Genomic tracking of SARS-CoV-2 evolution and spread
Trevor Bedford (@trvrb)
20 Oct 2020
VIDD Seminar
Fred Hutch
Slides at: bedford.io/talks
This talk
Start with an overview of what we've been up to since I last presented to the Division three years ago
A lot of our work is shaped by what's going on in the world, with previous focus on Ebola in West Africa and the Zika epidemic in the Americas
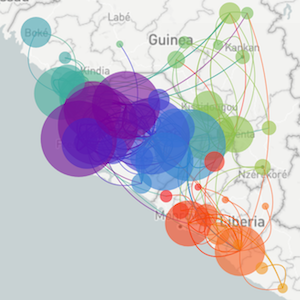

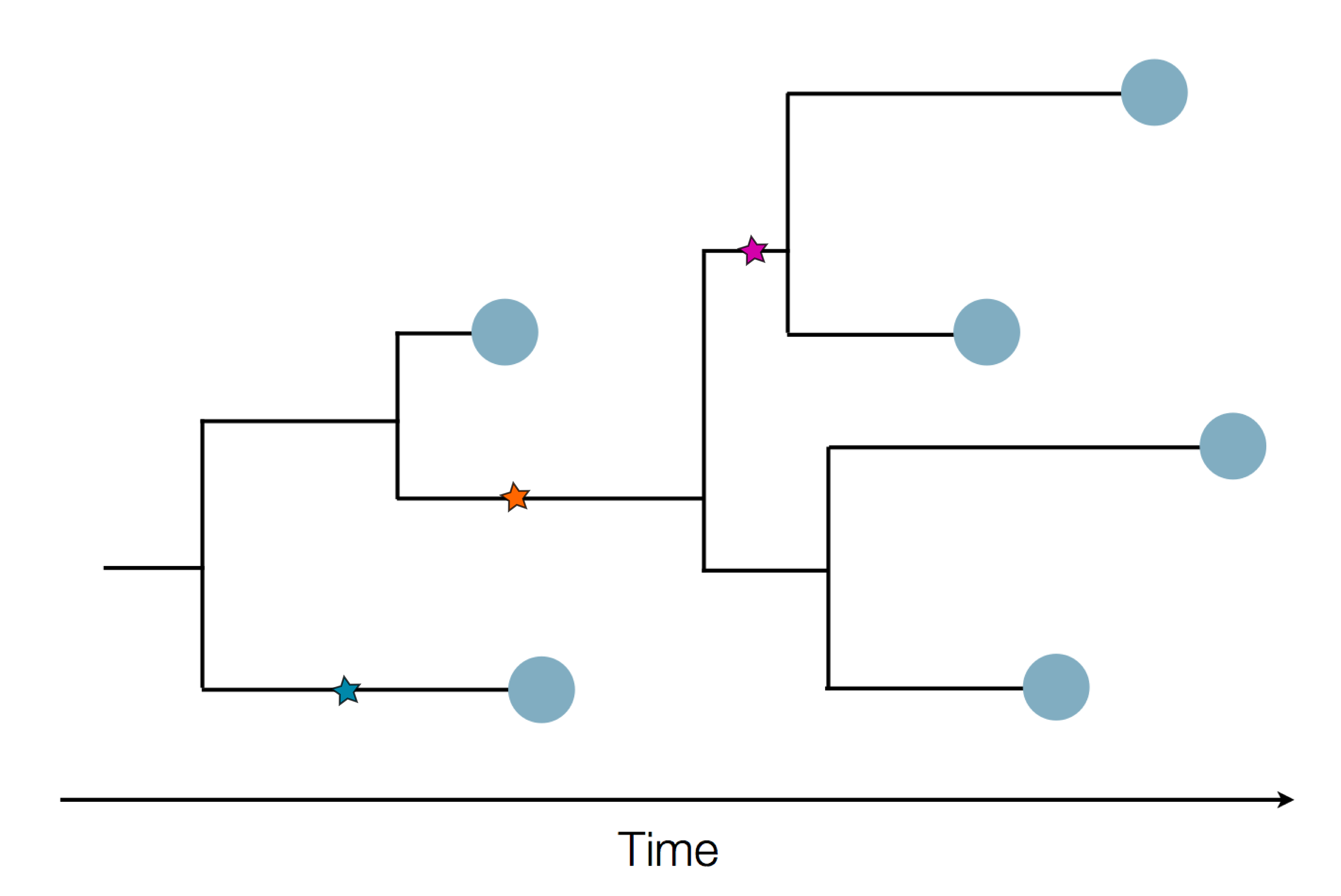
Epidemic process
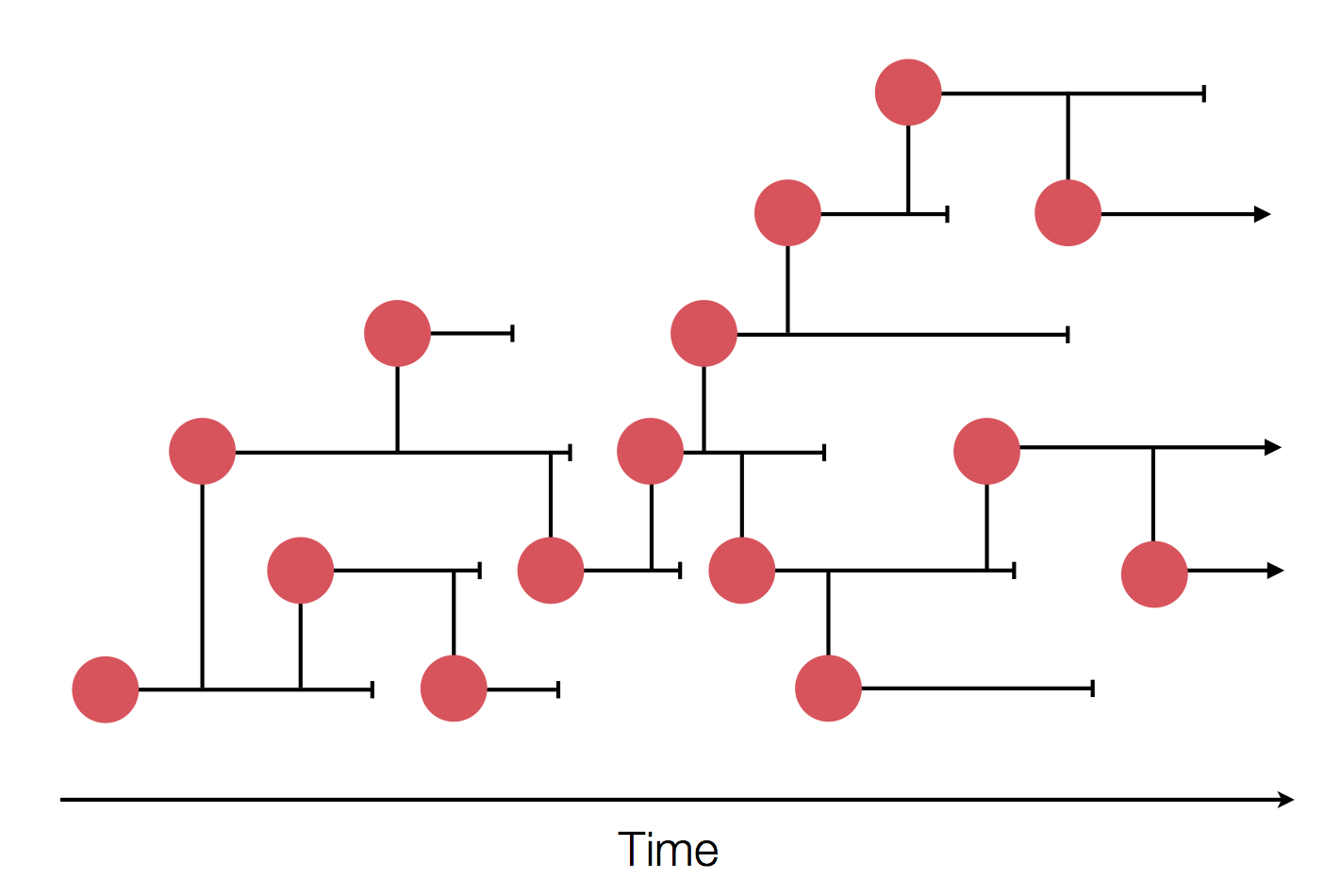
Sample some individuals
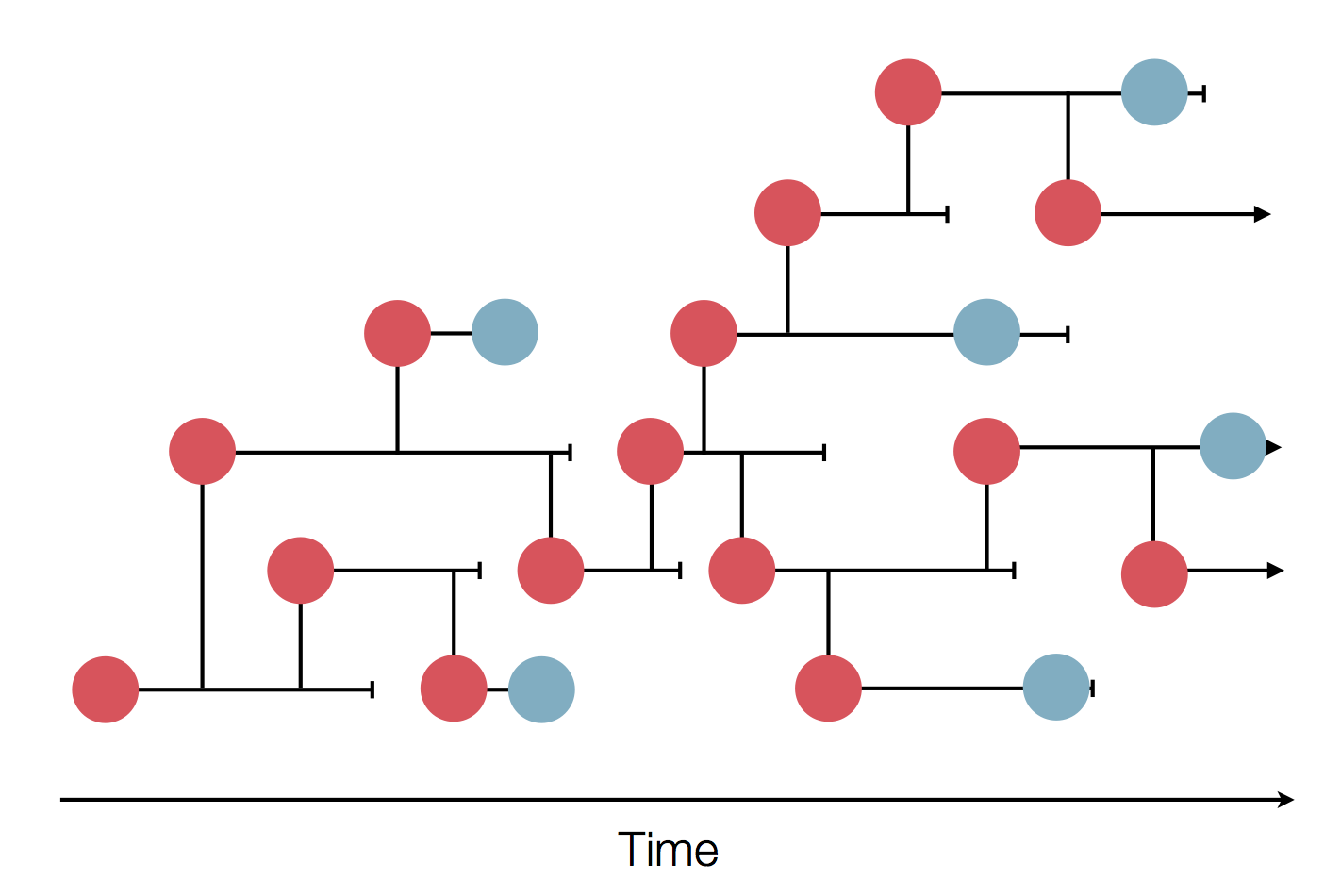
Sequence and determine phylogeny
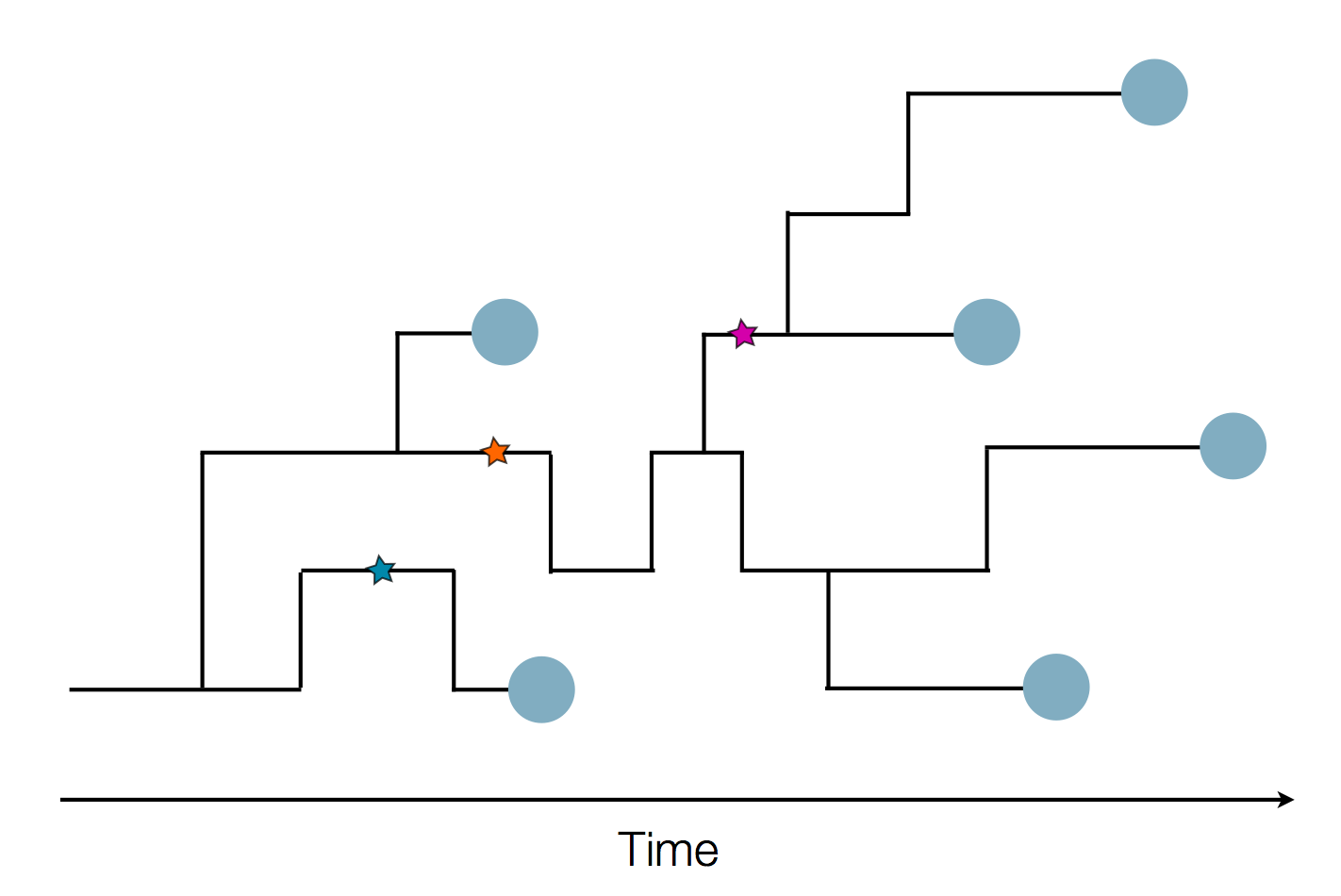
Sequence and determine phylogeny
Non-COVID "lab-lead" papers since last review in July 2017
 |
Dudas et al. 2018. MERS-CoV spillover at the camel-human interface. eLife. |  |
Hadfield et al. 2018. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. | |
 |
Lee et al. 2018. Deep mutational scanning of hemagglutinin helps predict evolutionary fates of human H3N2 influenza variants. PNAS. |  |
Bell et al. 2019. Dengue genetic divergence generates within-serotype antigenic variation, but serotypes dominate evolutionary dynamics. eLife. | |
 |
Hadfield et al. 2019. Twenty years of West Nile virus spread and evolution in the Americas visualized by Nextstrain. PLoS Pathog. |  |
Black et al. 2019. Genomic epidemiology supports multiple introductions and cryptic transmission of Zika virus in Colombia. BMC Infect Dis. | |
 |
Potter et al. 2019. Evolution and rapid spread of a reassortant A(H3N2) virus that predominated the 2017-2018 influenza season. Virus Evol. |  |
Dudas et al. 2019. The ability of single genes vs full genomes to resolve time and space in outbreak analysis. BMC Evol Biol. | |
 |
Moncla et al. 2020. Quantifying within-host evolution of H5N1 influenza in humans and poultry in Cambodia. PLoS Pathog. |  |
Black et al. 2020. Ten recommendations for supporting open pathogen genomic analysis in public health. Nat Med. | |
