Real-time tracking of virus evolution
Trevor Bedford (@trvrb)
Fred Hutchinson Cancer Center / Howard Hughes Medical Institute
5 Aug 2022
SURP Seminar
Fred Hutch
Slides at: bedford.io/talks
We work at the interface of virology, evolution and epidemiology

Segue through data vizualization
Data tells a story
Charles Minard and Napoleon's 1812 campaign
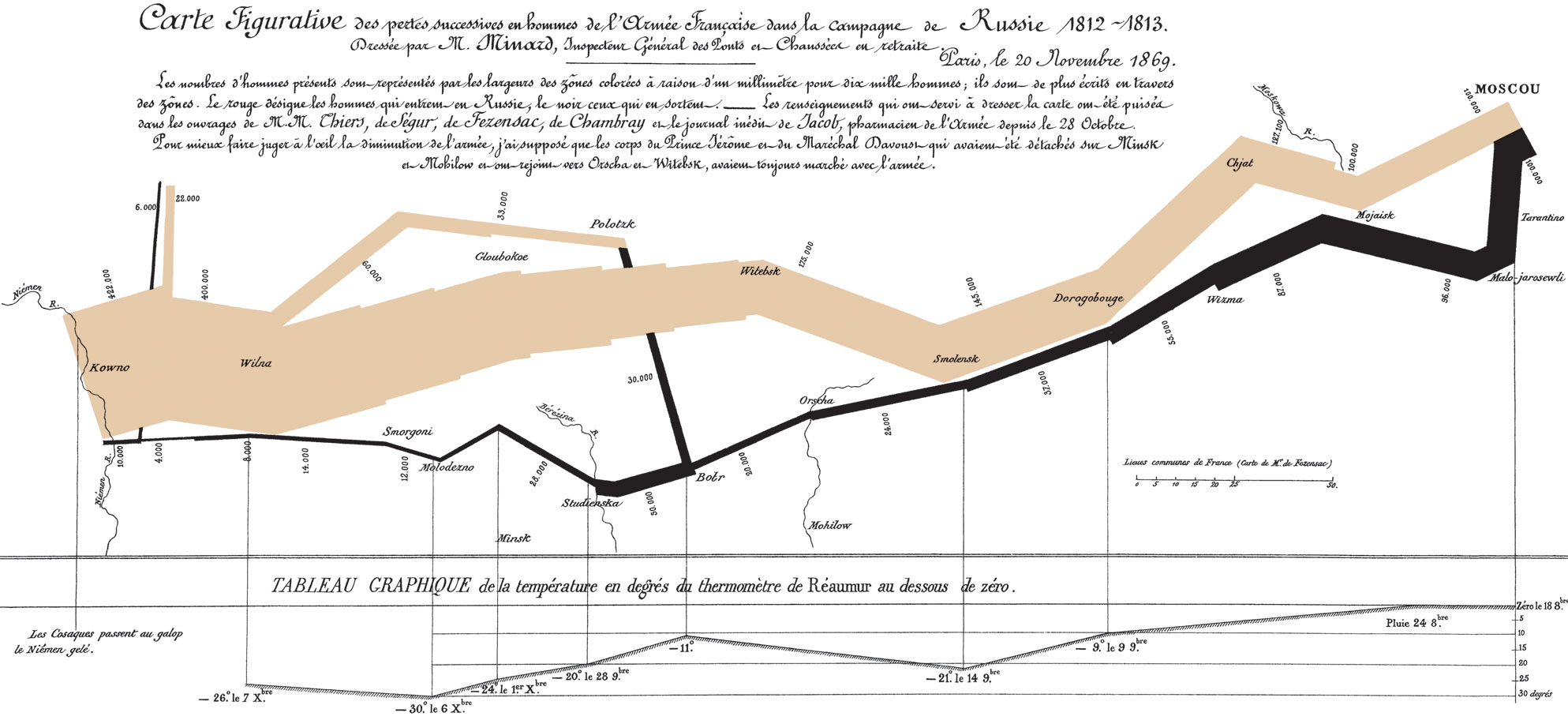
If data are looked at in the right way, answers become immediately clear
Edward Tufte




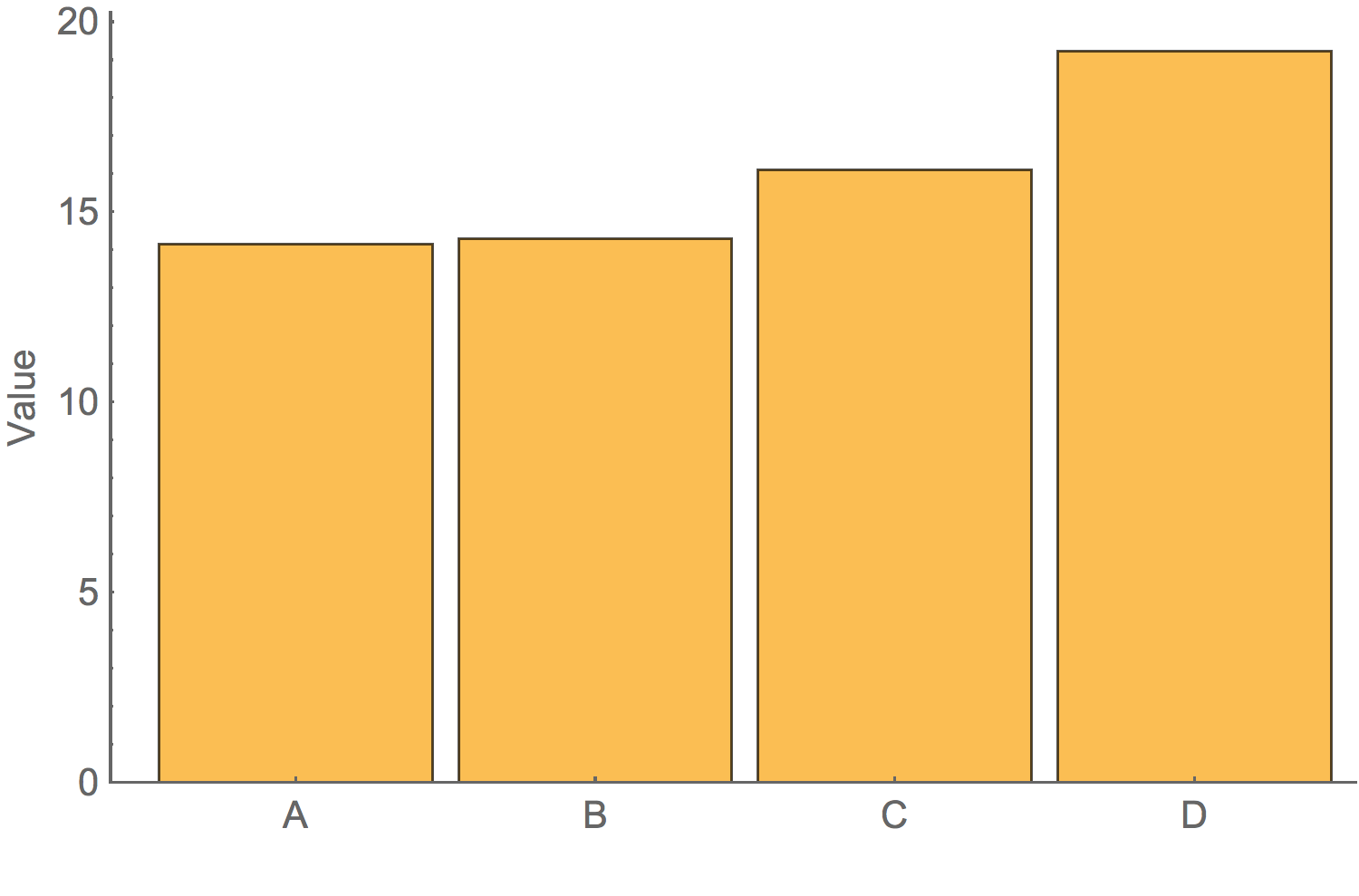
Above all, show the data
A good graphic is honestBarchart of average value
Box-and-whisker plot showing variation
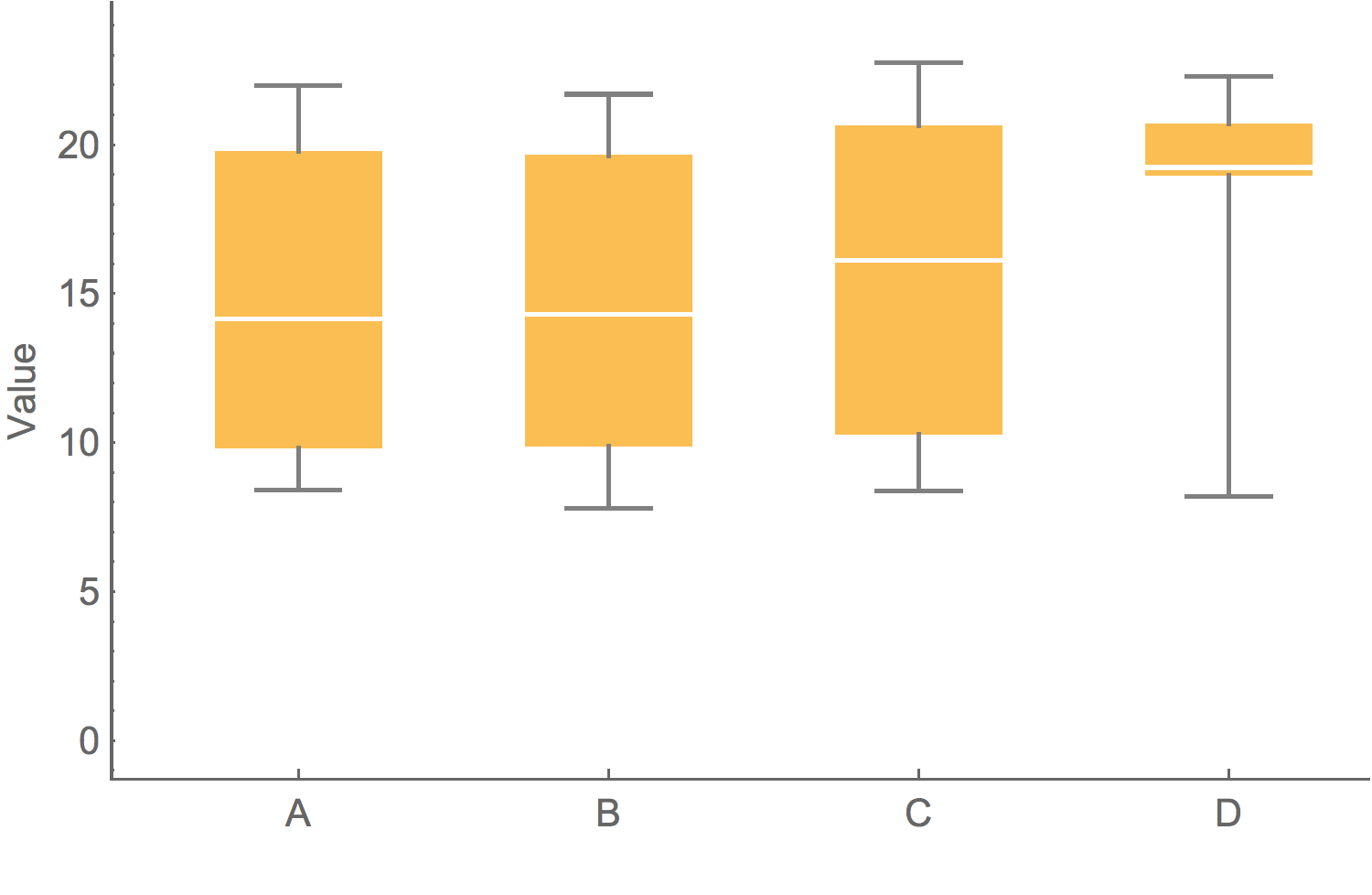
Showing the data
