Phylogenetic analyses of Ebola spread in West Africa and Zika spread in the Americas
Trevor Bedford
6 Mar 2017
MIDAS Steering Committee Call

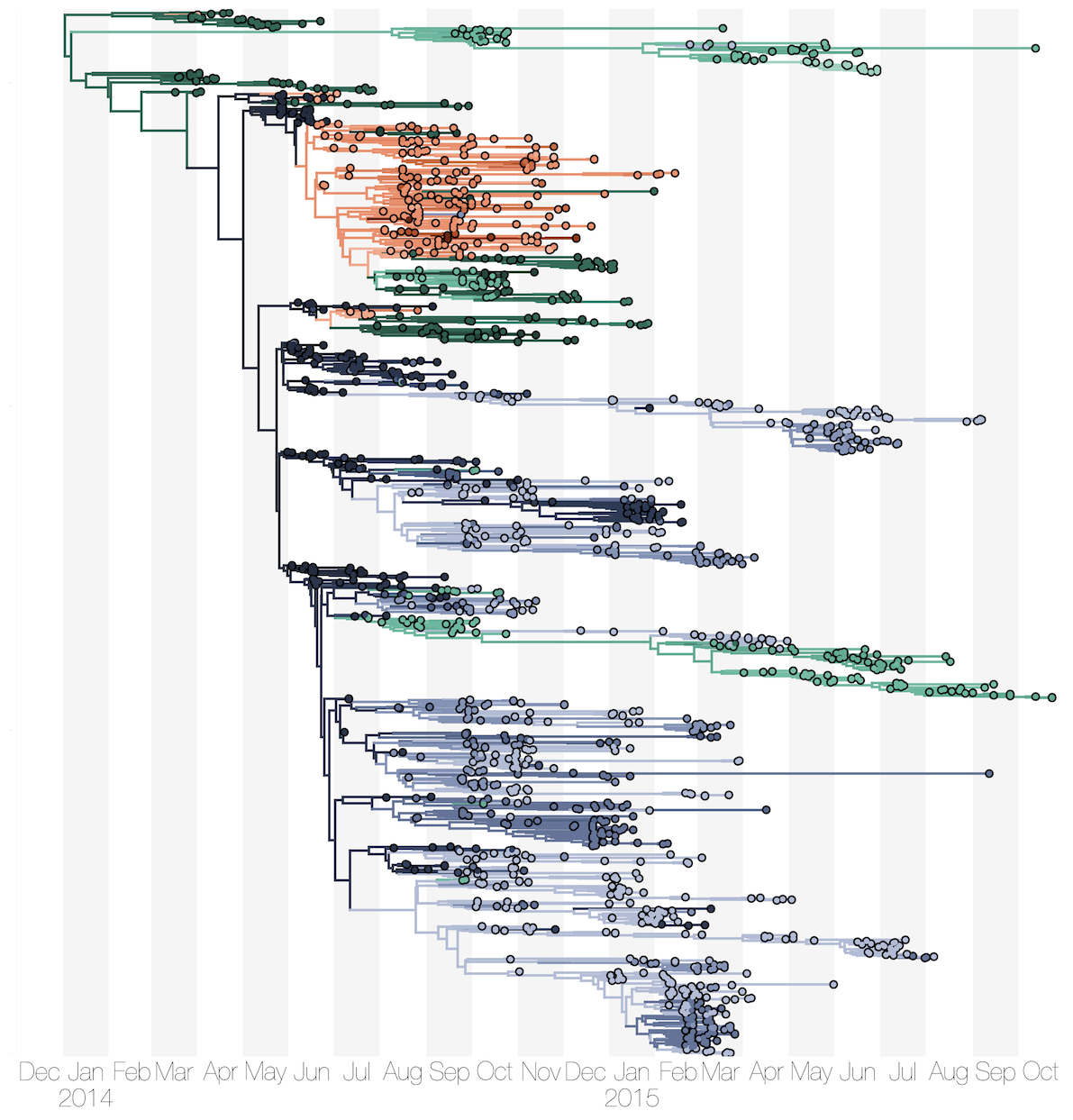
Dataset of 1610 Ebola virus genomes collected during the 2013-2016 epidemic
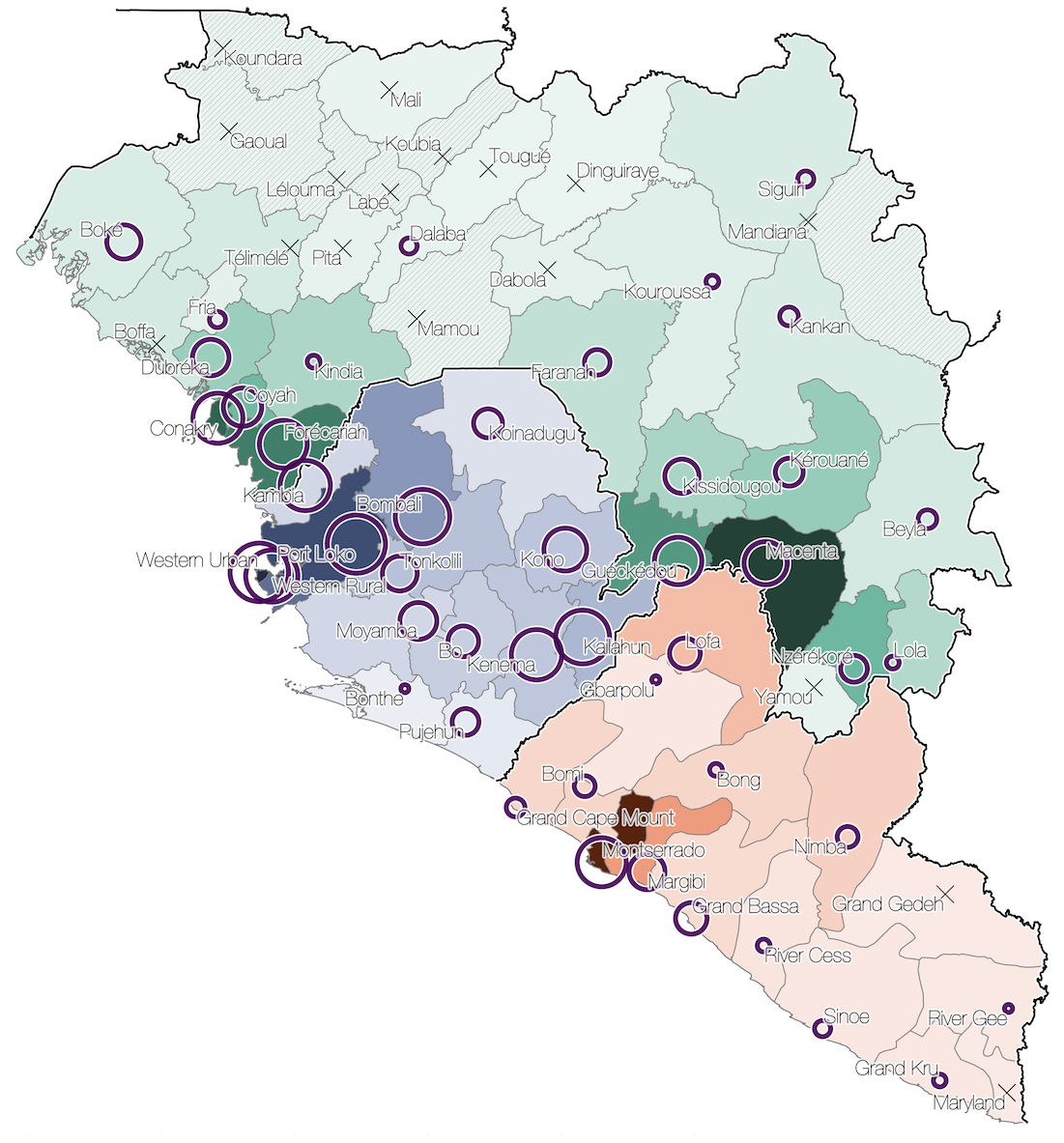
Phylogeographic reconstruction
