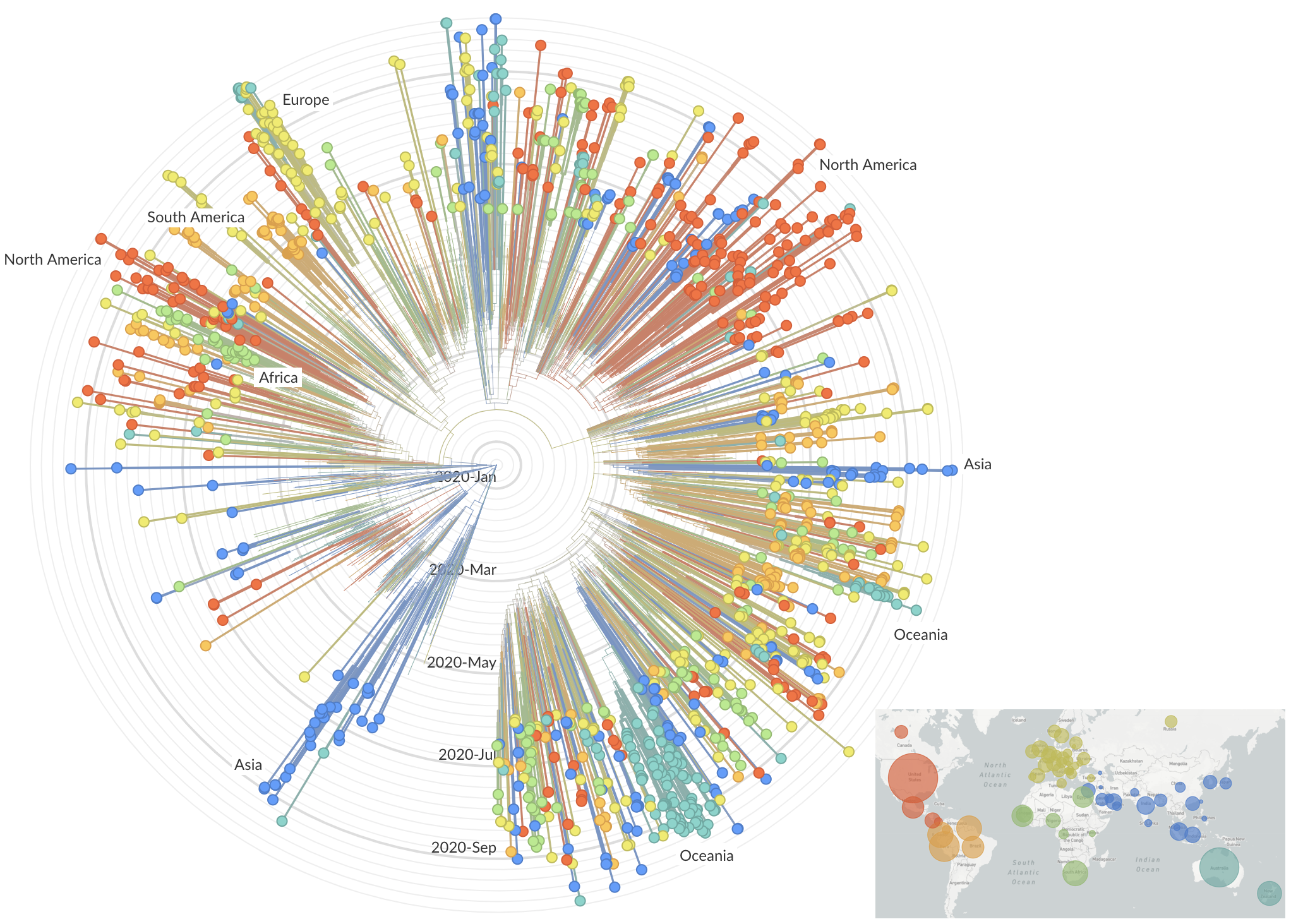
decreased international travel,
regional clades have emerged
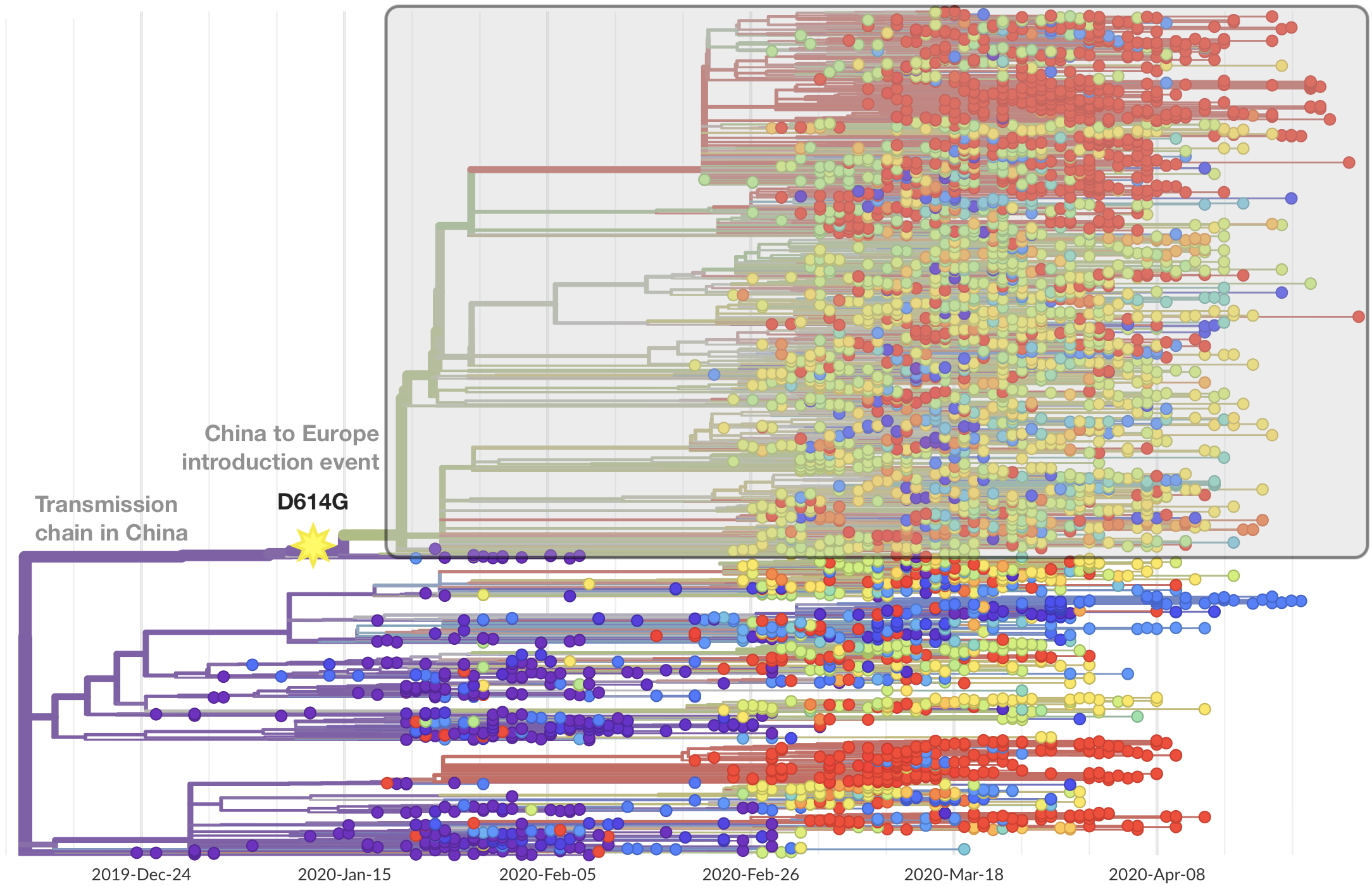
Limited early mutations like D614G