Genomic tracking of SARS-CoV-2 evolution and spread
Trevor Bedford (@trvrb)
17 Jun 2020
VIDD Mini-Symposium on Coronaviruses
Fred Hutch
Slides at: bedford.io/talks


Significant fog of war. Genomic approaches offer orthogonal data source to understand the pandemic.
Epidemic process
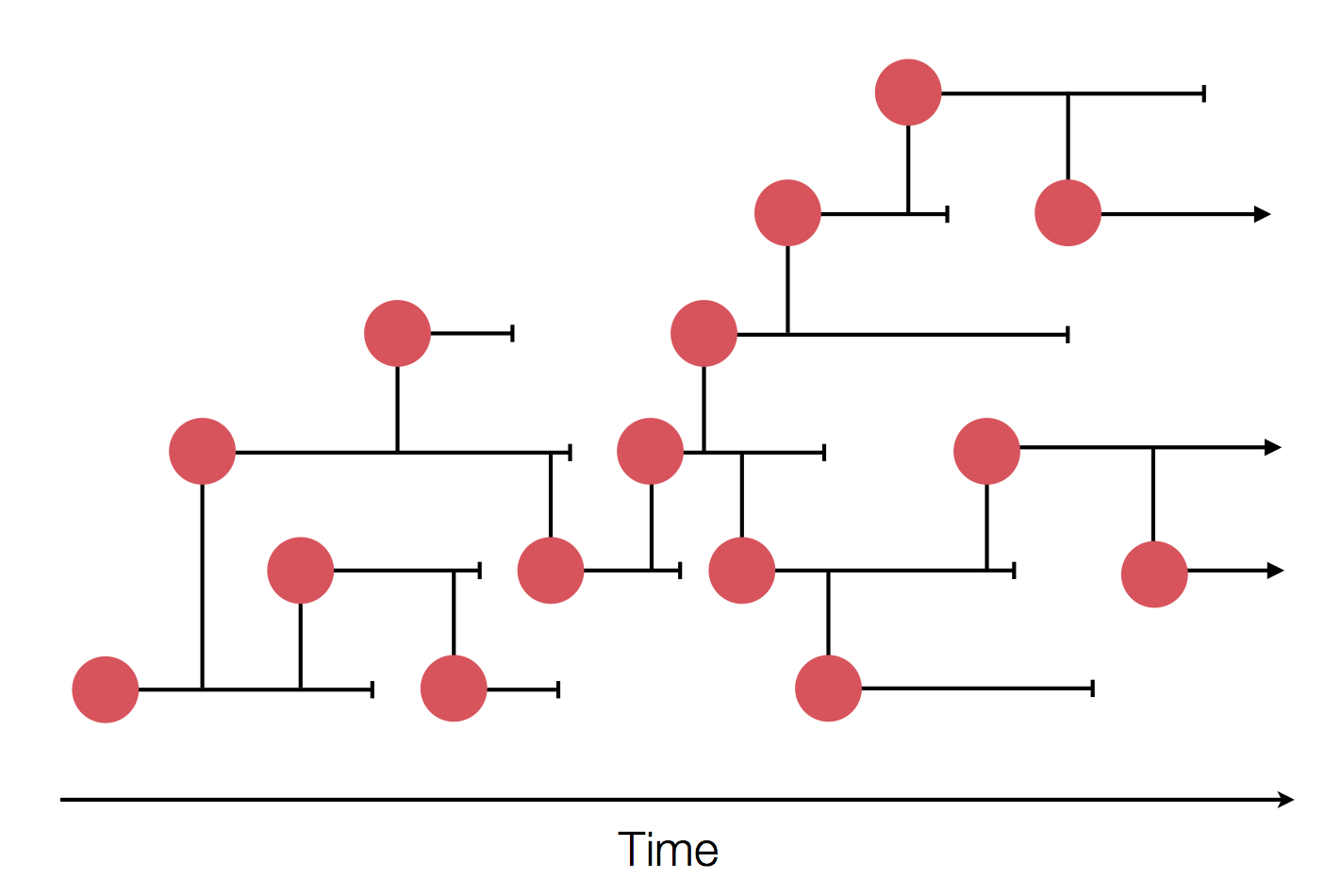
Sample some individuals
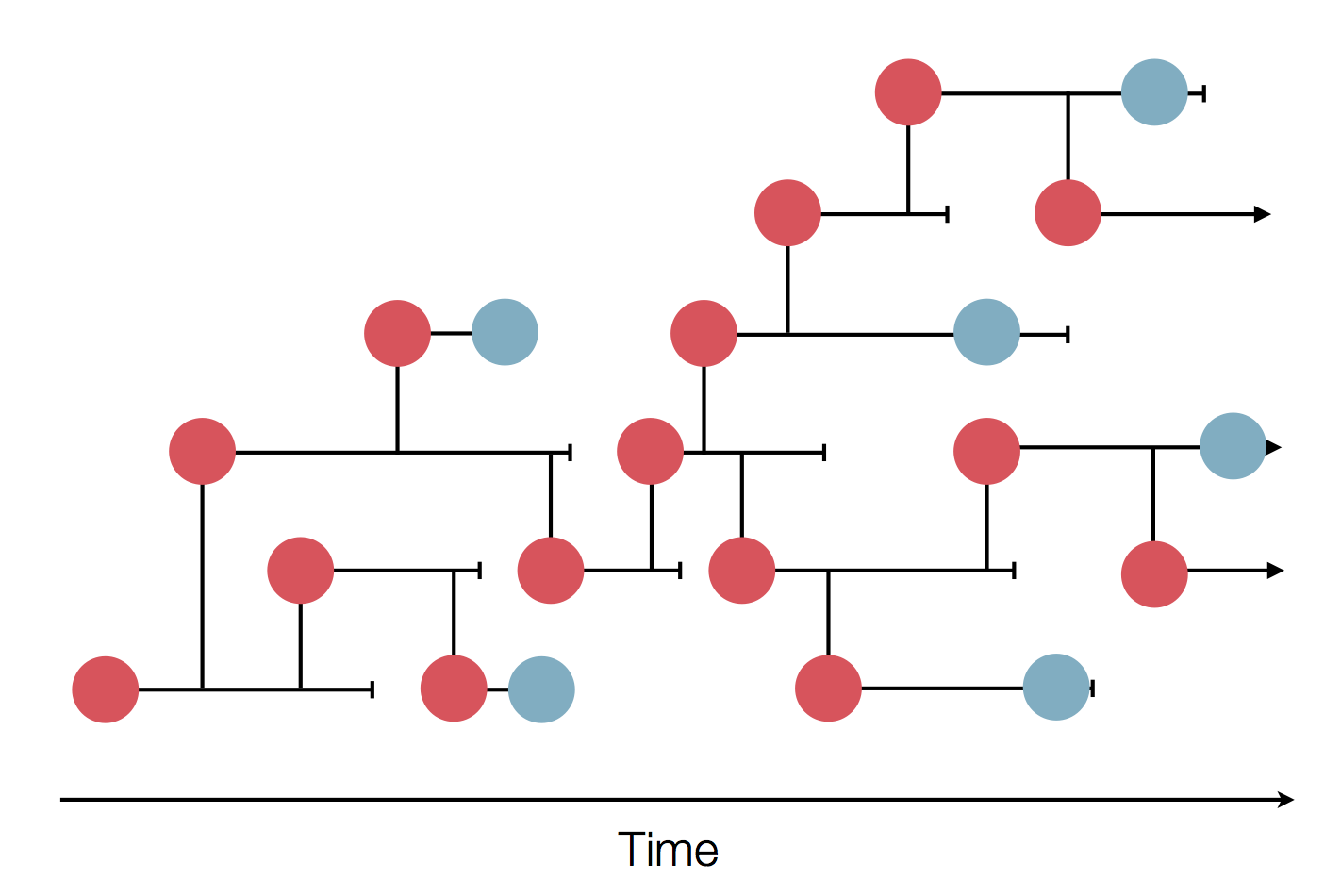
Sequence and determine phylogeny
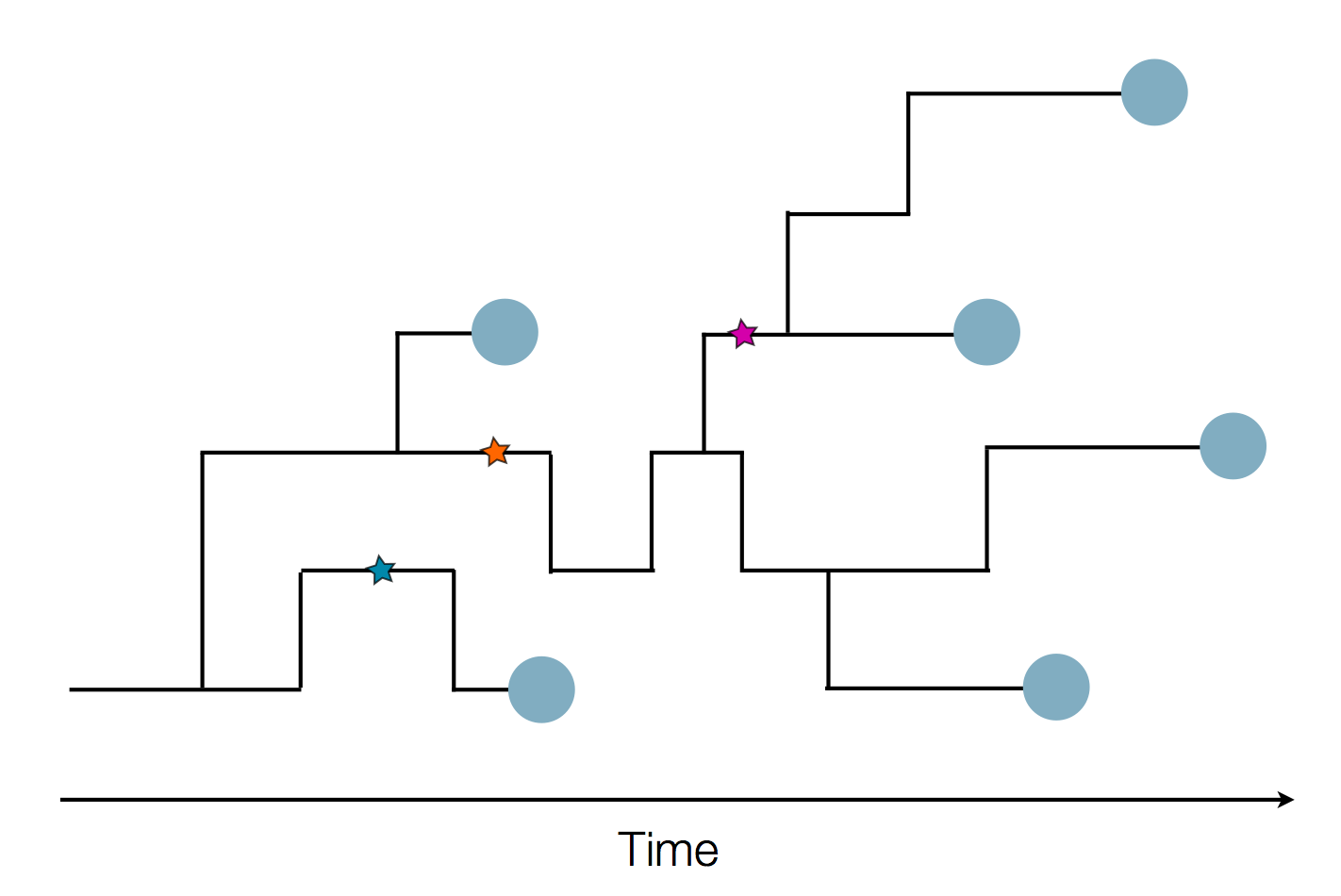
