Real-time tracking of virus evolution
Trevor Bedford (@trvrb)
23 May 2016
MIDAS Network Meeting
Washington, DC
Slides at bedford.io/talks/
Sequencing to reconstruct pathogen spread
Epidemic process
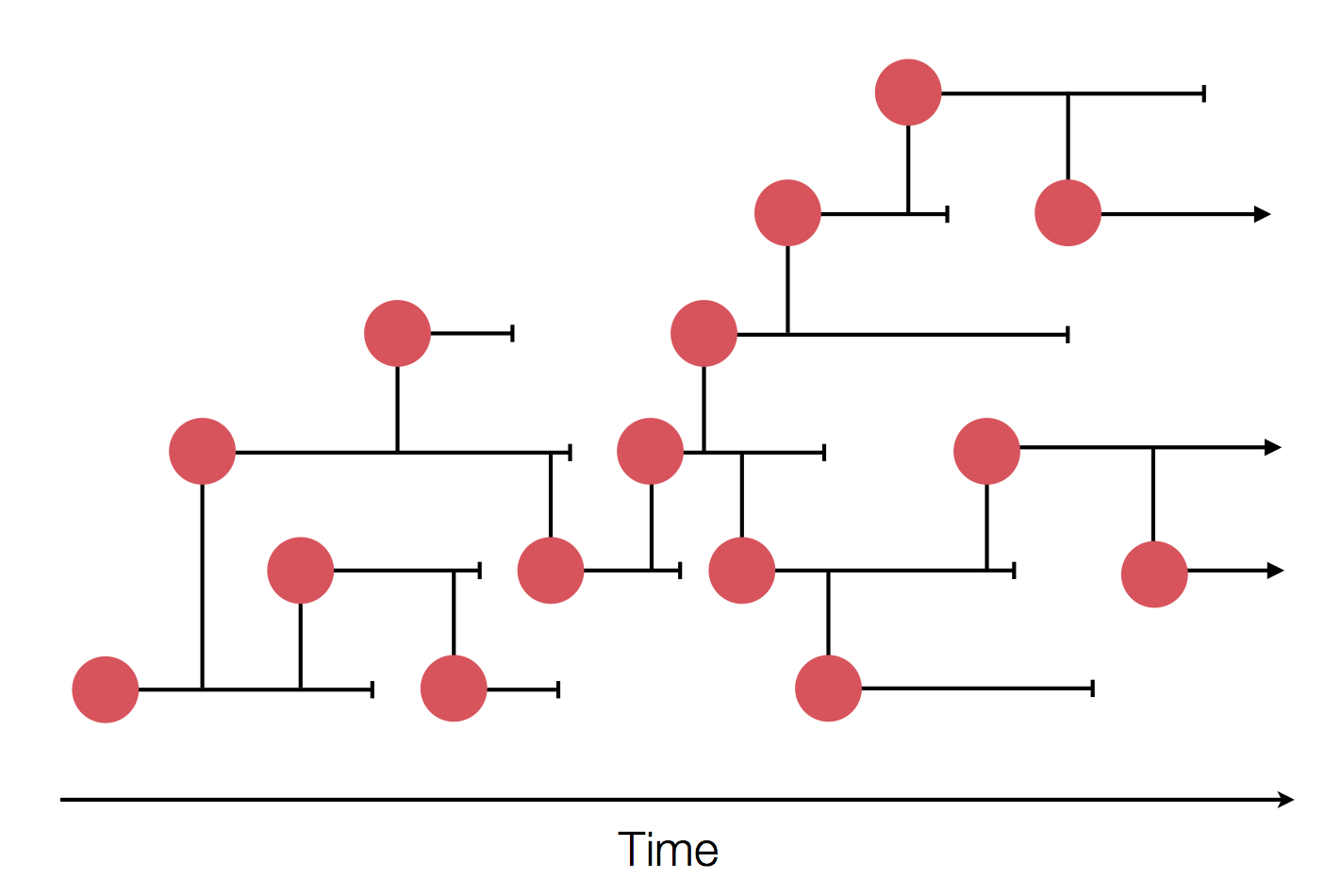
Sample some individuals
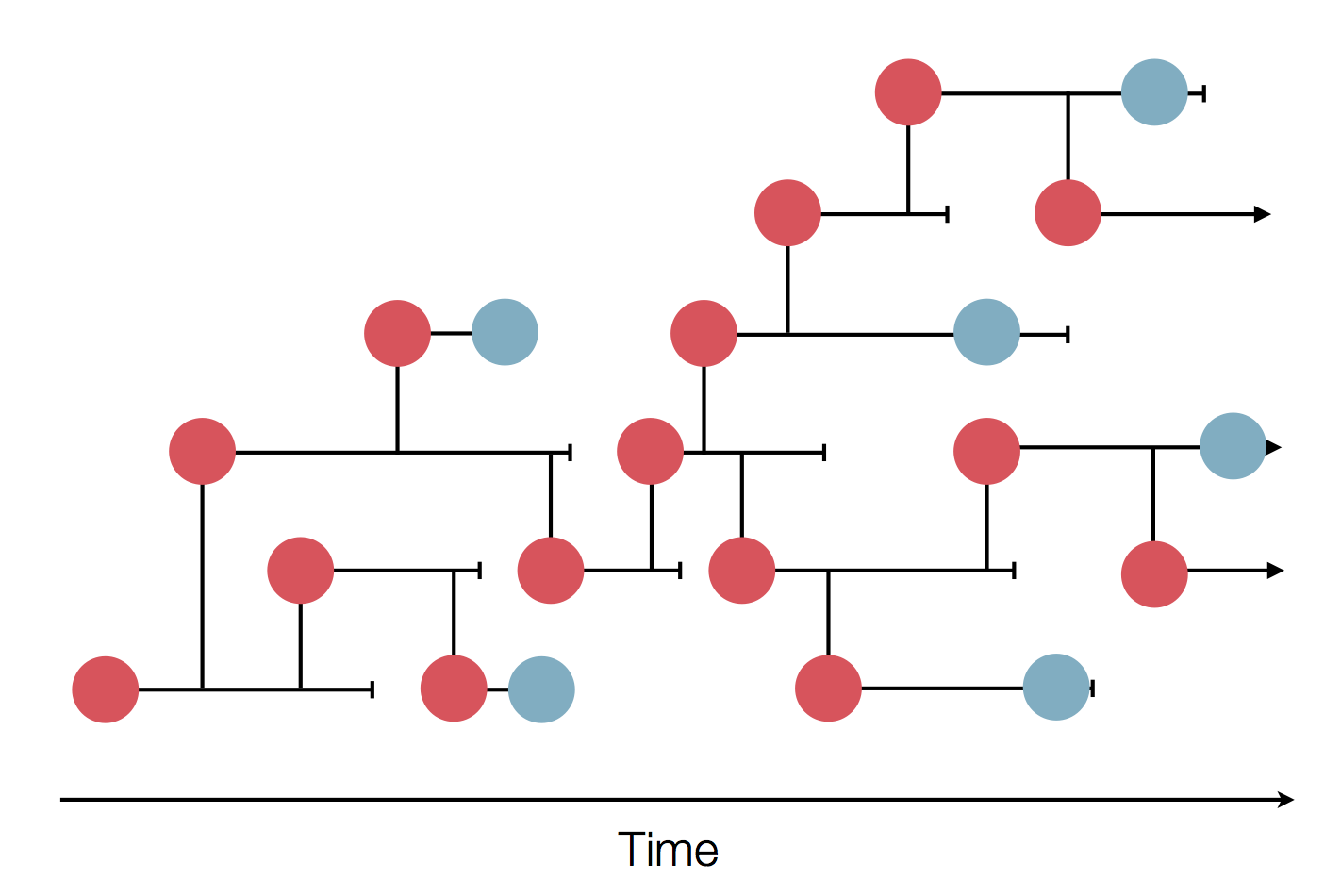
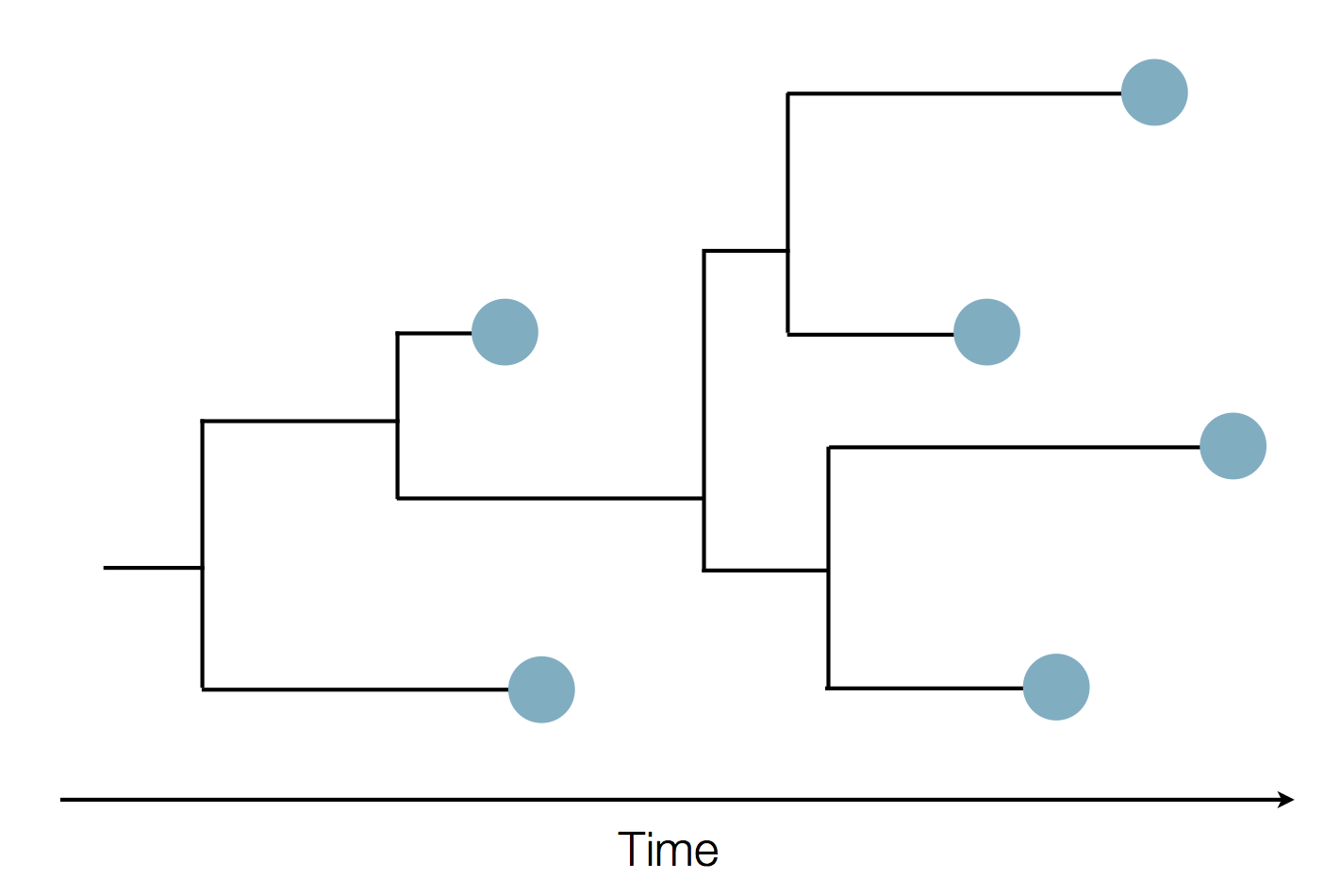
Sequence and determine phylogeny
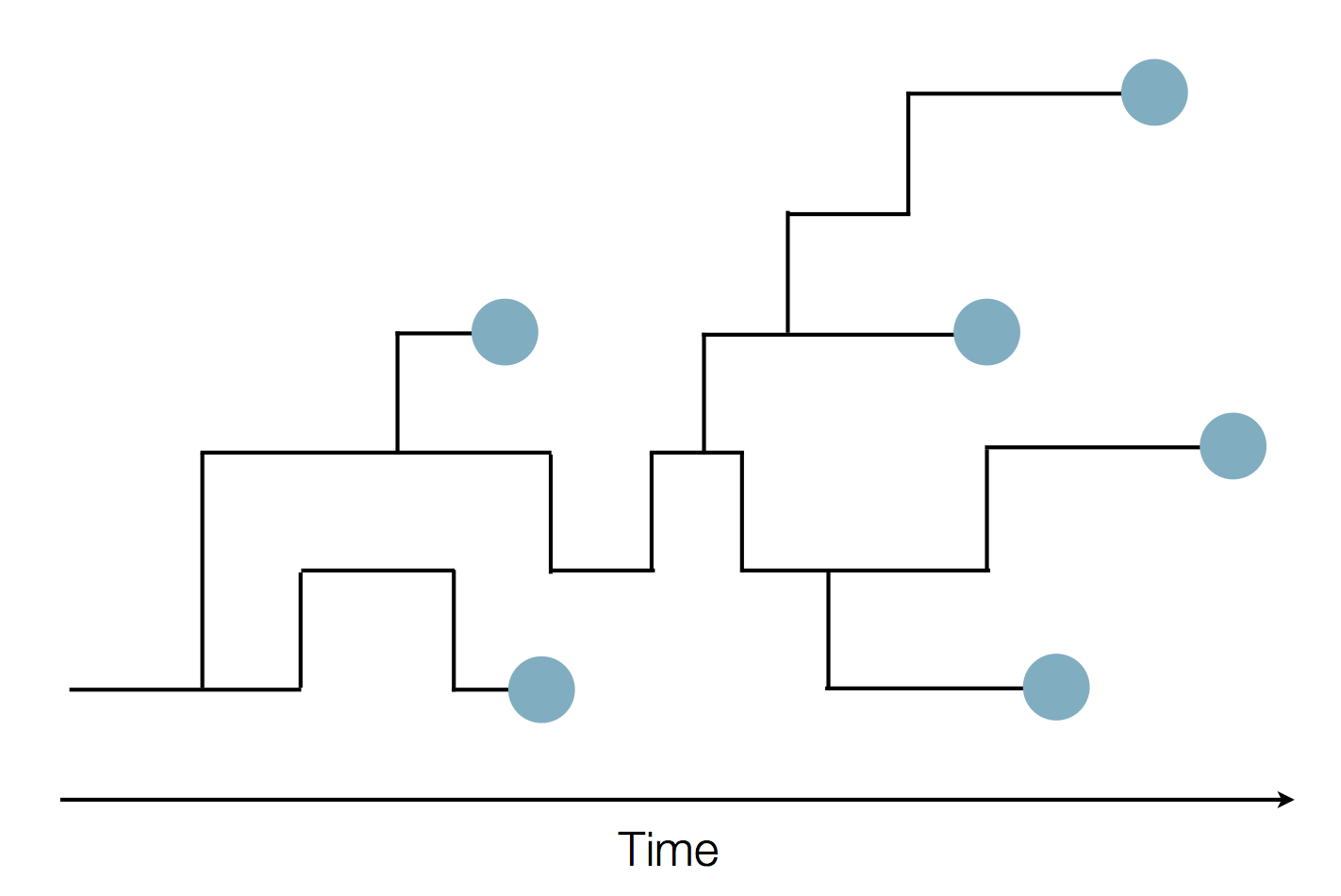
Sequence and determine phylogeny
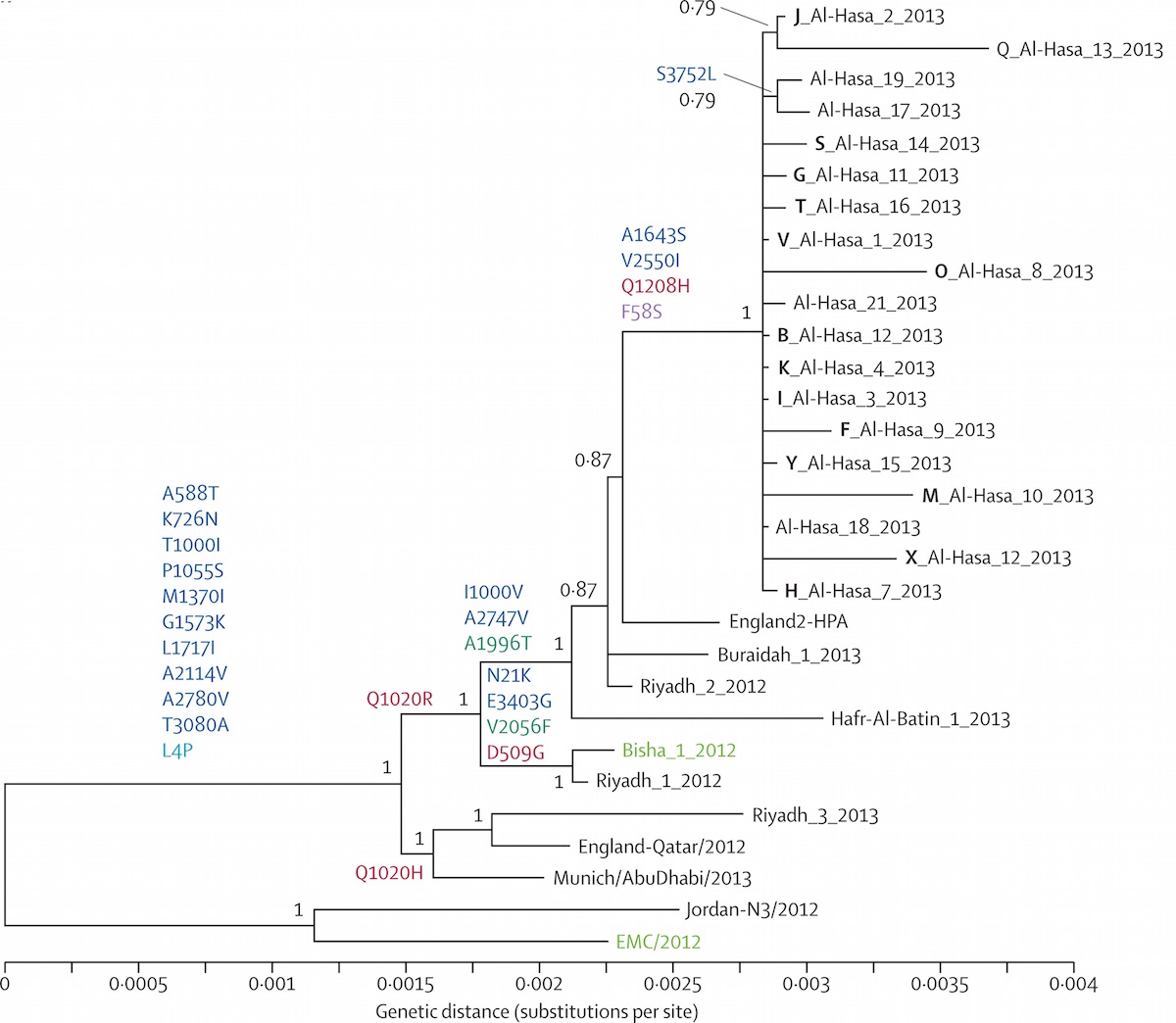
Localized Middle Eastern MERS-CoV phylogeny