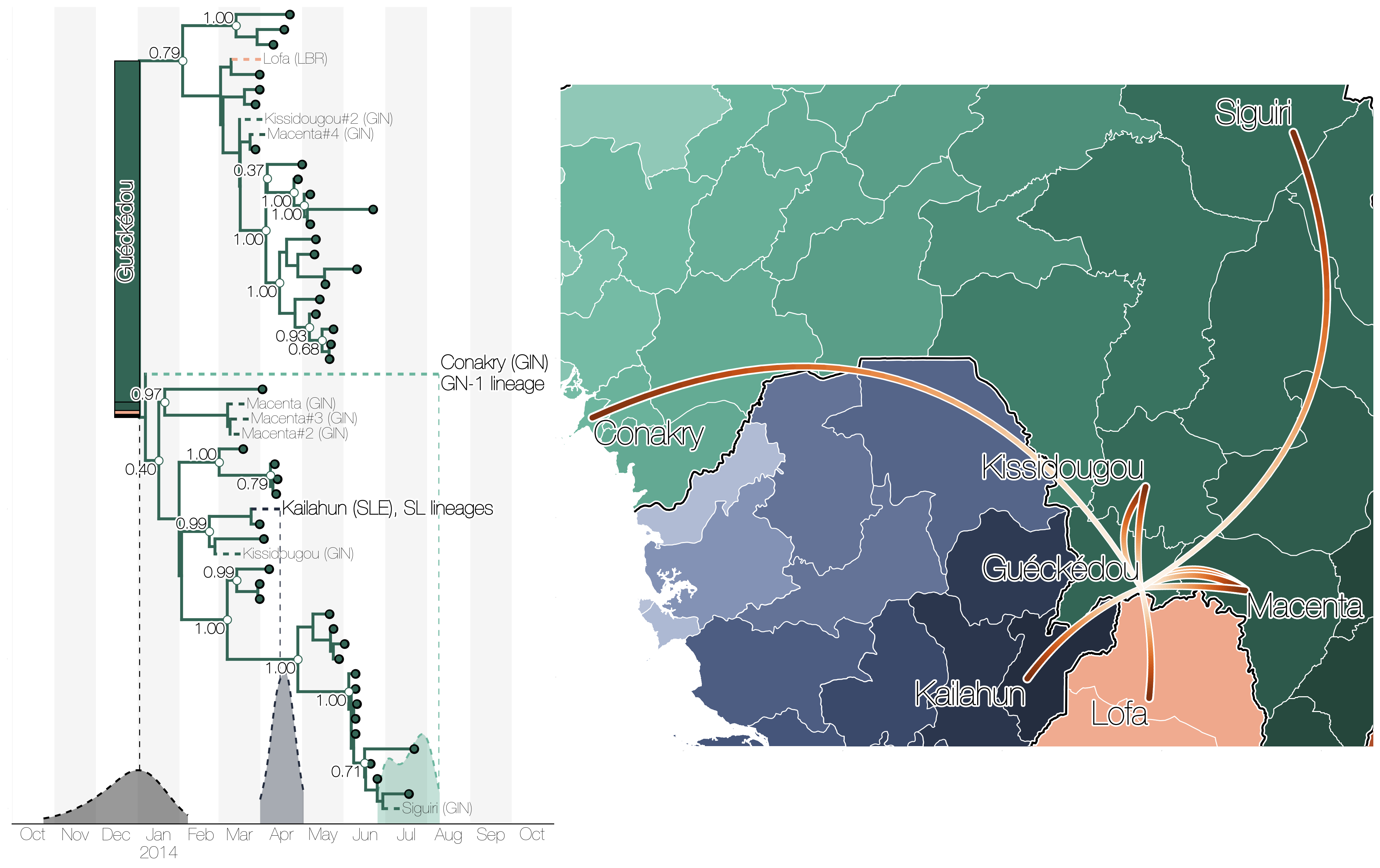
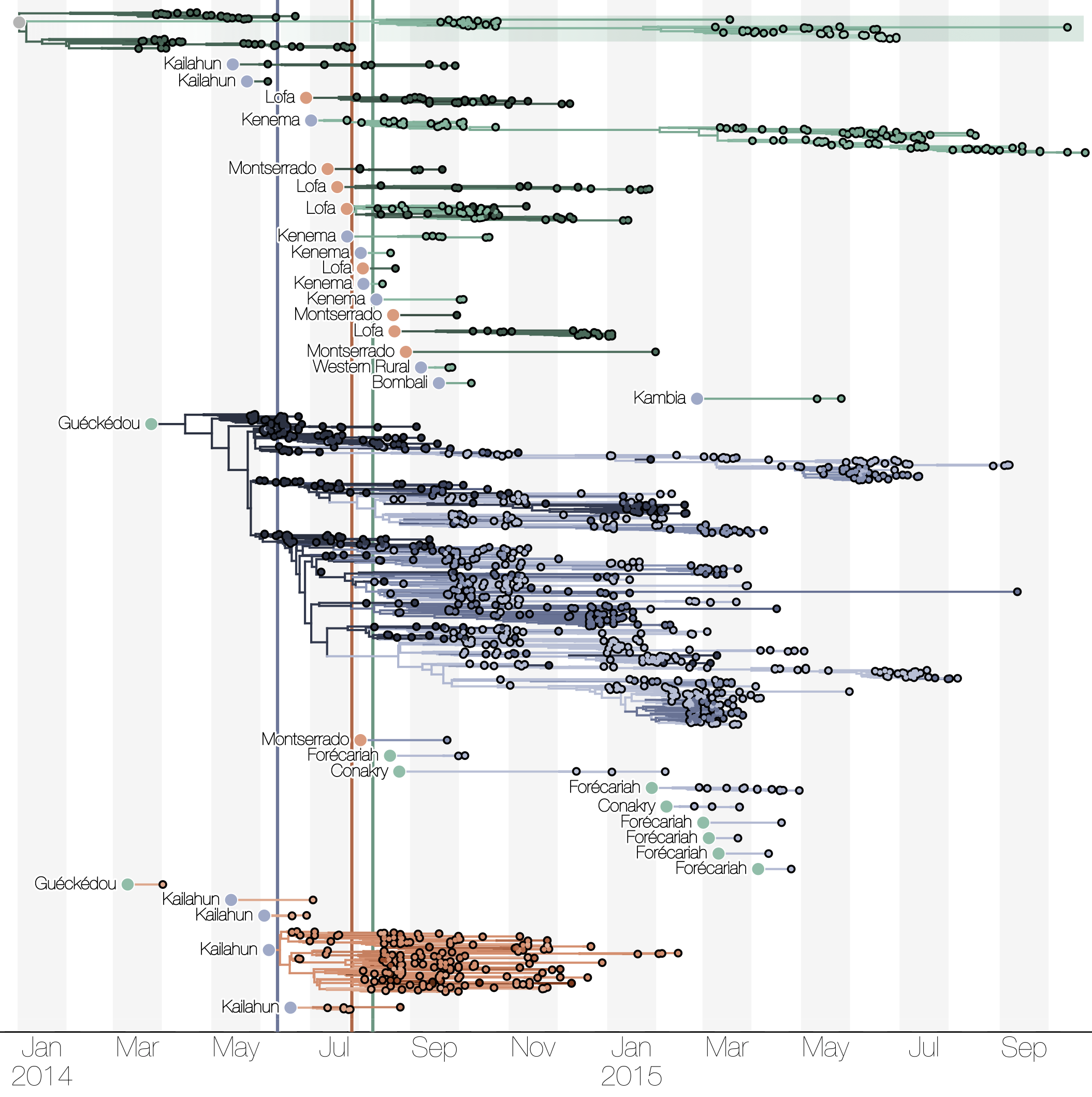
Initial emergence from Guéckédou
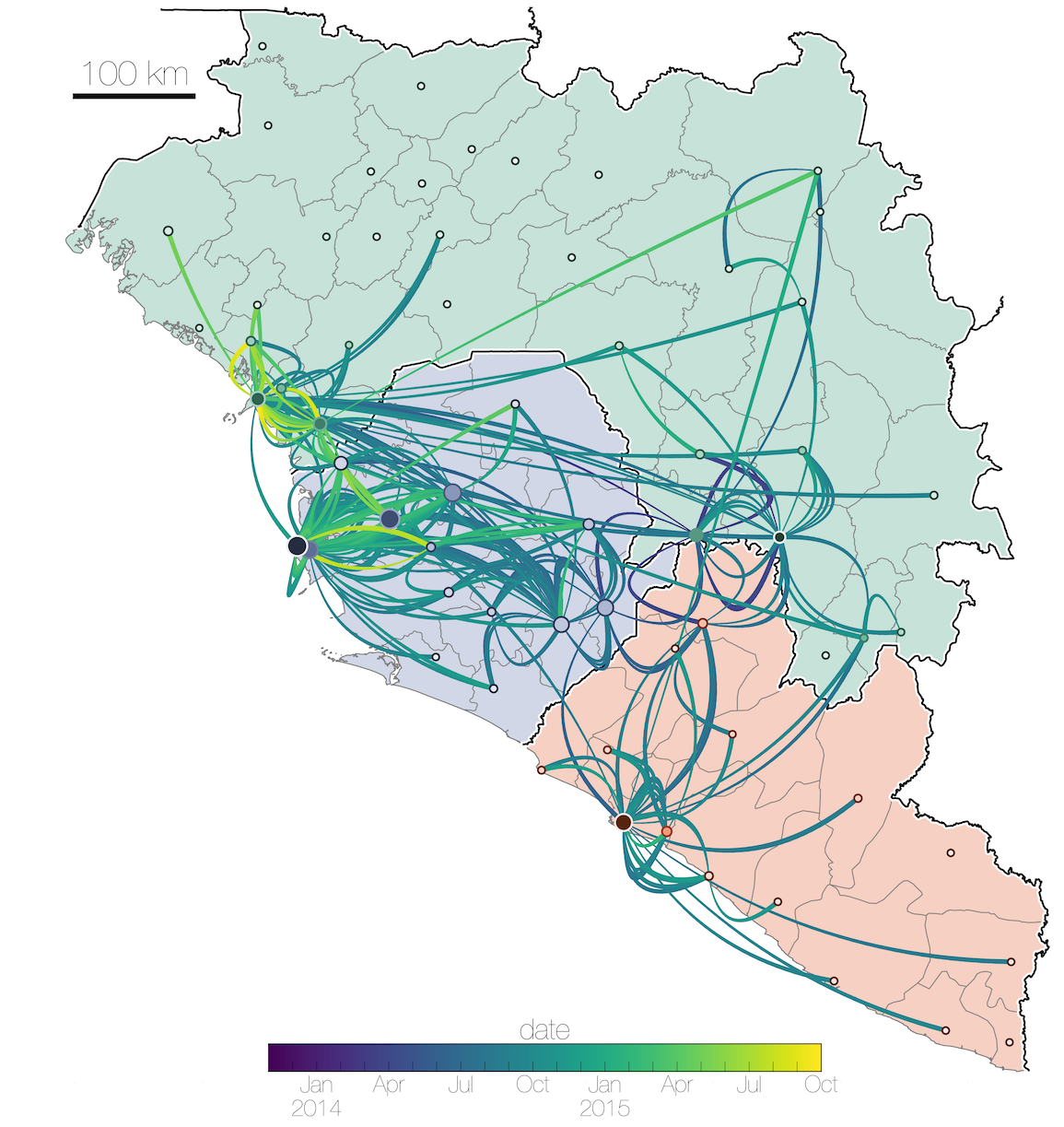
Tracking migration events
Factors influencing migration rates
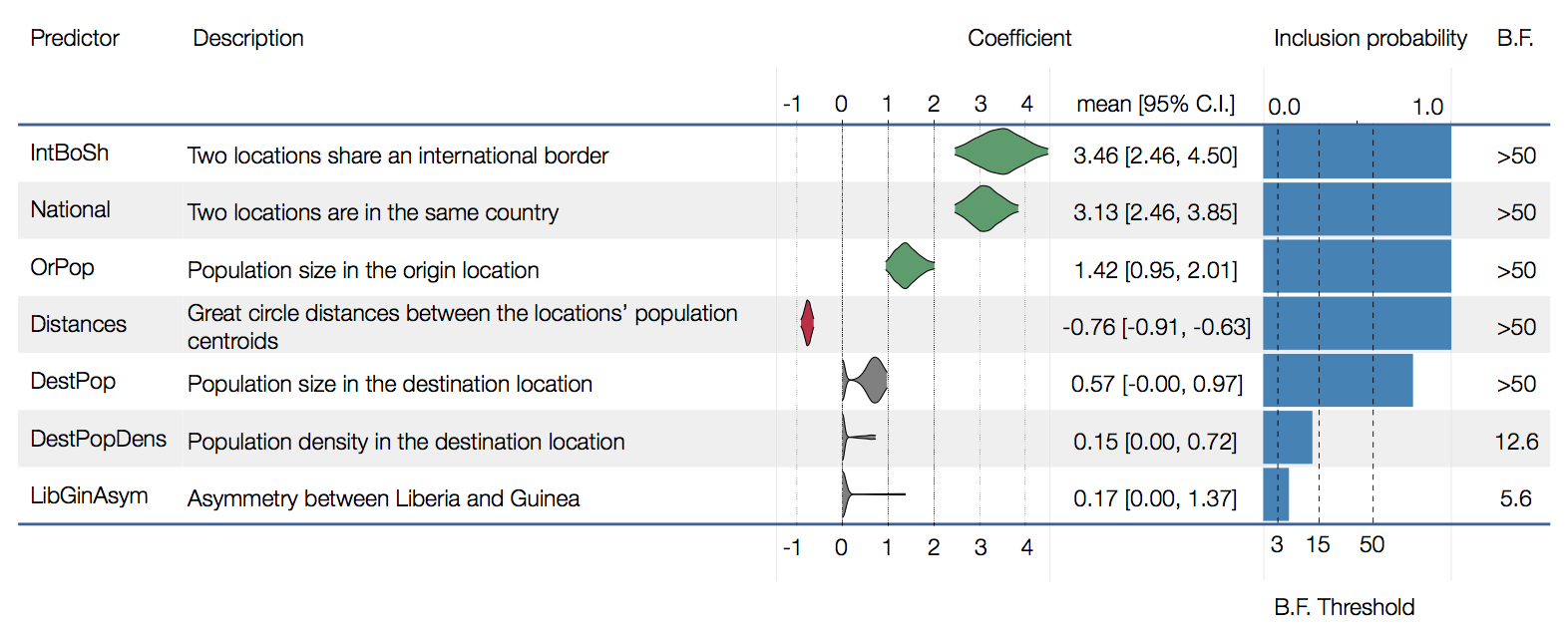
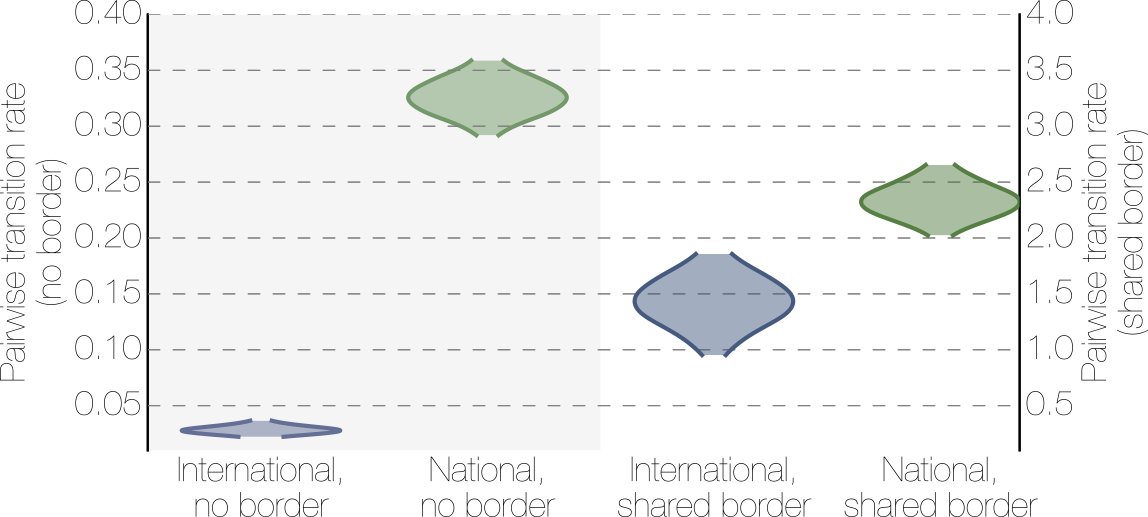
Effect of borders on migration rates