 |
Hilton et al. 2020. dms-view: Interactive visualization tool for deep mutational scanning data. J Open Source Softw. |  |
Huddleston et al. 2020. Integrating genotypes and phenotypes improves long-term forecasts of seasonal influenza A/H3N2 evolution. eLife. |
Non-COVID "lab-lead" papers since last review in July 2017
 |
Dudas et al. 2018. MERS-CoV spillover at the camel-human interface. eLife. |  |
Hadfield et al. 2018. Nextstrain: real-time tracking of pathogen evolution. Bioinformatics. | |
 |
Lee et al. 2018. Deep mutational scanning of hemagglutinin helps predict evolutionary fates of human H3N2 influenza variants. PNAS. |  |
Bell et al. 2019. Dengue genetic divergence generates within-serotype antigenic variation, but serotypes dominate evolutionary dynamics. eLife. | |
 |
Hadfield et al. 2019. Twenty years of West Nile virus spread and evolution in the Americas visualized by Nextstrain. PLoS Pathog. |  |
Black et al. 2019. Genomic epidemiology supports multiple introductions and cryptic transmission of Zika virus in Colombia. BMC Infect Dis. | |
 |
Potter et al. 2019. Evolution and rapid spread of a reassortant A(H3N2) virus that predominated the 2017-2018 influenza season. Virus Evol. |  |
Dudas et al. 2019. The ability of single genes vs full genomes to resolve time and space in outbreak analysis of Ebola. BMC Evol Biol. | |
 |
Moncla et al. 2020. Quantifying within-host evolution of H5N1 influenza in humans and poultry in Cambodia. PLoS Pathog. |  |
Black et al. 2020. Ten recommendations for supporting open pathogen genomic analysis in public health. Nat Med. | |
 |
Hilton et al. 2020. dms-view: Interactive visualization tool for deep mutational scanning data. J Open Source Softw. |  |
Huddleston et al. 2020. Integrating genotypes and phenotypes improves long-term forecasts of seasonal influenza A/H3N2 evolution. eLife. |
Recent work with seasonal flu and Ebola