Sequence and determine phylogeny
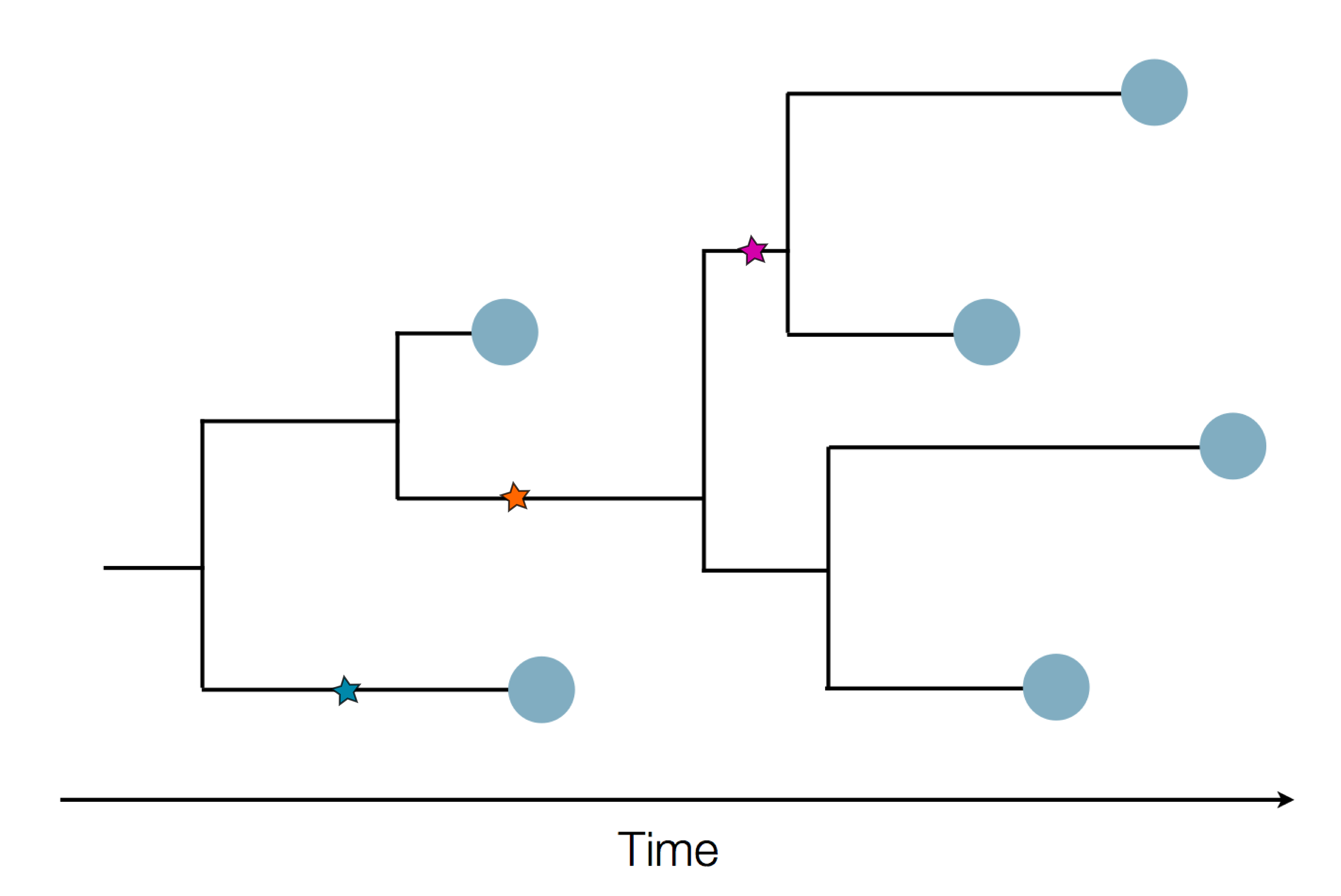
Nextstrain
Project to conduct real-time molecular epidemiology and evolutionary analysis of emerging epidemics
with
![]() Richard Neher,
Richard Neher,
![]() James Hadfield,
James Hadfield,
![]() Emma Hodcroft,
Emma Hodcroft,
![]() Thomas Sibley,
Thomas Sibley,
![]() John Huddleston,
John Huddleston,
![]() Louise Moncla,
Louise Moncla,
![]() Cassia Wagner,
Cassia Wagner,
![]() Miguel Paredes,