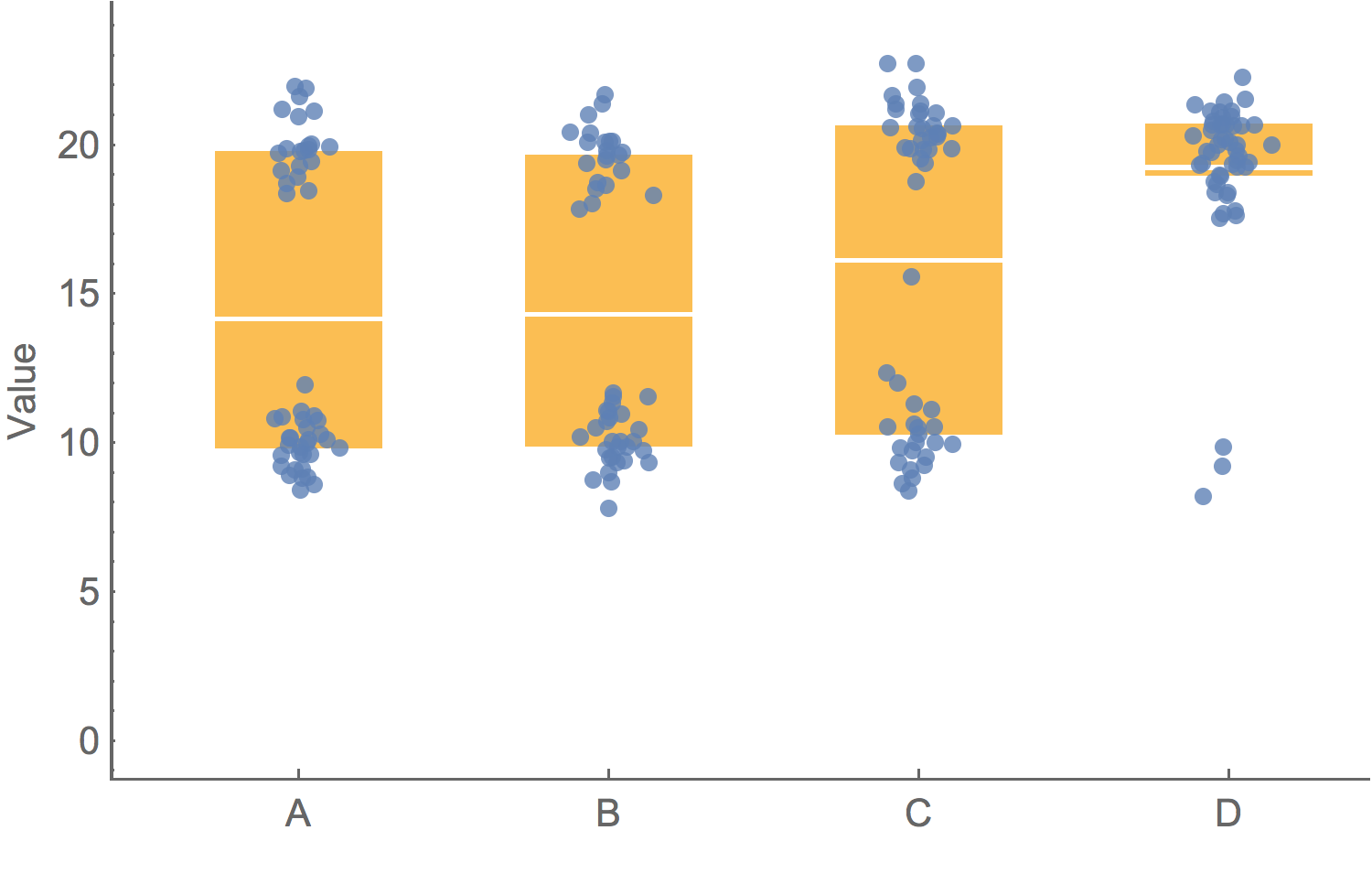
To clarify add detail
"To clarify, add detail... Clutter and overload are not attributes of information, they are failures of design. If the information is in chaos, don’t start throwing out information, instead fix the design."– Tufte
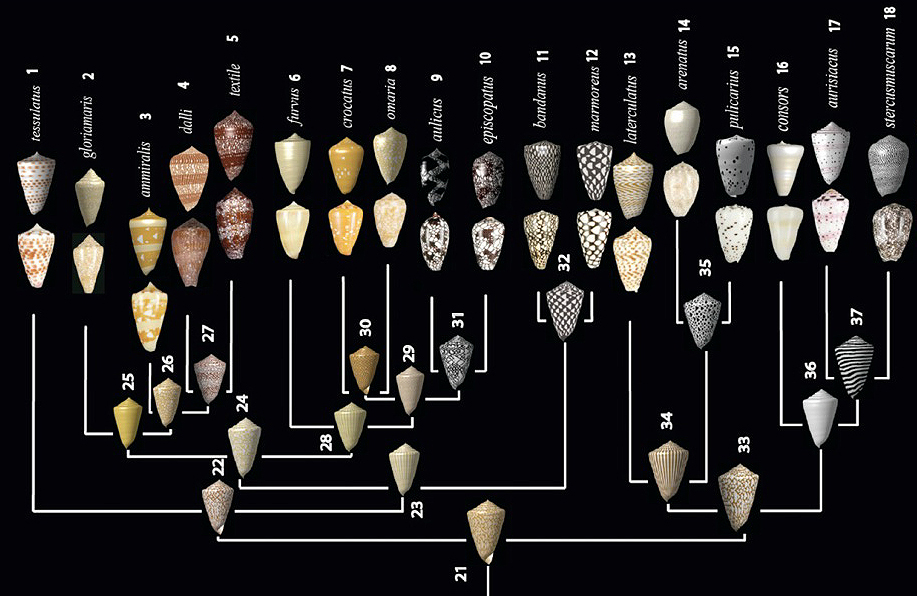
Evolution of Conus shells
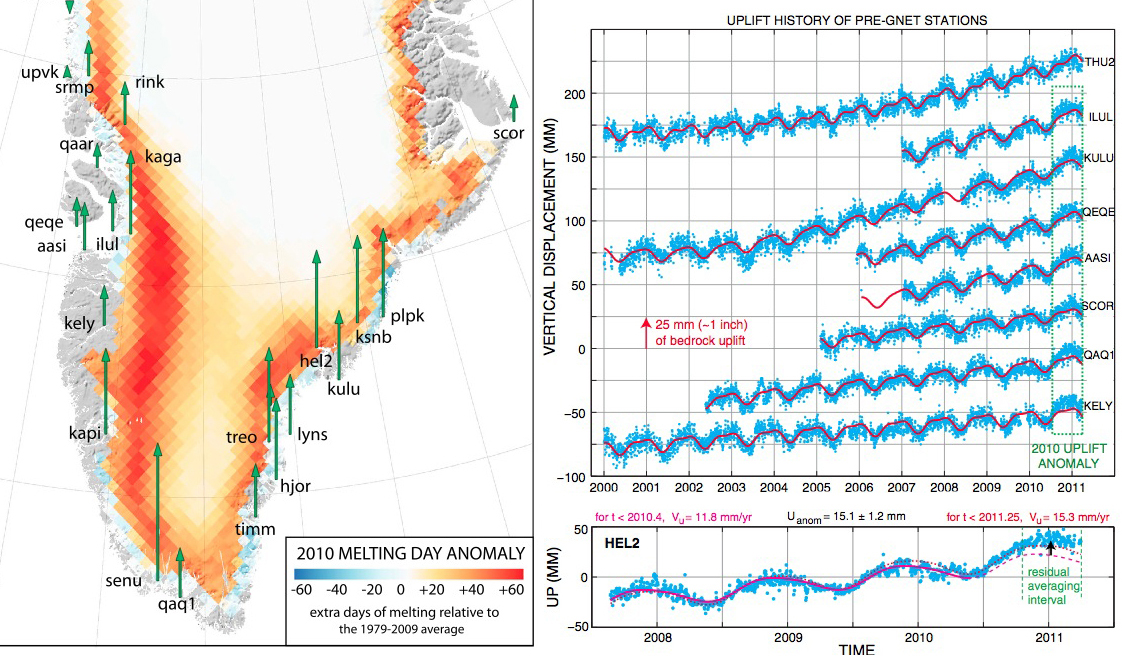
Greenland rising
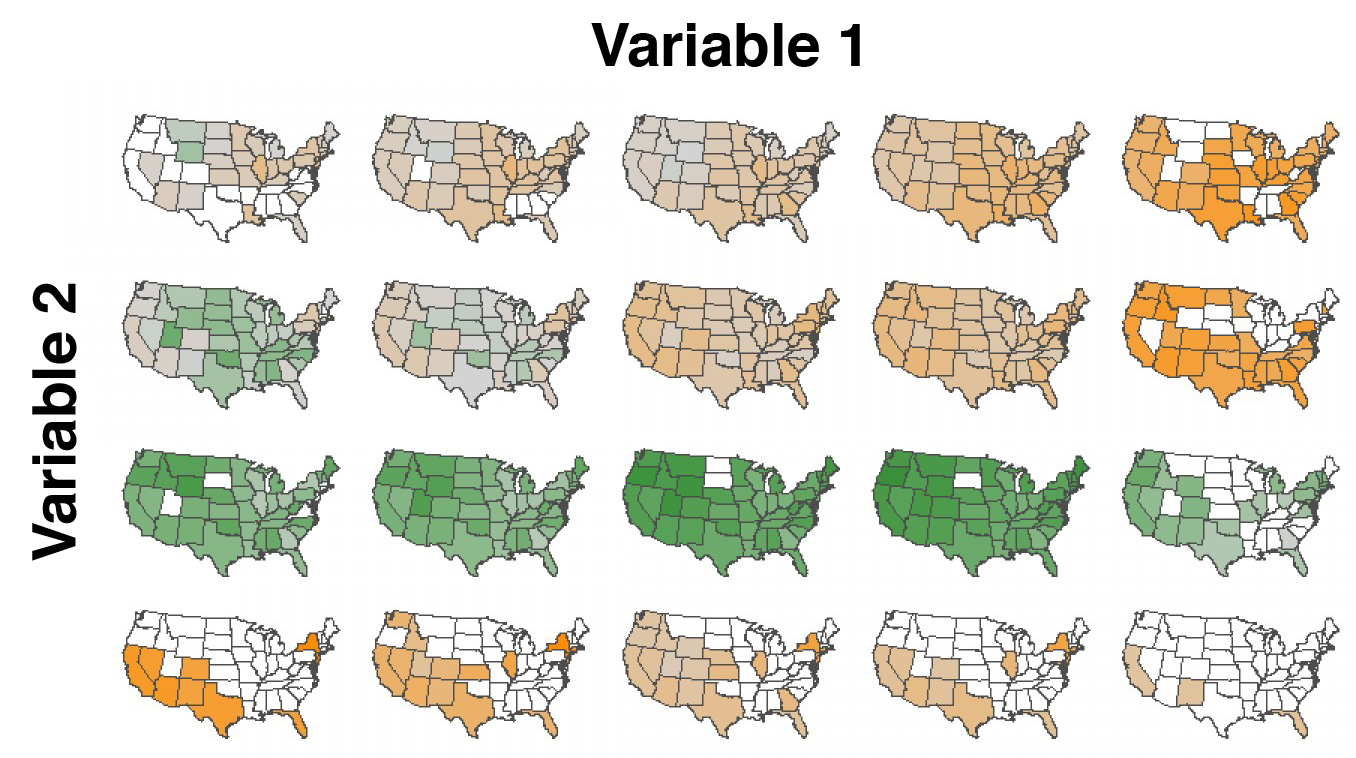
Small multiples and dimensionality
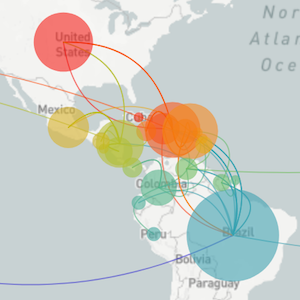
Mapping infection spread
John Snow and the founding of epidemiologyMy research
Methods focus on sequencing to reconstruct pathogen spread
Epidemic process
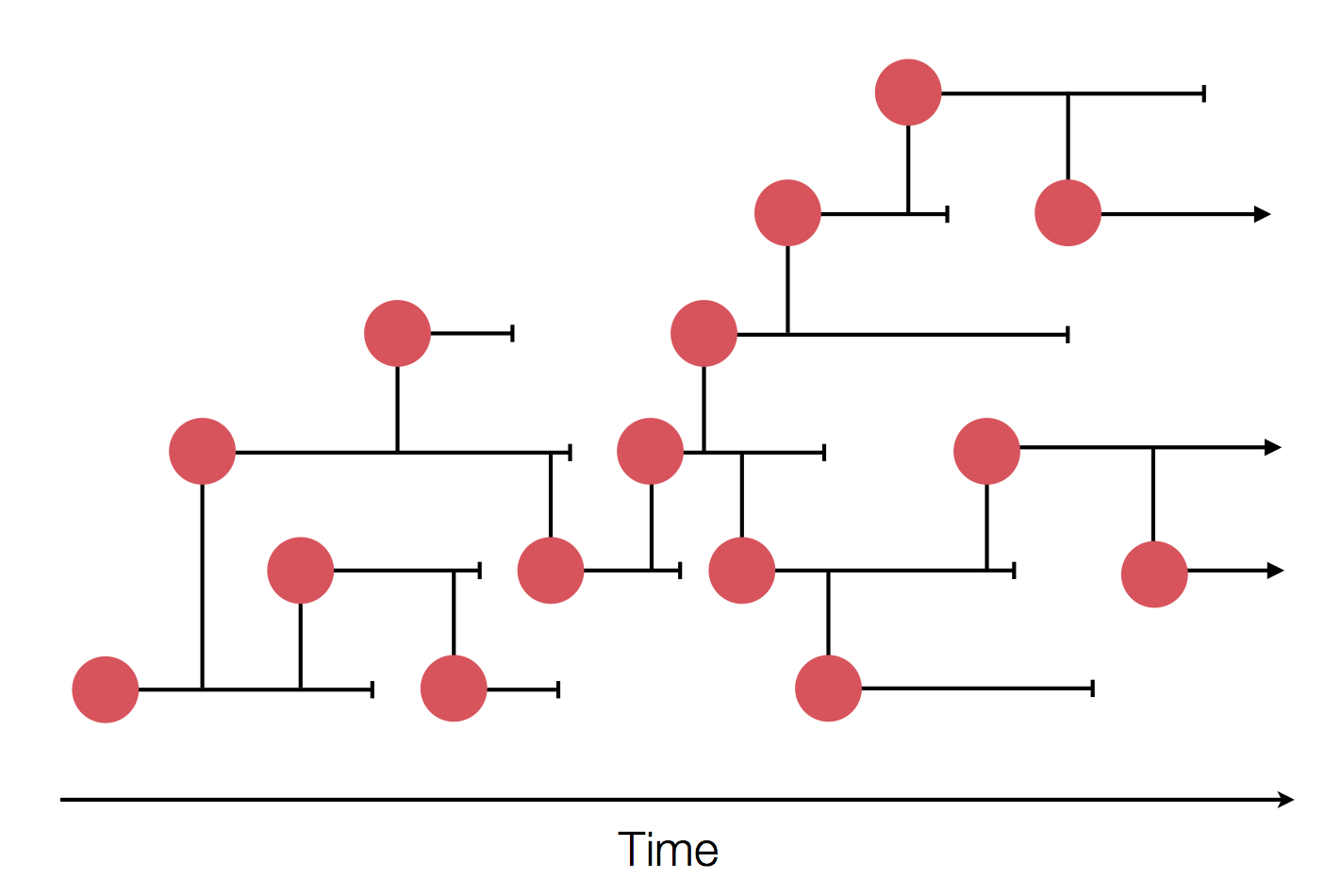