Spatial structure at the country level
Substantial mixing at the regional level
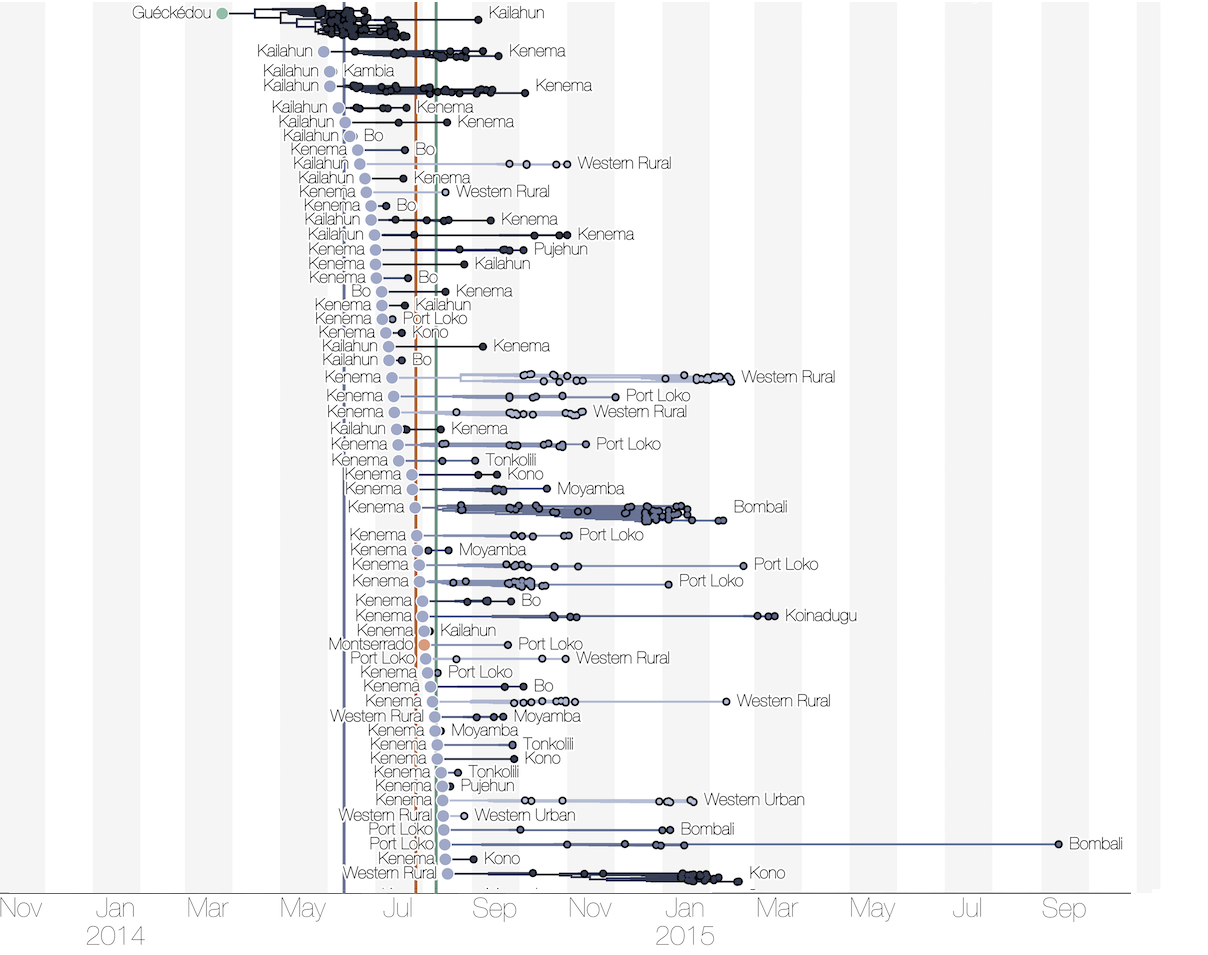
Regional outbreaks due to multiple introductions
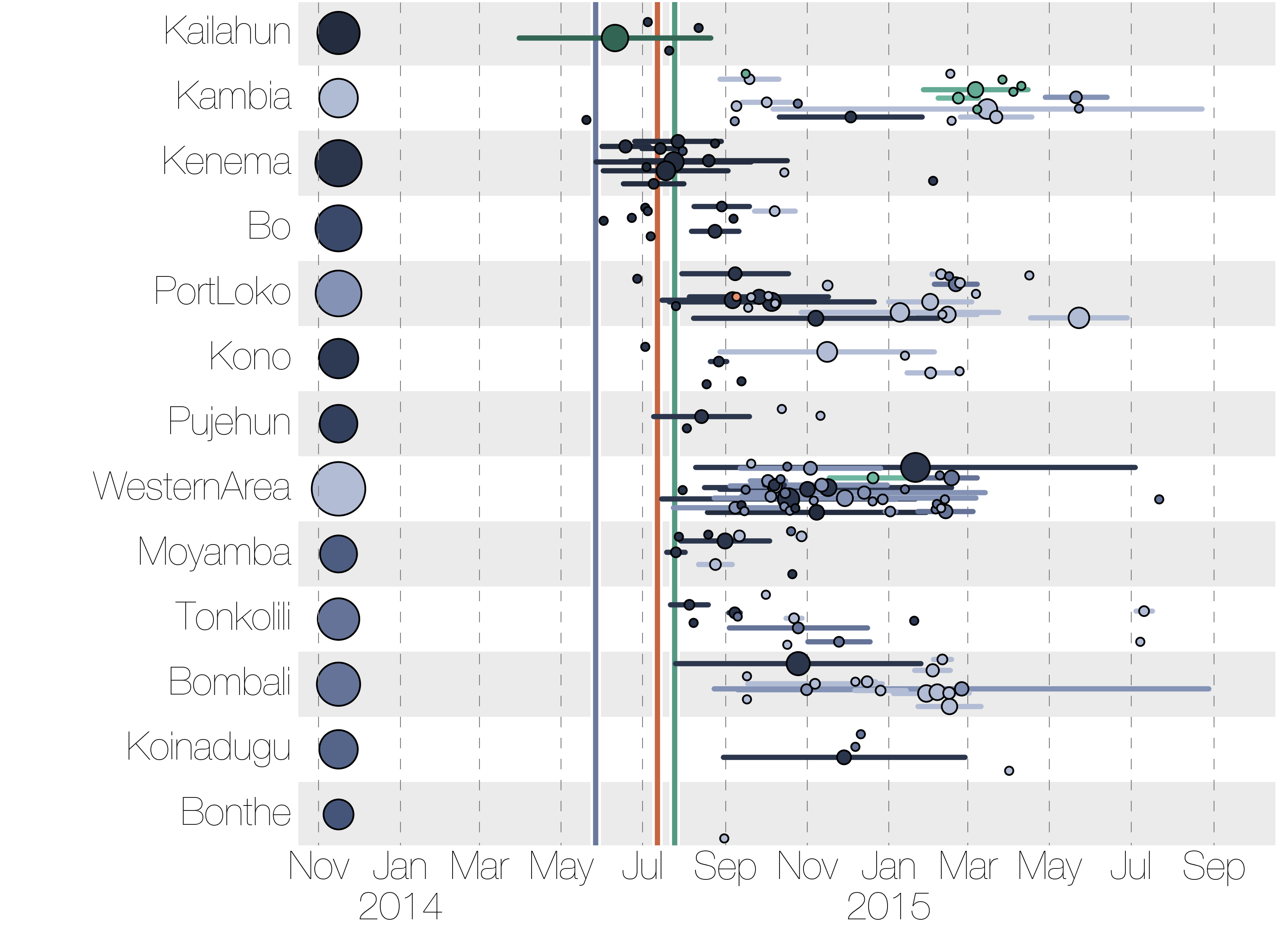
Each introduction results in a minor outbreak
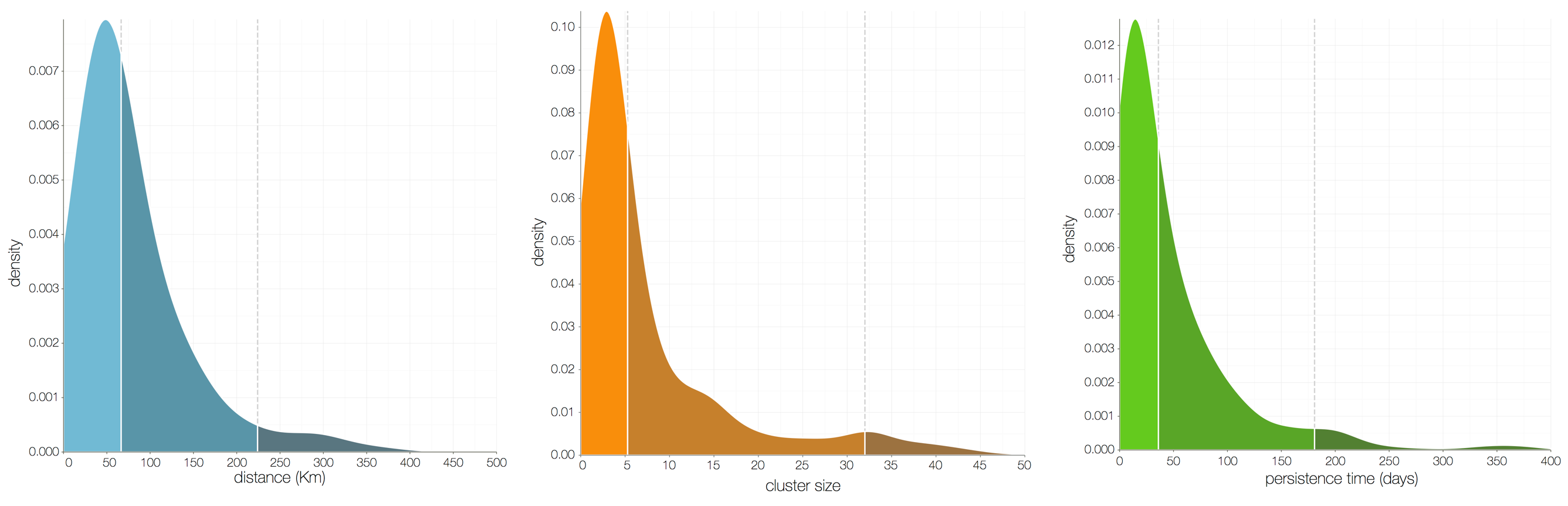