Miguel Paredes,
![]() Misja Ilcisin,
Misja Ilcisin,
![]() Kairsten Fay,
Kairsten Fay,
![]() Jover Lee,
Jover Lee,
![]() Allison Black,
Allison Black,
![]() Colin Megill,
Colin Megill,
![]() Sidney Bell,
Sidney Bell,
![]() Barney Potter,
Barney Potter,
![]() Charlton Callender
Charlton Callender
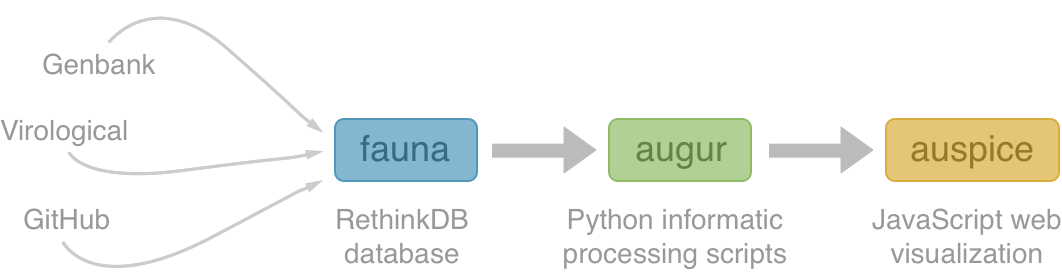
Nextstrain architecture
All code open source at github.com/nextstrain
Two central aims: (1) rapid and flexible phylodynamic analysis and