Sample some individuals
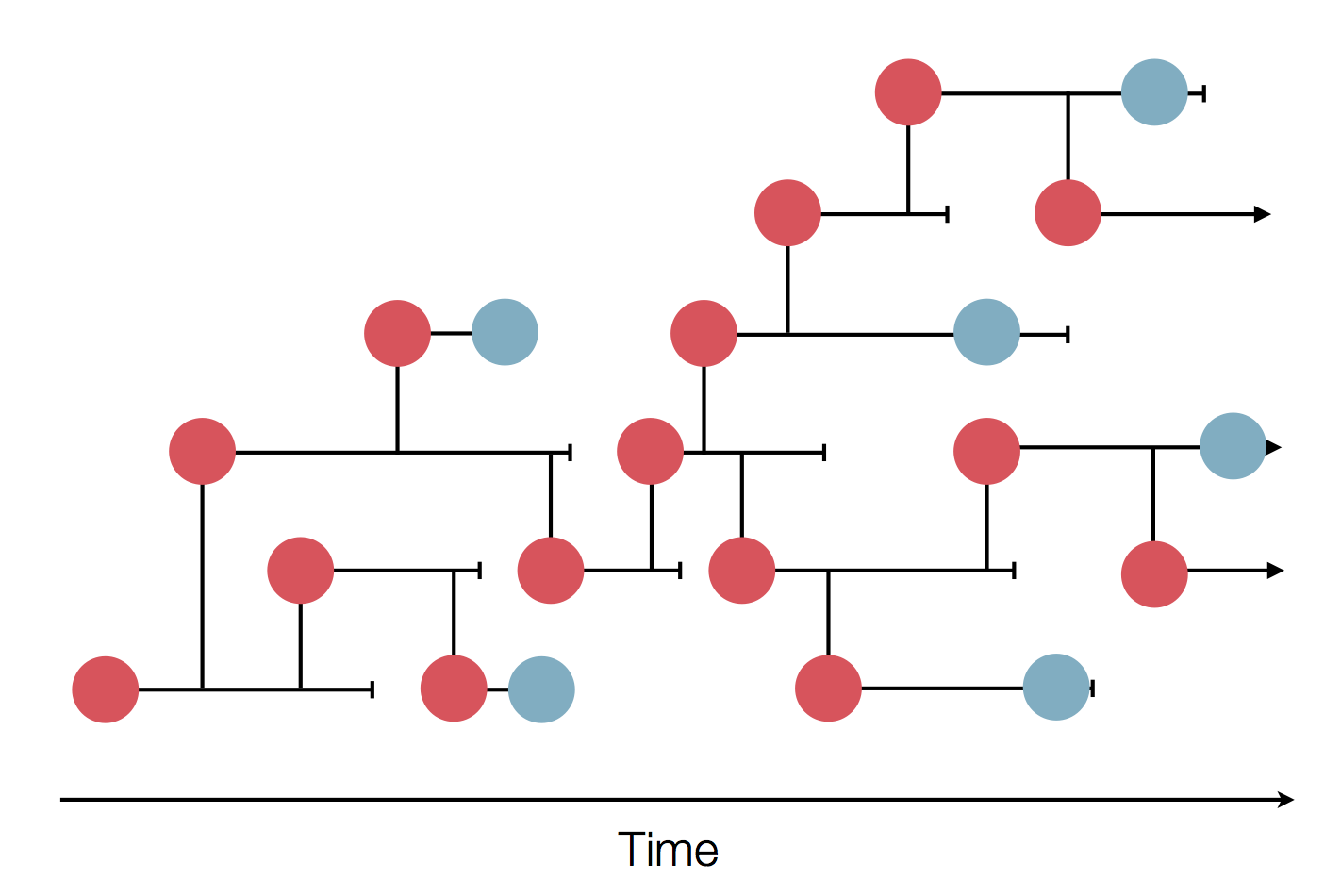
Sequence and determine phylogeny
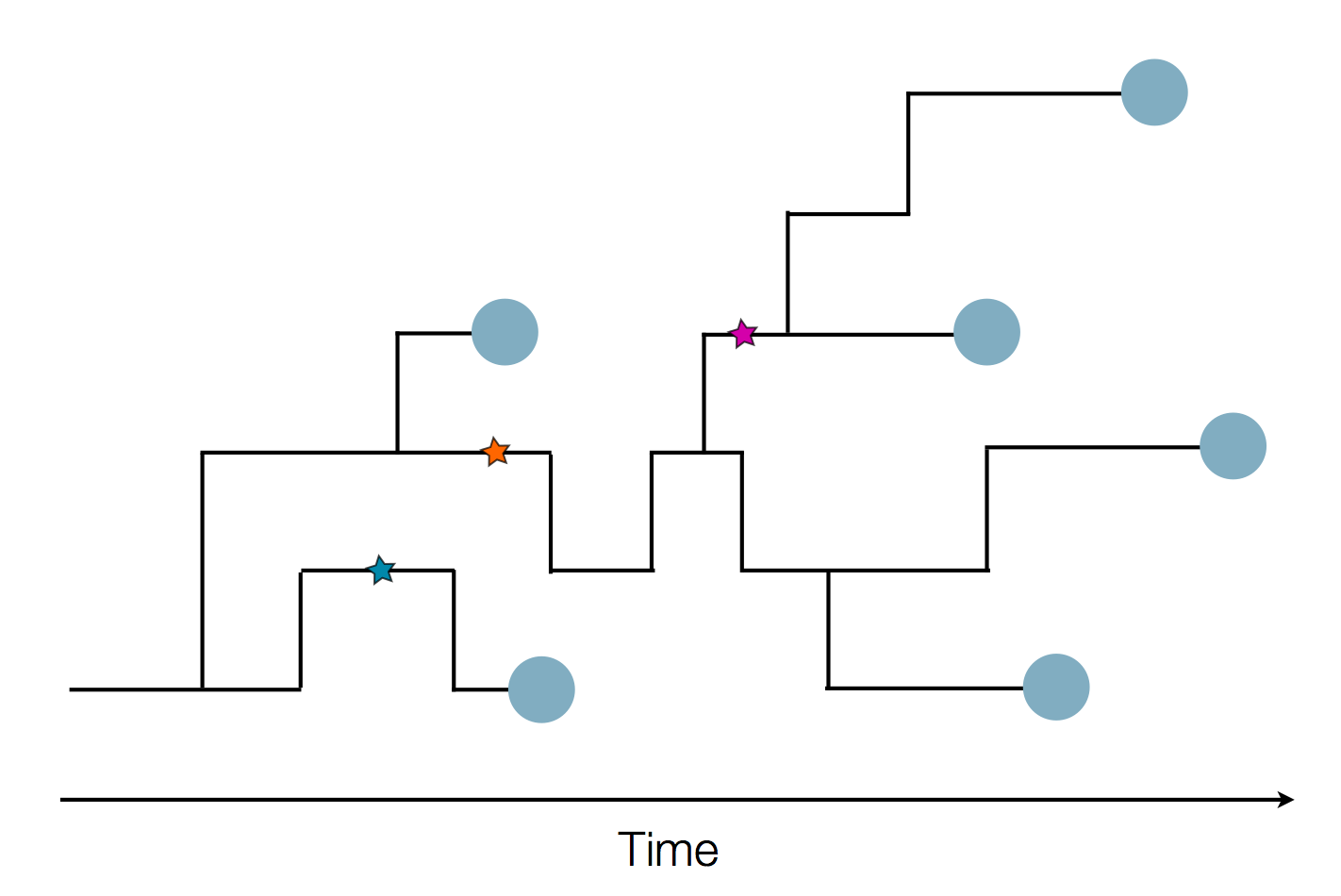
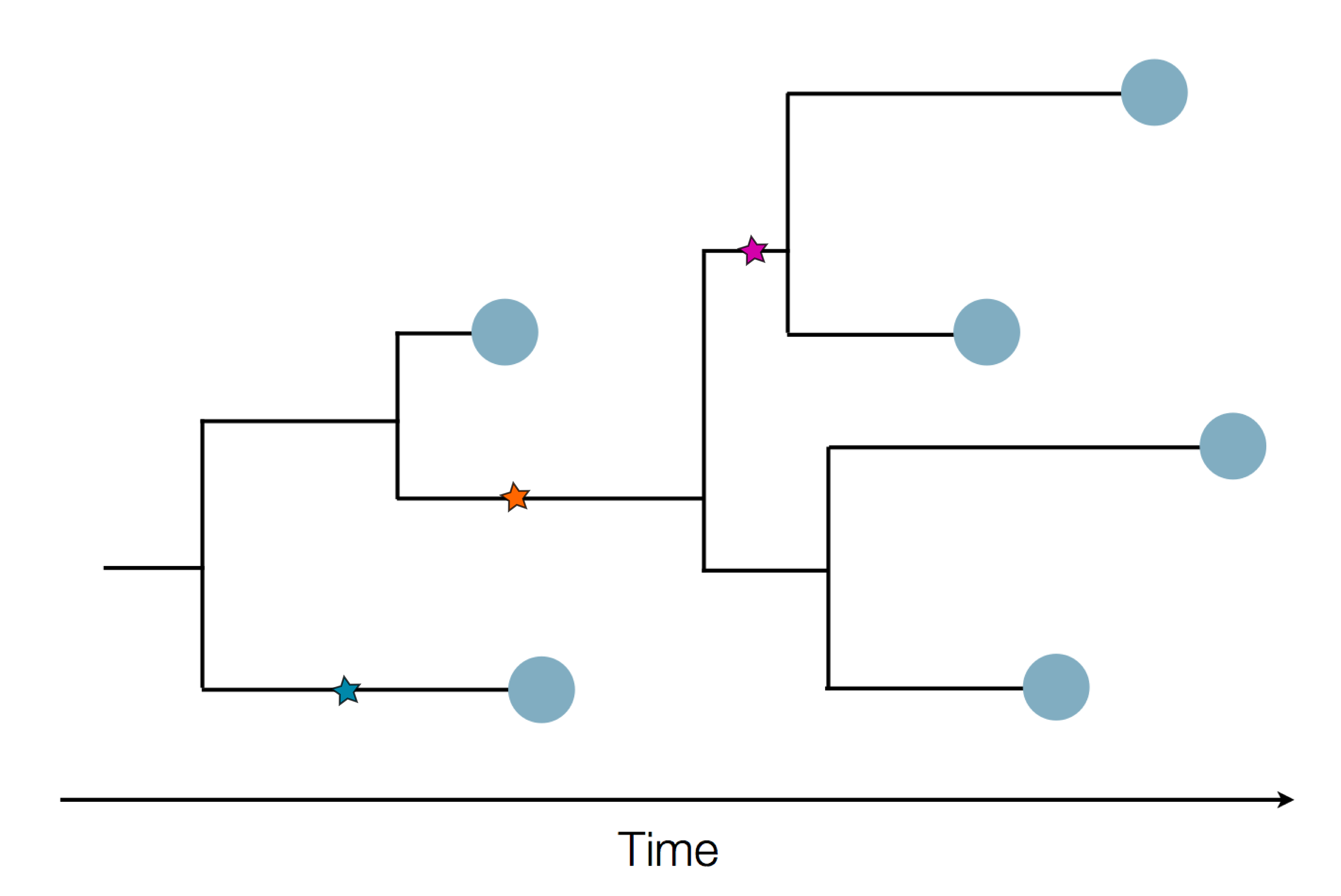
Sequence and determine phylogeny
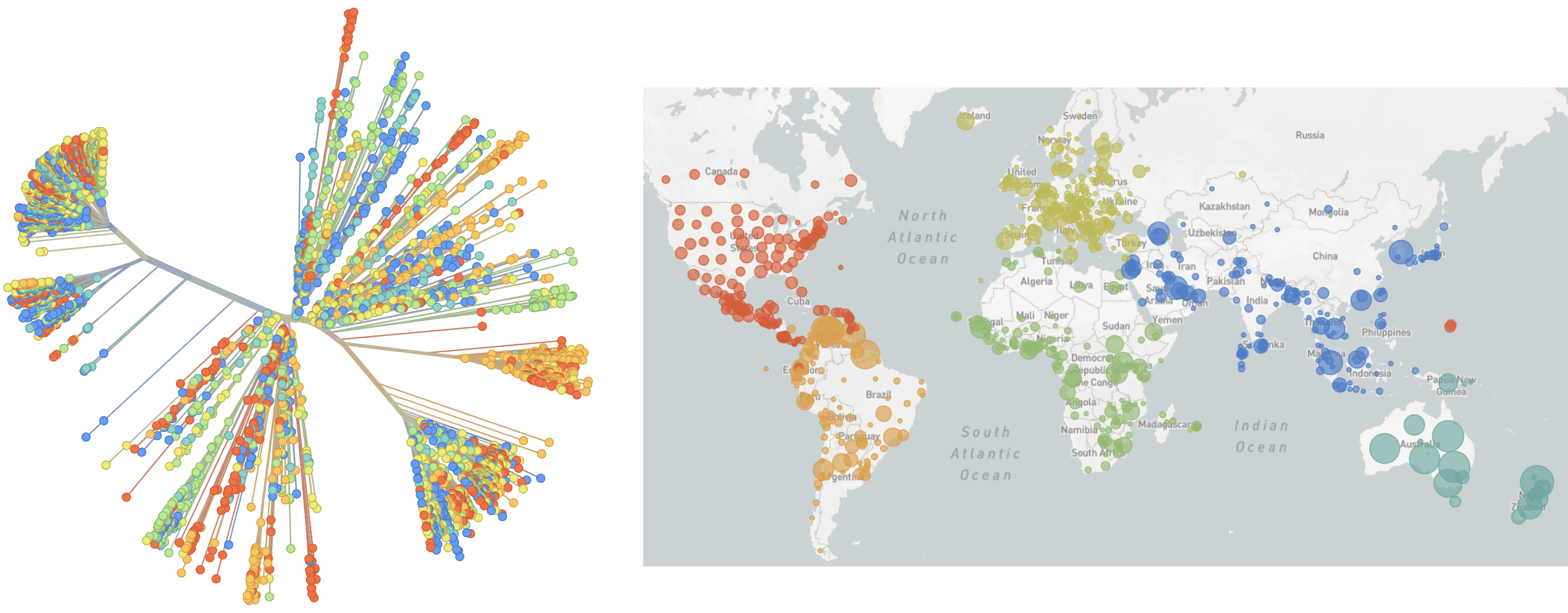
Genomic epidemiology during the COVID-19 pandemic
Over 12M SARS-CoV-2 genomes shared to GISAID and evolution tracked in real-time at nextstrain.org
![]() Richard Neher,
Richard Neher,
![]() Emma Hodcroft,
Emma Hodcroft,
![]() James Hadfield,
James Hadfield,
![]() Thomas Sibley,
Thomas Sibley,
![]() John Huddleston,
John Huddleston,
![]() Ivan Aksamentov,
Ivan Aksamentov,
![]() Moira Zuber,
Moira Zuber,
![]() Jover Lee,