Integrating genotypes and phenotypes improves long-term forecasts of seasonal influenza A/H3N2 evolution
with ![]() John Huddleston, Richard Neher, Dave Wentworth, Becky Kondor, John McCauley, Hideki Hasegawa, Kanta Subbarao and others
John Huddleston, Richard Neher, Dave Wentworth, Becky Kondor, John McCauley, Hideki Hasegawa, Kanta Subbarao and others
H3N2 vaccine updates occur every ~2 years
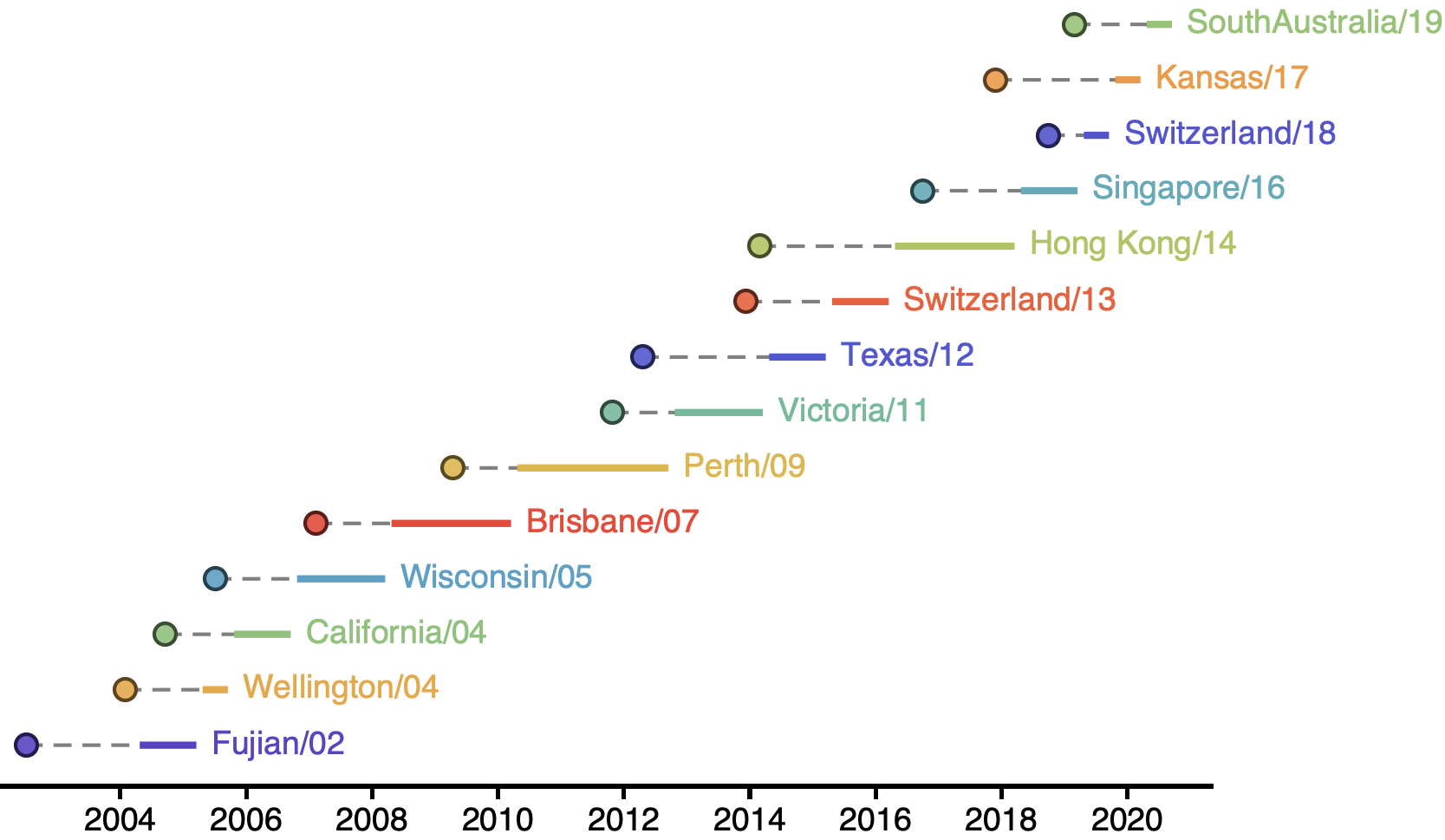
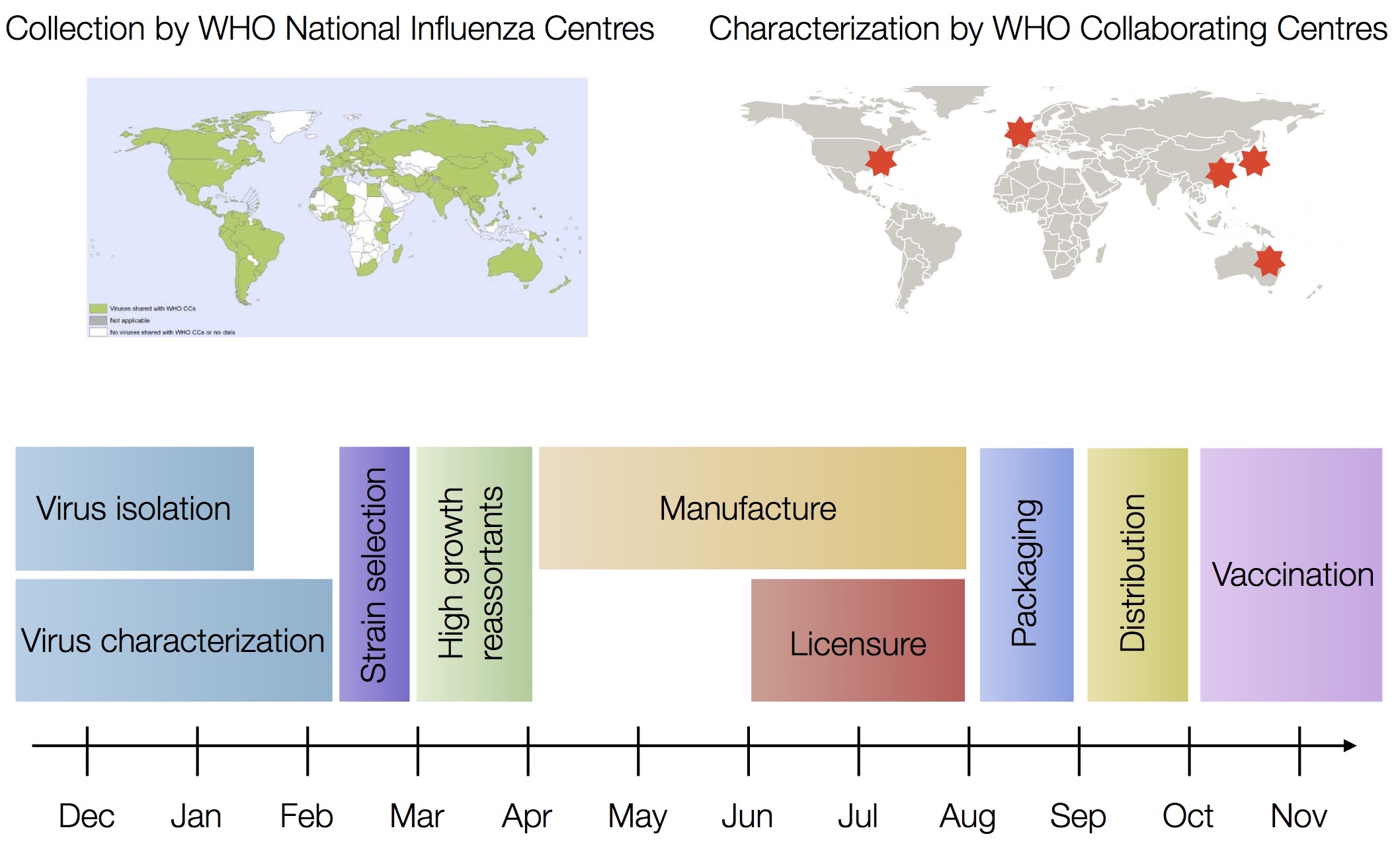
Vaccine strain selection by WHO
Select one strain from circulating diversity
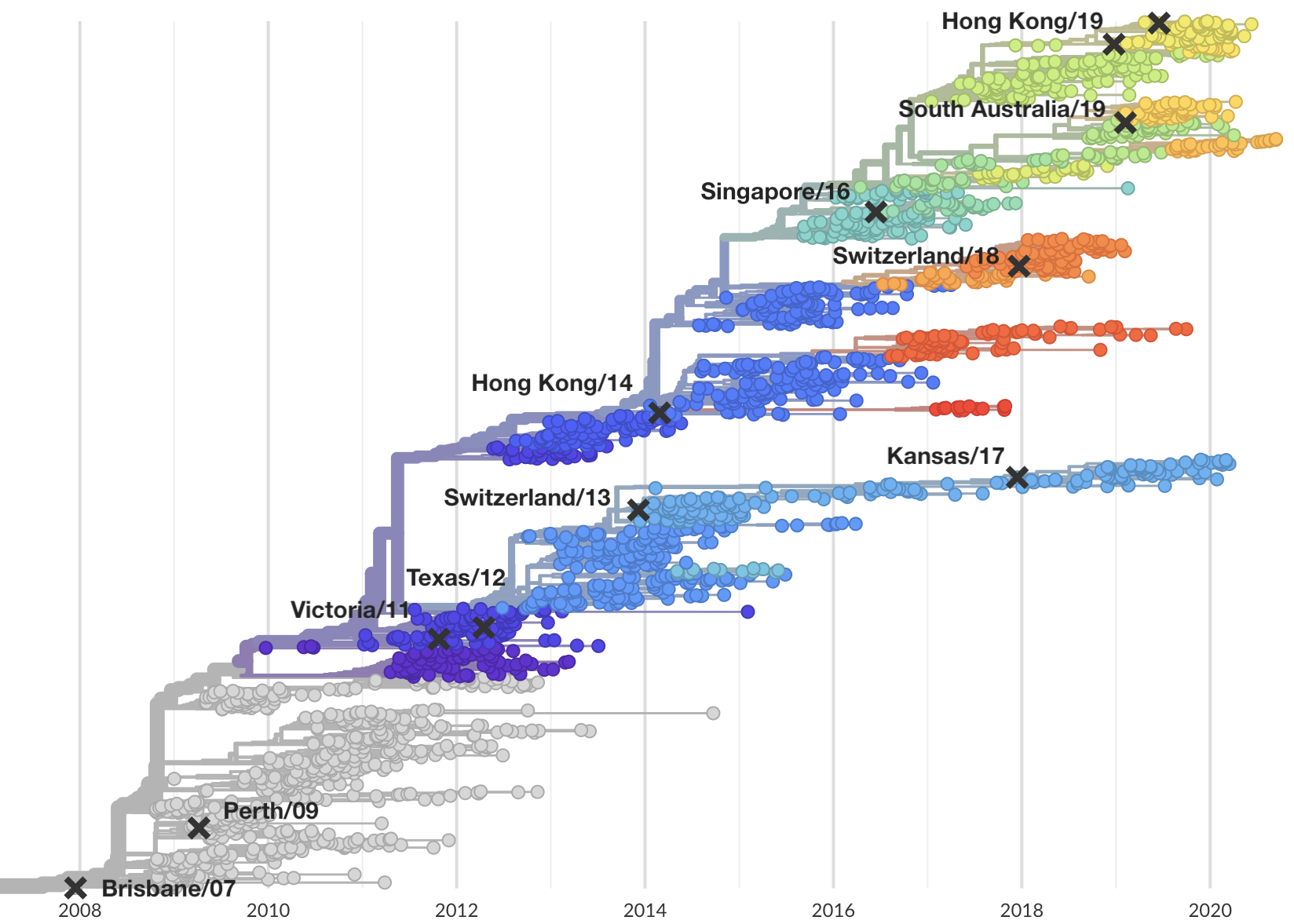
Fitness models project strain frequencies
Future frequency $x_i(t+\Delta t)$ of strain $i$ derives from strain fitness $f_i$ and present day frequency $x_i(t)$, such that
$$\hat{x}_i(t+\Delta t) = x_i(t) \, \mathrm{exp}(f_i \, \Delta t)$$
Total strain frequencies at each timepoint are normalized. This captures clonal interference between competing lineages.
Match strain forecast to retrospective circulation
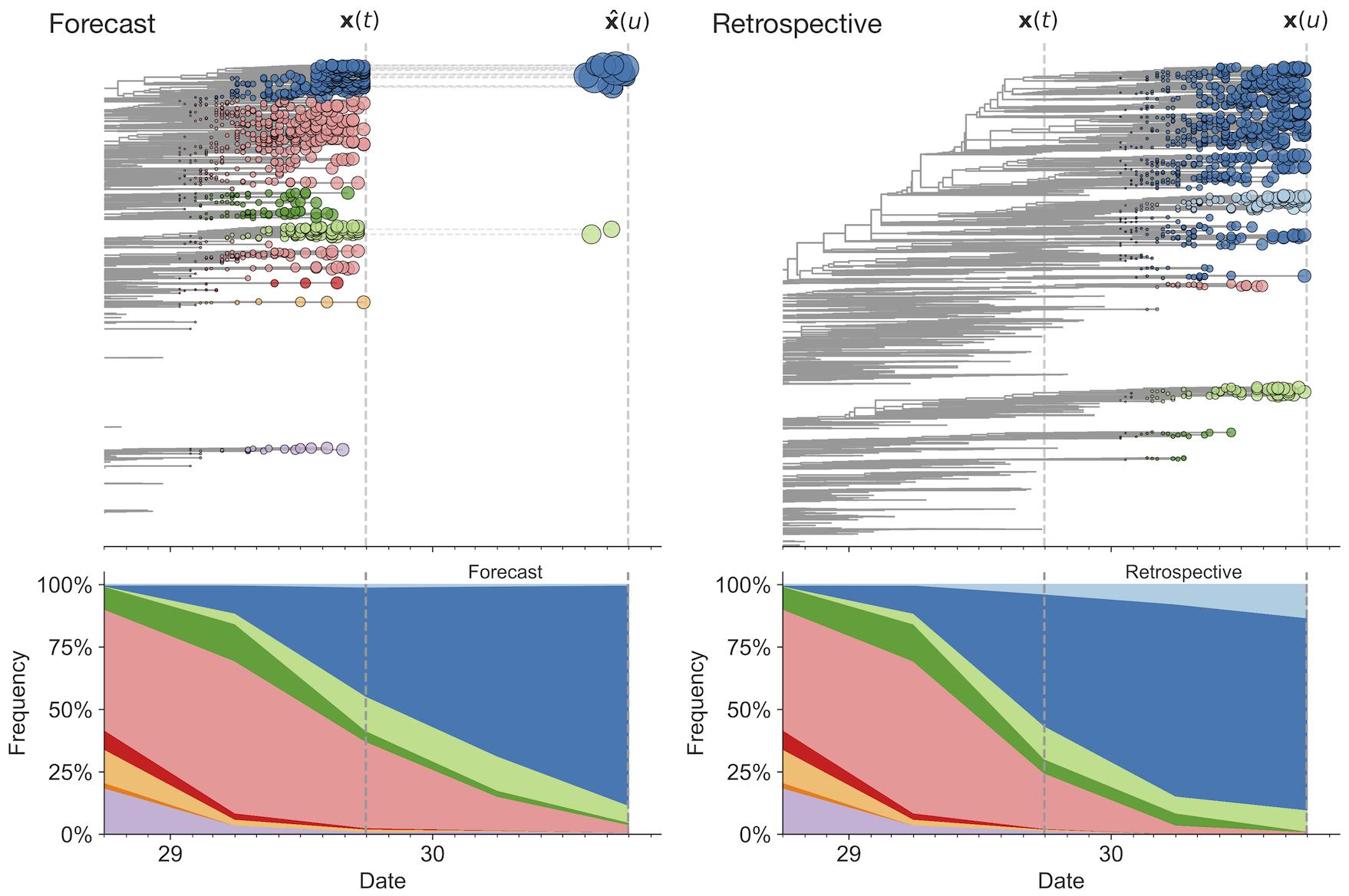
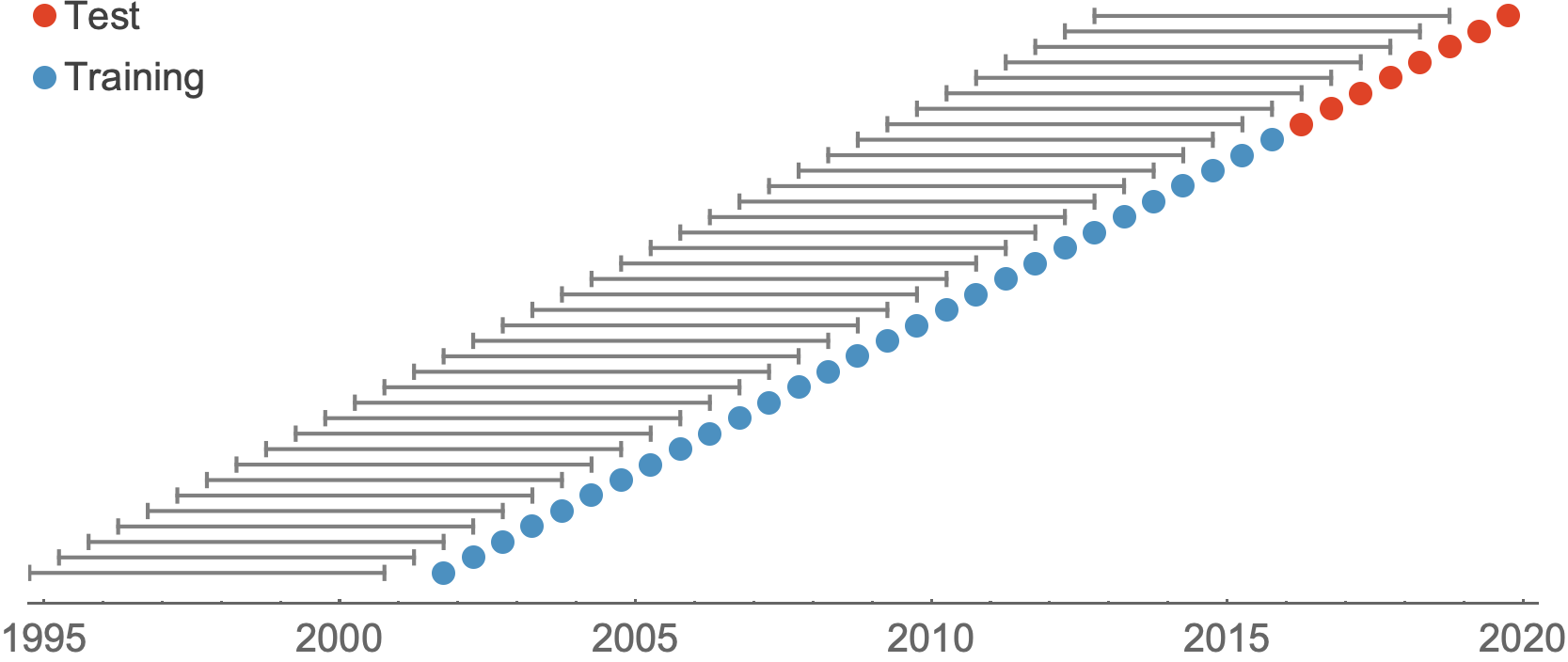
Train in 6-year sliding windows from 1995 to 2015 with most recent years held out as test
Composite models favor combinations of HI drift, local branching index and non-epitope fitness
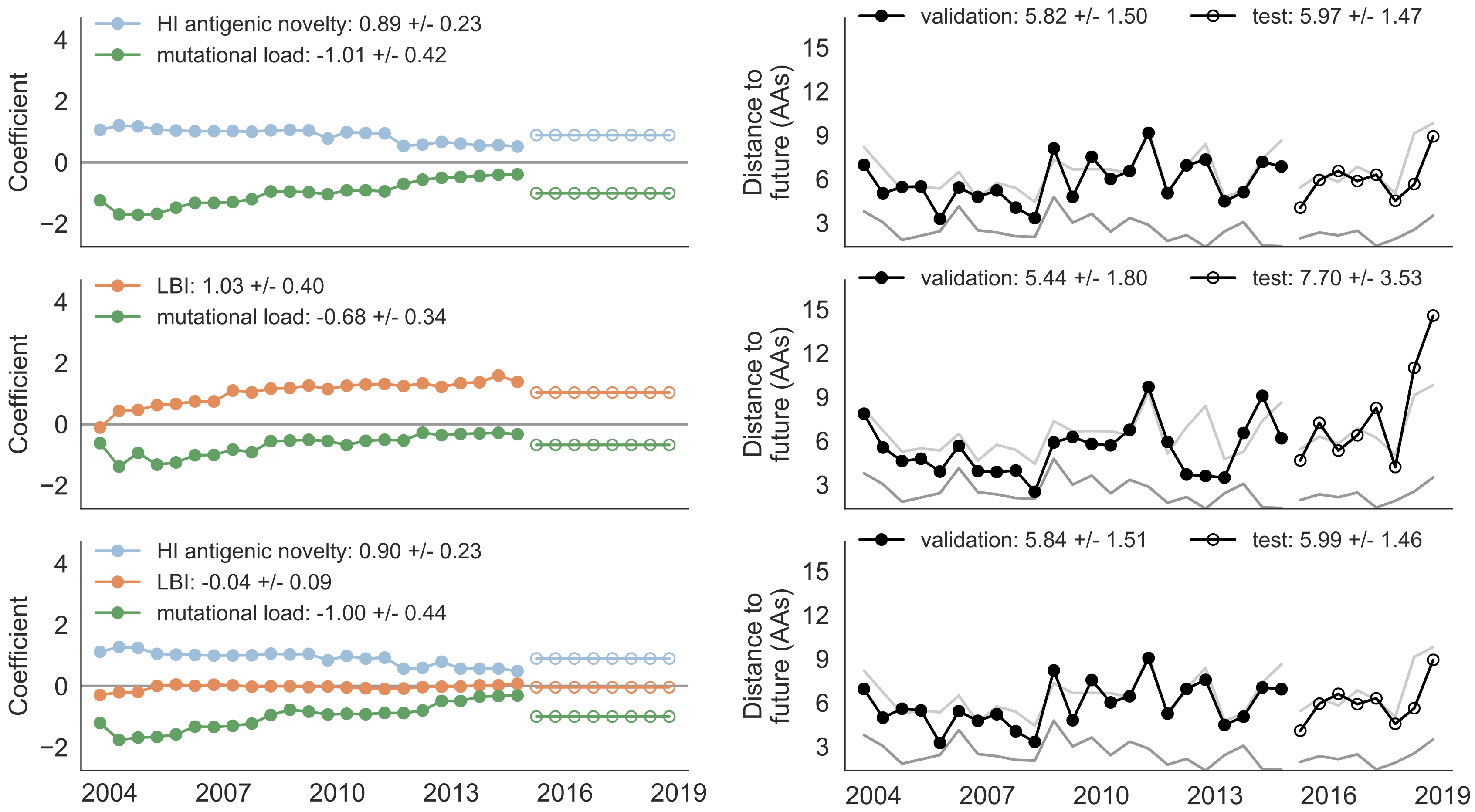
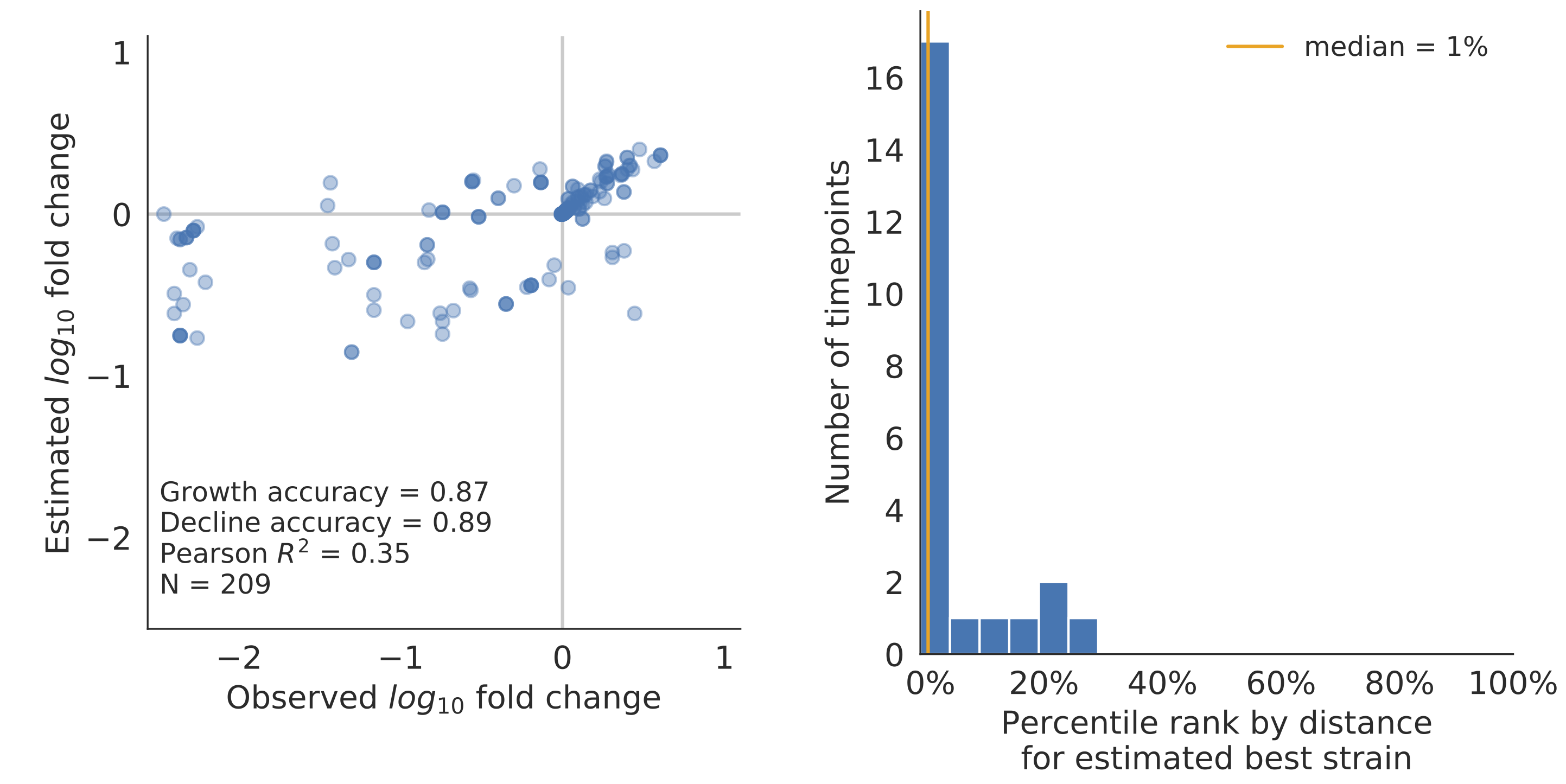
Model successfully predicts clade growth and best pick from model is generally close to best possible retrospective pick
Forecasts are live at nextstrain.org/flu, but are currently fraught due to lack of recent isolates
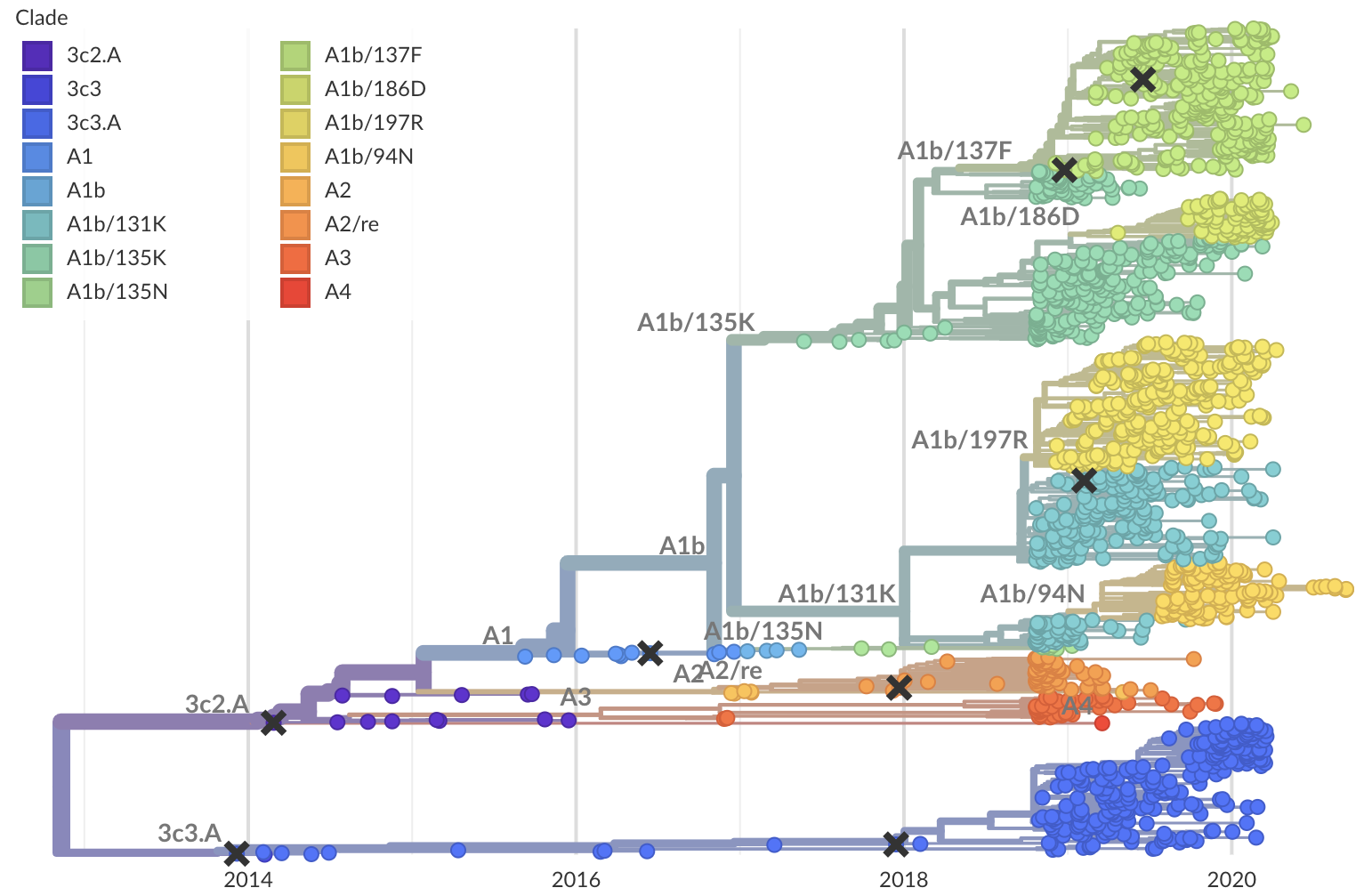
Operationalizing genomic epidemiology during the Nord-Kivu Ebola outbreak, Democratic Republic of the Congo
with Eddy Kinganda-Lusamaki, ![]() Allison Black, Daniel Mukadi,
Allison Black, Daniel Mukadi, ![]() James Hadfield, Placide Mbala-Kingebeni, Catherine Pratt, Jean-Jacques Muyembe Tamfum, Steve Ahuka-Mundeke, Michael Wiley, Martine Peeters, Amadou Alpha Sall, Eric Delaporte, Gustavo Palacios and many others