spread globally during initial wave
More recent variants are confined to regional dominance
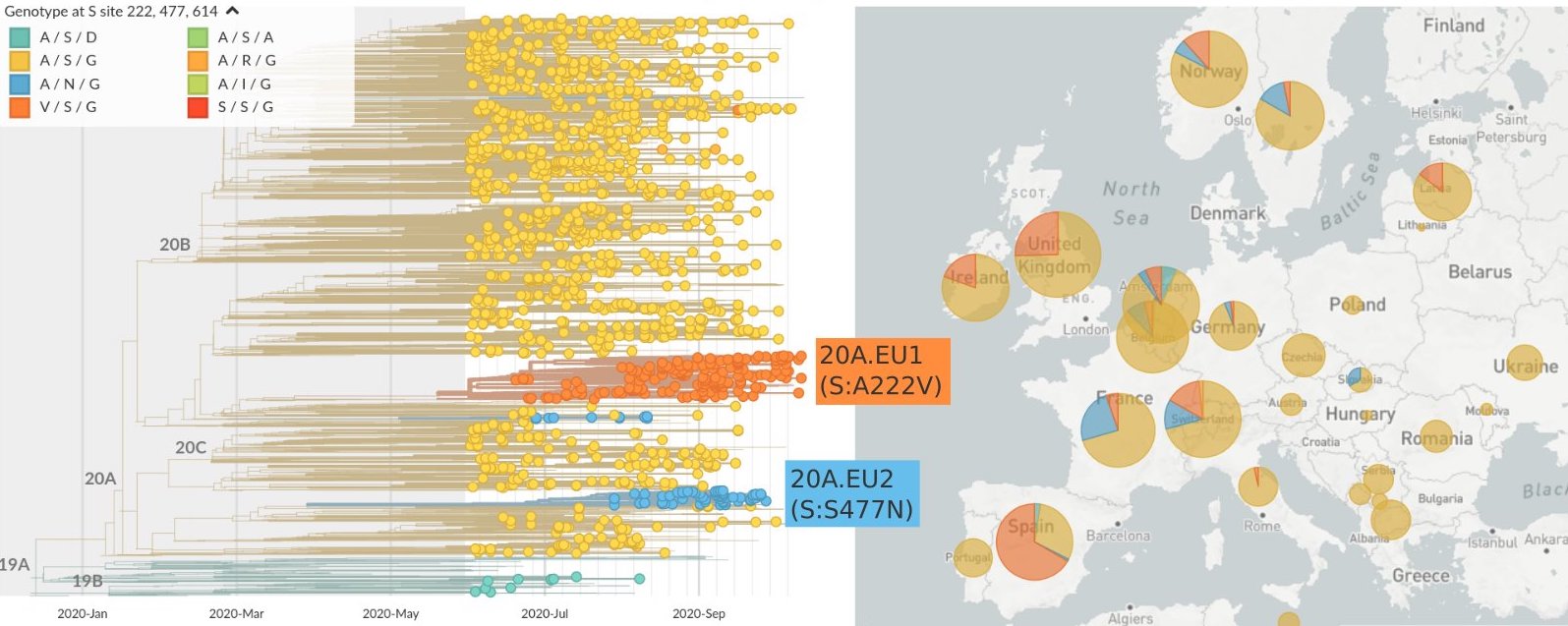
Starting to see circulating spike variants