Jover Lee,
![]() Cassia Wagner,
Cassia Wagner,
![]() Denisse Sequeira,
Denisse Sequeira,
![]() Cornelius Roemer,
Cornelius Roemer,
![]() Victor Lin,
Victor Lin,
![]() Jennifer Chang
Jennifer Chang
Three key insights that genomic epi provided during pandemic
- Rapid human-to-human spread in Wuhan beyond initial market outbreak
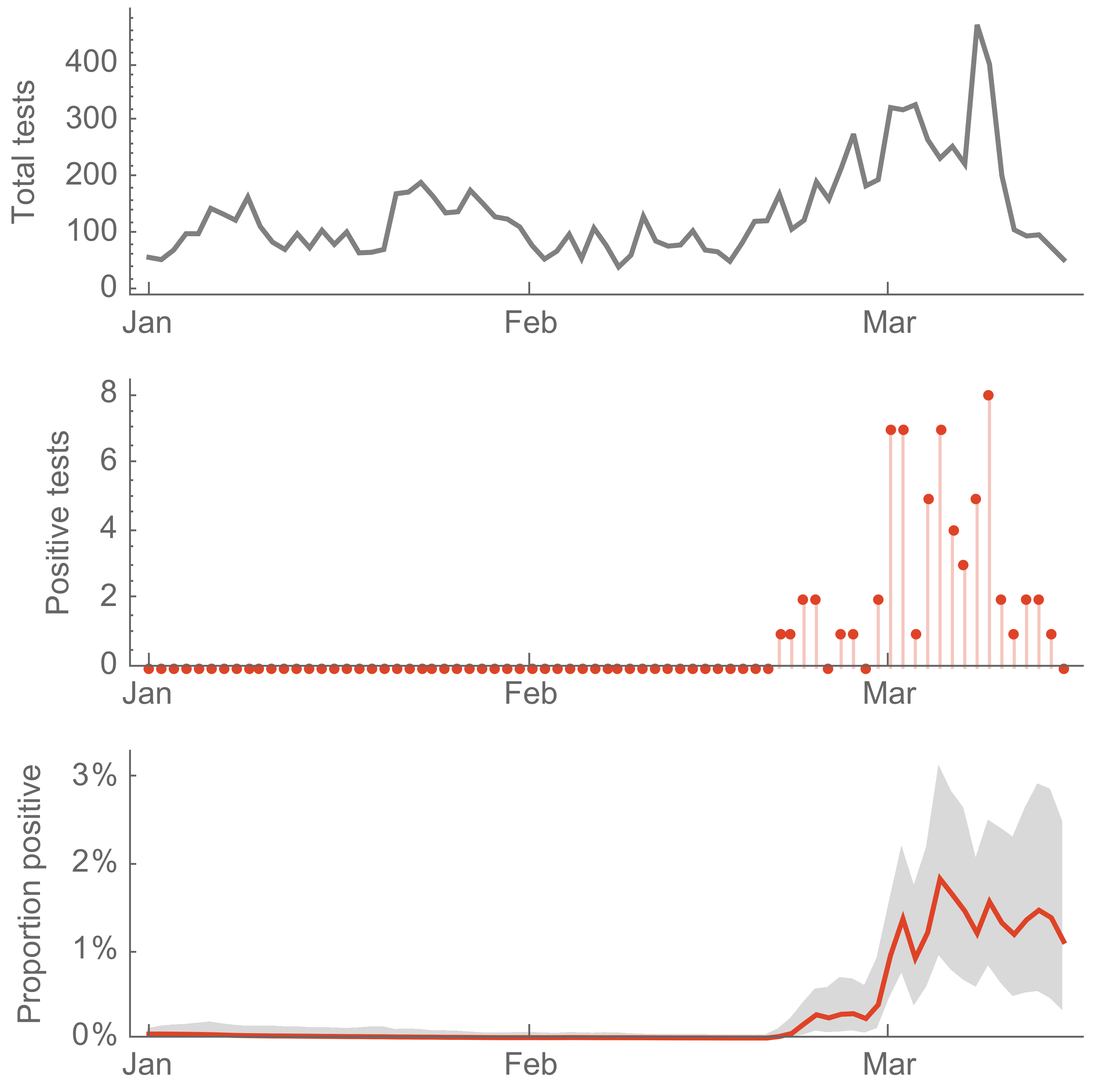
- Extensive local transmission while testing was rare
- Identification of variants of concern and mapping of increased transmission
Jan 11: First five genomes from Wuhan showed a novel SARS-like coronavirus
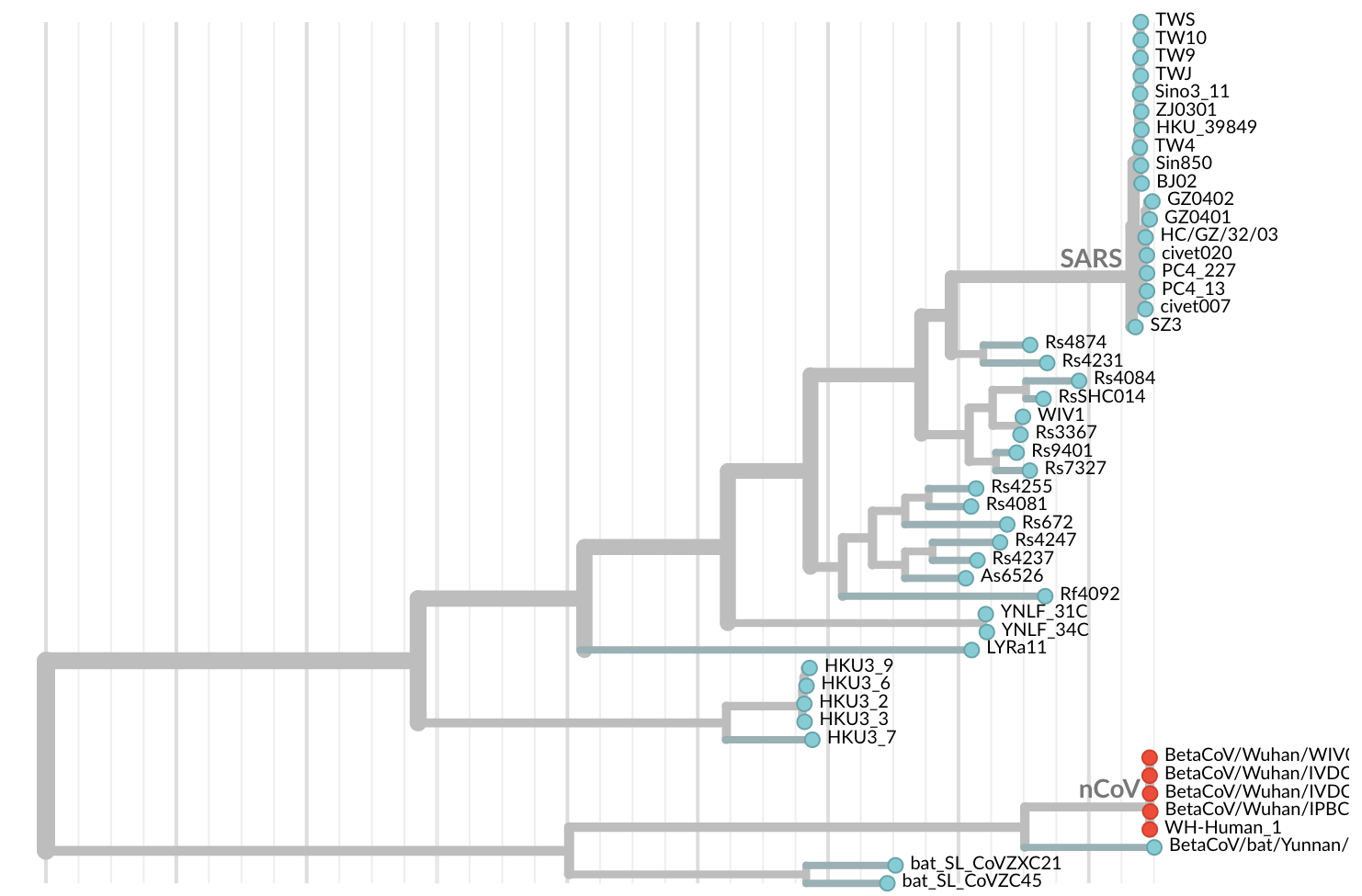
Jan 19: First 12 genomes from Wuhan and Bangkok showed lack of genetic diversity