James Hadfield, Placide Mbala-Kingebeni, Catherine Pratt, Jean-Jacques Muyembe Tamfum, Steve Ahuka-Mundeke, Michael Wiley, Martine Peeters, Amadou Alpha Sall, Eric Delaporte, Gustavo Palacios and many others
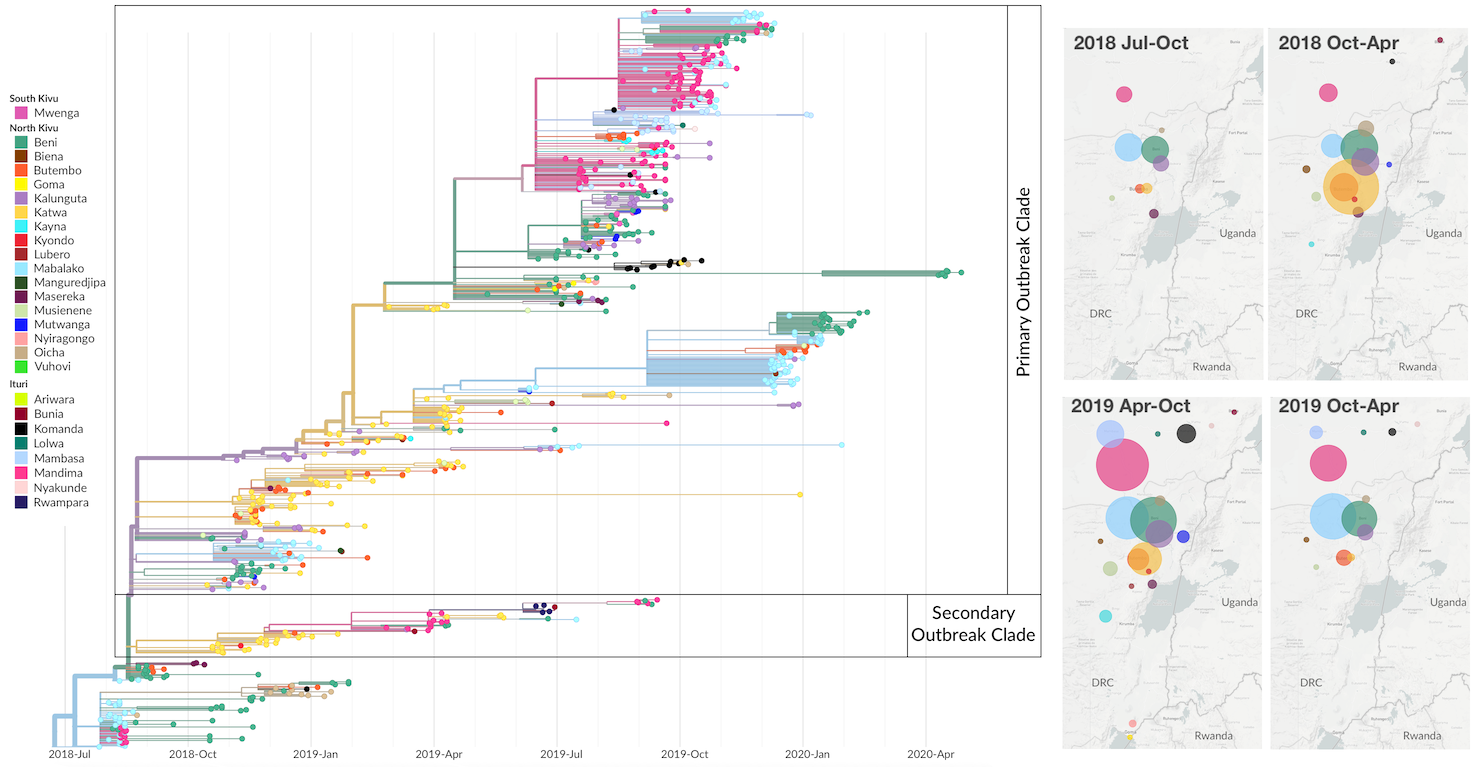
Genomic epidemiology used to track North Kivu outbreak
- Institut National de Recherche Biomédicale (INRB) sequenced samples in Kinshasa and Katwa
- Data and pipeline housed at github.com/inrb-drc/ebola-nord-kivu
- Resulting analyses deployed live to nextstrain.org/community/inrb-drc/ebola-nord-kivu
792 Ebola virus genomes sequenced generally within 4 weeks of sample collection, representing 24% of confirmed cases
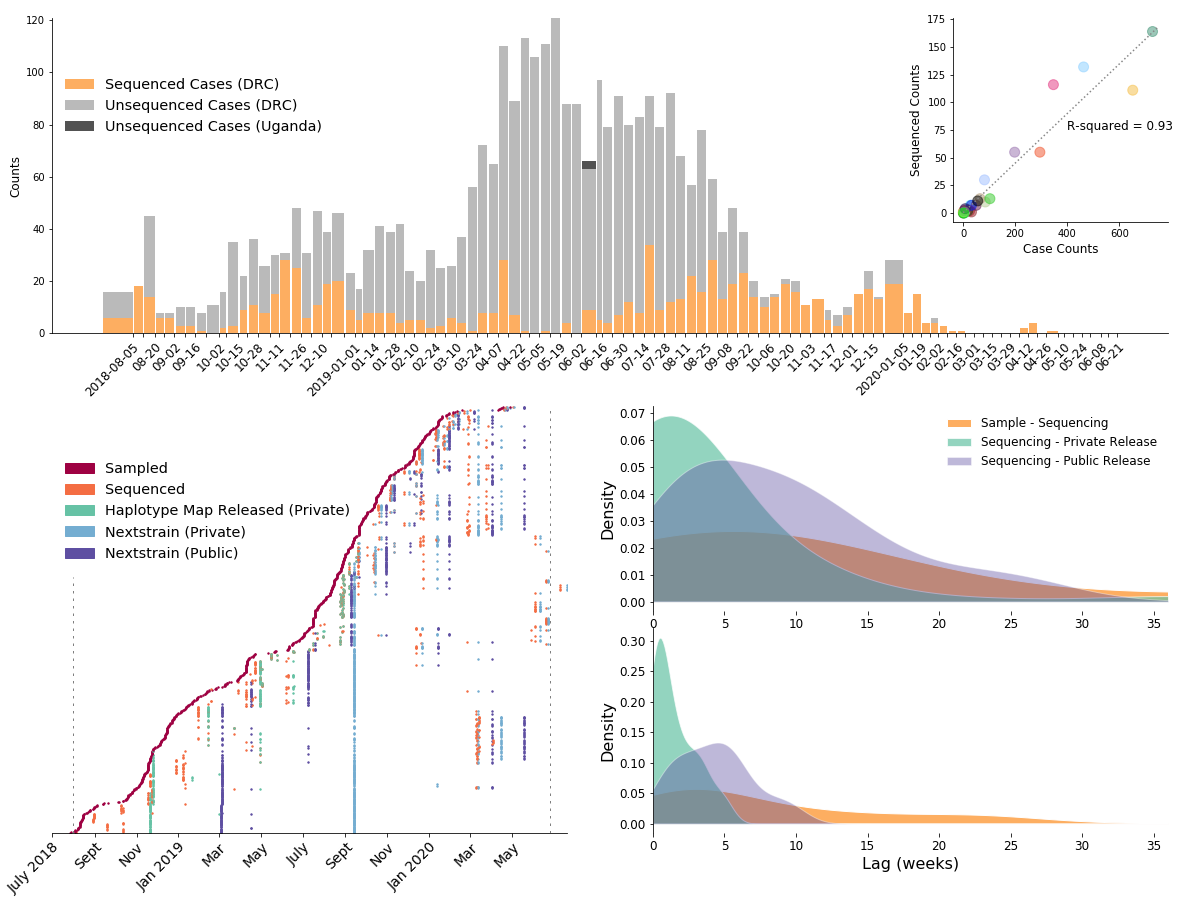
Outbreak sustained by locally transitory transmission chains
Situation reports deployed as Nextstrain Narratives
- Narratives are Markdown posts that allow pairing of narrative text to visualization state
- Situation reports describing new batches of sequenced cases shared as narratives and discussed at EOC meetings
- Example narrative for Ebola in the DRC here: nextstrain.org/narratives/inrb-ebola-example-sit-rep
- Focus on epidemiological inferences important for contact tracing
SARS-CoV-2
Detection and sequencing of SARS-CoV-2 in January
Jan 11: First five genomes showed that the outbreak was caused by a novel SARS-like coronavirus
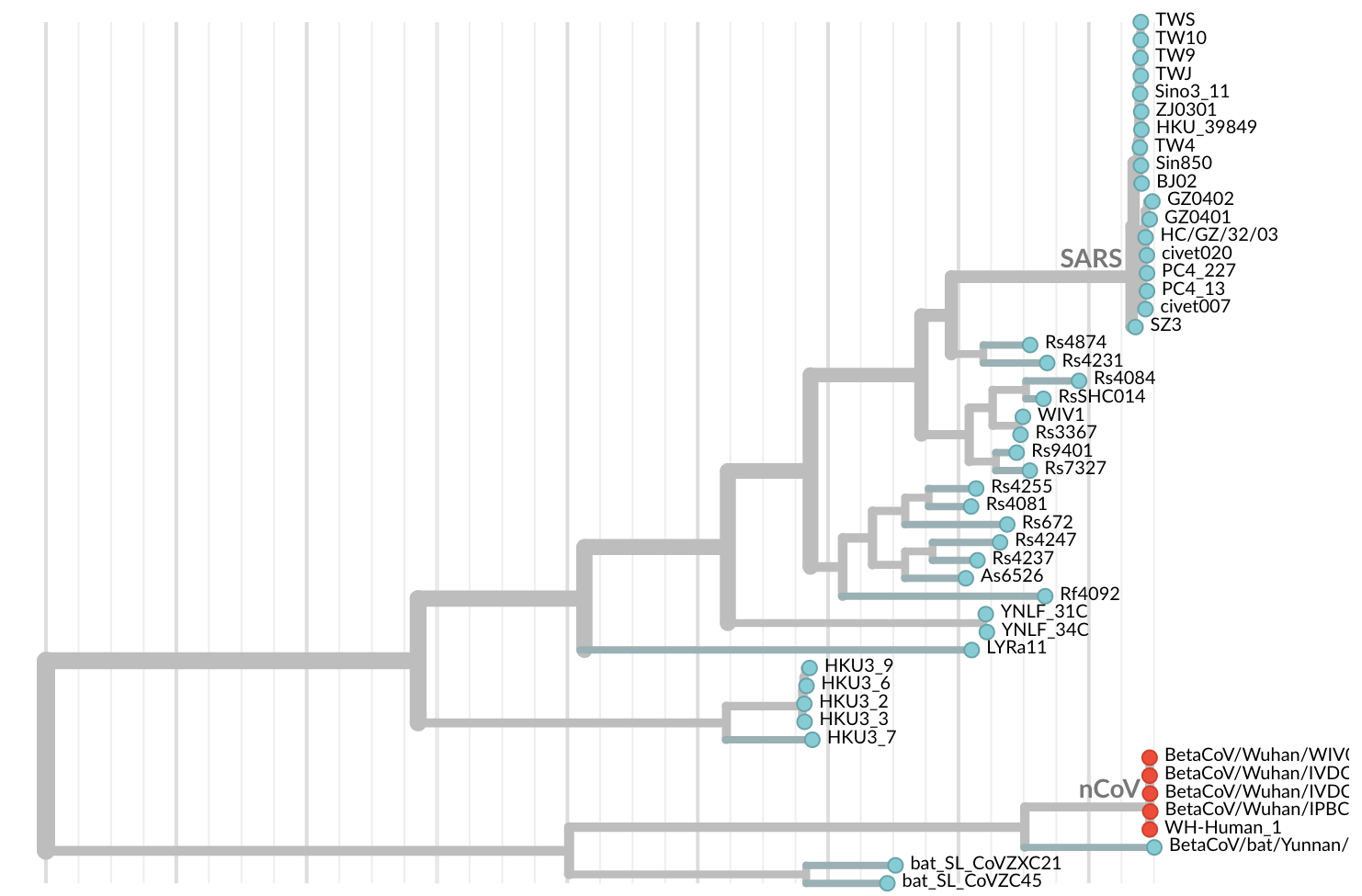
Jan 19: First 12 genomes from Wuhan and Bangkok lack genetic diversity
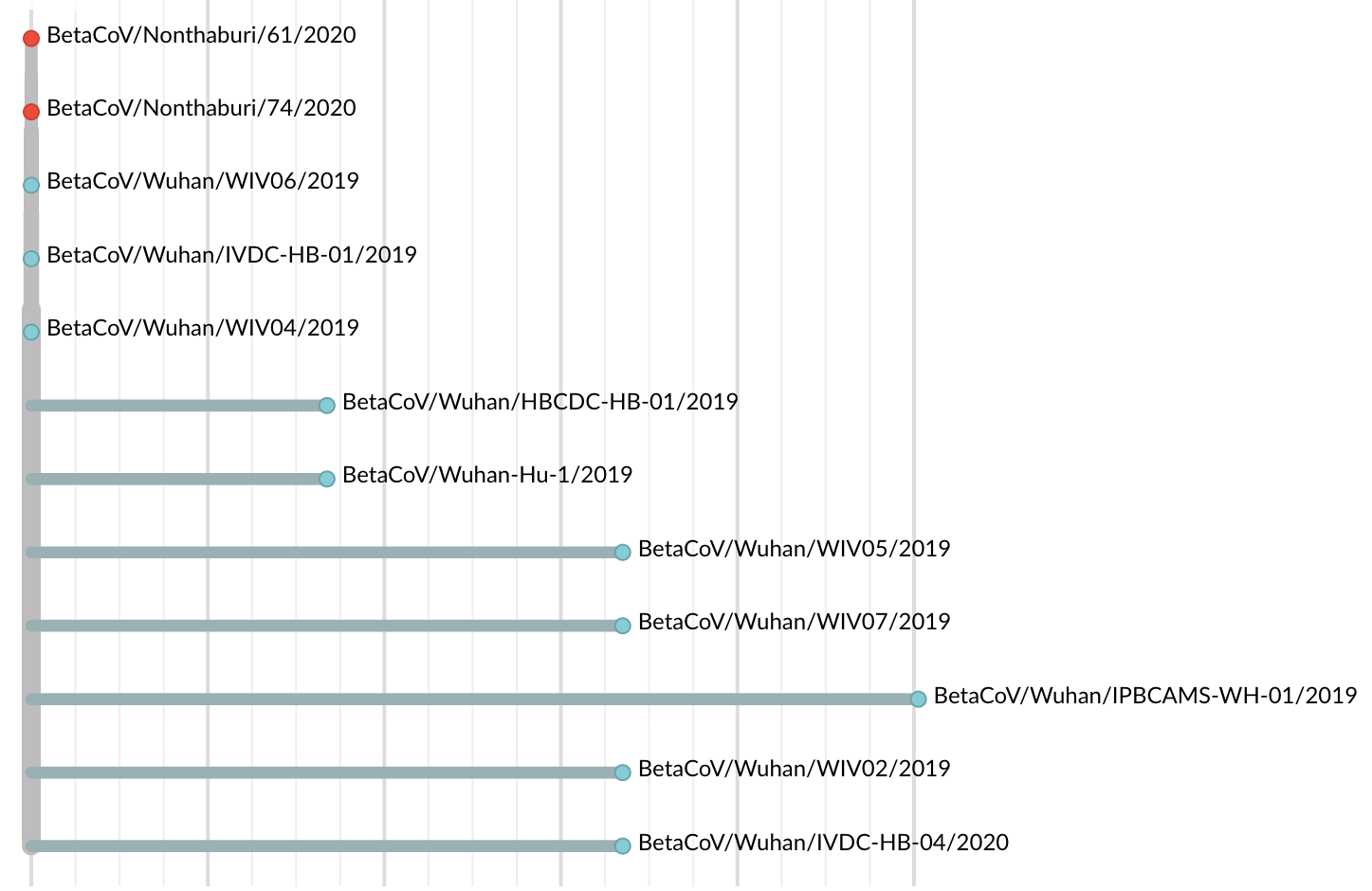
Single introduction into the human population between Nov 15 and Dec 15 and human-to-human epidemic spread from this point forward
😧
Spent the week of Jan 20 alerting public health officials, and since then have aimed to keep nextstrain.org up-to-date
Nextstrain
Project to conduct real-time genomic epidemiology and evolutionary analysis of emerging epidemics
with
![]() Richard Neher,
Richard Neher,