Ebola spread in West Africa followed a gravity model with moderate slowing by international borders, in which spread is driven by short-lived migratory clusters

Road trip through northeast Brazil to collect samples and sequence
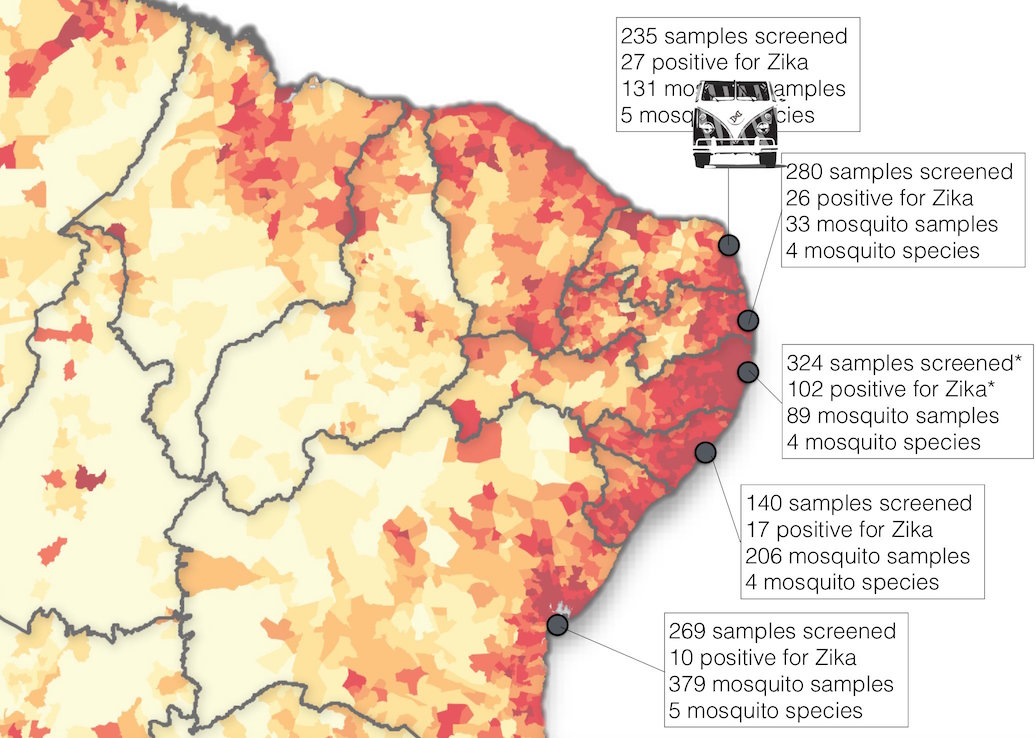
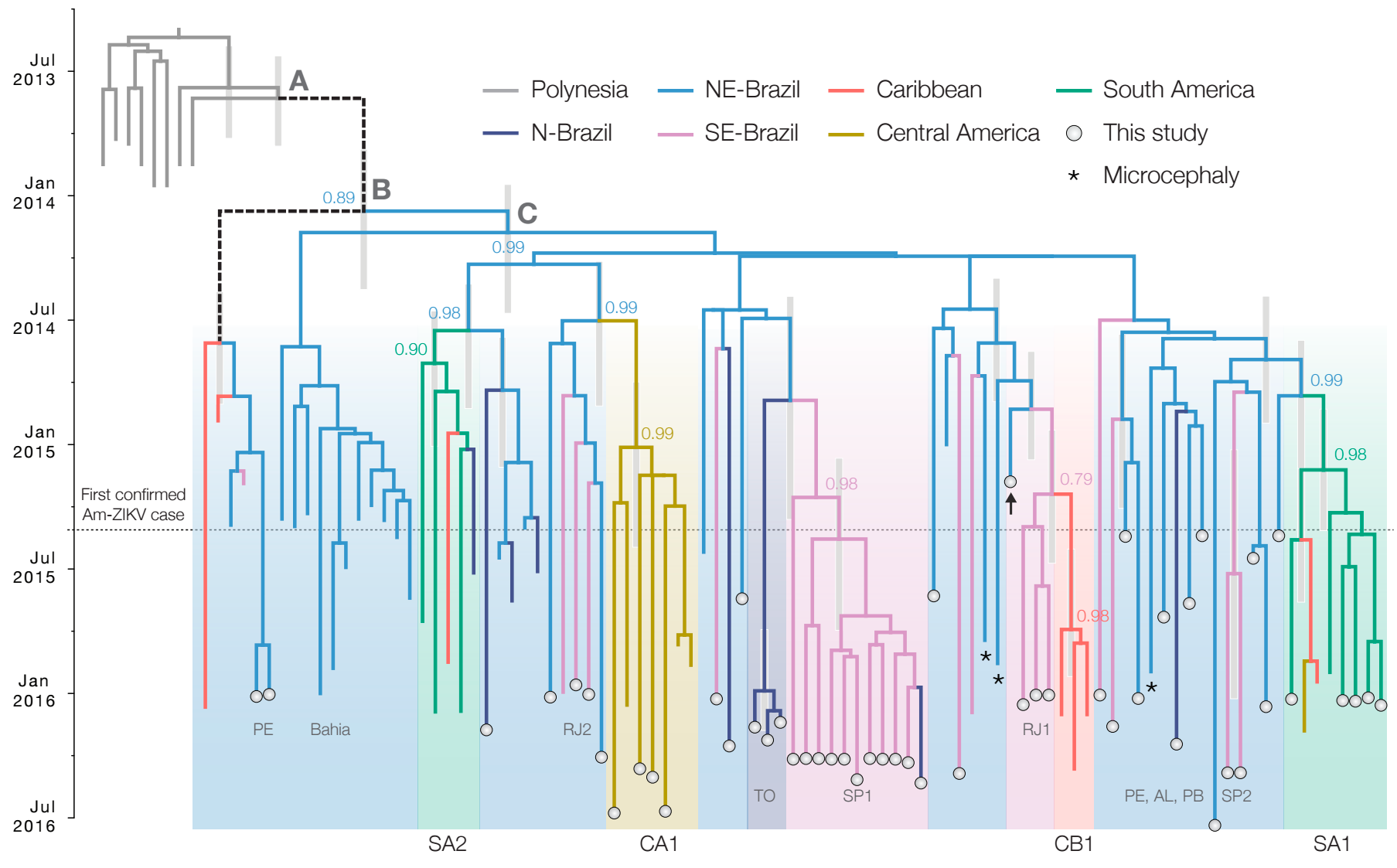
Case reports suggest initiation in northeast Brazil