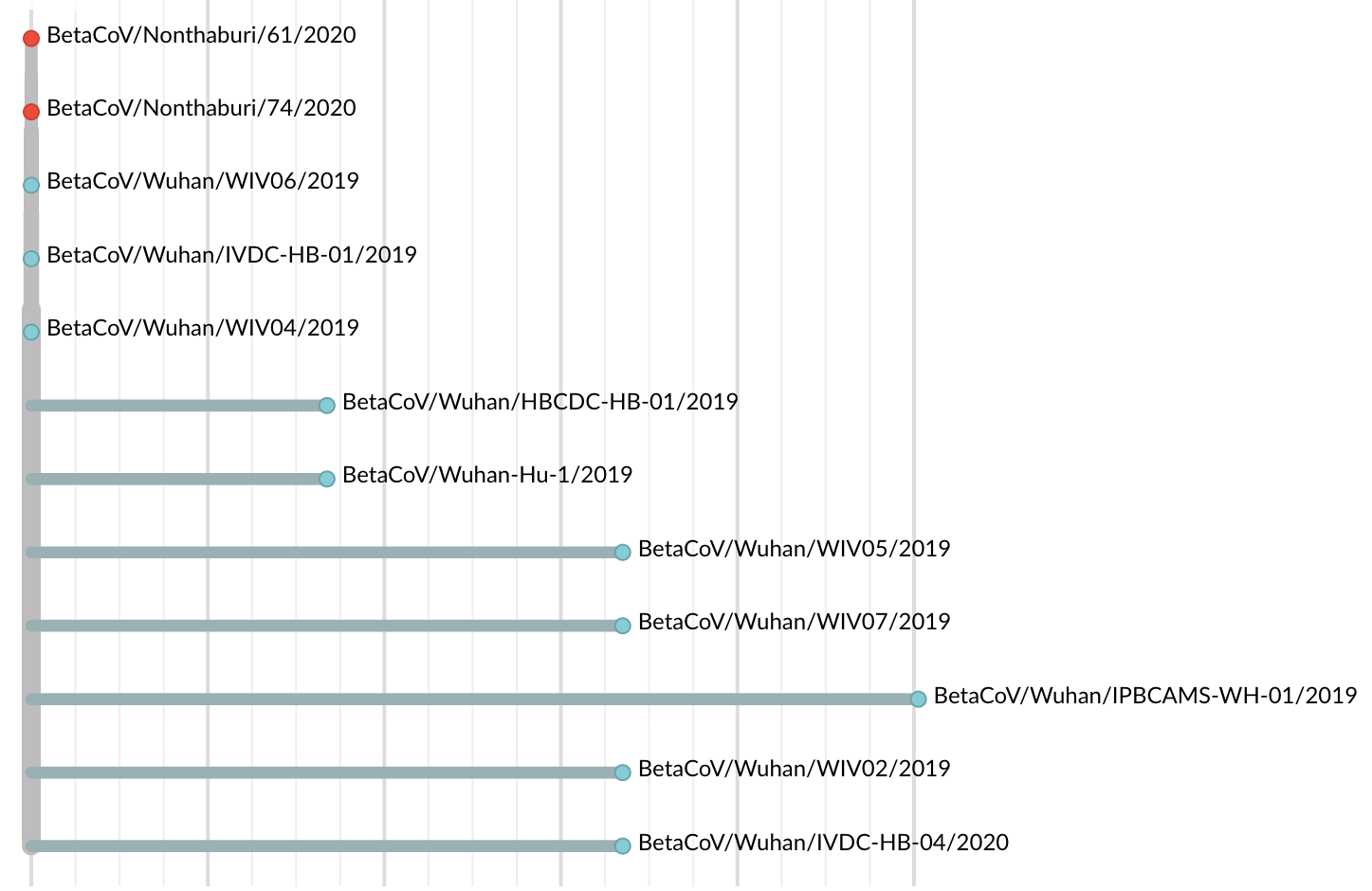
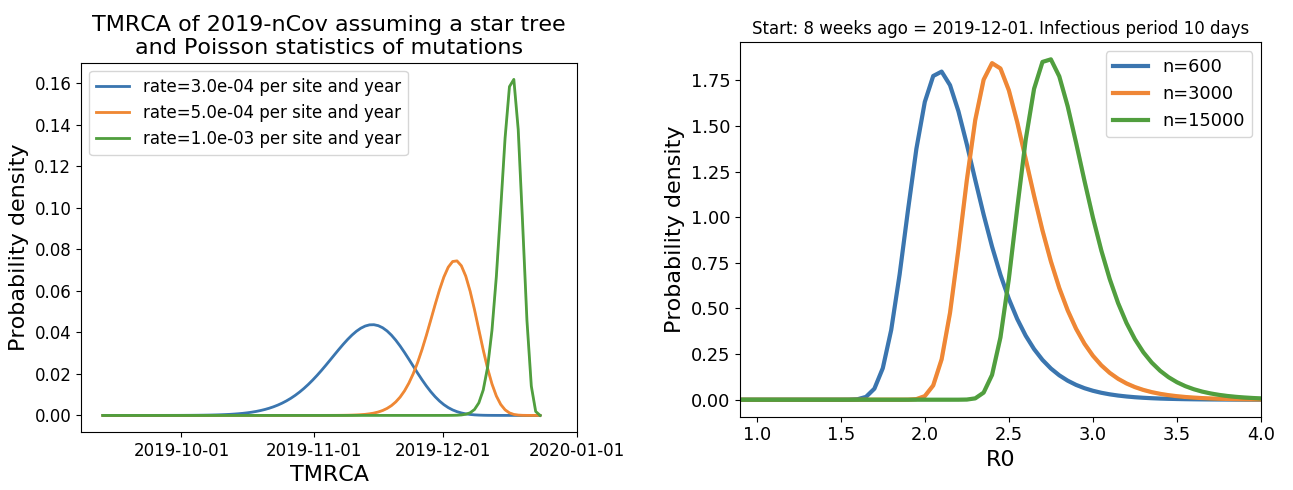
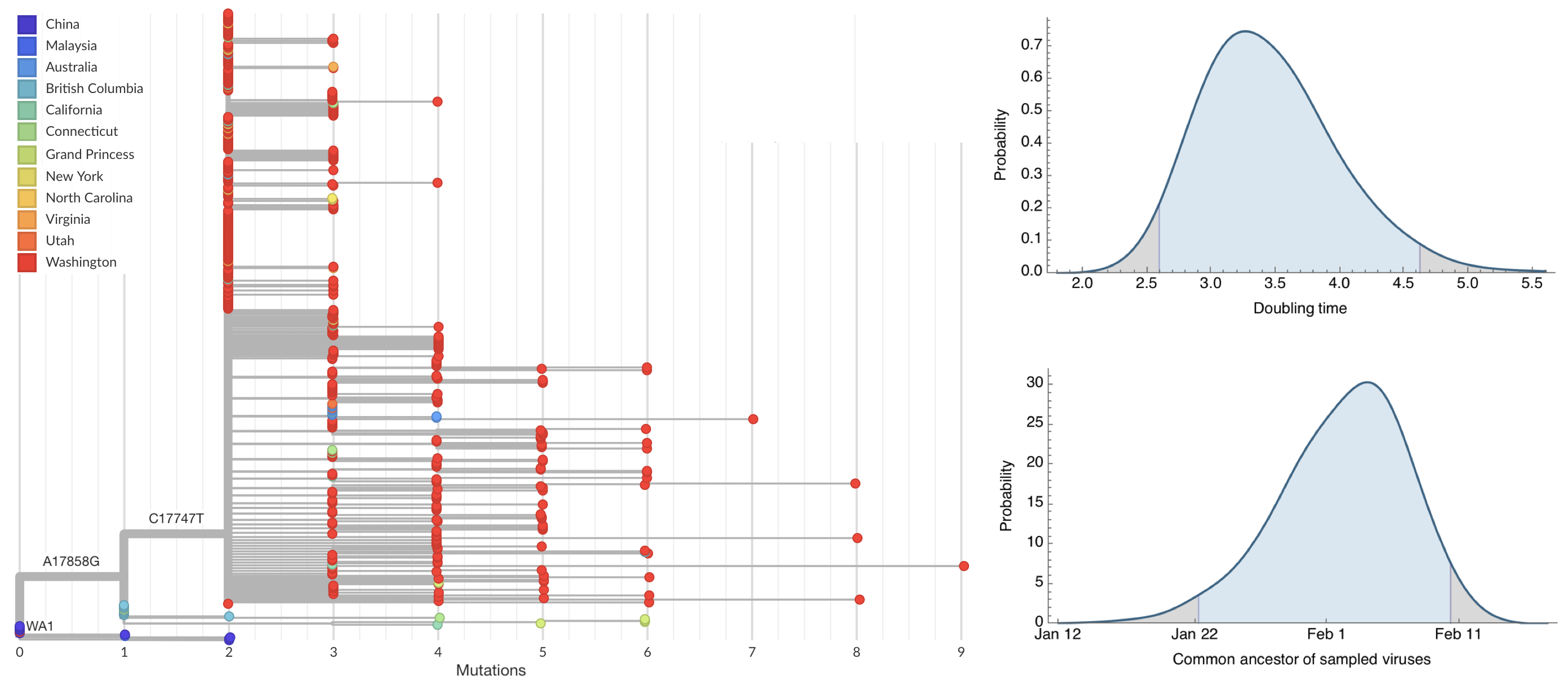
Single introduction into the human population between Nov 15 and Dec 15 and subsequent rapid human-to-human spread
Rapid global epidemic spread from China
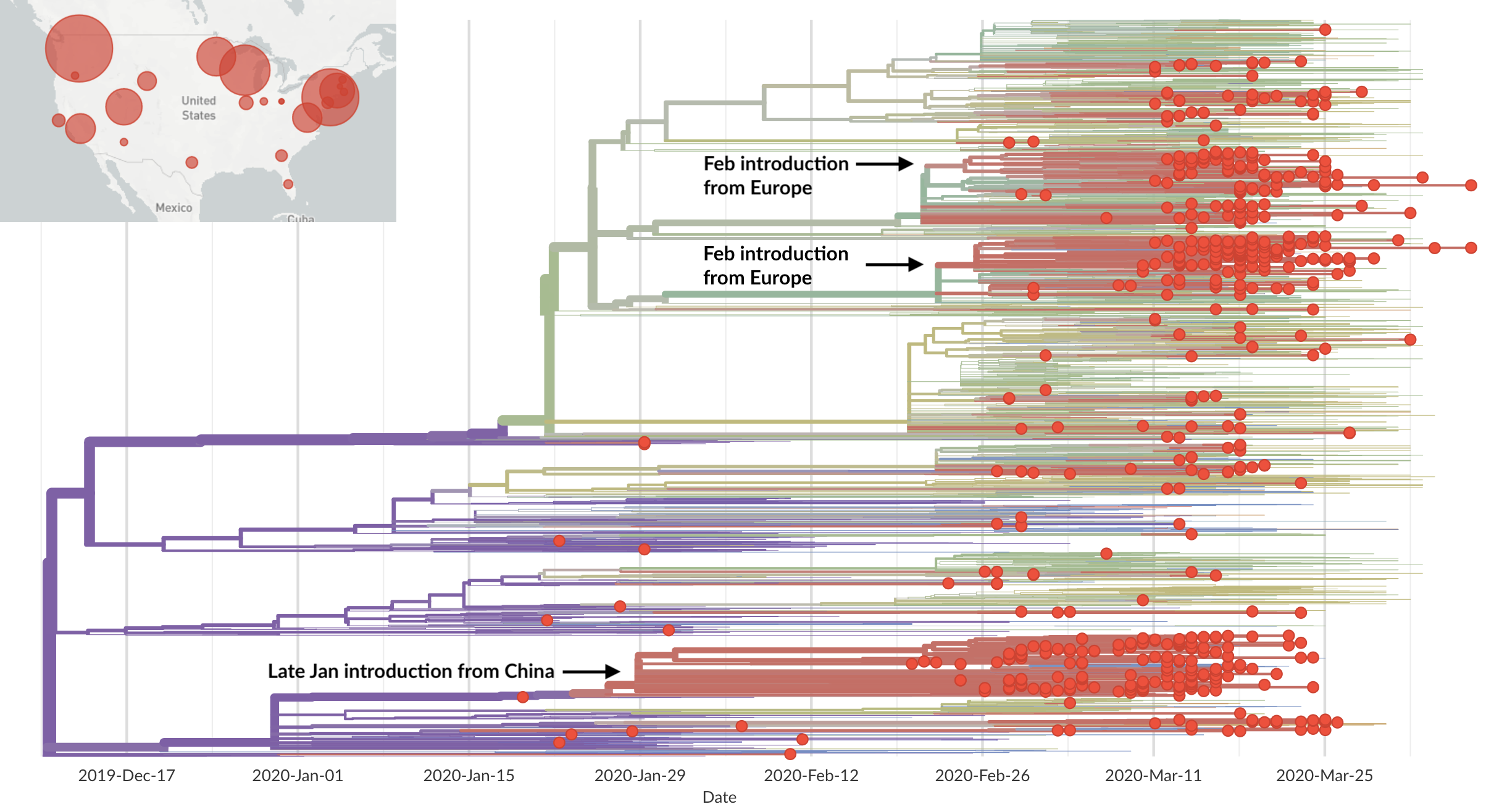
Epidemic in the USA was introduced from China in late Jan and from Europe during Feb
Seattle Flu Study detects local circulation and charts early epidemic in Washington State
Direct introduction from China ~Feb 1 responsible for the majority of the epidemic


Mitigation efforts visible in virus genetic diversity
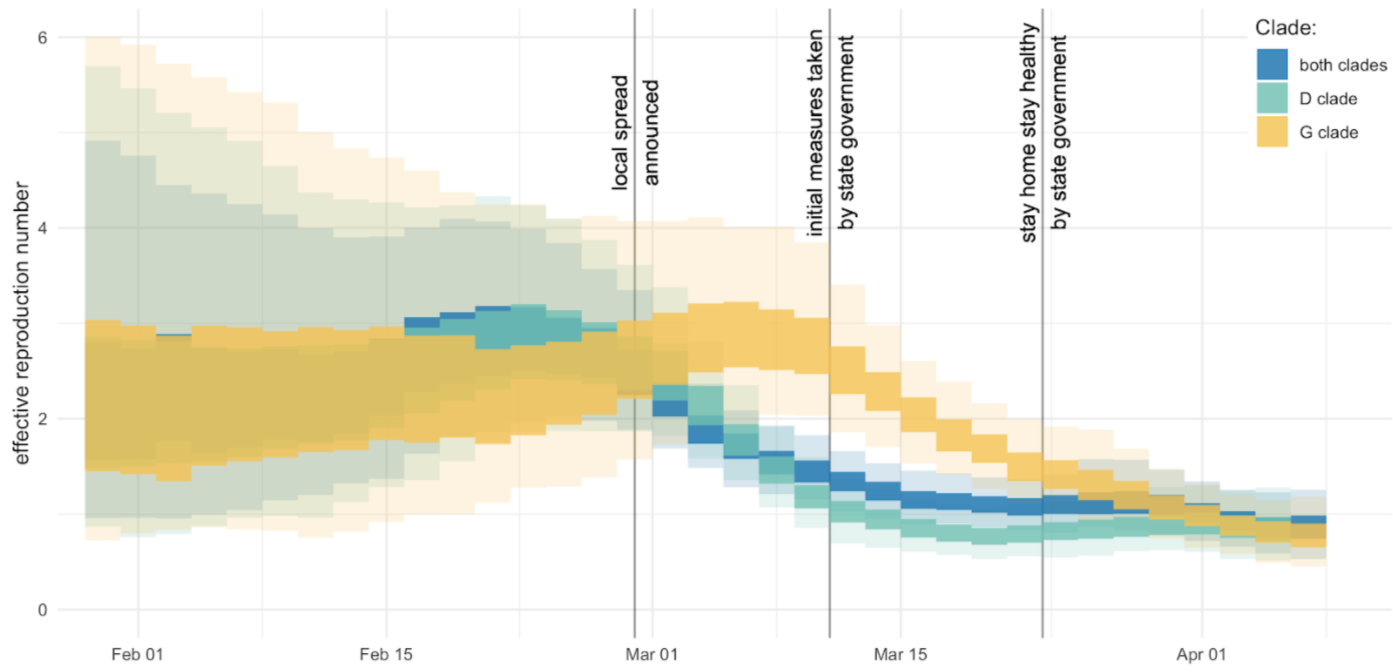
After initial wave, with mitigation