![]() James Hadfield,
James Hadfield,
![]() Emma Hodcroft,
Emma Hodcroft,
![]() Thomas Sibley,
Thomas Sibley,
![]() John Huddleston,
John Huddleston,
![]() Louise Moncla,
Louise Moncla,
![]() Cassia Wagner,
Cassia Wagner,
![]() Ivan Aksamentov,
Ivan Aksamentov,
![]() Moira Zuber,
Moira Zuber,
![]() Eli Harkins,
Eli Harkins,
![]() Misja Ilcisin,
Misja Ilcisin,
![]() Kairsten Fay,
Kairsten Fay,
![]() Jover Lee,
Jover Lee,
![]() Allison Black,
Allison Black,
![]() Miguel Paredes,
Miguel Paredes,
![]() Sidney Bell,
Sidney Bell,
![]() Colin Megill
Colin Megill
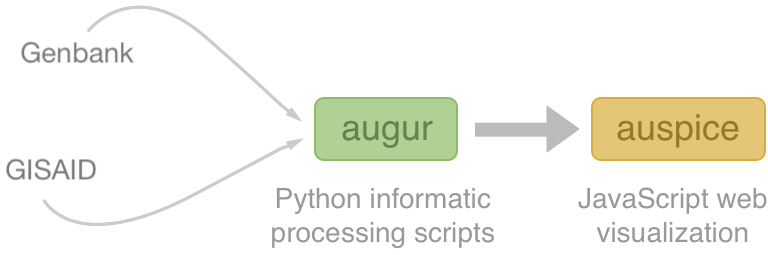
Nextstrain architecture
All code open source at github.com/nextstrain
Two central aims: (1) rapid and flexible phylodynamic analysis and
(2) interactive visualization
Rapid build pipeline for 3000 SARS-CoV-2 genomes (timings are for a laptop)
- Align with MAFFT (~20 min)
- Build ML tree with IQTREE (~40 min)
- Temporally resolve tree and geographic ancestry with TreeTime (~50 min)
- Total pipeline (~2 hr)
Current data flow for SARS-CoV-2
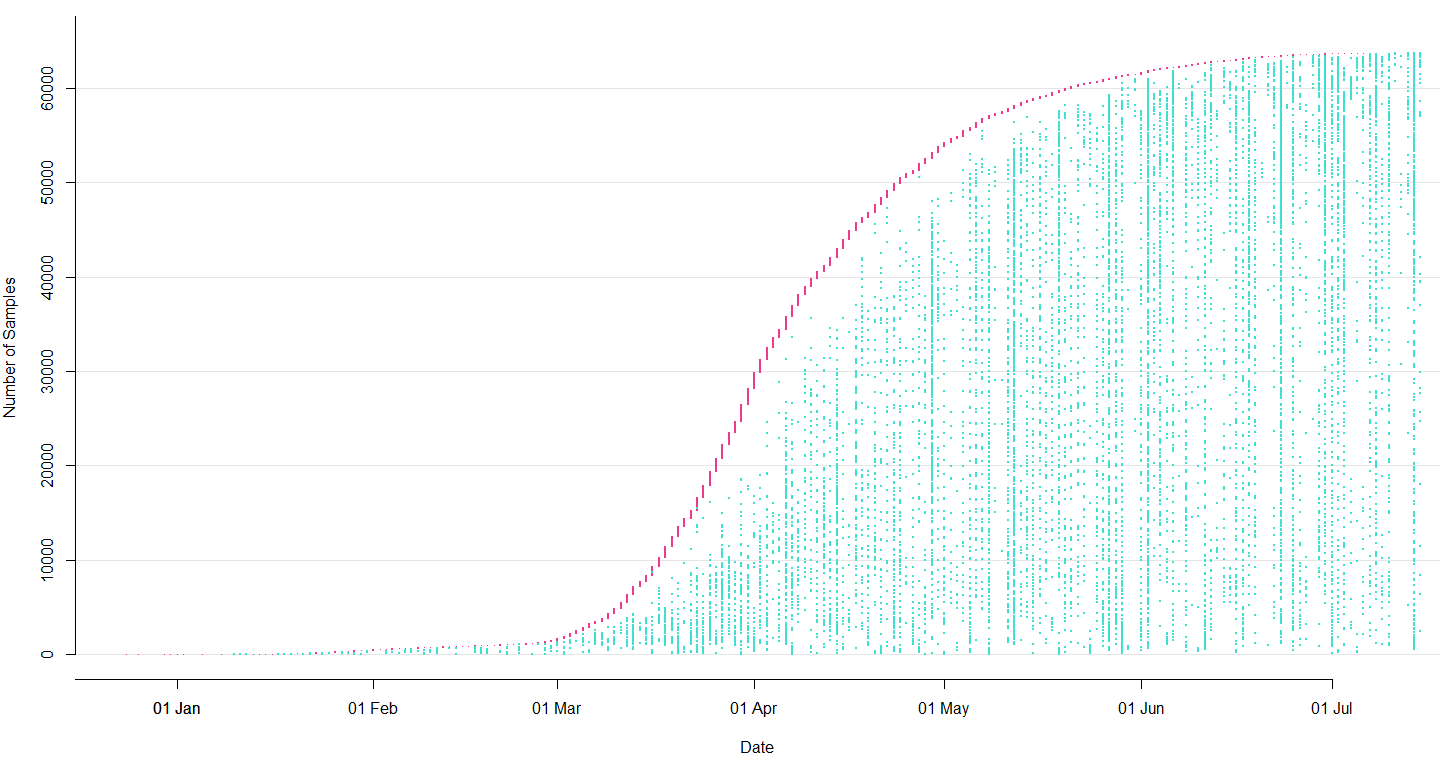
- Labs contribute directly to GISAID (now have >150k full genomes)
- Nextstrain pulls a complete dataset from GISAID every 24 hours
- This triggers an automatic rebuild on Amazon Web Services
- We manually update new lat/longs, etc...
- We push this build online to nextstrain.org and tweet the update from @nextstrain
Sequencing and data sharing in almost real-time
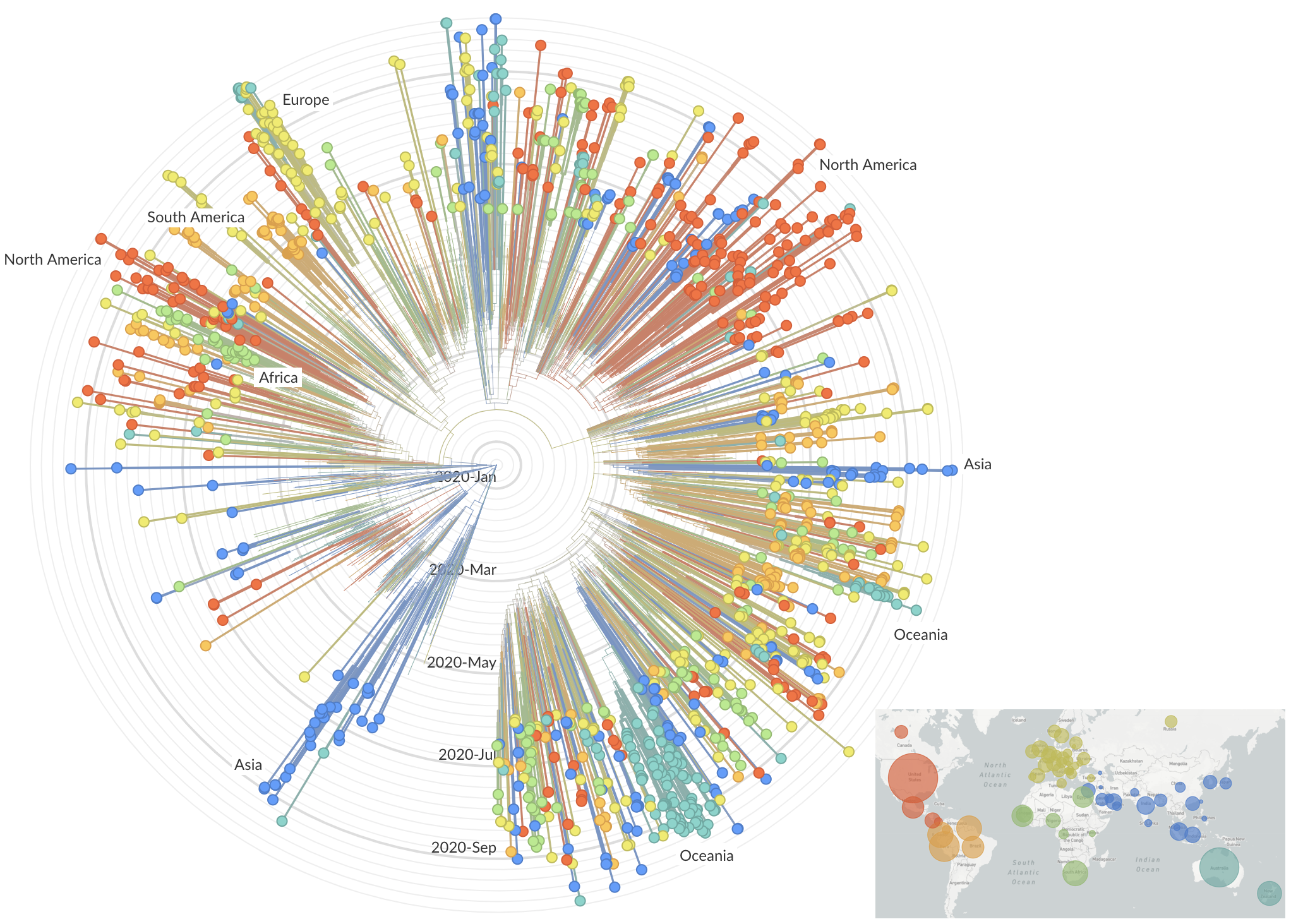
Dec/Jan: Emergence from Wuhan in ~Nov 2019
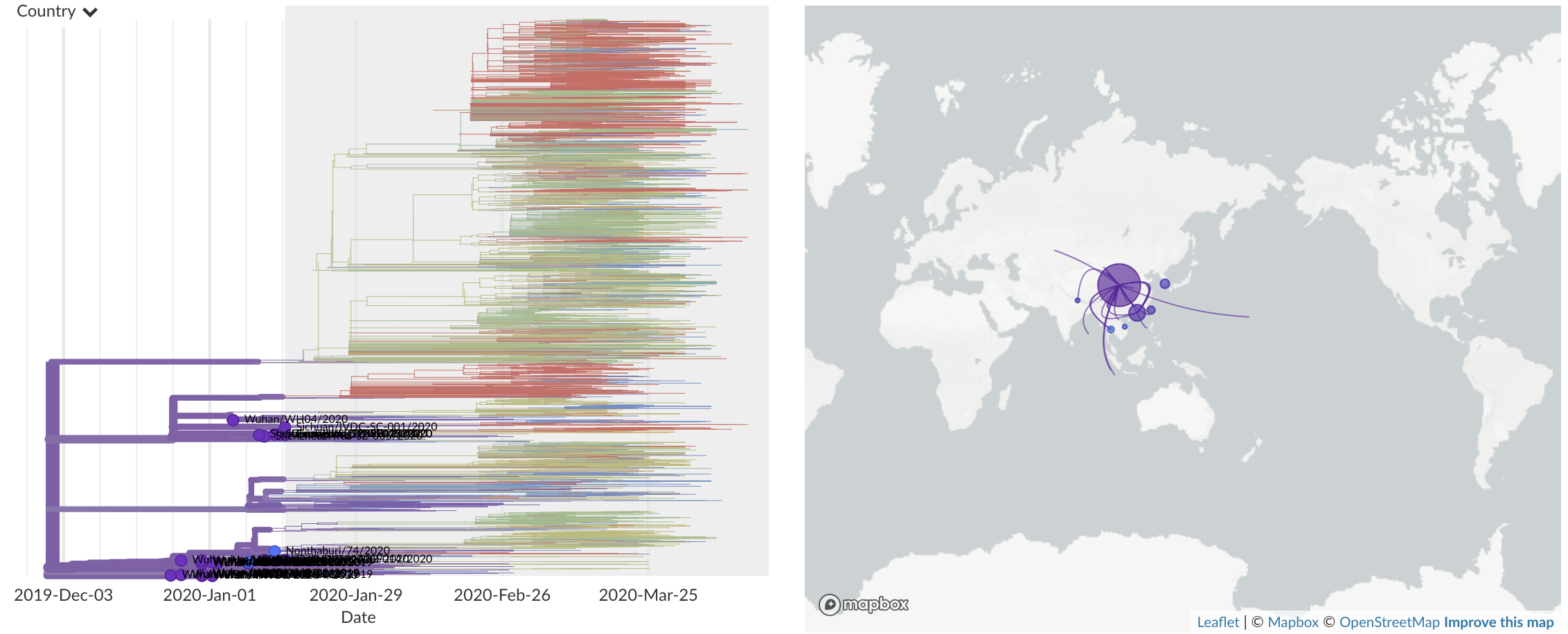
Jan/Feb: Spread within China and seeding elsewhere
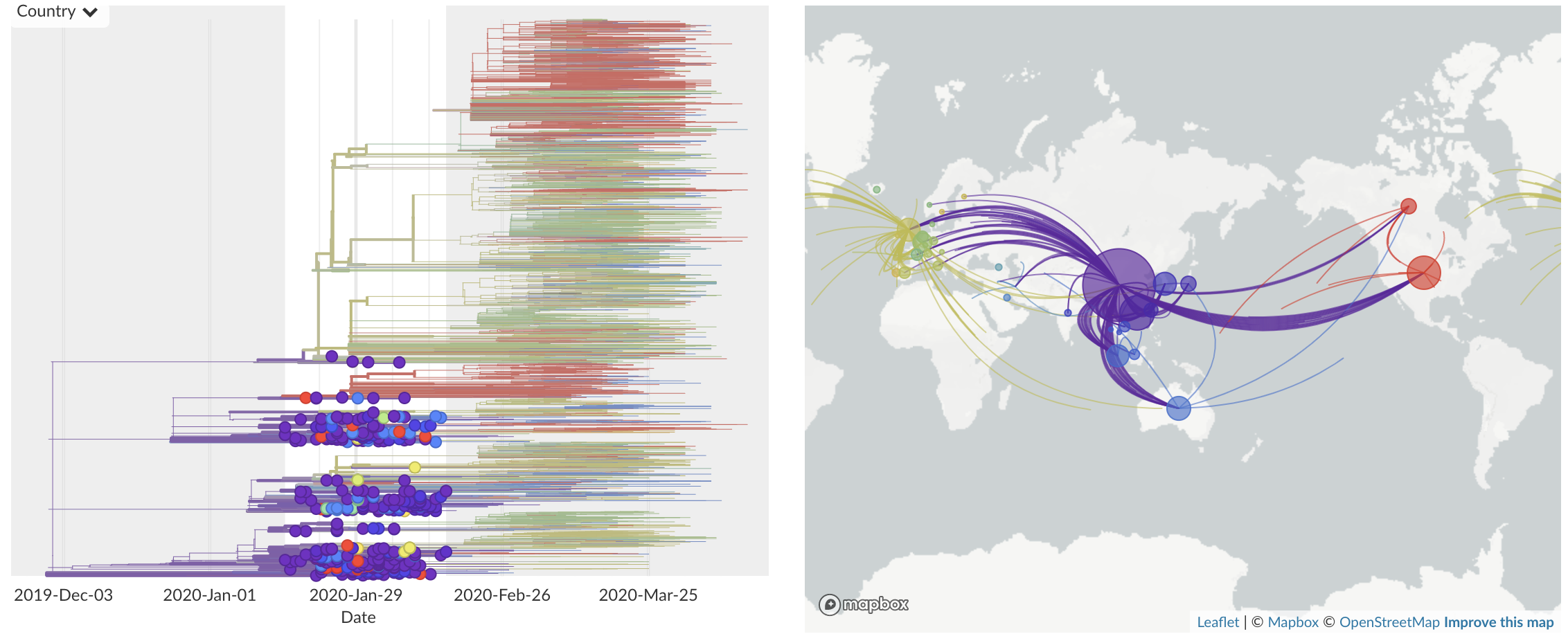
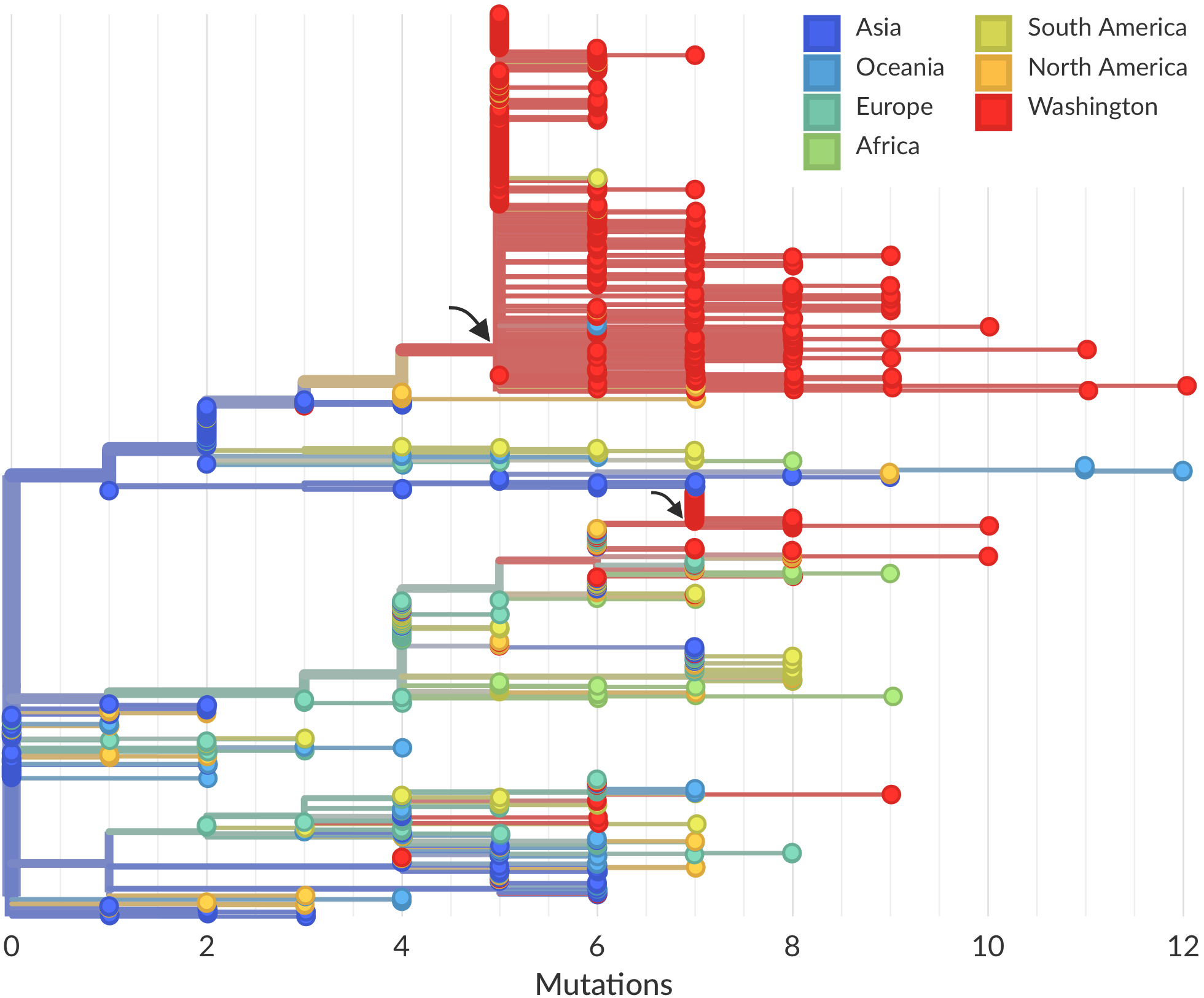
Feb/Mar: Epidemic spread within North America and Europe
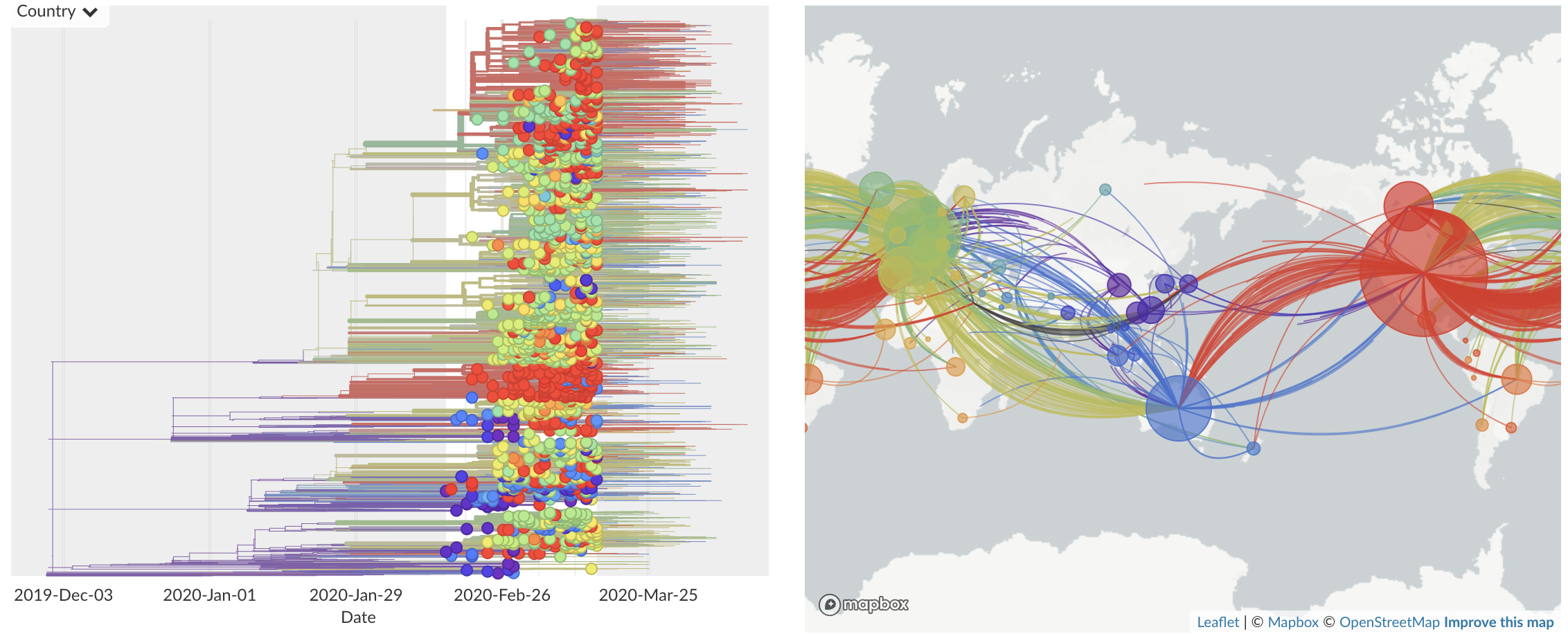
Mar/Apr: Decreasing transmission with social distancing
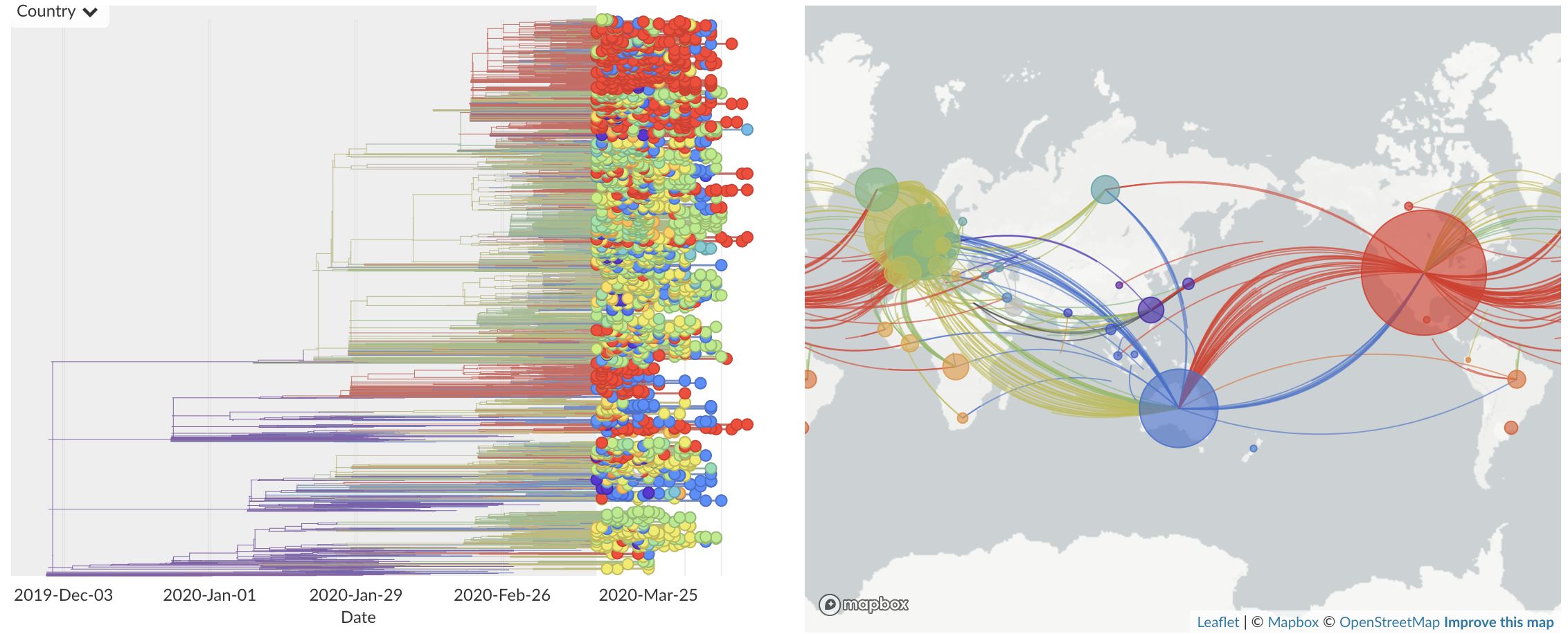
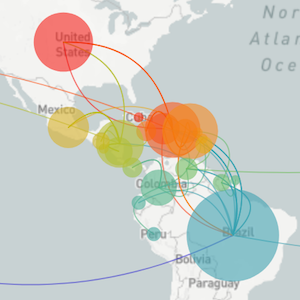