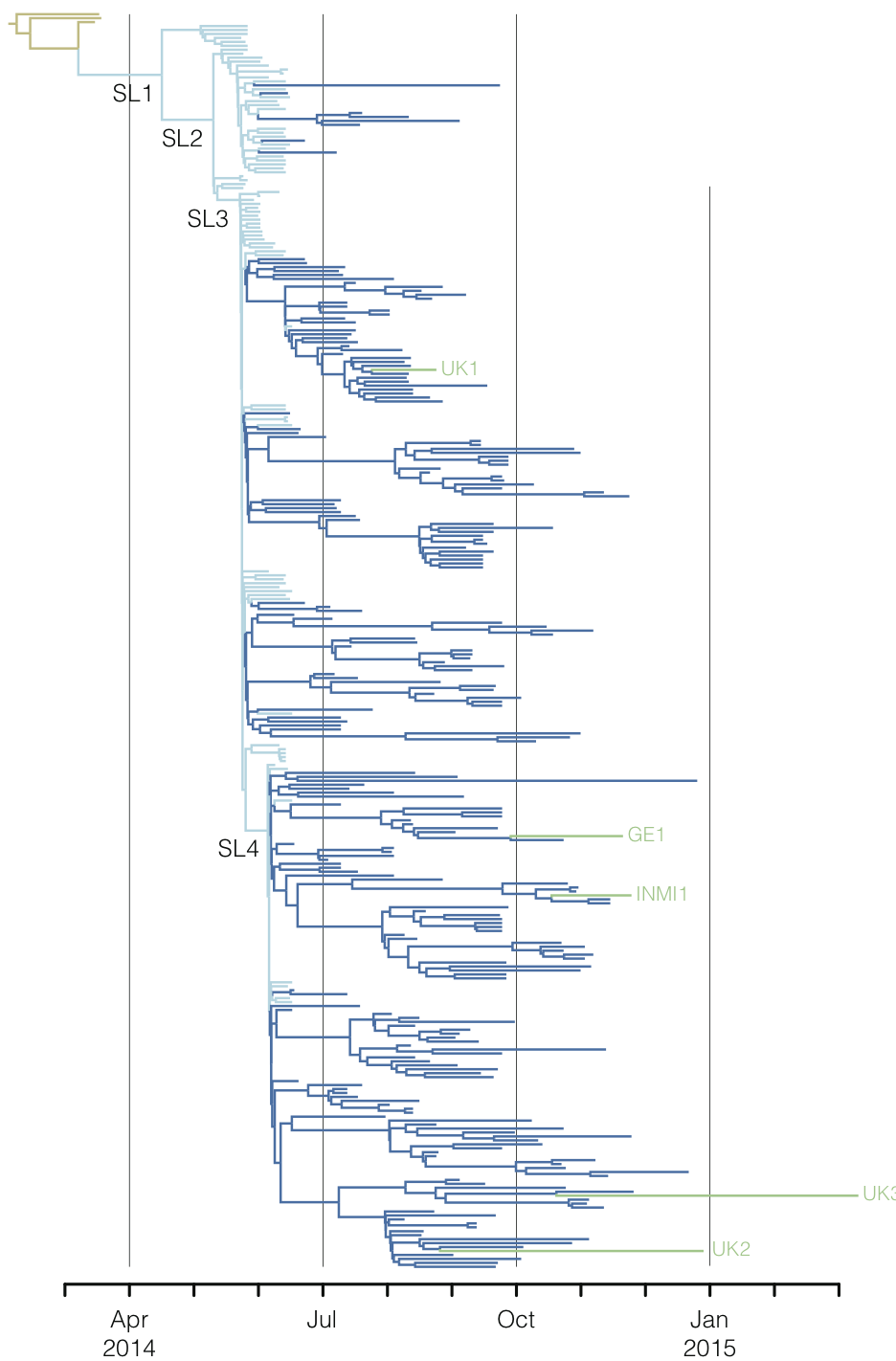
Regional West African Ebola phylogeny
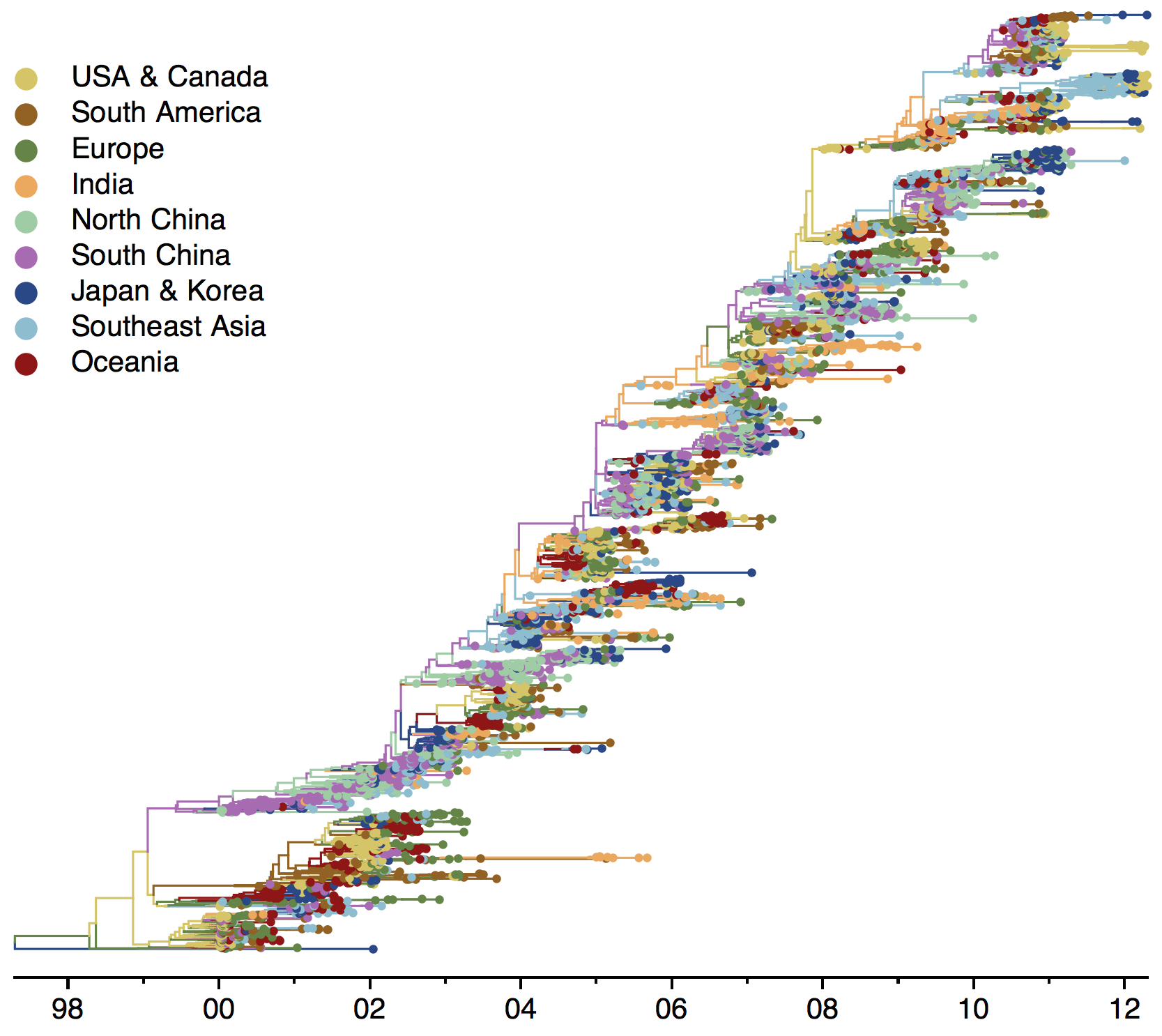
Global influenza phylogeny
Applications of evolutionary analysis for vaccine strain selection and charting outbreak spread
Influenza
Influenza H3N2 vaccine updates
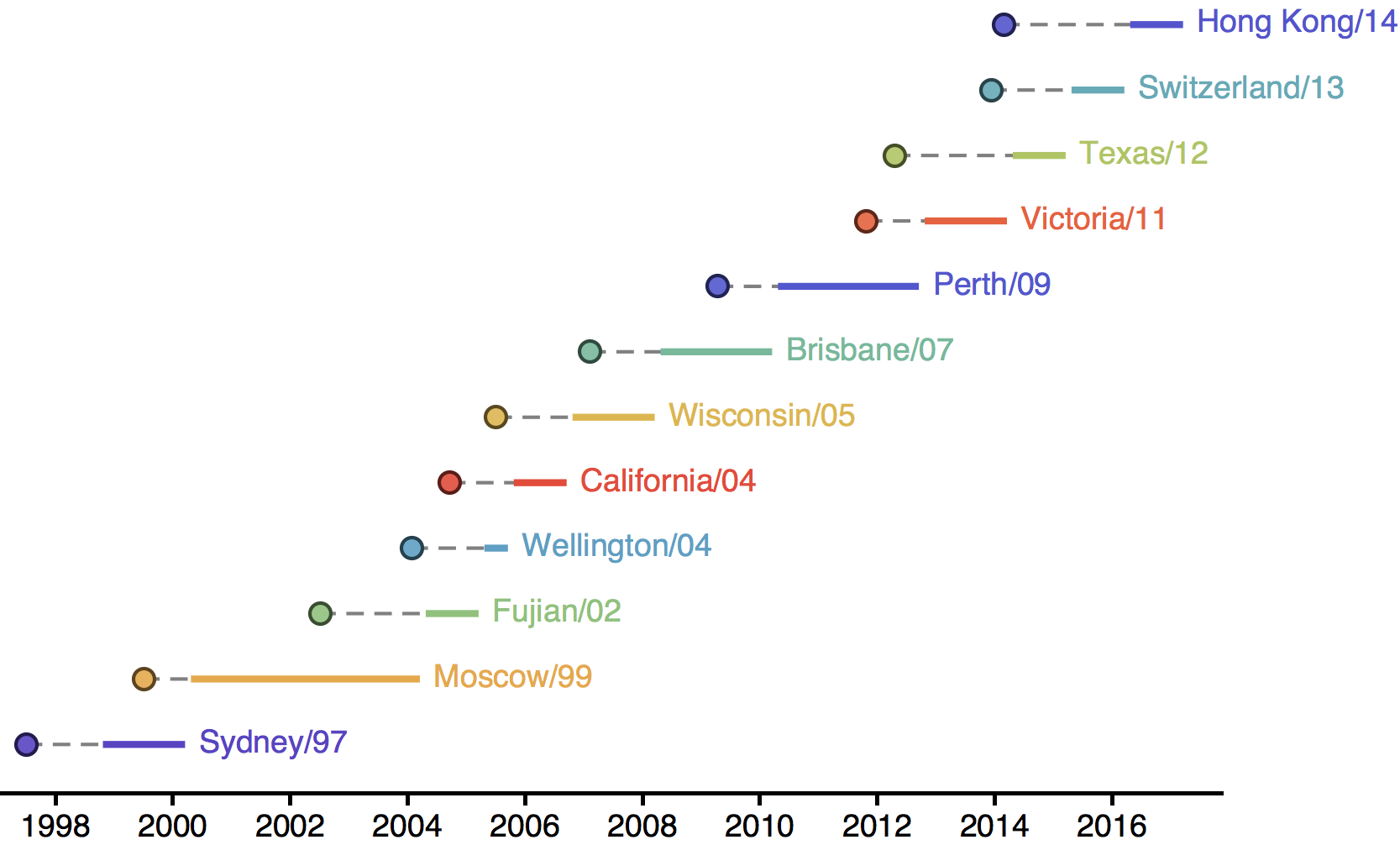
H3N2 phylogeny showing antigenic drift
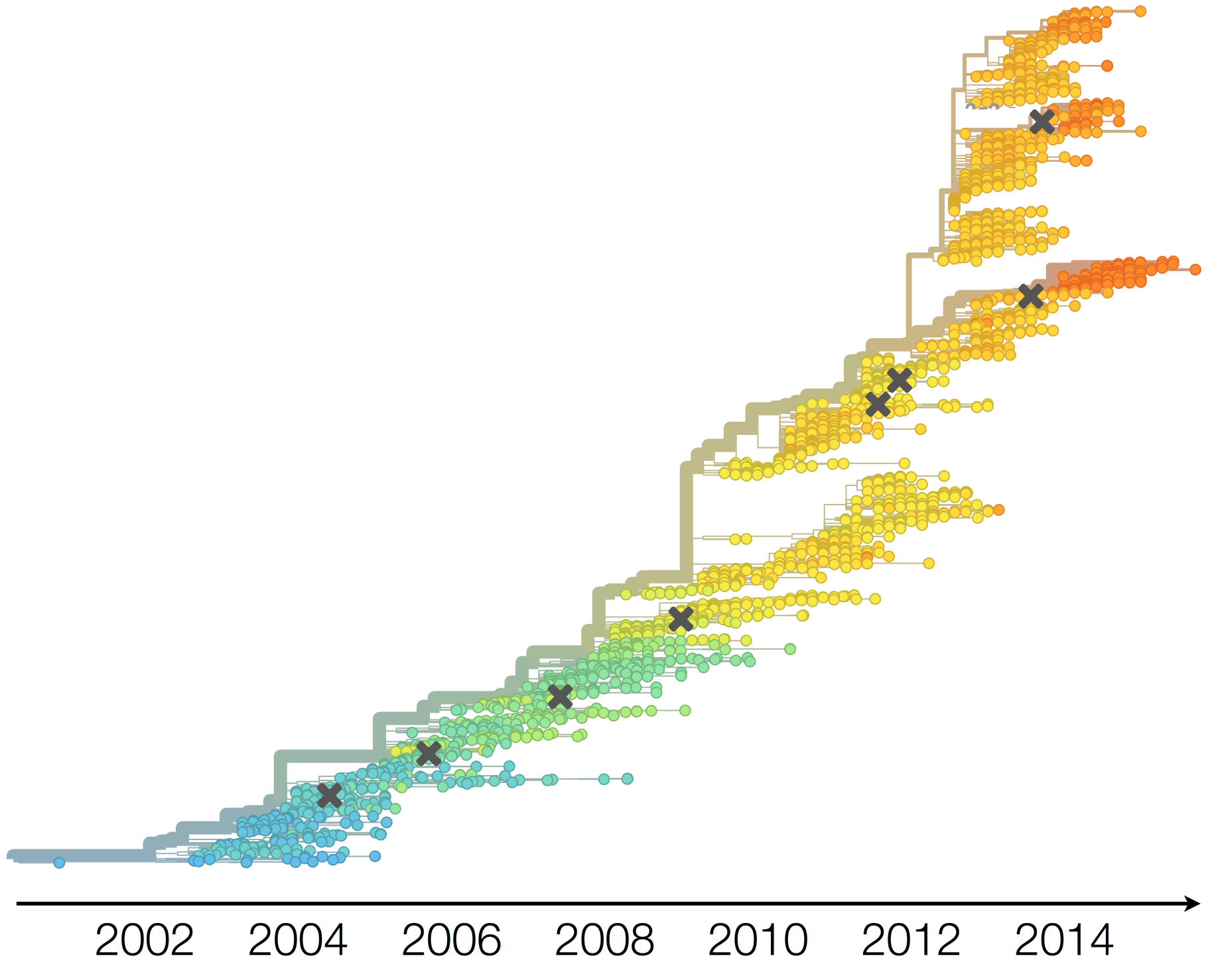
H3N2 phylogeny showing antigenic drift
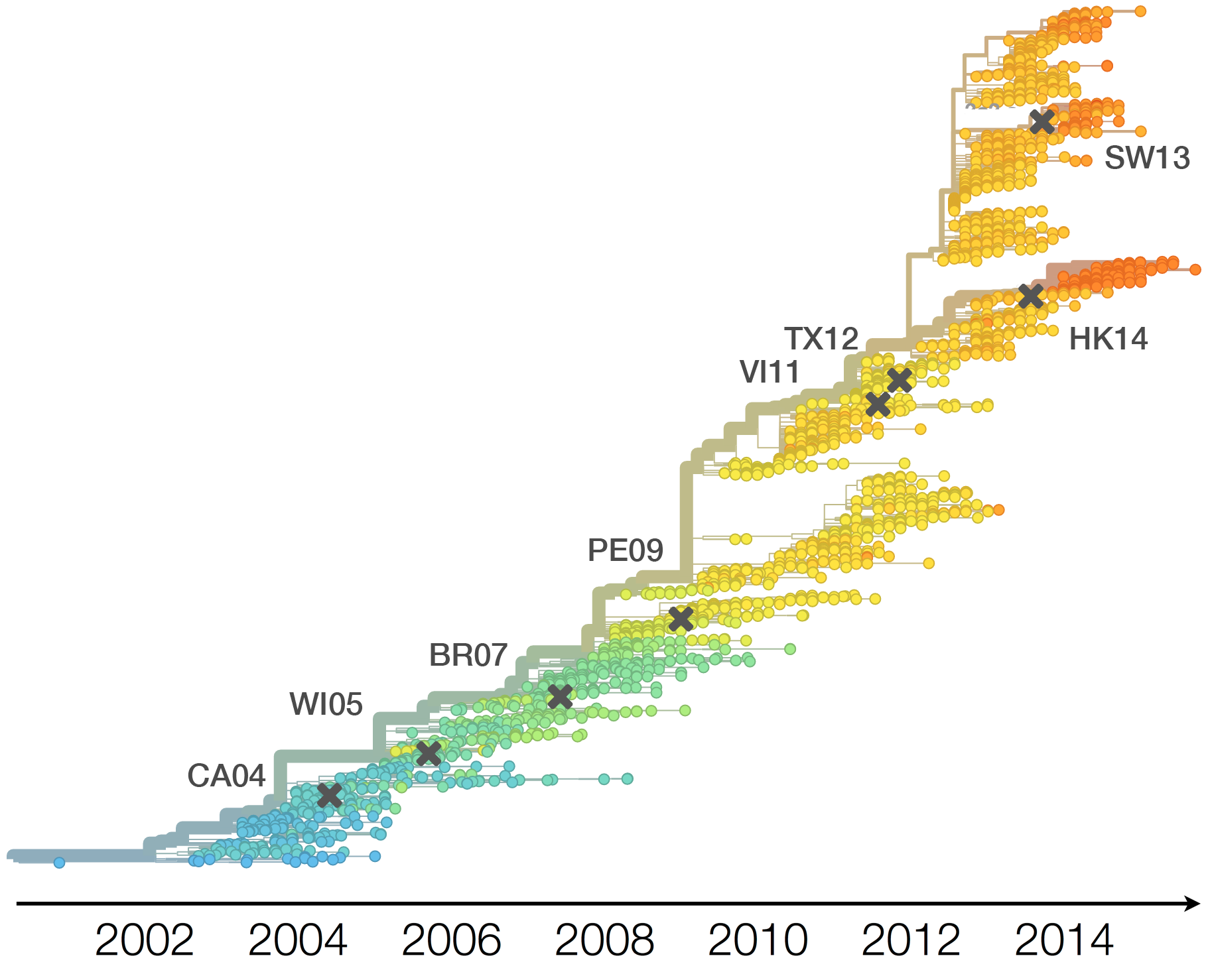
Timely surveillance and rapid analysis essential to understand ongoing influenza evolution