efforts and decreased travel,
regional clades emerge
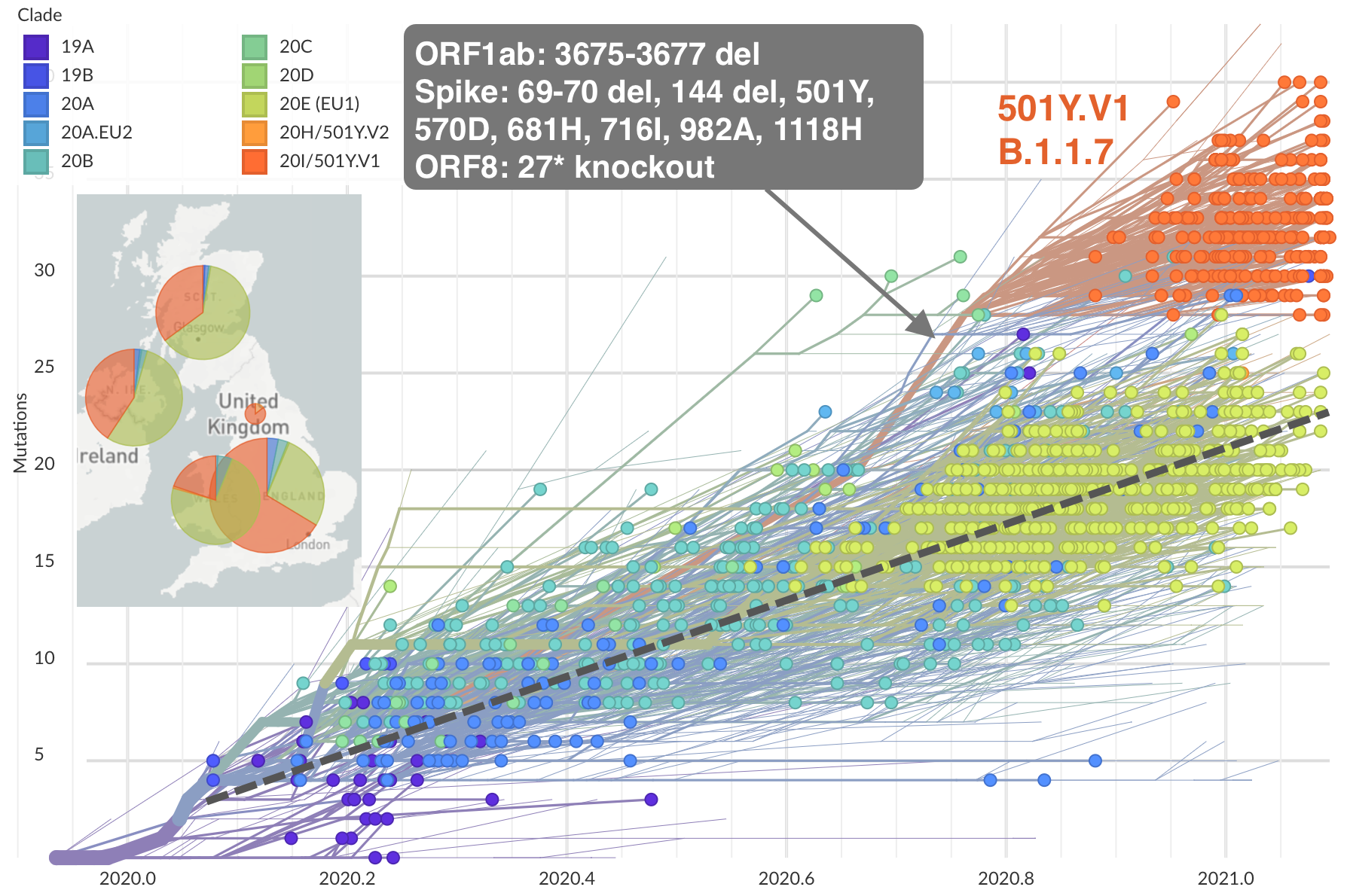
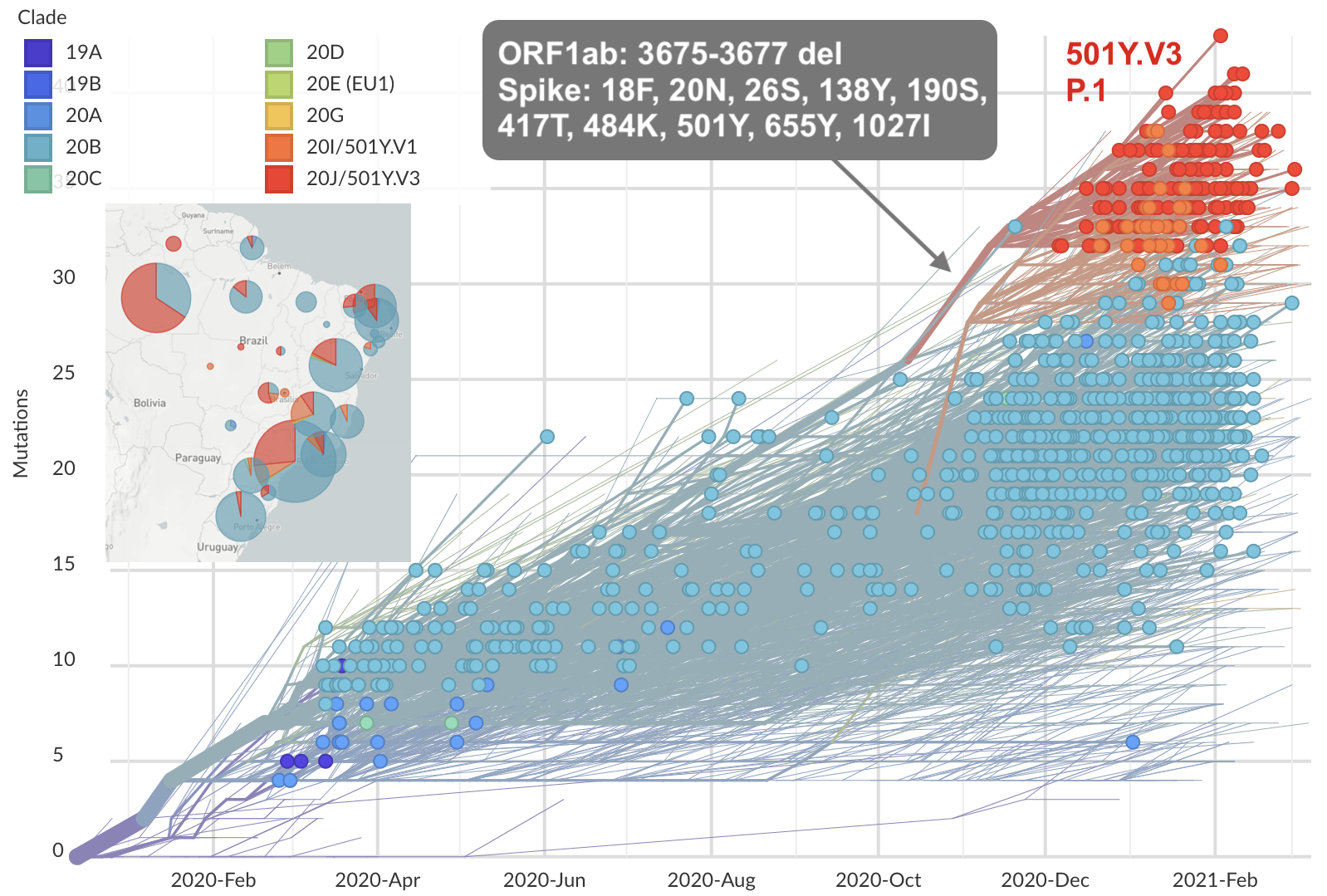
Repeated emergence of variant of concern viruses
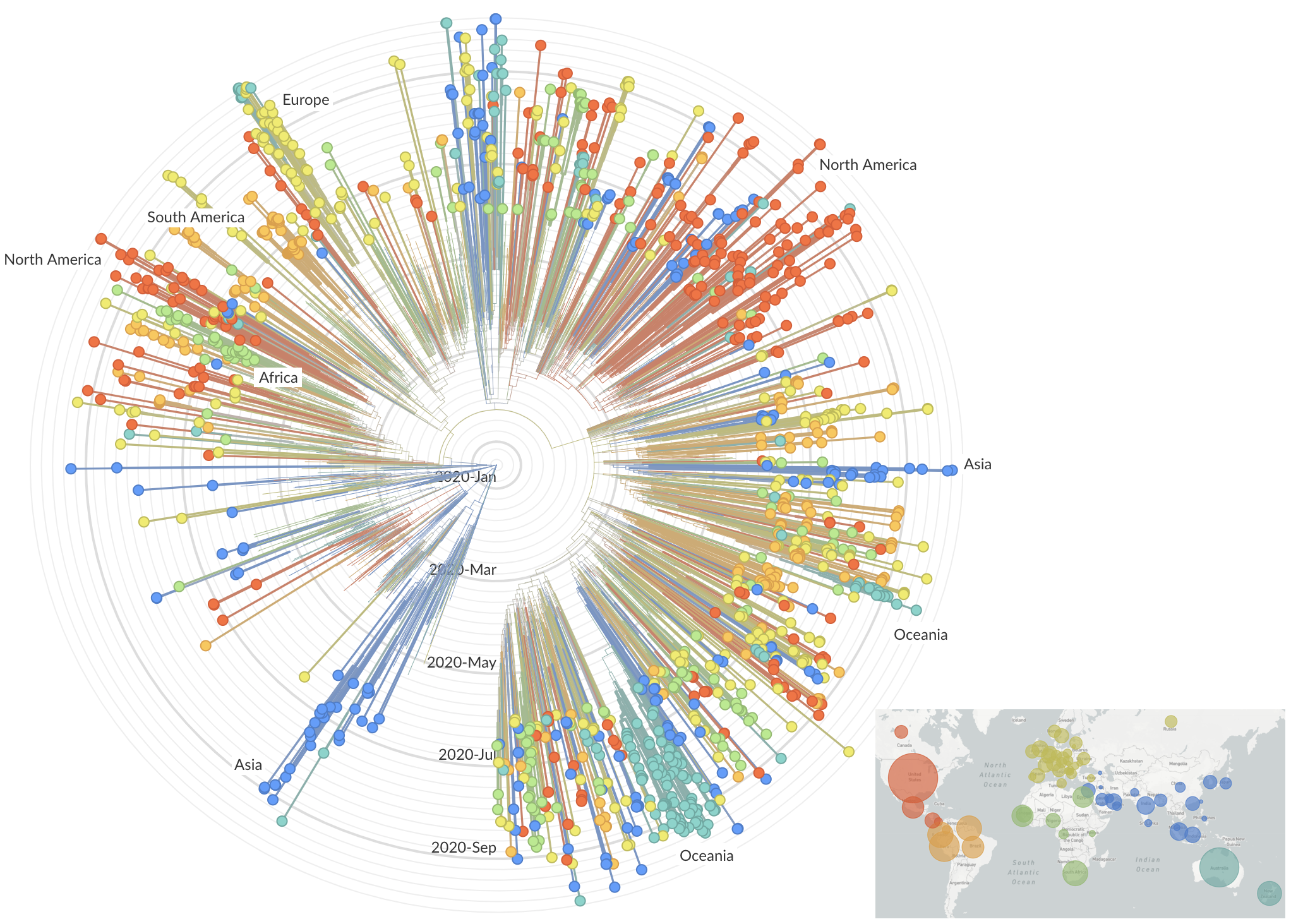
Genetic relationships of globally sampled SARS-CoV-2 to present
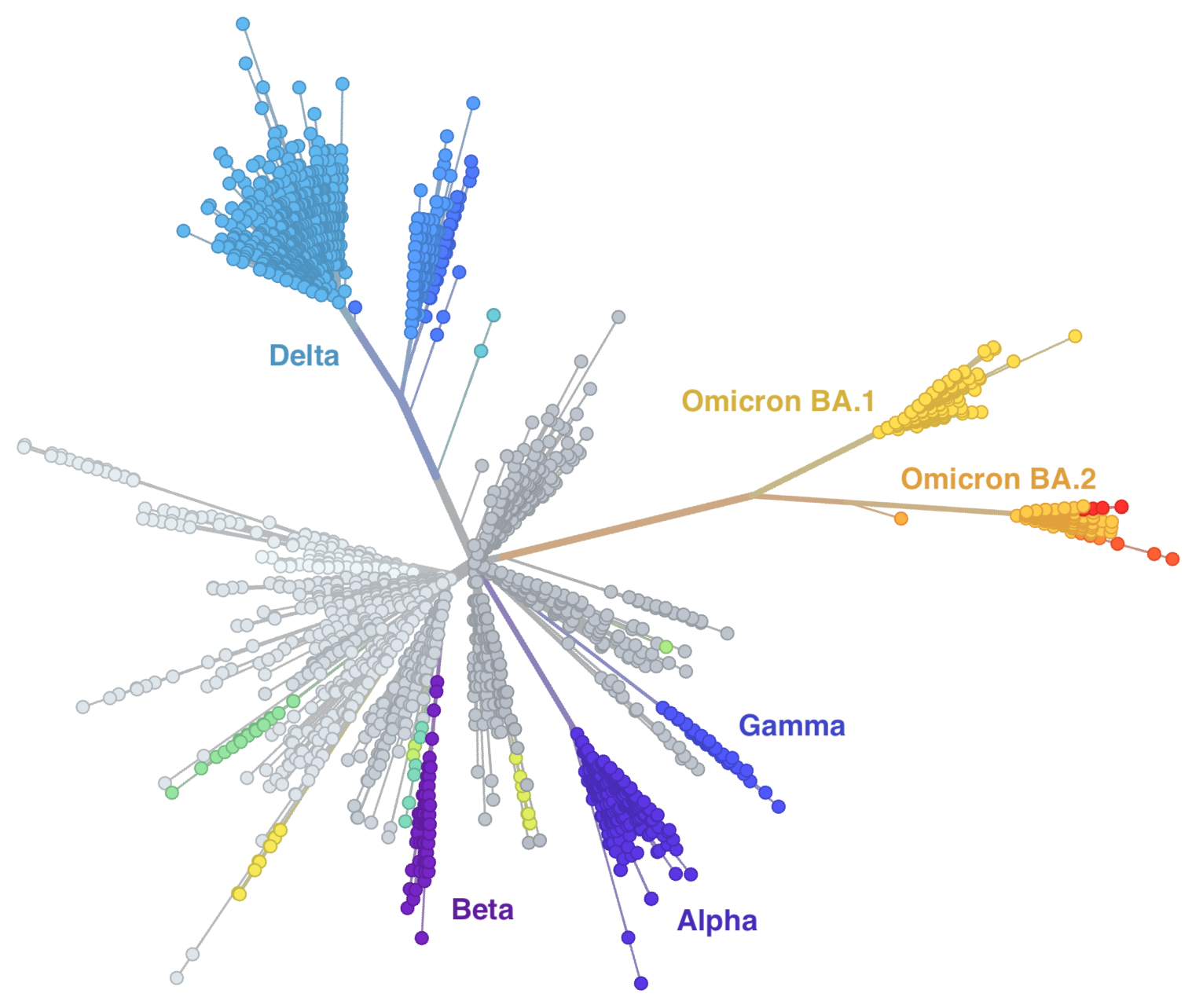
Rapid displacement of existing diversity by emerging variants

Rapid accumulation of spike mutations driving displacement

S1 evolution remarkably fast relative to seasonal influenza

BA.4 and BA.5 emerging from South Africa with spike mutations 452R and 486V
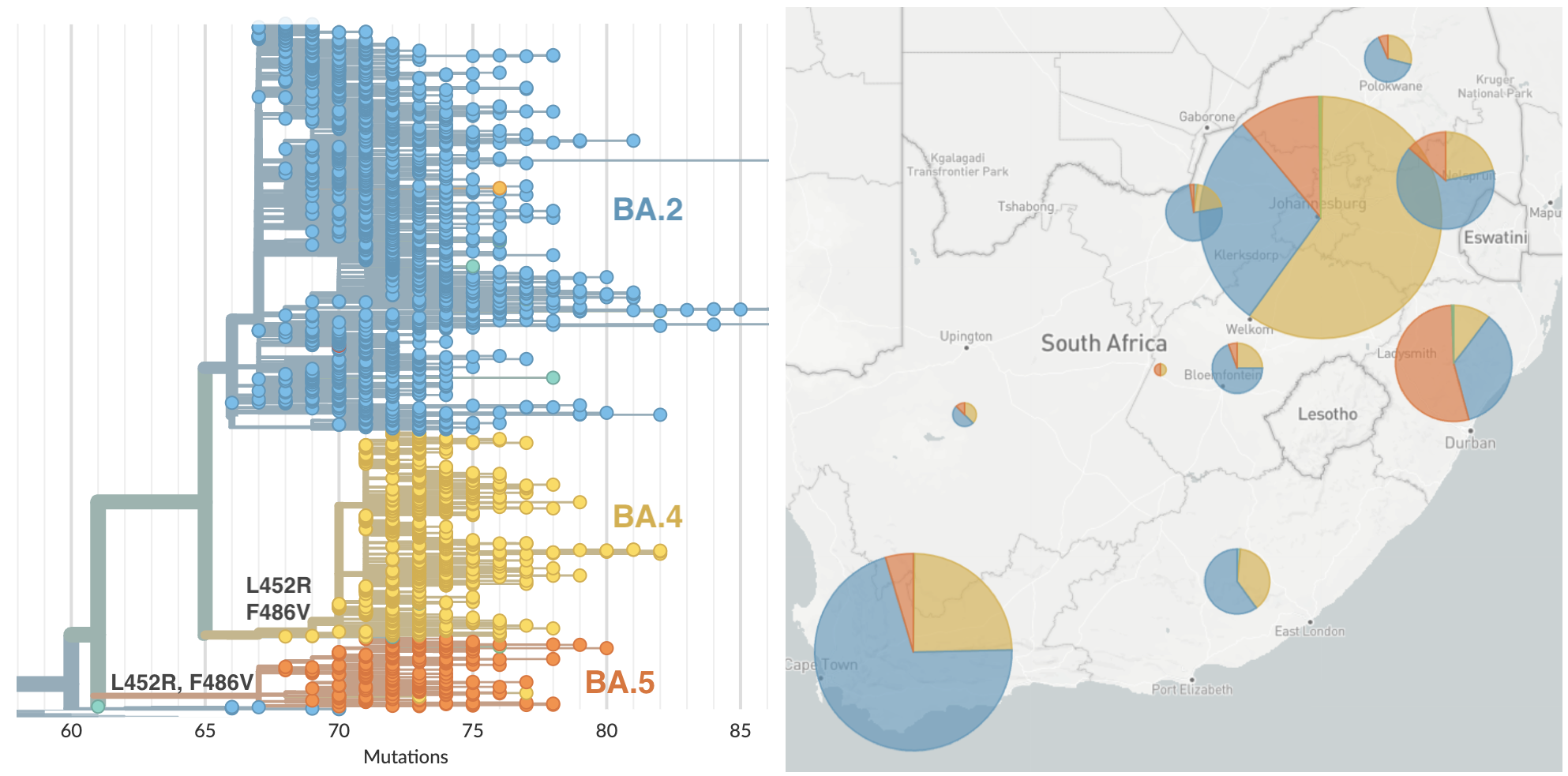
BA.5 is now dominant in the US (and most of the world)