Epidemic in the USA was introduced from China in late Jan and from Europe during Feb
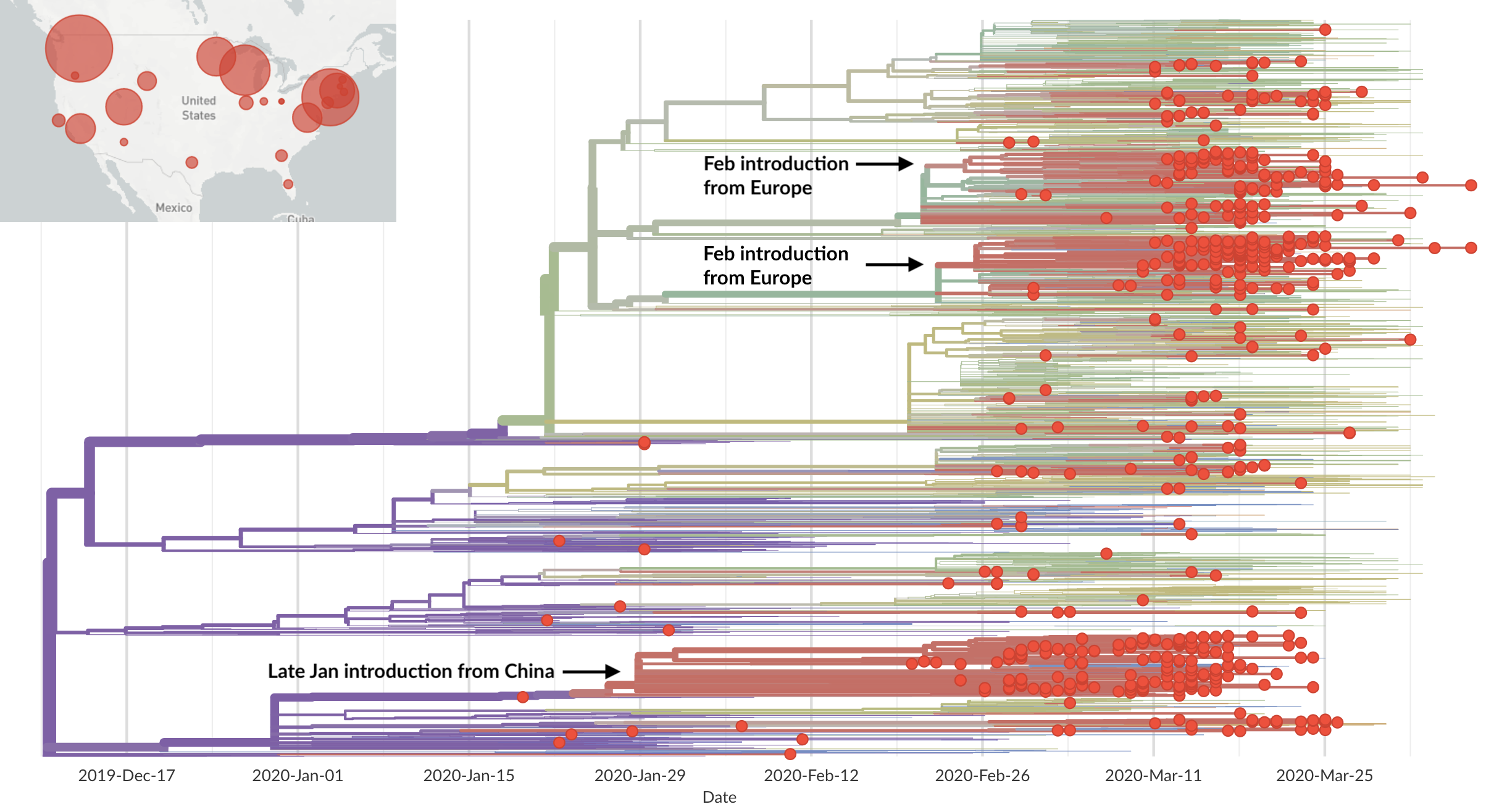
Once in the US, virus spread rapidly
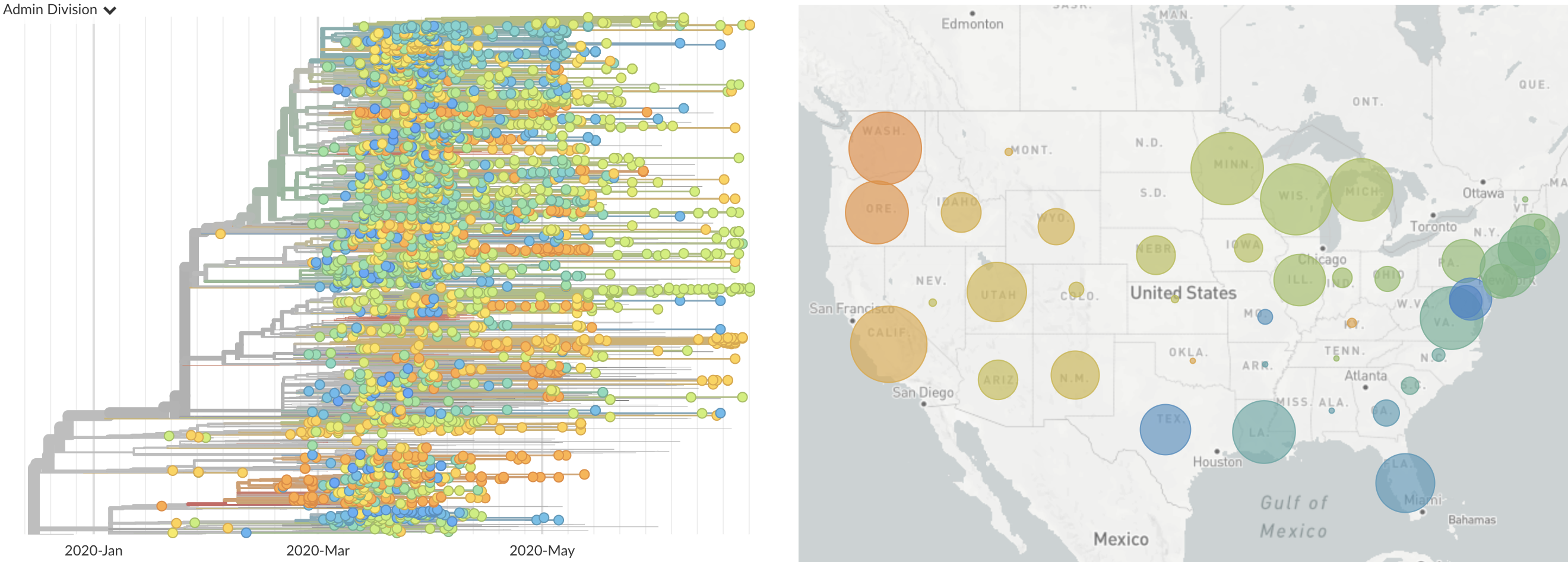
Single introduction at the beginning of Feb quickly shows up throughout the country
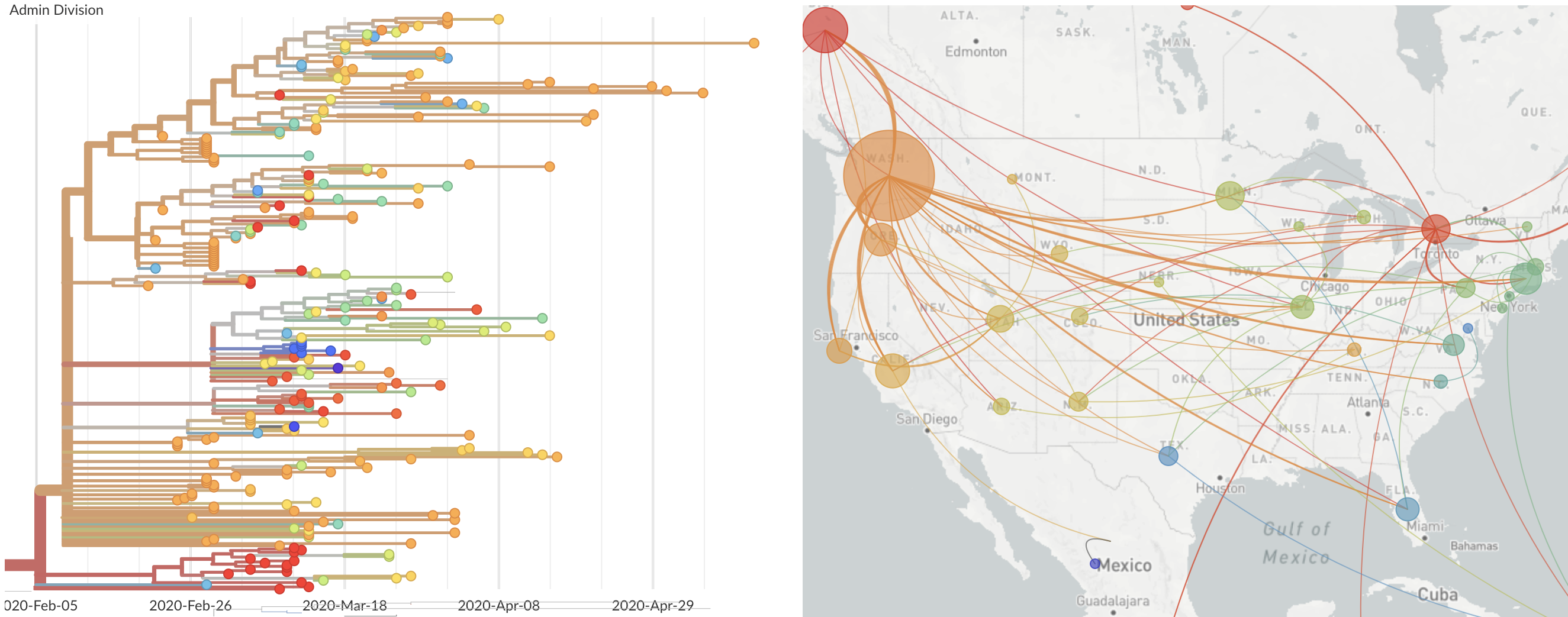
and decreased international travel,
regional clades have emerged
and decreased international travel,
regional clades have emerged
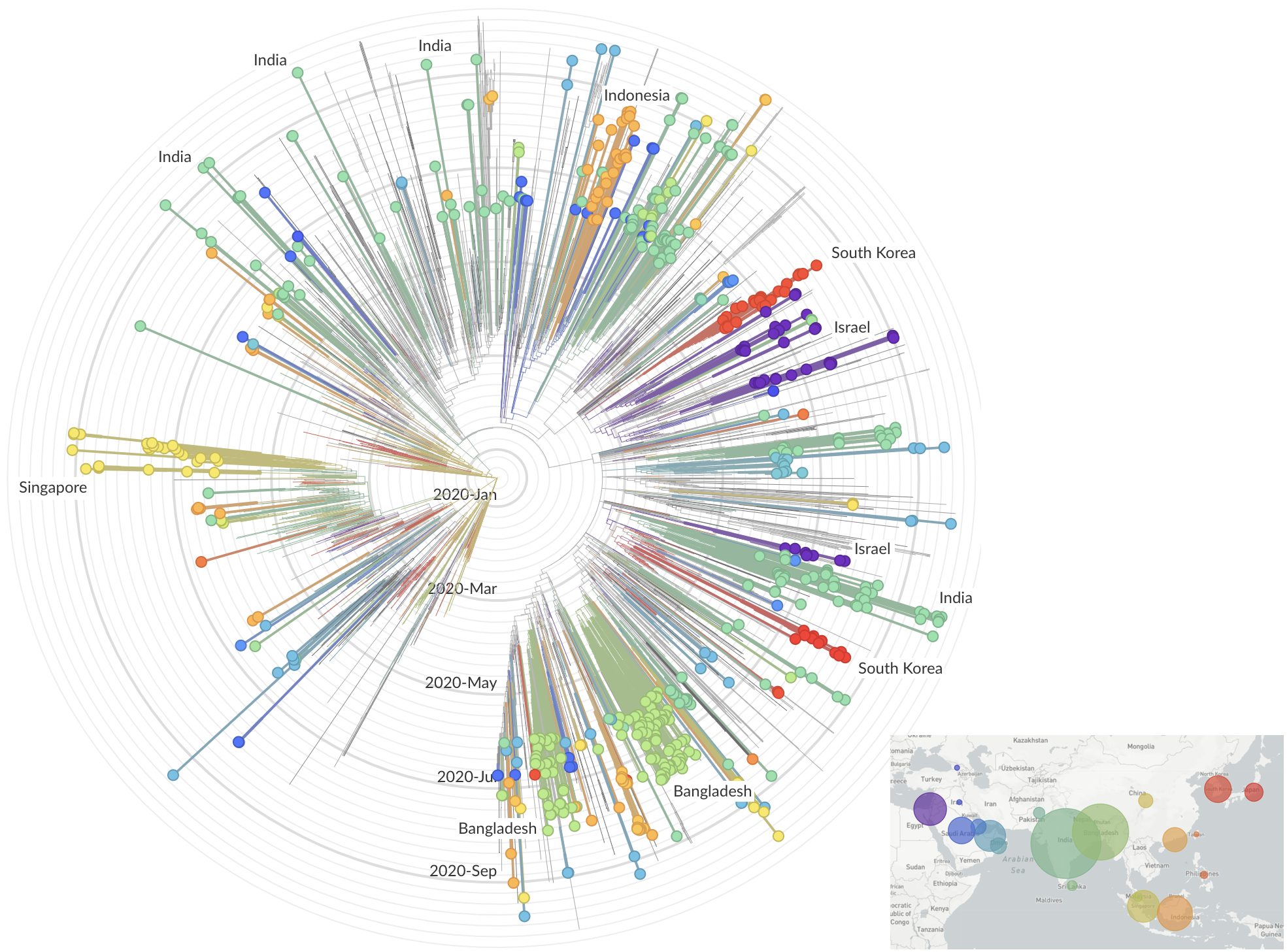
Sequencing immediately useful for epidemiological understanding, but selection and functional impacts should also be studied
Significant interest in spike mutation D614G
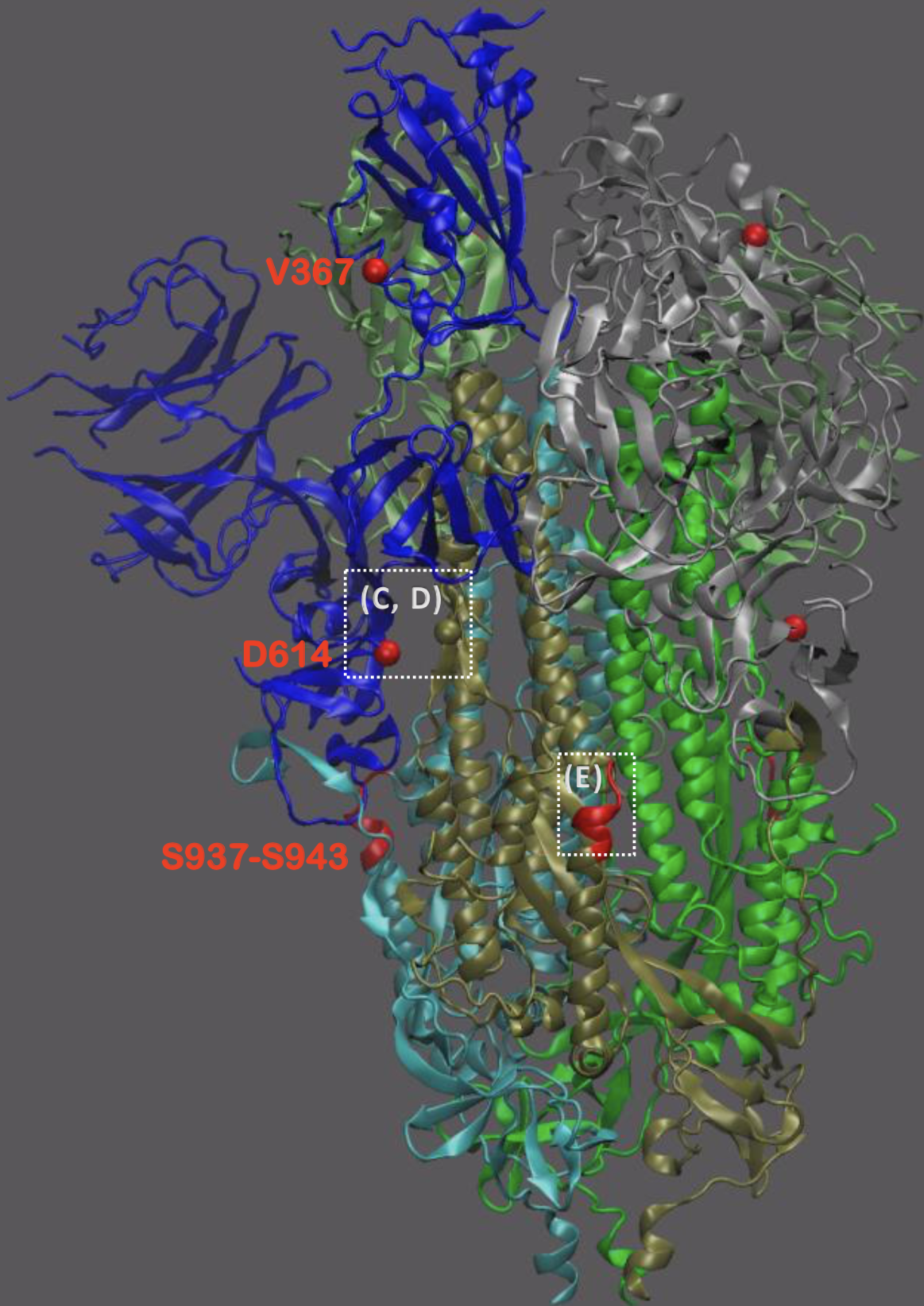
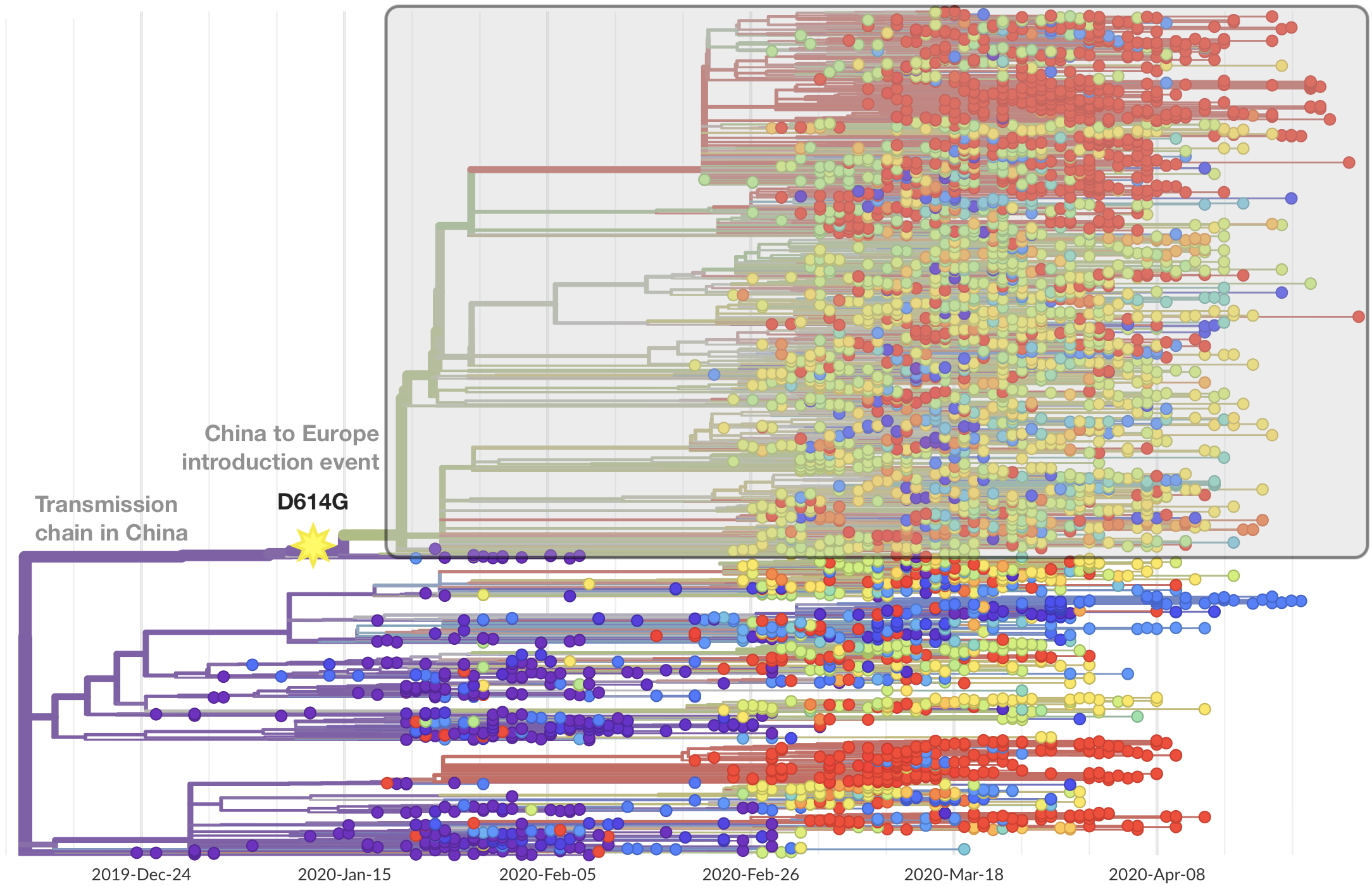
This mutation occurred in the initial European introduction
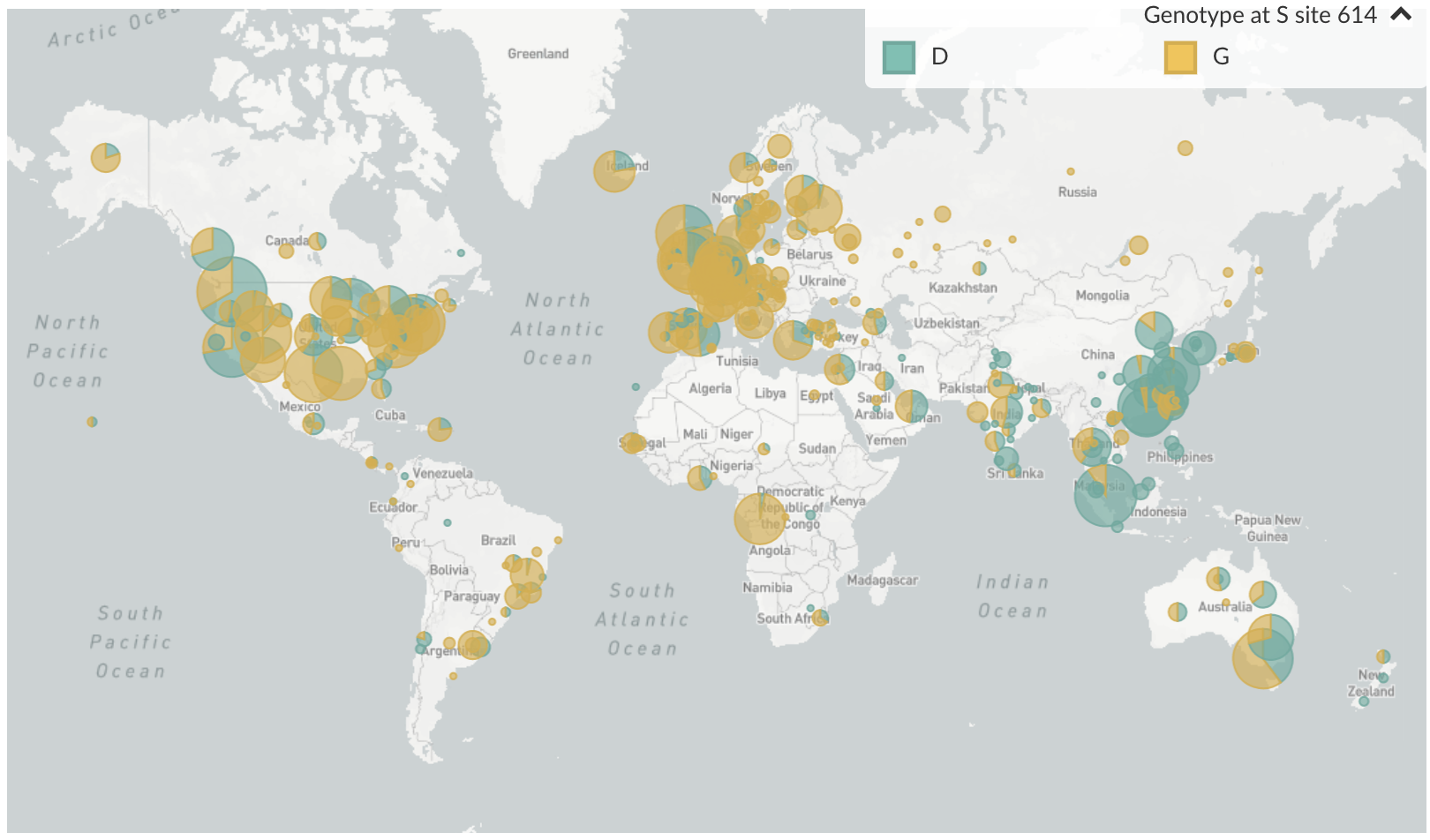
D614G is prevalent throughout Europe and mixed in US and Australia
D614G is increasing in frequency across states in US and Australia
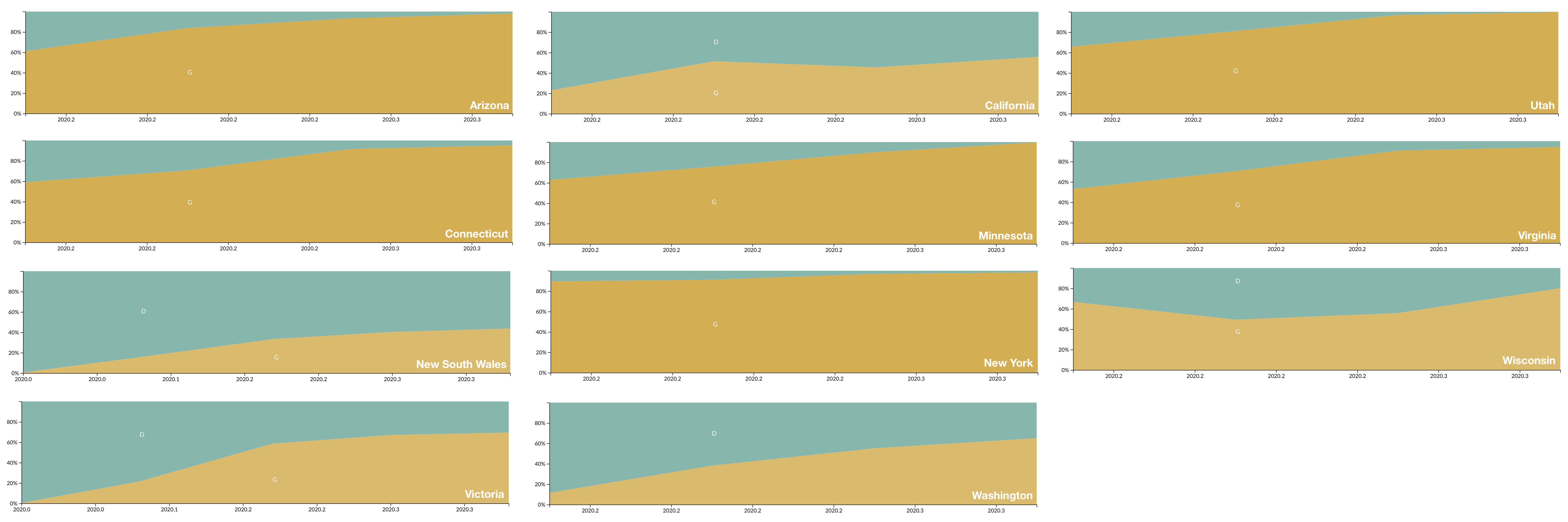
D614G is increasing in frequency across states in US and Australia
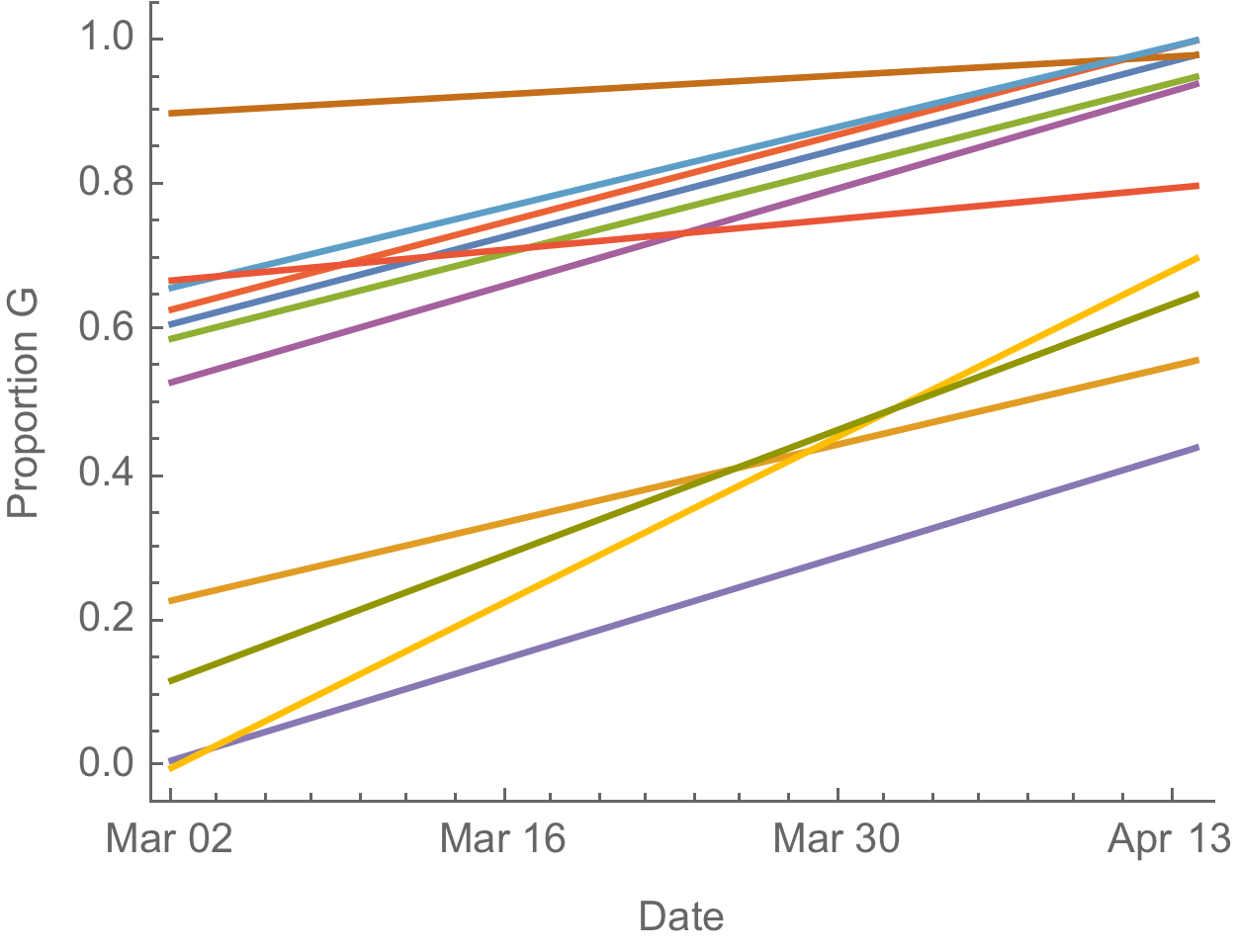
The success of D614G can be explained by either:
- D614G is more transmissible and has higher $R_0$
- founder effects and epidemiological confounding
Additional evidence from Ct values of clinical specimens
Sheffield, UK Seattle, USA
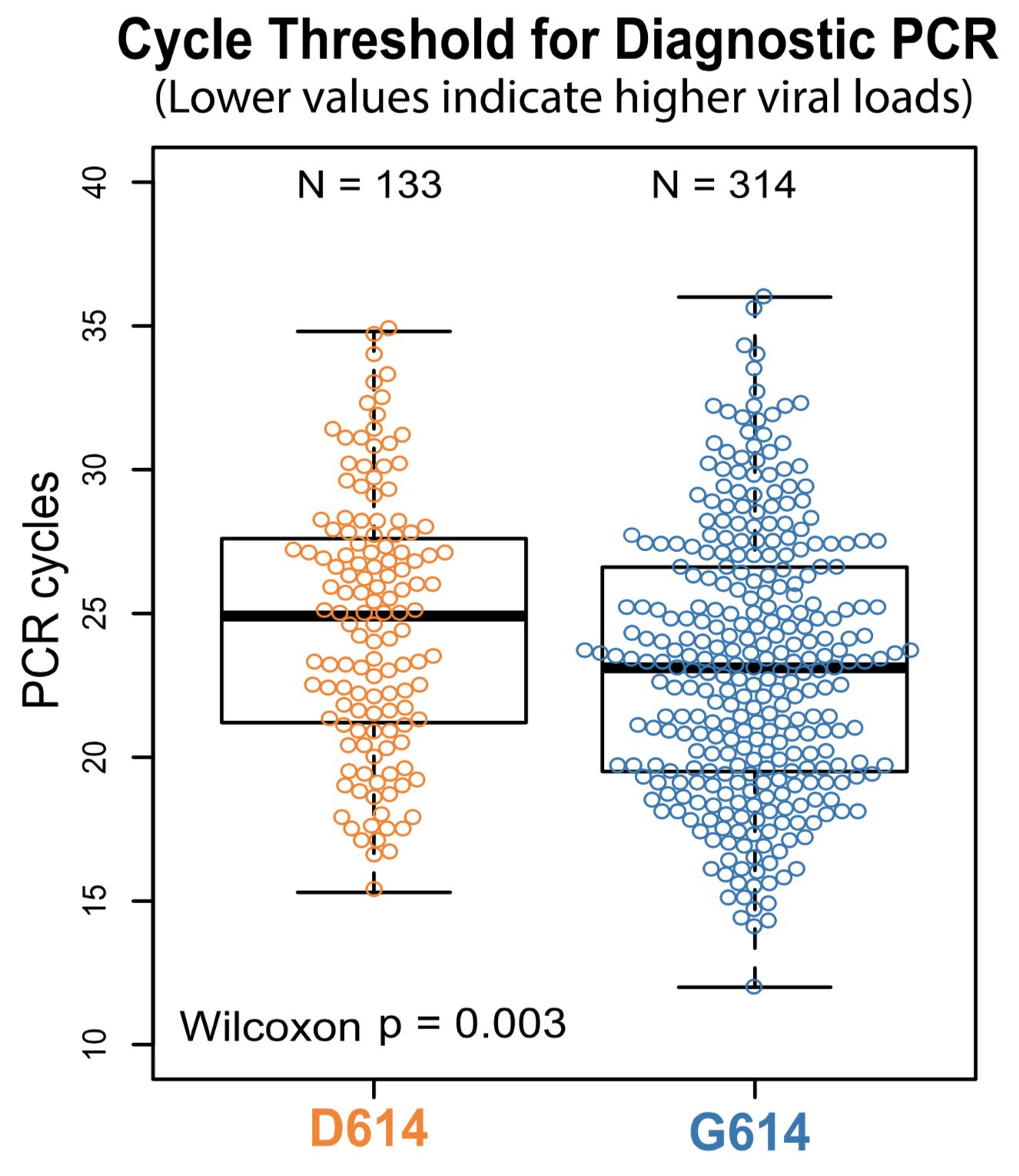
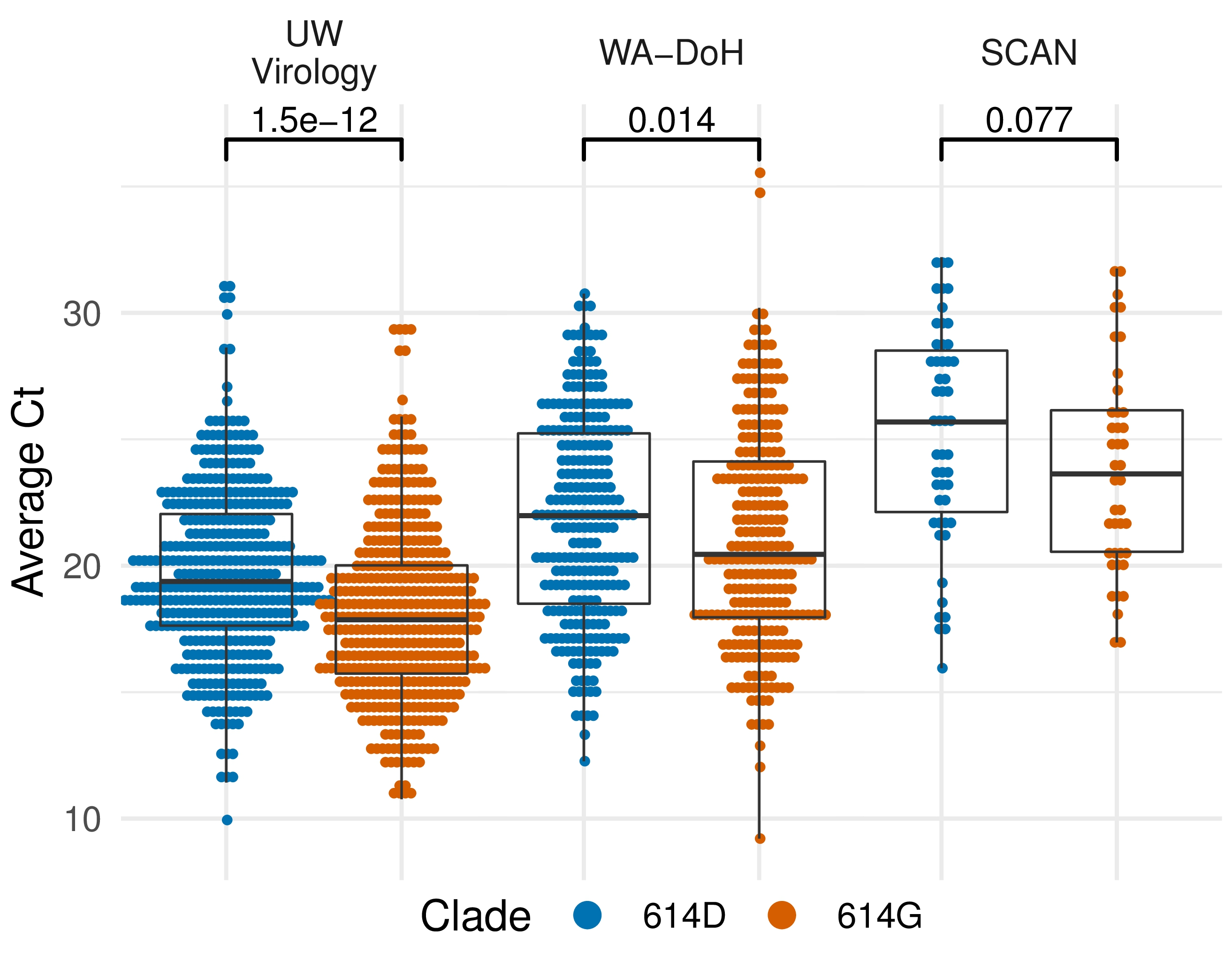
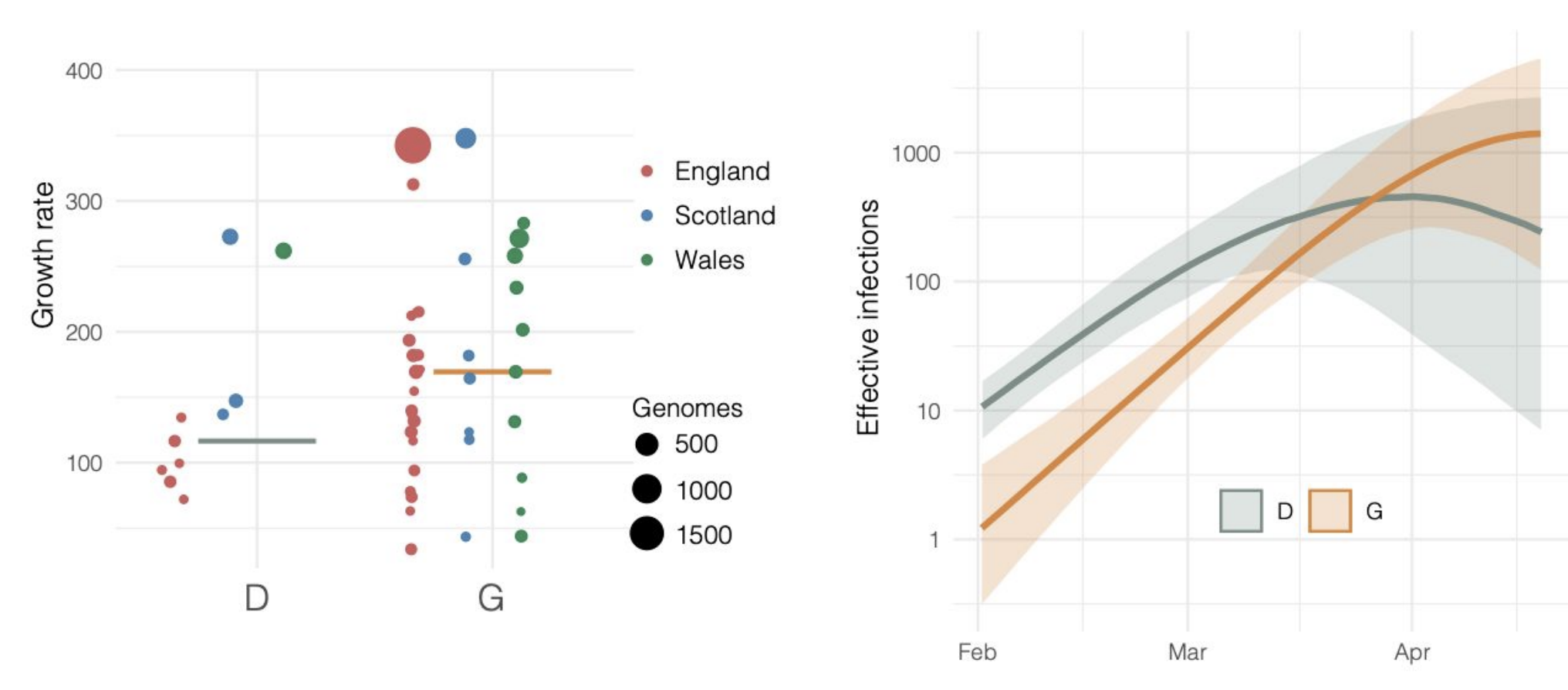
Repeated introductions to the UK suggest transmission advantage of D614G
Advancing genomic epidemiology
- Better methods for large datasets
- Distinguishing endogenous spread from importations
- Tying genomic epidemiology together with richer epi data to better understand local transmission
- Incorporating within-host variation to improve phylogenetic resolution
- Integrating clinical data to look for mutations that impact clinical outcomes
Surveillance is critically important to our ability to combat epidemics and pandemics