nextflu
Project to provide a real-time view of the evolving influenza population
All in collaboration with Richard Neher

nextflu pipeline
- Download all recent HA sequences from GISAID
- Filter to remove outliers
- Subsample across time and space
- Align sequences
- Build tree
- Estimate frequencies
- Export for visualization
Up-to-date analysis publicly available at:
nextflu.org
Forecasting
Vaccine strain selection timeline
Fitness models can project strain dynamics
Clade frequencies $X$ derive from the fitnesses $f$ and frequencies $x$ of constituent viruses, such that
$$\hat{X}_v(t+\Delta t) = \sum_{i:v} x_i(t) \, \mathrm{exp}(f_i \, \Delta t)$$
This captures clonal interference between competing lineages
We implement a model based on two predictors
- Clade frequency change
- Antigenic advancement
Predicted clade frequency trajectories show congruence
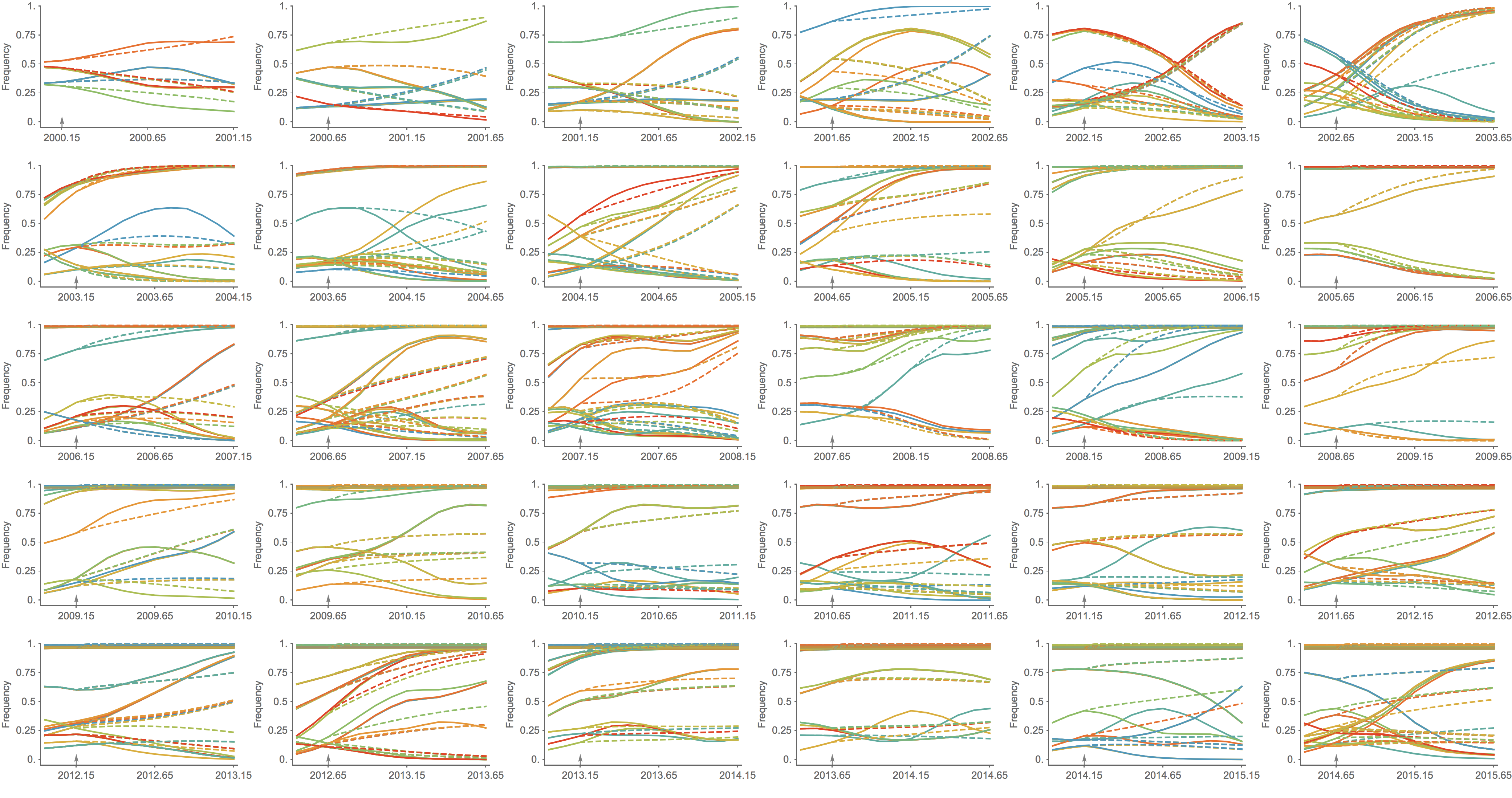
Real-time analyses are actionable and may inform influenza vaccine strain selection
Outbreak analysis
Ebola
Early sequencing showed single origin of epidemic
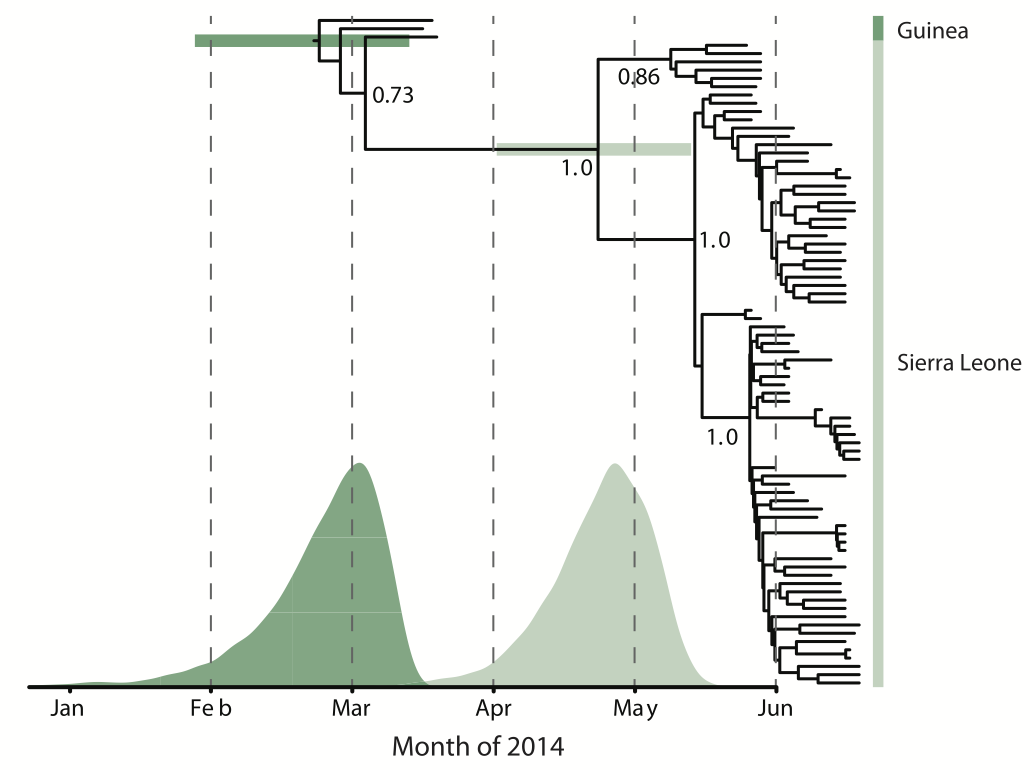