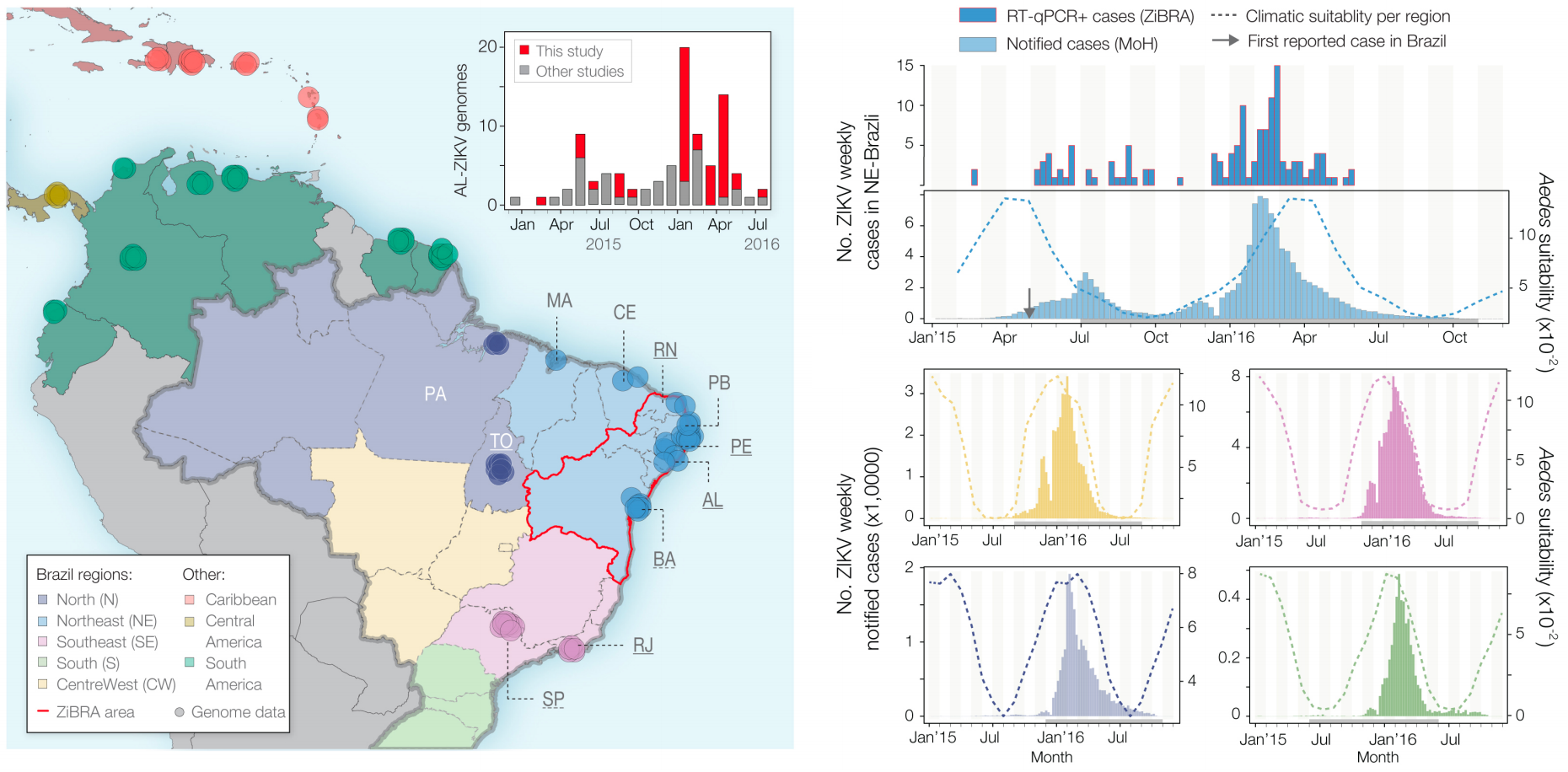
Phylogeny infers an origin in northeast Brazil

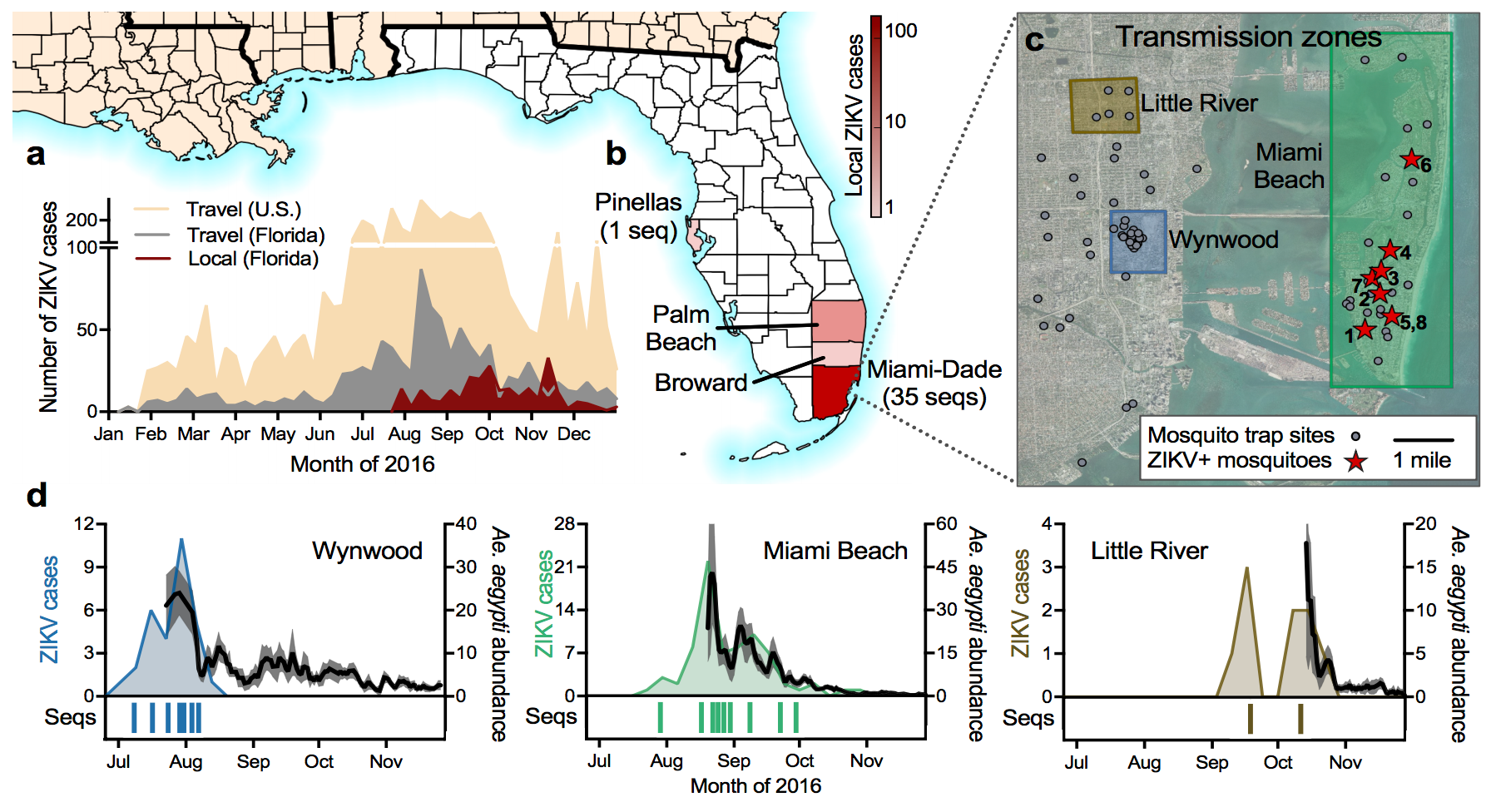
Outbreak of locally-acquired infections focused in Miami-Dade county
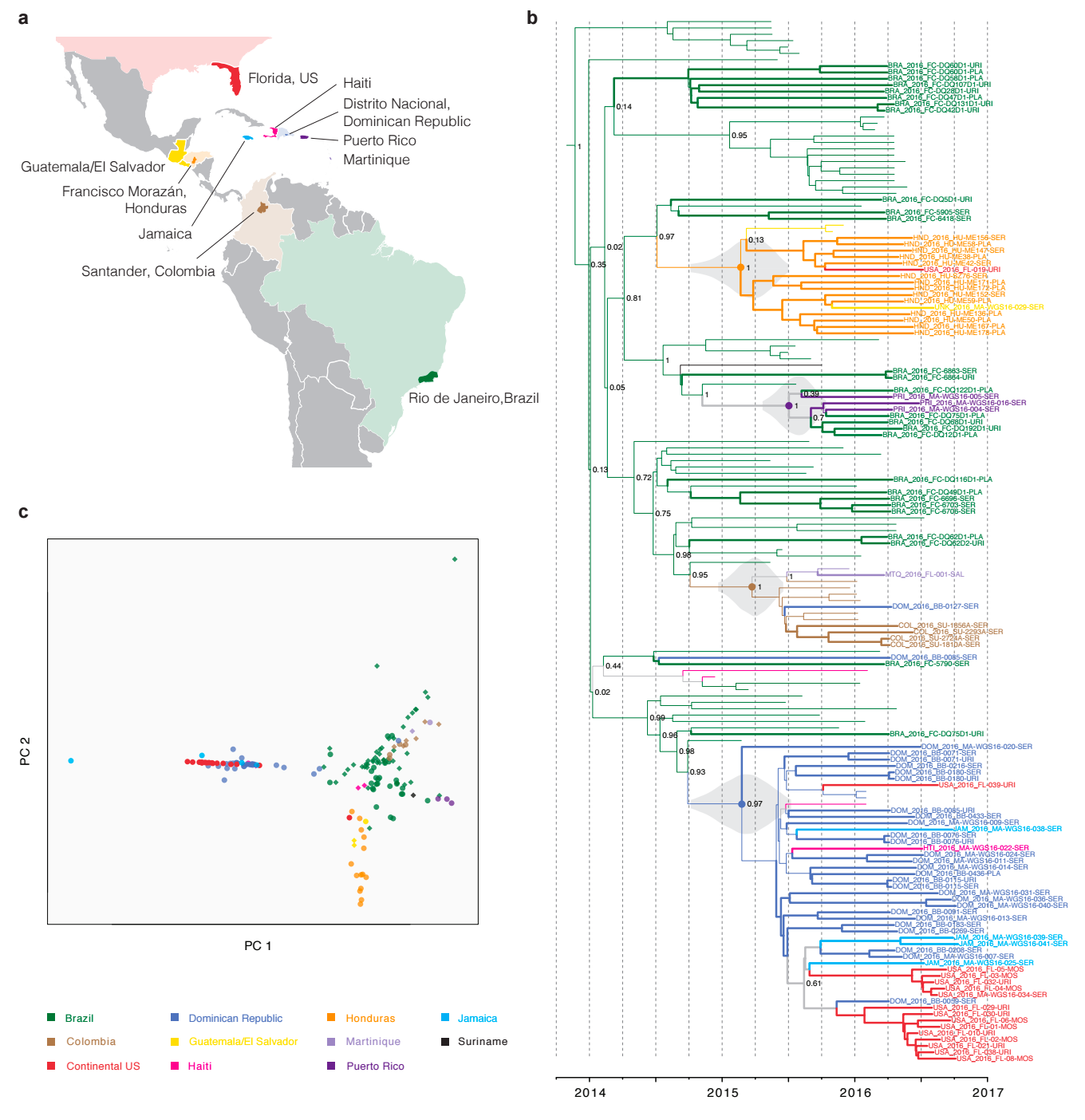
Phylogeny shows a surprising degree of clustering