 Seattle Flu Study
Seattle Flu Study
Lead investigators: Helen Chu, Michael Boeckh, Janet Englund, Michael Famulare, Barry Lutz, Deborah Nickerson, Mark Rieder, Lea Starita, Matthew Thompson, Jay Shendure, Trevor Bedford
Co-investigators: Amanda Adler, Elisabeth Brandstetter, Roy Burstein, Shari Cho, Anne Emanuels, Kairsten Fay, Chris Frazar, Rachel Geyer, Peter Han, James Hadfield, Jessica Heimonen, Misja Ilcisin, Michael Jackson, Anahita Kiavand, Ashley Kim, Louise Kimball, Jack Henry Kotnik, Kirsten Lacombe, Jover Lee, Jennifer Logue, Victoria Lyon, Denise McCulloch, Jessica O’Hanlon, Matthew Richardson, Julia Rogers, Thomas Sibley, Monica Zigman Suchsland, Melissa Truong, Caitlin Wolf, Weizhi Zhong




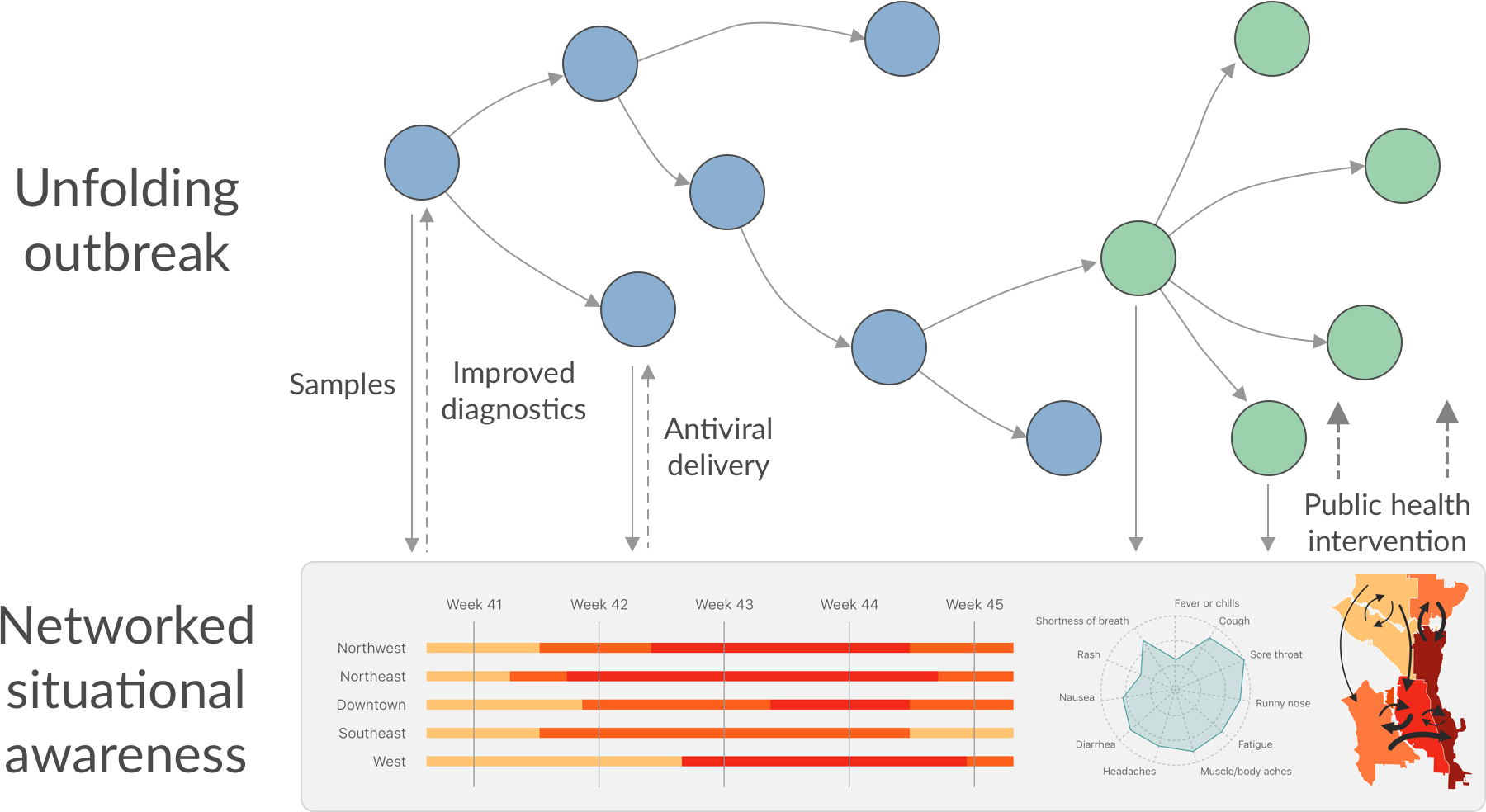
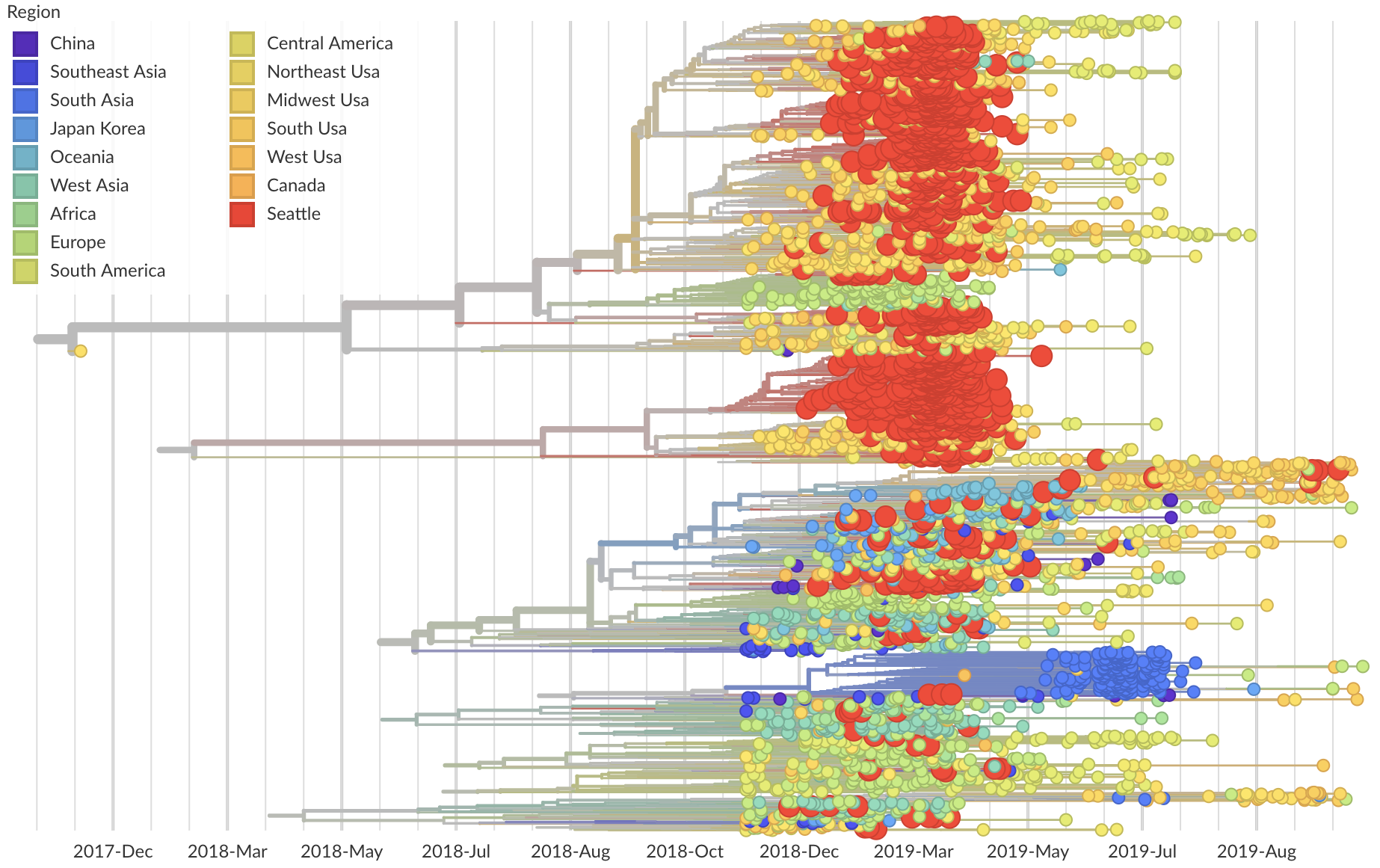
Phylogeny of 1150 Seattle H3N2 viruses
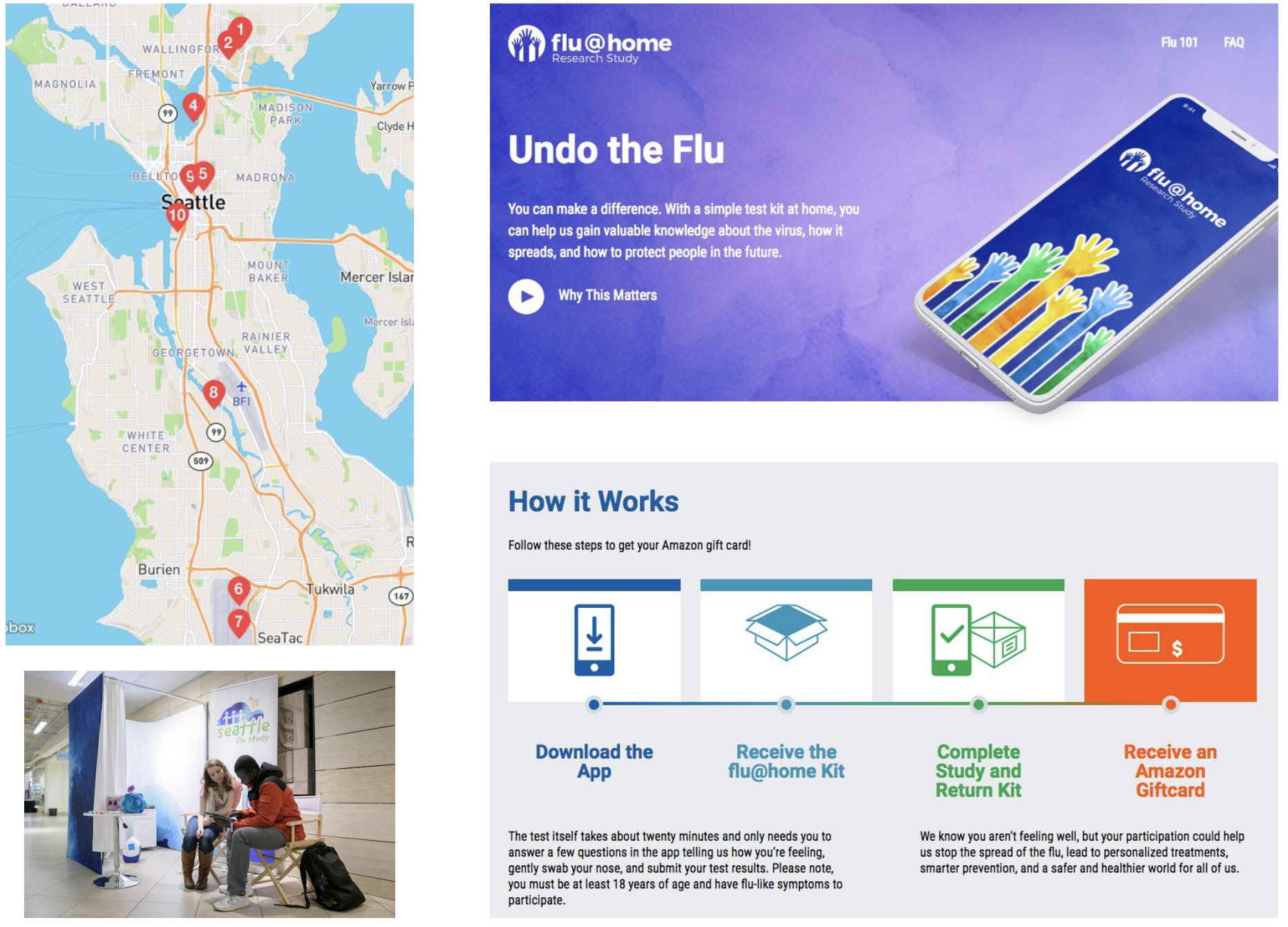
Screening of acute respiratory infections for SARS-CoV-2
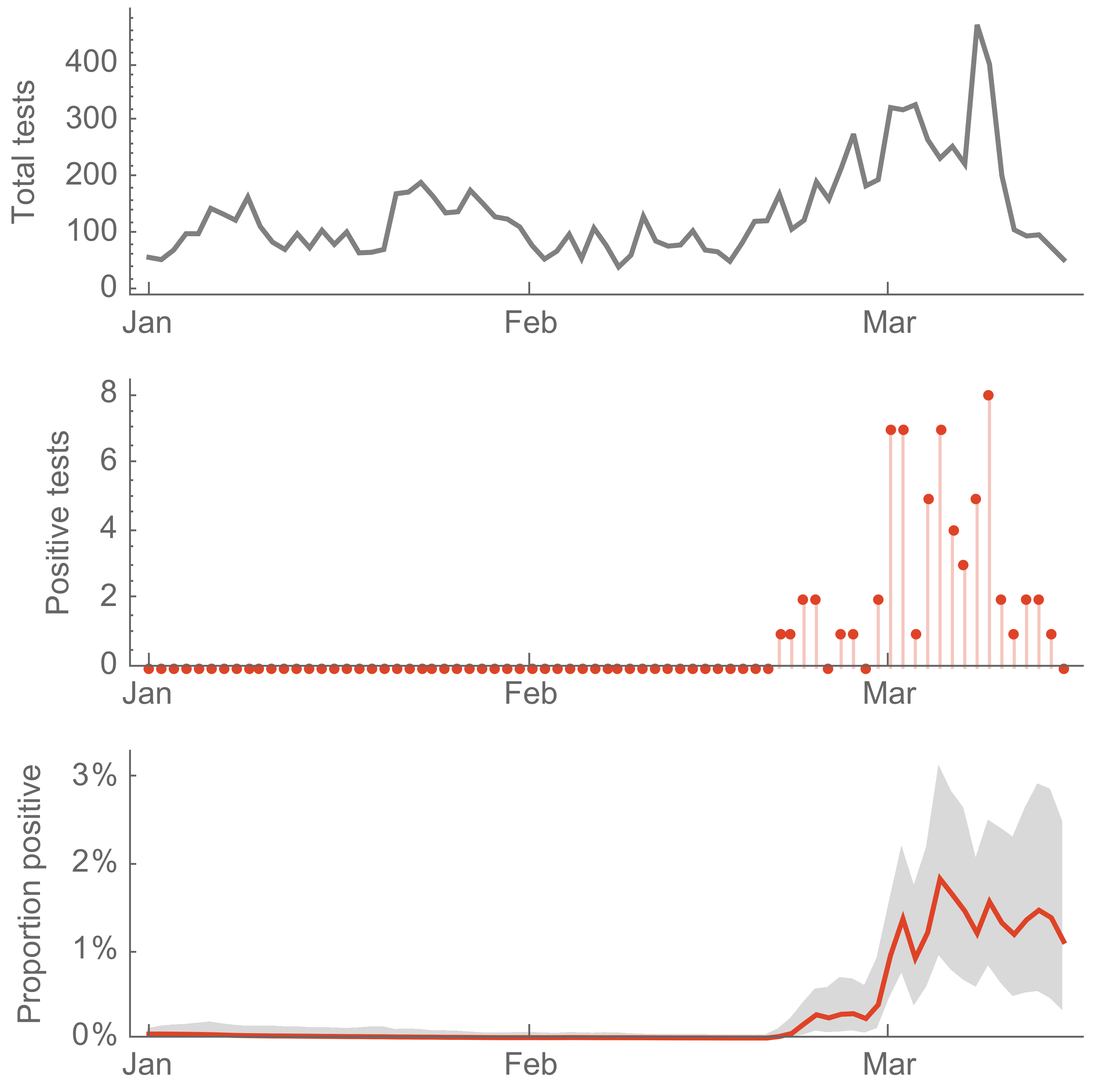
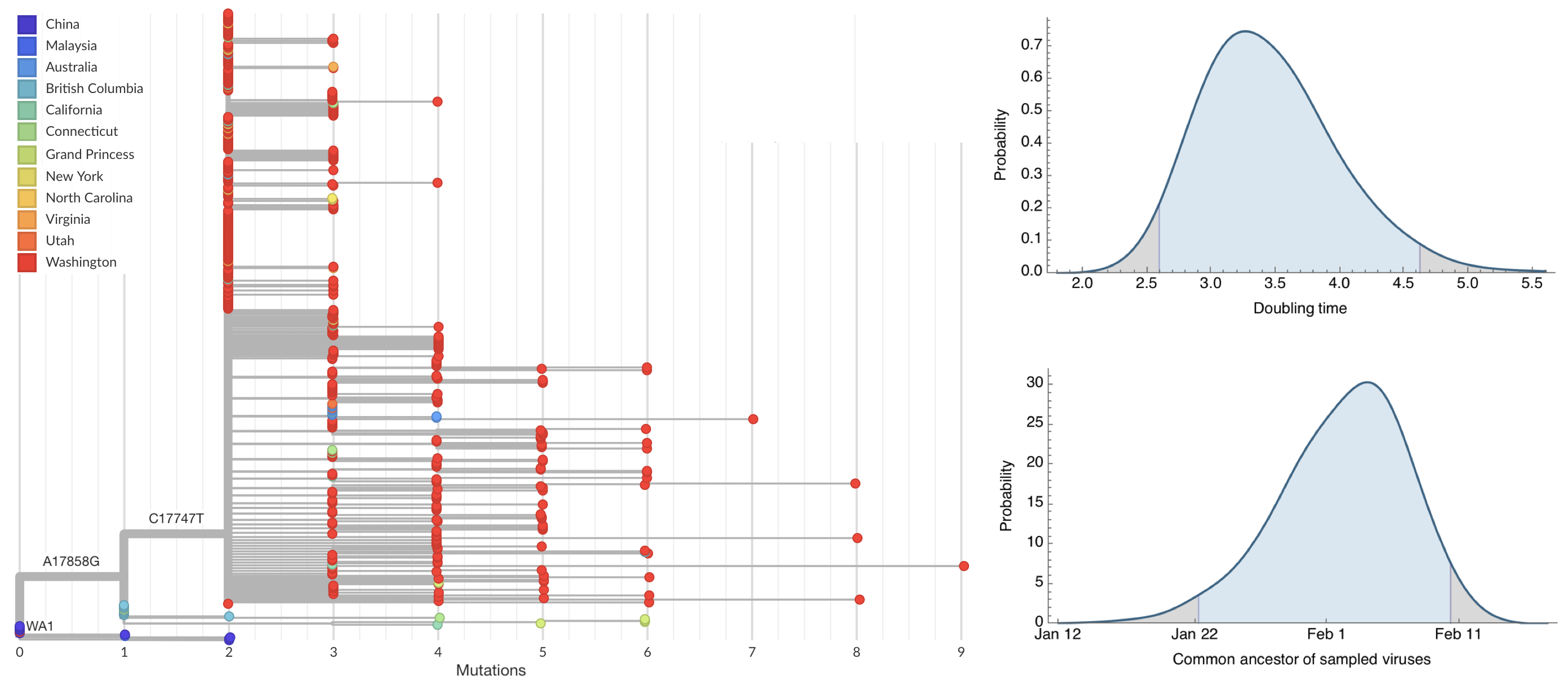
Sequencing of viruses collected prior to March 15 detects origins and rate of local spread
Sequencing of viruses collected prior to March 15 detects origins and rate of local spread



Prevalence estimates and supplemental testing
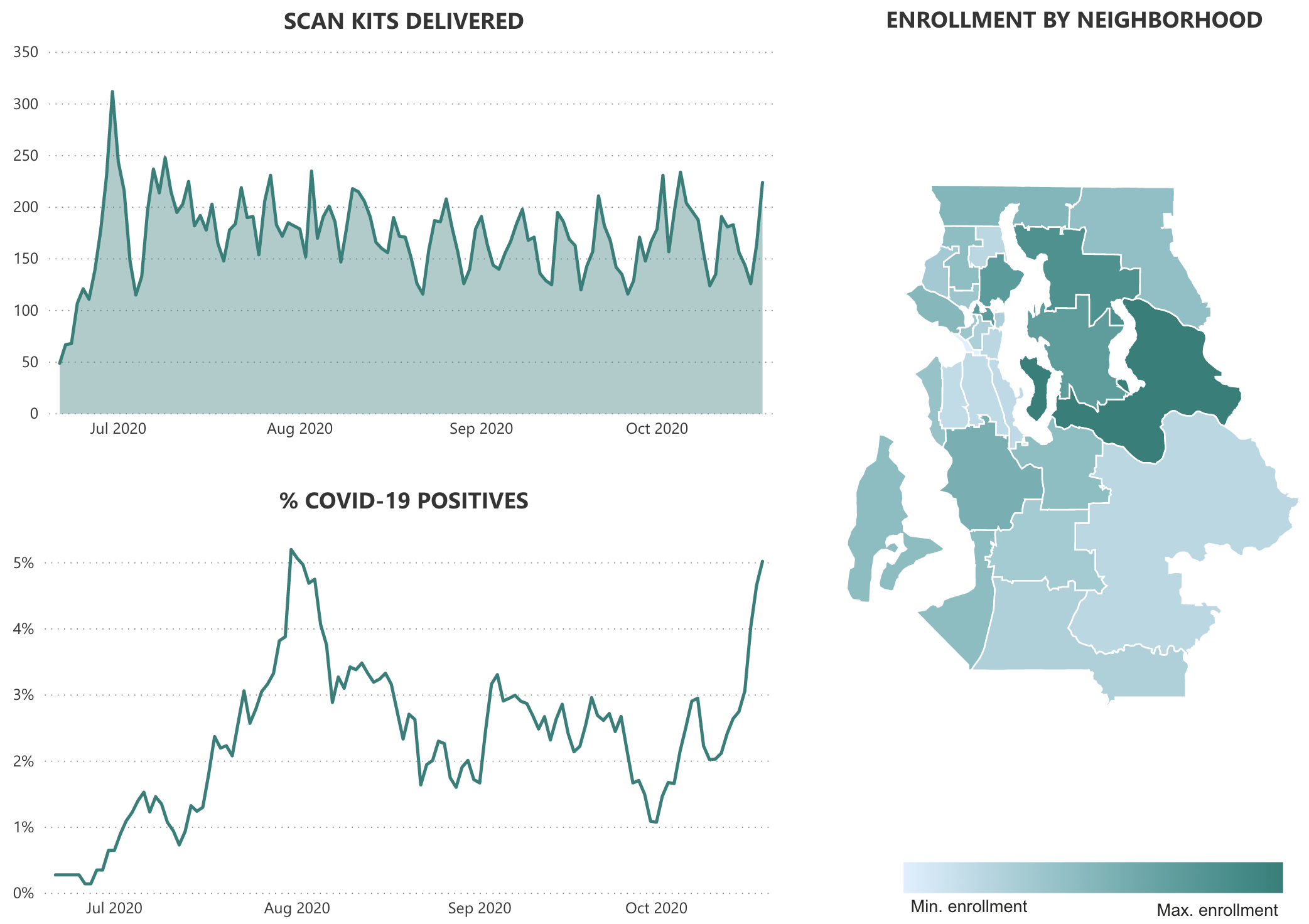
Continued SFS activities
- SCAN platform for distributed testing
- Improvements to cost and throughput of SARS-CoV-2 testing
- Testing for mitigation with Husky Coronavirus Testing
- Continued investigation of circulation patterns across common respiratory pathogens
- Further integration of genome sequencing for epidemiological insights
Circulation of everything except COVID-19 and rhinovirus basically disappeared
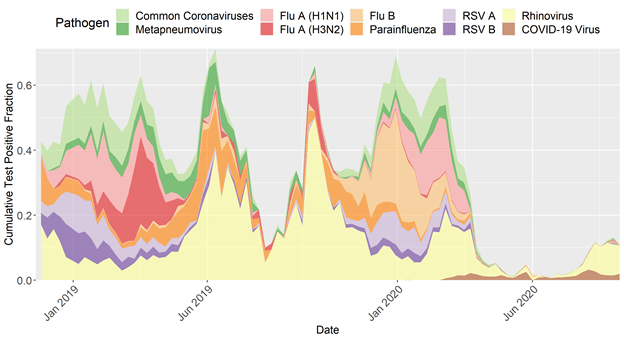
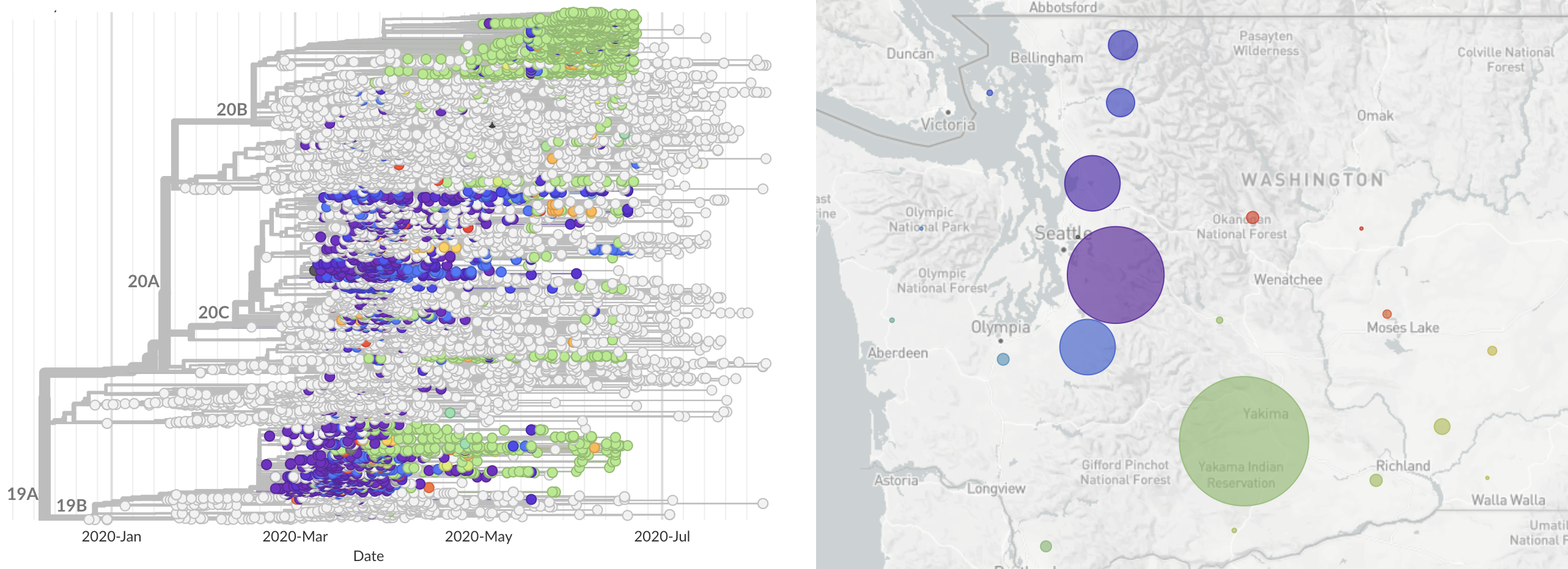
State-level transmission patterns, moving towards more detailed epi metadata
Closing thoughts
- Pandemic warning systems need to be constructed on top of burden of endemic / seasonal pathogens