Continued spread through Dec 2014
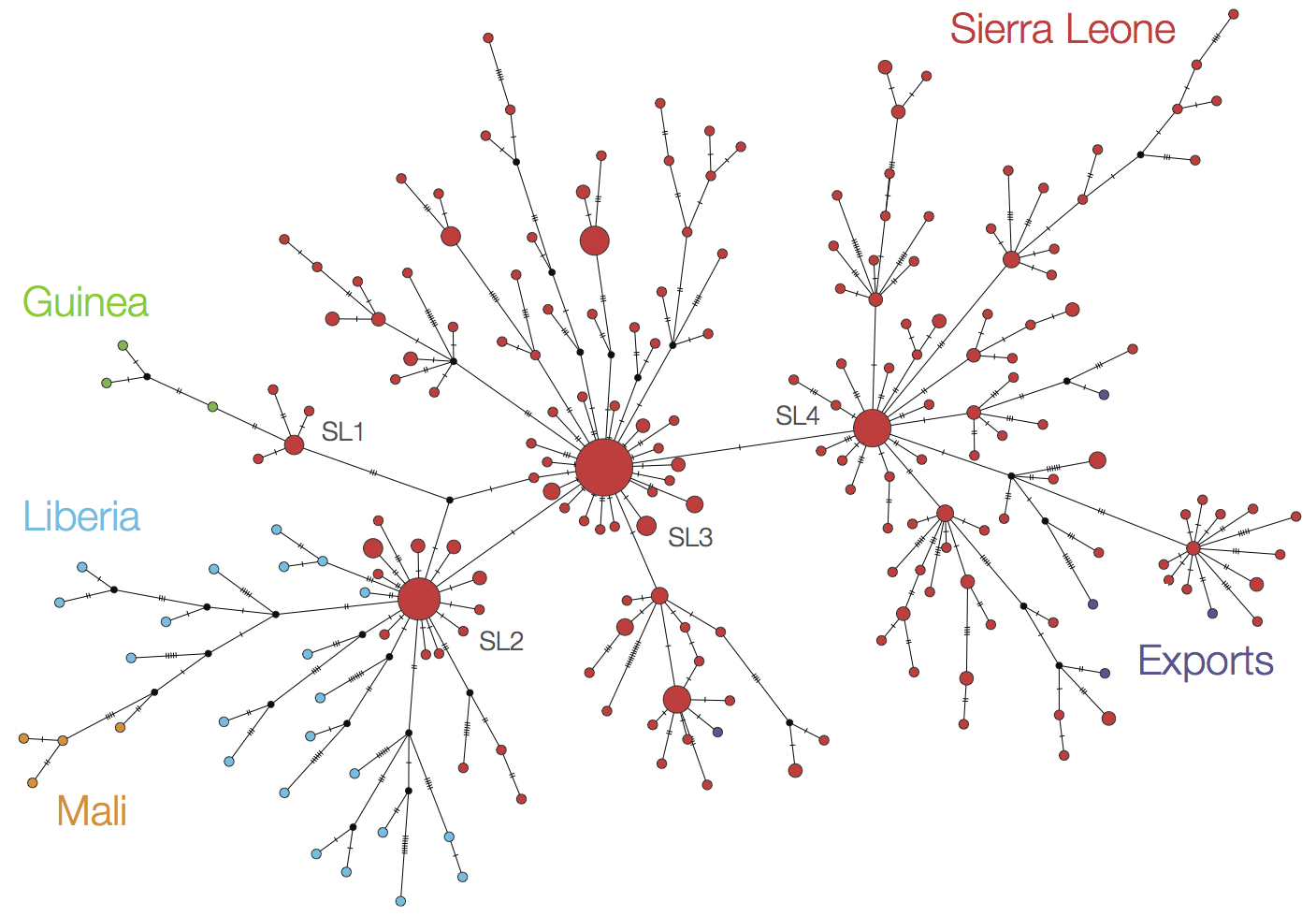
At epidemic height, geographic spread of particular interest
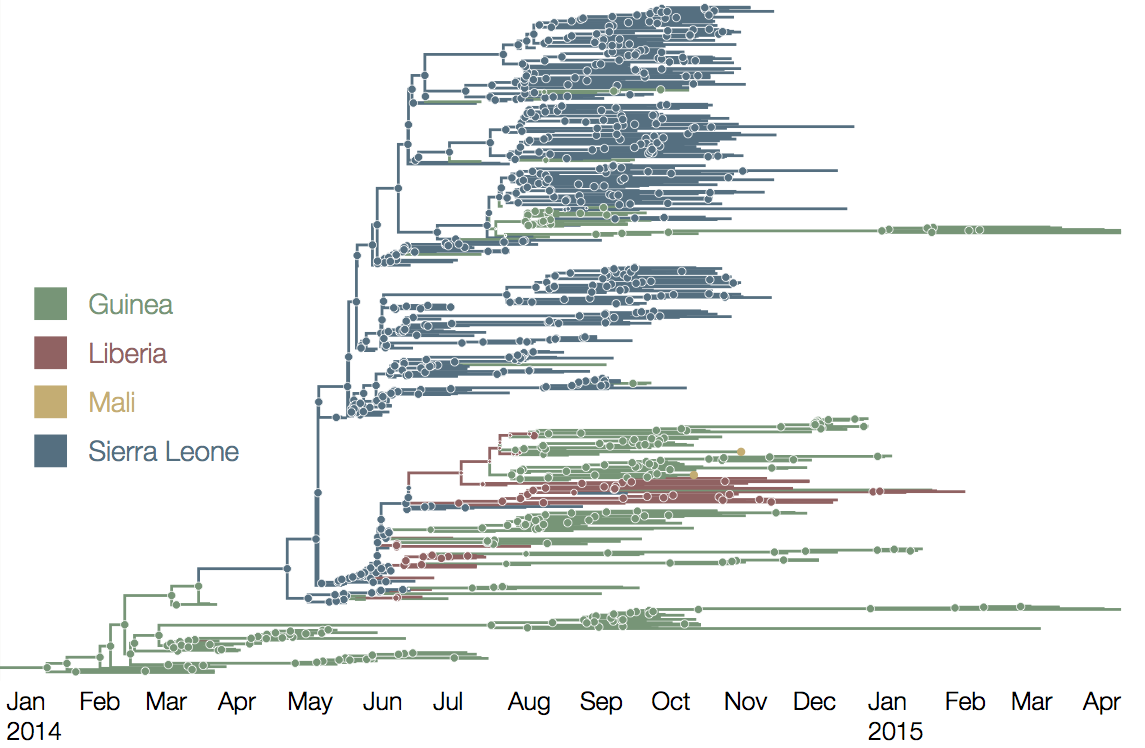
Later on, tracking transmission clusters of primary importance
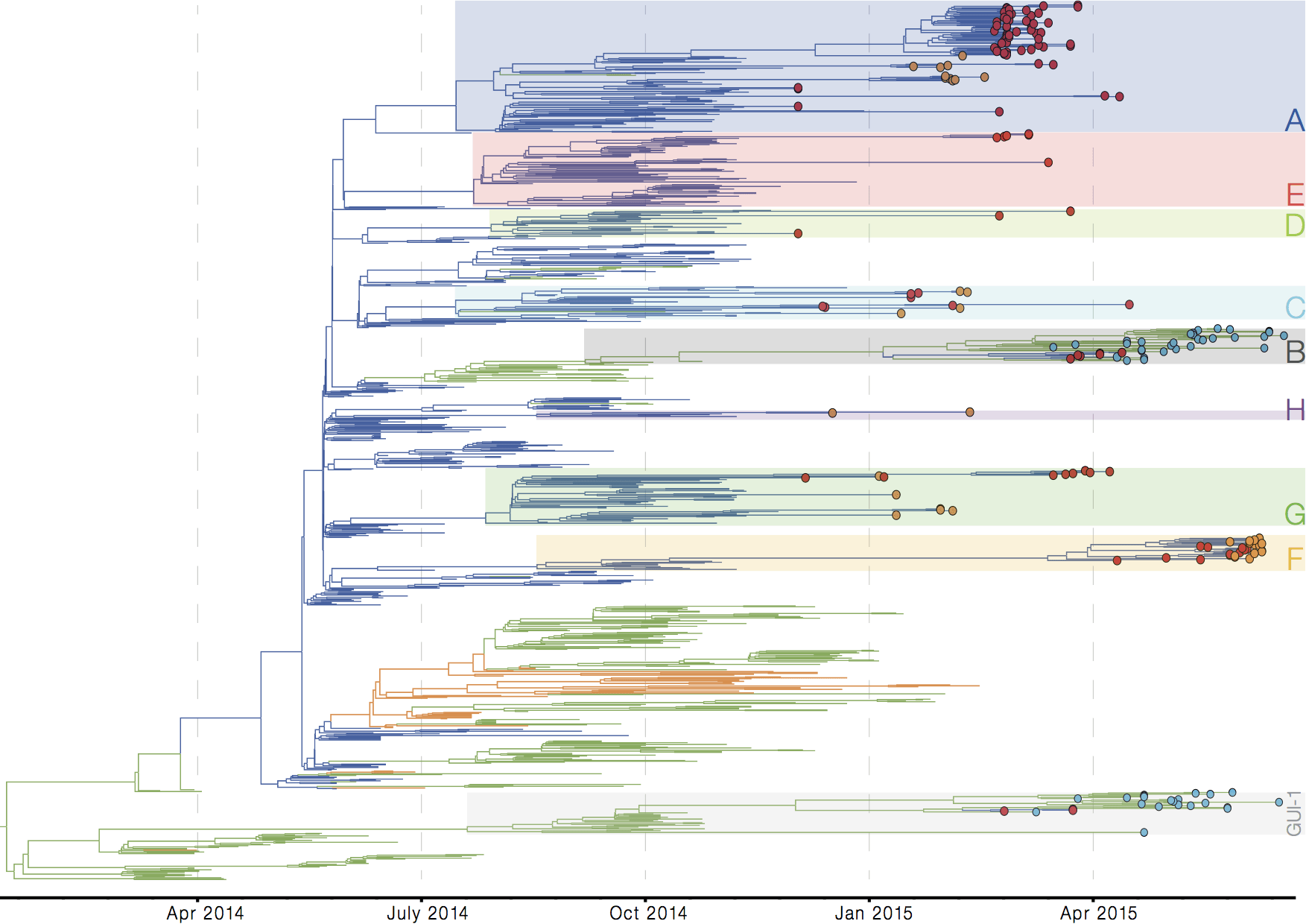
Important analyses, let's make them more rapid and more automated
Tracking epidemic spread in real-time:
ebola.nextstrain.org
Rapid on-the-ground sequencing by Ian Goodfellow, Matt Cotten and colleagues
Deployment of MinION sequencing to Guinea by Nick Loman, Josh Quick, Lauren Cowley and colleagues