A222V, N439K and S477N spreading in Europe (although may be largely explained by epidemiology)

S477N also spreading in Oceania, having arisen independently

N439K shows evidence of reduced binding and neutralization

Y453F shows associated with passage in farmed mink

I think SARS-CoV-2 will undergo antigenic drift
Strong immune response to mRNA vaccines may help here

Monitoring for antigenic drift
- Test neutralization of circulating variants against serum panels from
recovered and vaccinated individuals - Combine vaccination records with sequencing of clinical samples
Acknowledgements
Seasonal flu: WHO Global Influenza Surveillance Network, GISAID, John Huddleston, Richard Neher, Jover Lee, Dave Wentworth, Becky Kondor
Seasonal CoVs: Katie Kistler
SARS-CoV-2 genomic epi: Data producers from all over the world, GISAID and the Nextstrain team
Bedford Lab:
![]() Alli Black,
Alli Black,
![]() John Huddleston,
John Huddleston,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Louise Moncla,
Louise Moncla,
![]() Maya Lewinsohn,
Maya Lewinsohn,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Kairsten Fay,
Kairsten Fay,
![]() Misja Ilcisin,
Misja Ilcisin,
![]() Cassia Wagner,
Cassia Wagner,
![]() Miguel Paredes,
Miguel Paredes,
![]() Nicola Müller,
Nicola Müller,
![]() Marlin Figgins,
Marlin Figgins,
![]() Eli Harkins
Eli Harkins