- Addressing need of endemic / seasonal diseases provides sample flows for detection of novel pathogens
- I don't think we need to be sequencing every acute respiratory infection, but we should be sequencing clusters and/or infections of unknown etiology
- Will require global surveillance / reporting to achieve
Acknowledgements
Seasonal flu: WHO Global Influenza Surveillance Network, GISAID, John Huddleston, Richard Neher, Jover Lee, Barney Potter, Dave Wentworth, Becky Garten
Ebola in DRC: James Hadfield, Allison Black, Eddy Kinganda Lusamaki, Placide Mbala-Kingebeni, Catherine Pratt, Mike Wiley, Jean-Jacques Muyembe Tamfum, Steve Ahuka-Mundeke, Daniel Mukadi, Gustavo Palacios, Amadou Sall, Ousmane Faye, Eric Delaporte, Martine Peeters, David Blazes, Cecile Viboud, David Spiro
SARS-CoV-2 genomic epi: Data producers from all over the world, GISAID and the Nextstrain team
Seattle Flu Study: Helen Chu, Michael Boeckh, Janet Englund, Michael Famulare, Barry Lutz, Deborah Nickerson, Mark Rieder, Lea Starita, Matthew Thompson, Jay Shendure, Amanda Adler, Jeris Bosua, Elisabeth Brandstetter, Kairsten Fay, Chris Frazar, Peter Han, Reena Gulati, James Hadfield, ShiChu Huang, Misja Ilcisin, Michael Jackson, Anahita Kiavand, Louise Kimball, Enos Kline, Kirsten Lacombe, Jover Lee, Jennifer Logue, Victoria Lyon, Kira Newman, Miguel Paredes, Thomas Sibley, Monica Zigman Suchsland, Cassia Wagner, Caitlin Wolf
Bedford Lab:
![]() Alli Black,
Alli Black,
![]() John Huddleston,
John Huddleston,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Louise Moncla,
Louise Moncla,
![]() Maya Lewinsohn,
Maya Lewinsohn,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Kairsten Fay,
Kairsten Fay,
![]() Misja Ilcisin,
Misja Ilcisin,
![]() Cassia Wagner,
Cassia Wagner,
![]() Miguel Paredes,
Miguel Paredes,
![]() Nicola Müller,
Nicola Müller,
![]() Marlin Figgins,
Marlin Figgins,
![]() Eli Harkins
Eli Harkins