(2) interactive visualization
Rapid build pipeline for 3000 SARS-CoV-2 genomes (timings are for a laptop)
- Align with MAFFT (~20 min)
- Build ML tree with IQTREE (~40 min)
- Temporally resolve tree and geographic ancestry with TreeTime (~50 min)
- Total pipeline (~2 hr)
Current data flow for SARS-CoV-2
- Labs contribute directly to GISAID (now have >17k full genomes)
- Nextstrain pulls a complete dataset from GISAID every 60 minutes
- This triggers an automatic rebuild on Amazon Web Services
- We manually update new lat/longs, etc...
- We push this build online to nextstrain.org and tweet the update from @nextstrain
Dec/Jan: Emergence from Wuhan in ~Nov 2019
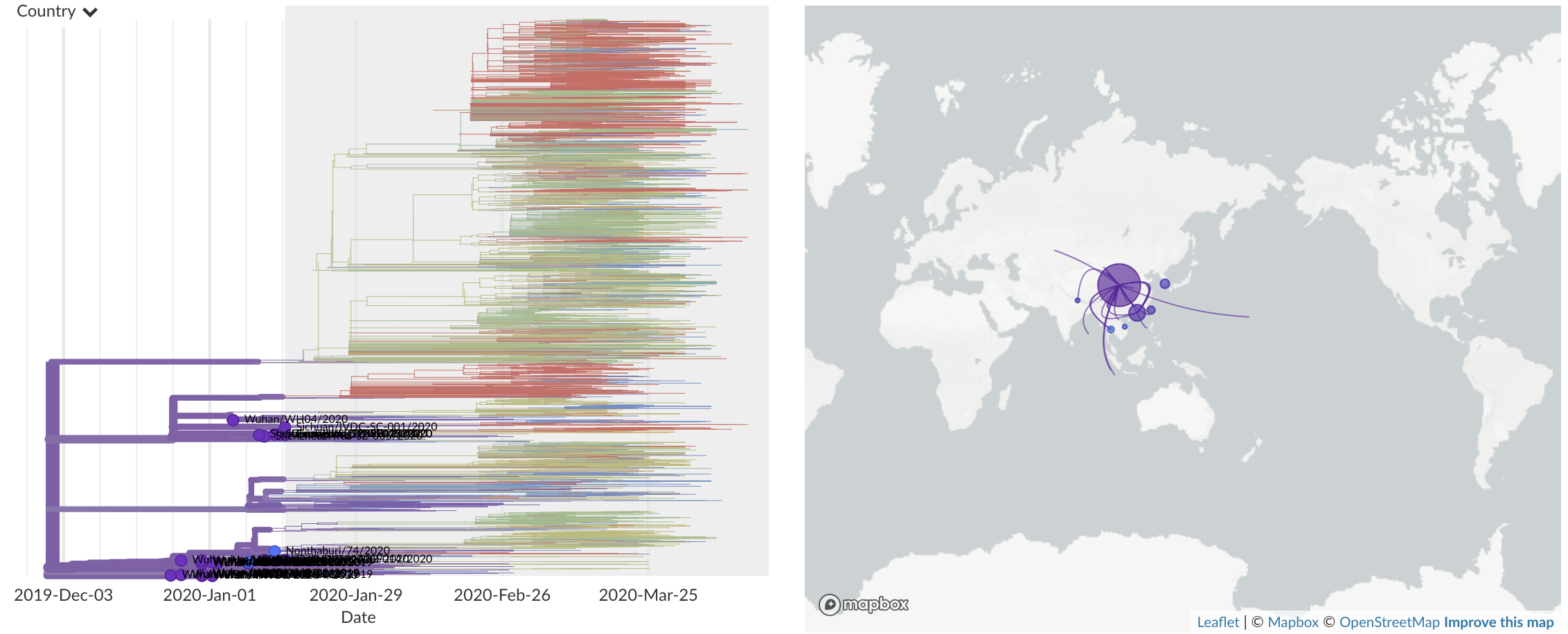
Jan/Feb: Spread within China and seeding elsewhere
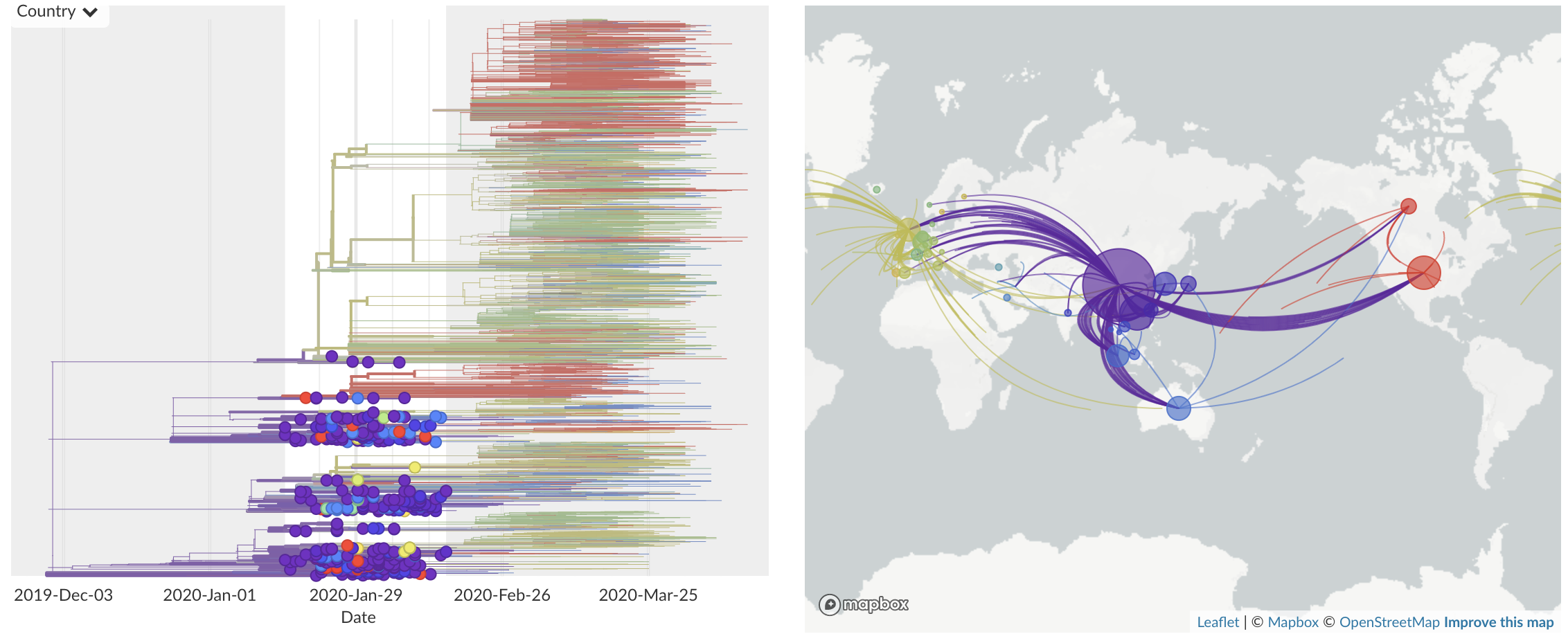
Feb/Mar: Epidemic spread within North America and Europe
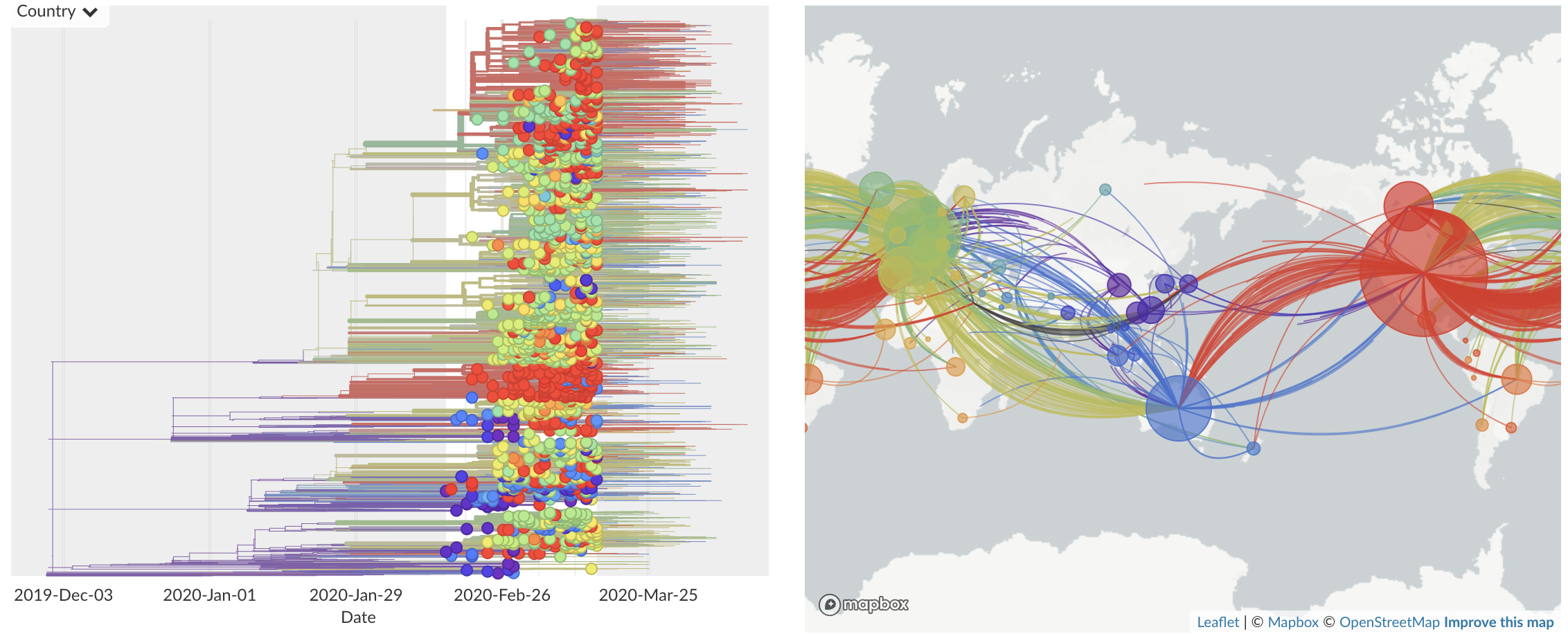