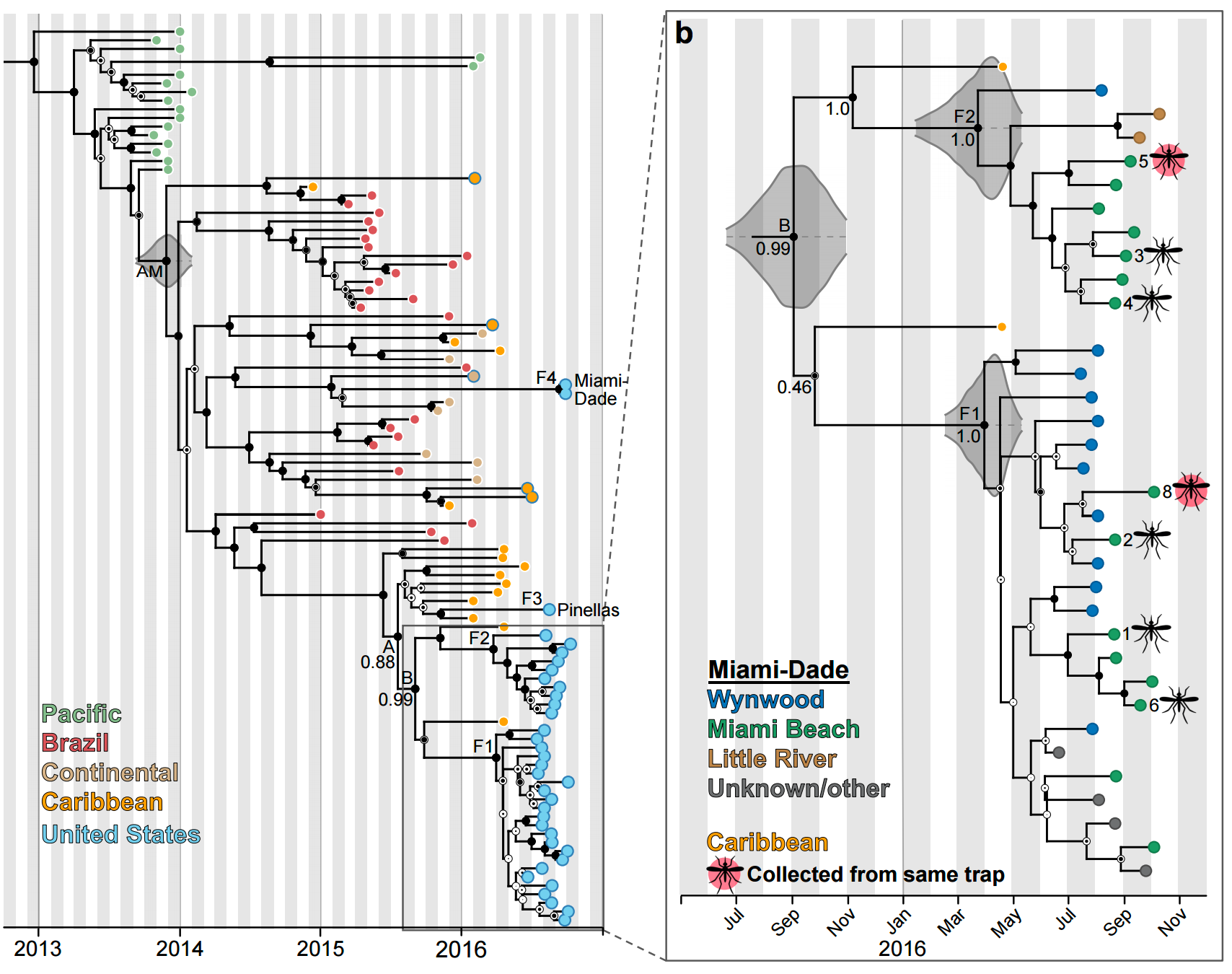
Clustering suggests fewer, longer transmission chains and higher R0
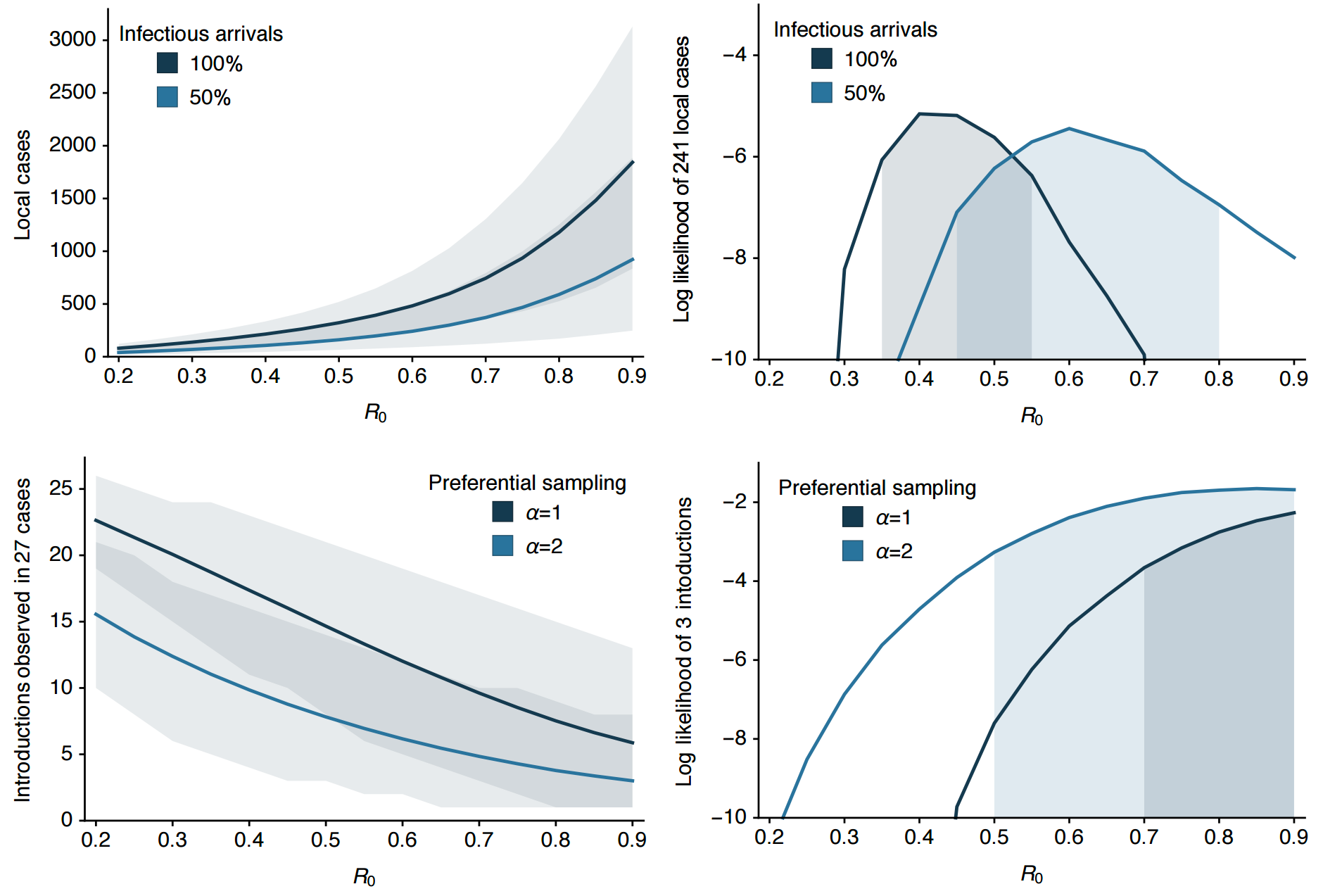
Flow of infected travelers greatest from Caribbean
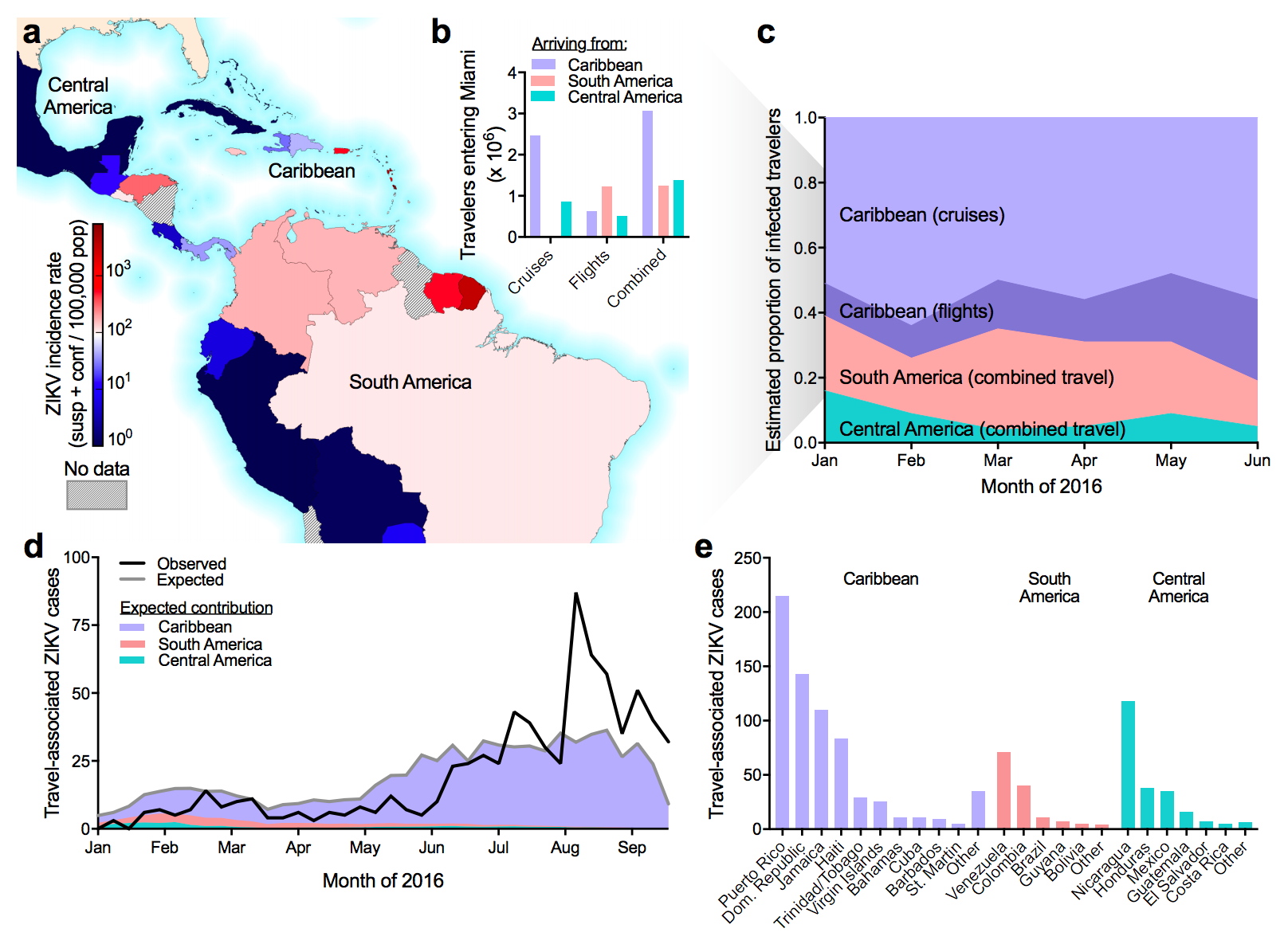
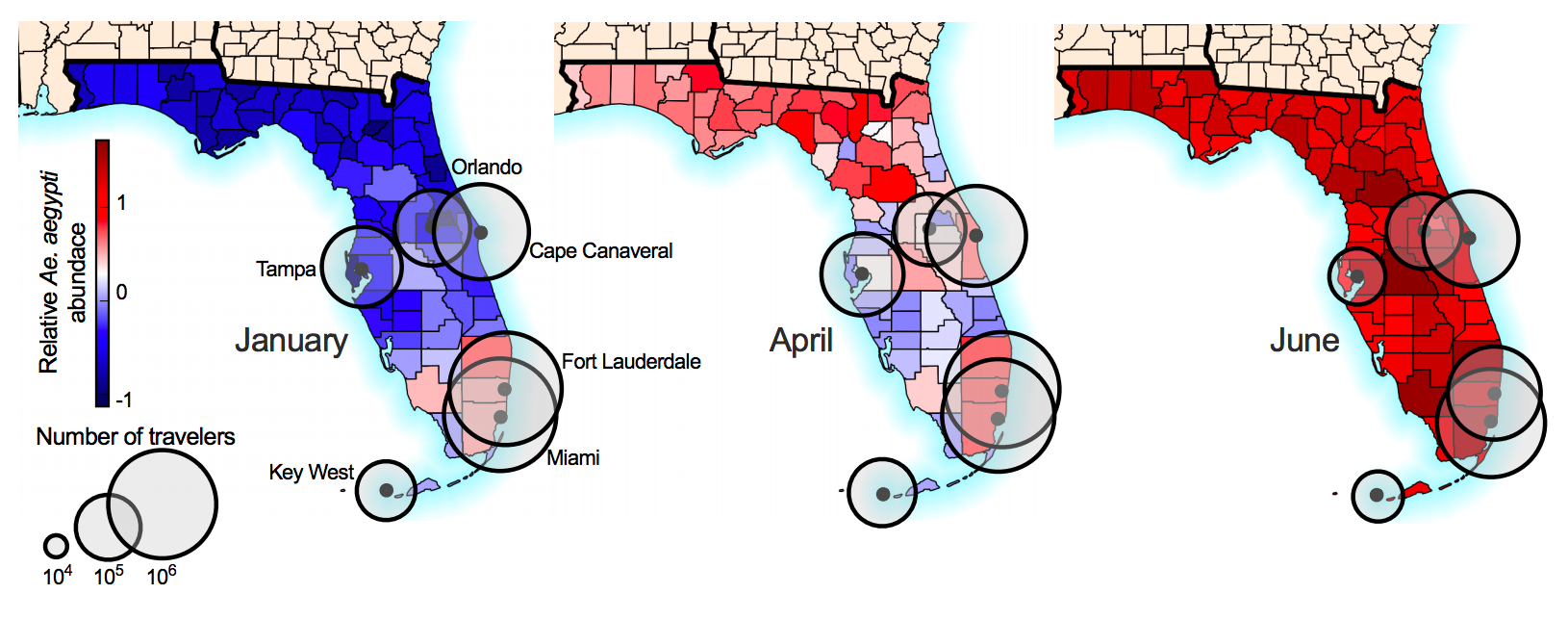
Southern Florida has high potential for Aedes borne outbreaks

Diverse phylogeny
nextstrain.org
Project to conduct real-time molecular epidemiology and evolutionary analysis of emerging epidemics
Acknowledgements
Ebola: Gytis Dudas, Andrew Rambaut, Luiz Carvalho, Philippe Lemey, Marc Suchard, Andrew Tatem, many others
Zika in Brazil: Nick Loman, Nuno Faria, Oliver Pybus, Josh Quick, Ingra Morales, Allison Black, the rest of the ZiBRA team
Zika in Florida: Kristian Andersen, Nathan Grubaugh, Oliver Pybus, Gustavo Palacios, Sharon Isern, Gytis Dudas, Moritz Kraemer, many others
Nextstrain: Richard Neher, Colin Megill, James Hadfield, Charlton Callender, Sarah Murata, Sidney Bell, Barney Potter