Zika
Virus endemic to Africa, emergence in Southeast Asia in the last century
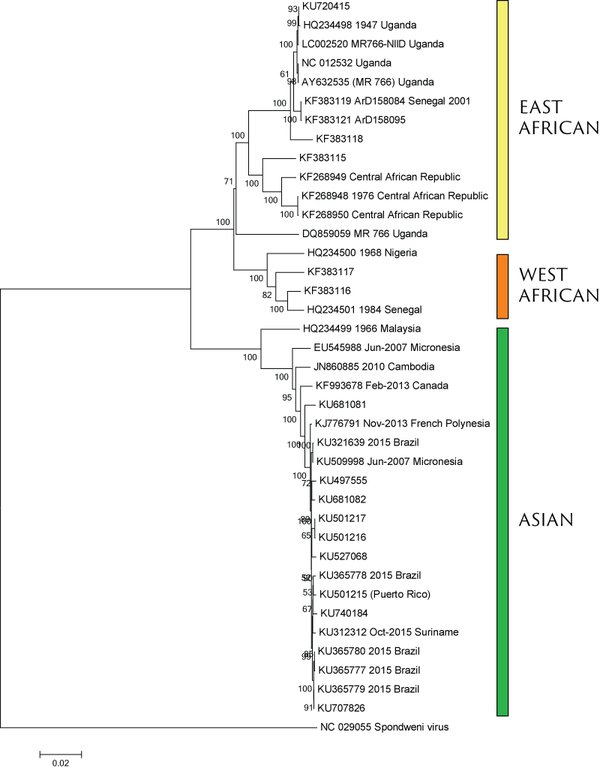
Spread eastward through the South Pacific
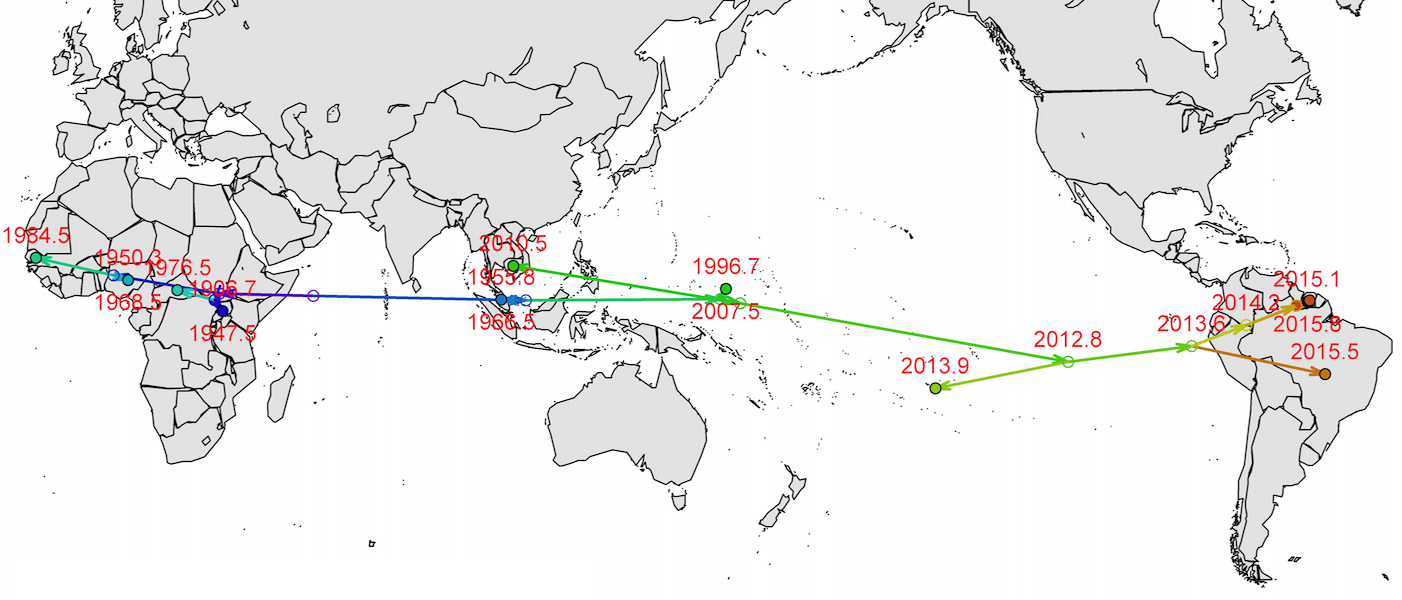
Single arrival into the Americas in early 2014
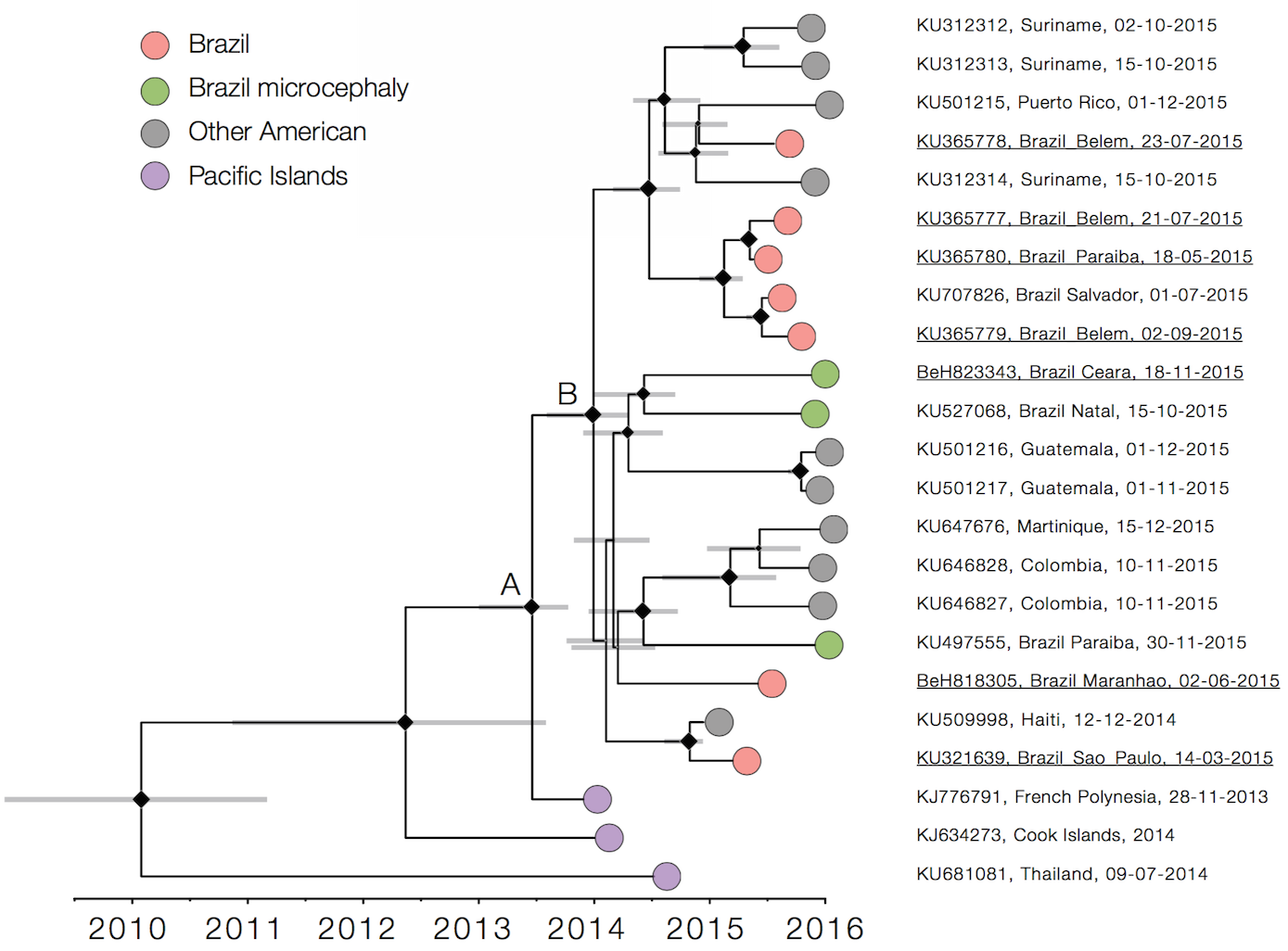
Working on analysis of ongoing evolution:
nextstrain.org/zika/
Discussion points for breakout session
Discussion points
- Mechanisms to promote data sharing (genetic data, epi data) and proper scientific attribution
- Key insights from phylodynamic analyses for understanding outbreak spread and aiding epidemiological investigation
- Interface of modeling results with public health policy
Acknowledgements
Influenza: WHO Global Influenza Surveillance Network, GISAID, Worldwide Influenza Centre at the Francis Crick Institute, Richard Neher, Colin Russell, Andrew Rambaut
Ebola: data producers, Gytis Dudas, Andrew Rambaut, Philipe Lemey, Richard Neher, Nick Loman, Ian Goodfellow, Matt Cotten, Paul Kellam, Danny Park, Kristian Andersen, Pardis Sabeti
Zika: data producers, Nick Loman, Nuno Faria, Oliver Pybus, Andrew Rambaut, Richard Neher, Charlton Callender



Contact
- Website: bedford.io
- Twitter: @trvrb
- Slides: bedford.io/talks/real-time-tracking-midas/
Discussion points for breakout session
Options for genetic data sharing
- Genbank: General purpose, wild-west
- Virological: Discussion oriented, control of reuse
- Lab websites: Control of reuse
- GISAID: Influenza only, redistributed restricted
Incentives for genetic data sharing
Statements of support from major journals and from the WHO
Confusion over behavioral norms