Mar/Apr: Continued growth, but decreasing transmission with social distancing measures
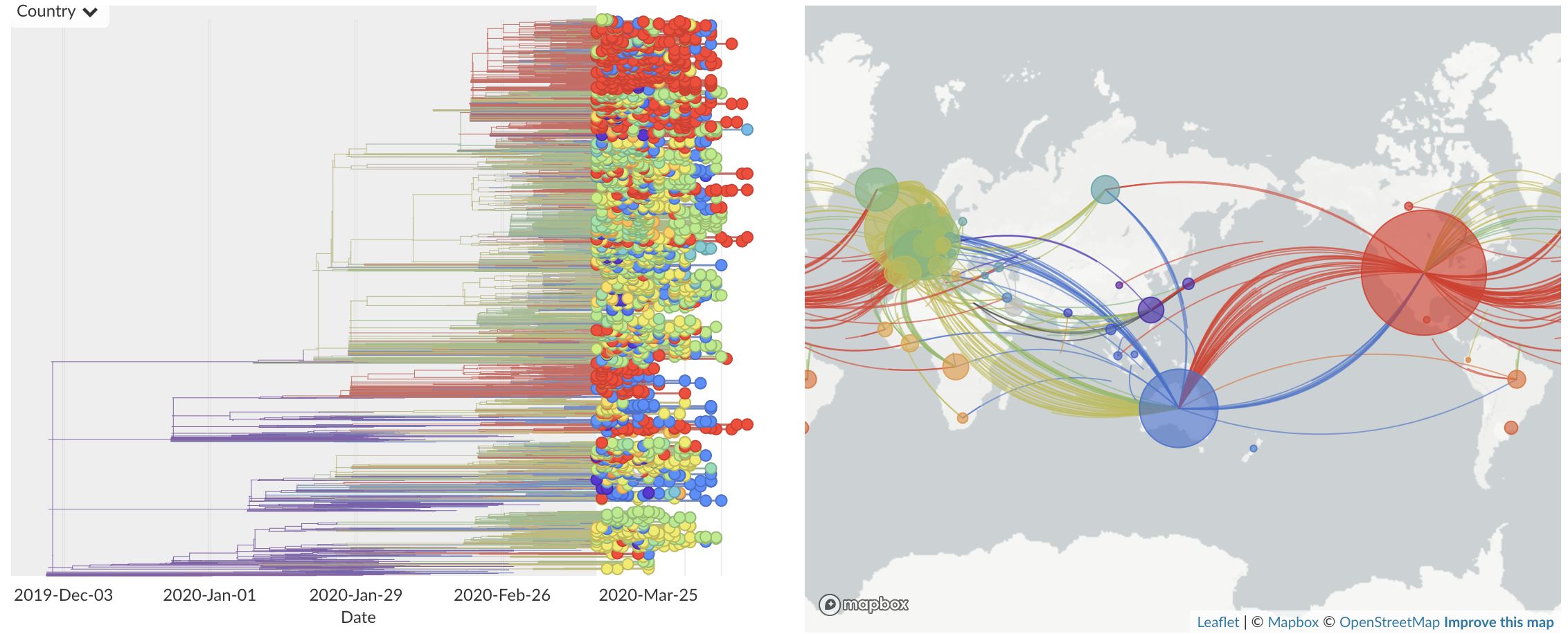
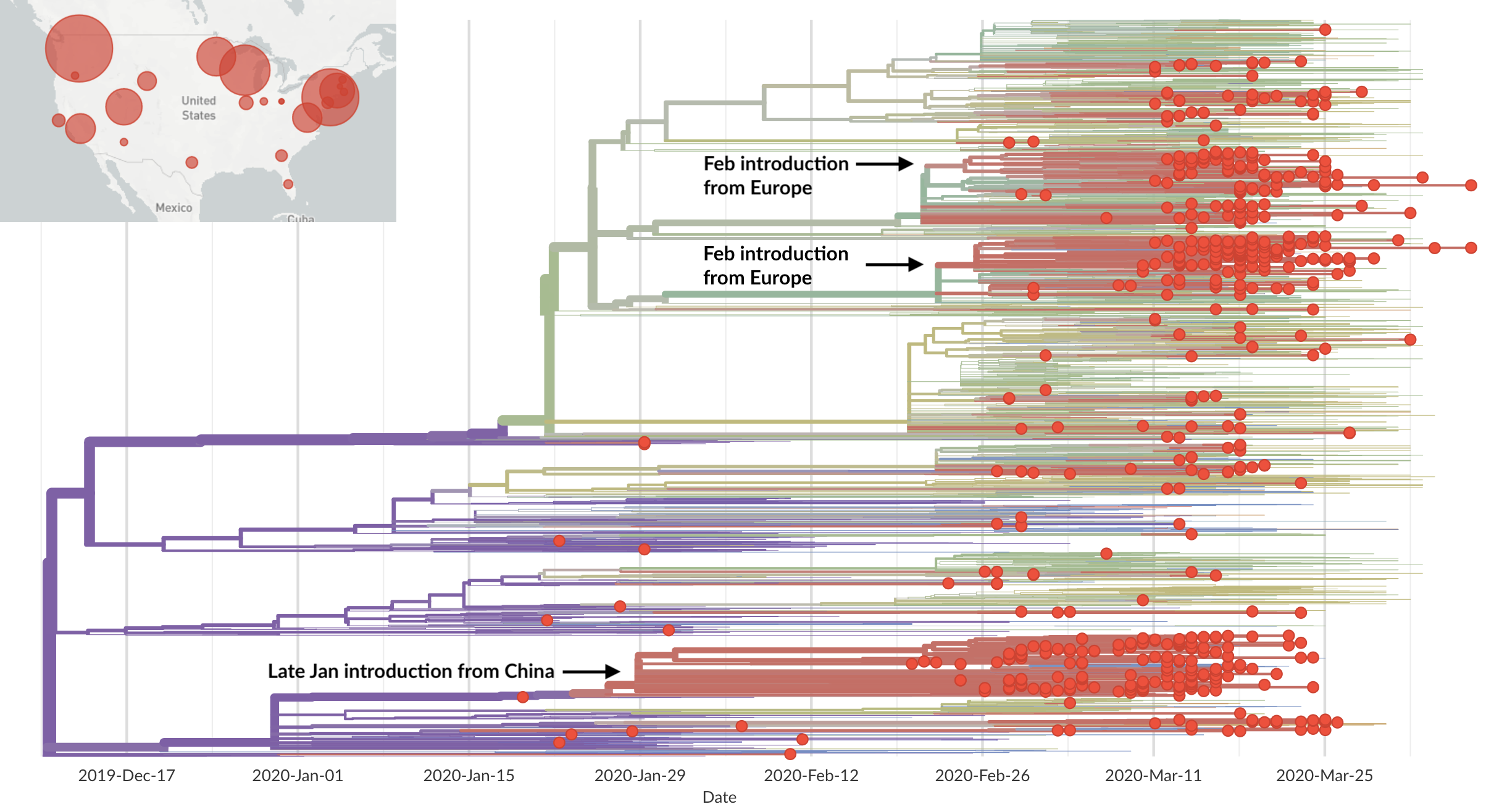
Epidemic in the USA was introduced from China in late Jan and from Europe during Feb
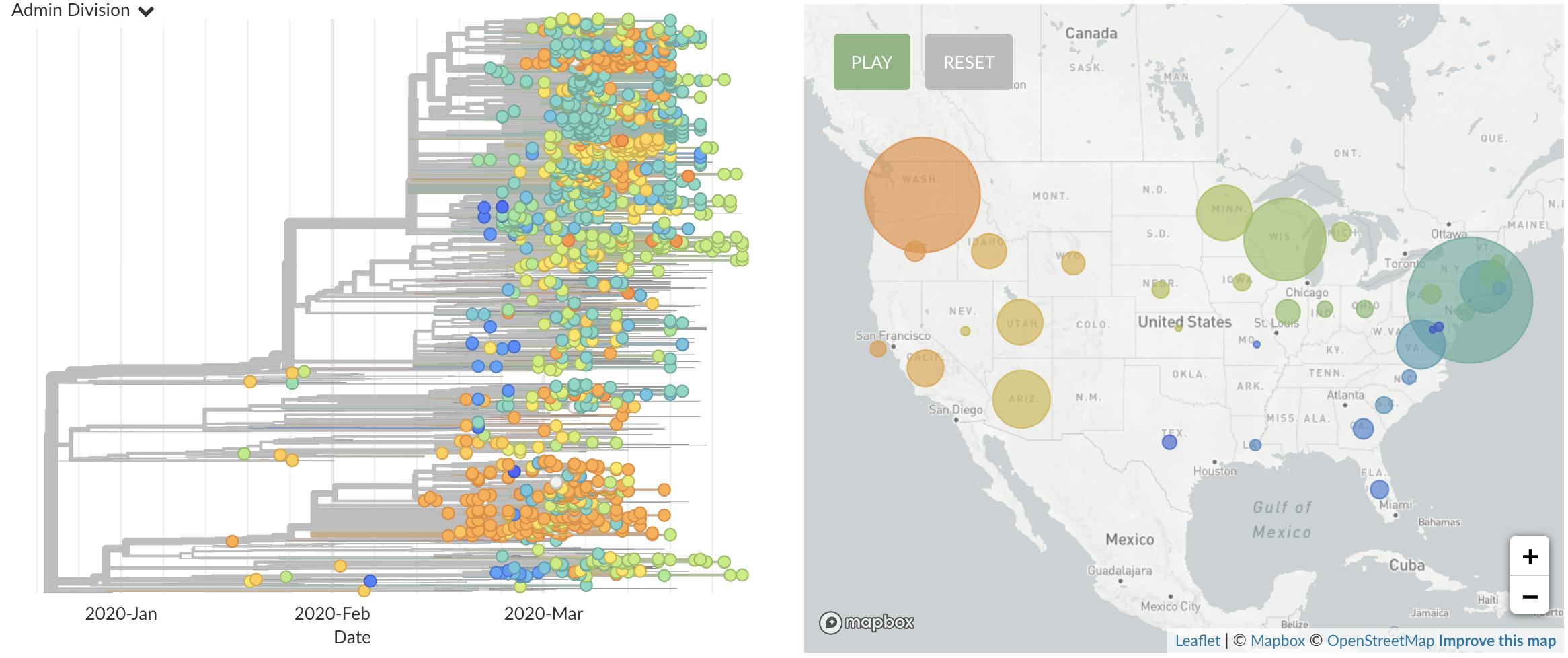
Once in the US, virus spread rapidly
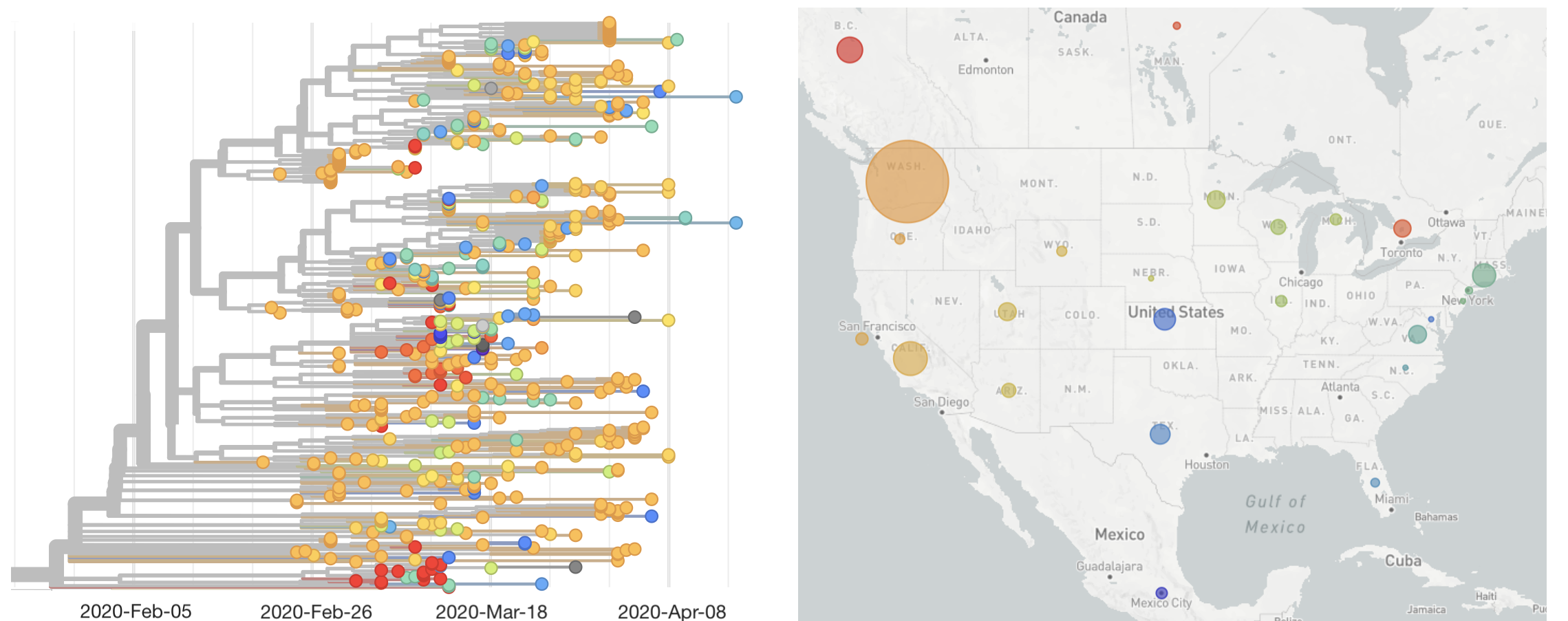
Single introduction ~Feb 1 quickly shows up throughout the country
There are now >45k publicly available SARS-CoV-2 genomes sequenced globally. Intractable to do a single global analysis. Best path forward is for different groups to analyze their local outbreaks.
Local Nextstrain instances
- US: California (CA DPH / CZ BioHub), Connecticut (CT DPH / Yale), Massachusetts (MA DPH / Broad), Michigan (MI HHS), Minnesota (MN PHL), Oregon (OHA / OHSU), Utah (UT PHL), Wisconsin (WI DPH / UW)
- Europe: ECDC, Austria (AAS), Israel (TAU), Spain (FISABIO)
- Elsewhere: India (CSIR IGIB), New Zealand (PMI), South Africa (SANGS), Uruguay (IPM)
Moving forward
- Better methods for large datasets
- Tying genomic epidemiology together with richer epi data to better understand local transmission
- Incorporating within-host variation to improve phylogenetic resolution
- Integrating clinical data to look for mutations that impact clinical outcomes
Acknowledgements
Genomic epi: Data producers from all over the world, GISAID and the Nextstrain team