Variant-specific epidemics shown by partitioning case counts by variant frequency

Variant-specific Rt decreasing as immunity accumulates

Similar dynamics seen in other countries

BA.2.75 emerging from India that possesses a number of spike mutations on top of BA.2

Frequency growth outpacing BA.5 in India

Inferred initial Rt ~1.9, but rapidly decreasing due to lack of growing case counts in India

Lab directions
- Outbreak investigations (mumps in Washington, Ebola in DRC, monkeypox, etc...)
- Genomic surveillance in King County and Washington State
- Phylodynamics methods development
- Evolutionary forecasting methods development
- Software infrastructure for Nextstrain and genomic epidemiology
- Other applications of evolutionary dynamics (within-host malaria and cancer evolution)
Acknowledgements
SARS-CoV-2 genomic epi: Data producers from all over the world, GISAID and the Nextstrain team
Bedford Lab:
![]() John Huddleston,
John Huddleston,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Louise Moncla,
Louise Moncla,
![]() Maya Lewinsohn,
Maya Lewinsohn,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Cassia Wagner,
Cassia Wagner,
![]() Miguel Paredes,
Miguel Paredes,
![]() Nicola Müller,
Nicola Müller,
![]() Marlin Figgins,
Marlin Figgins,
![]() Denisse Sequeira,
Denisse Sequeira,
![]() Victor Lin,
Victor Lin,
![]() Jennifer Chang
Jennifer Chang






