Genomic insights during initial viral emergence and continued surveillance of viral evolution
Trevor Bedford (@trvrb)
Fred Hutchinson Cancer Center / Howard Hughes Medical Institute
23 Jun 2022
VIDD Faculty Retreat
Fred Hutch
Slides at: bedford.io/talks
1. Initial viral emergence (SARS-CoV-2 and MPXV)
2. Continuing viral evolution (SARS-CoV-2)
SARS-CoV-2 emergence
Jan 11: First five genomes from Wuhan showed a novel SARS-like coronavirus
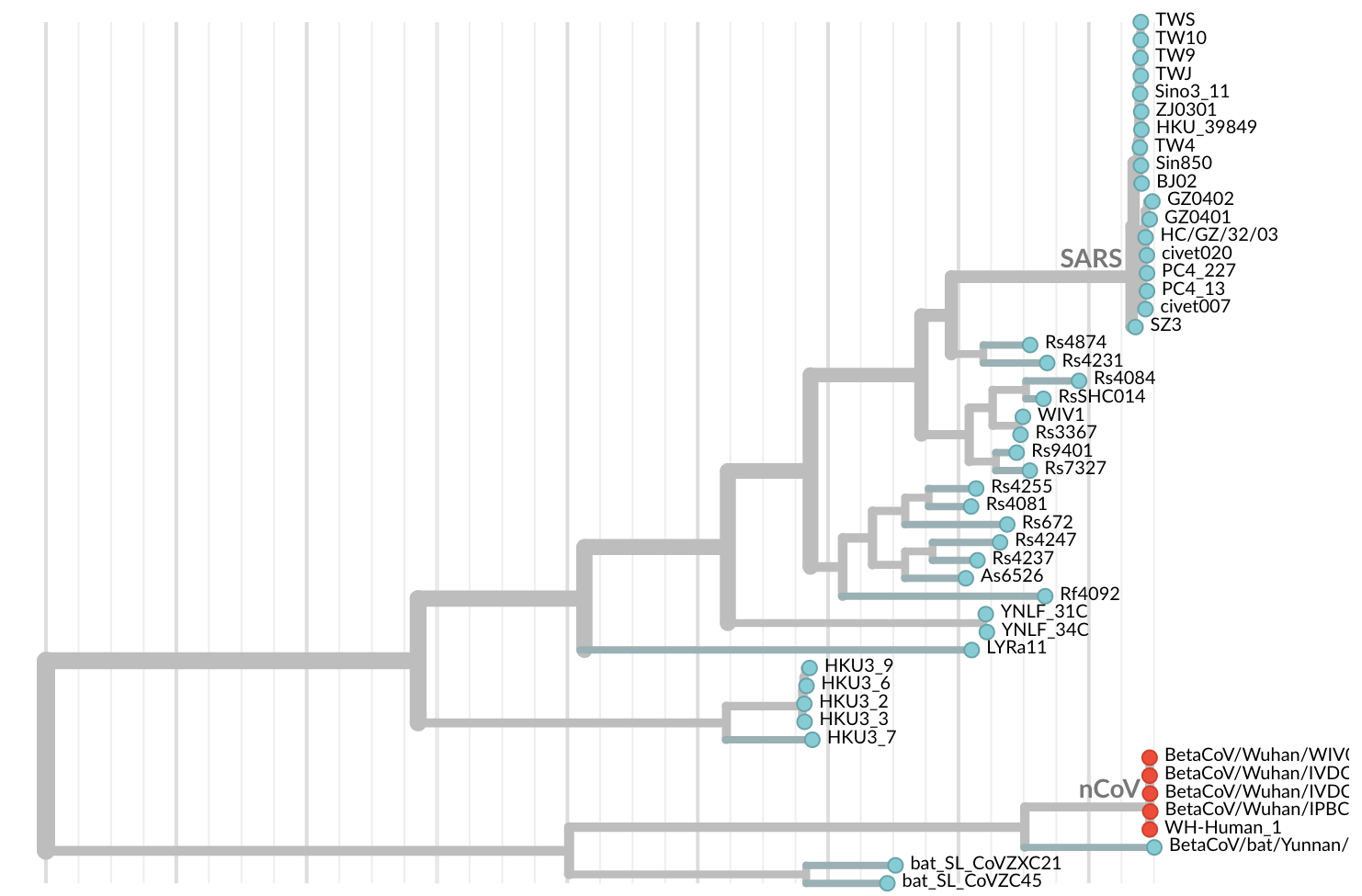
Jan 19: First 12 genomes from Wuhan and Bangkok showed lack of genetic diversity
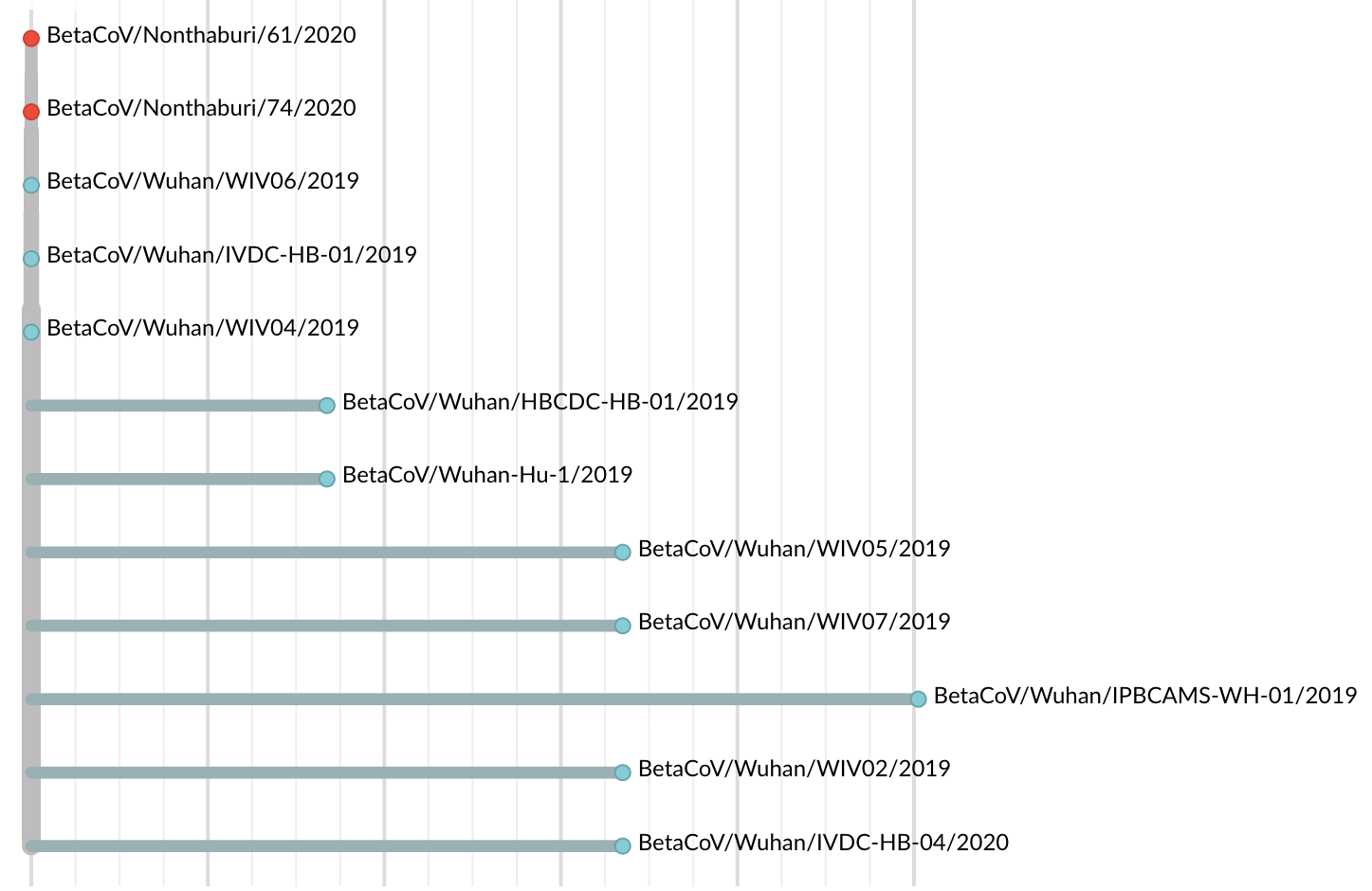
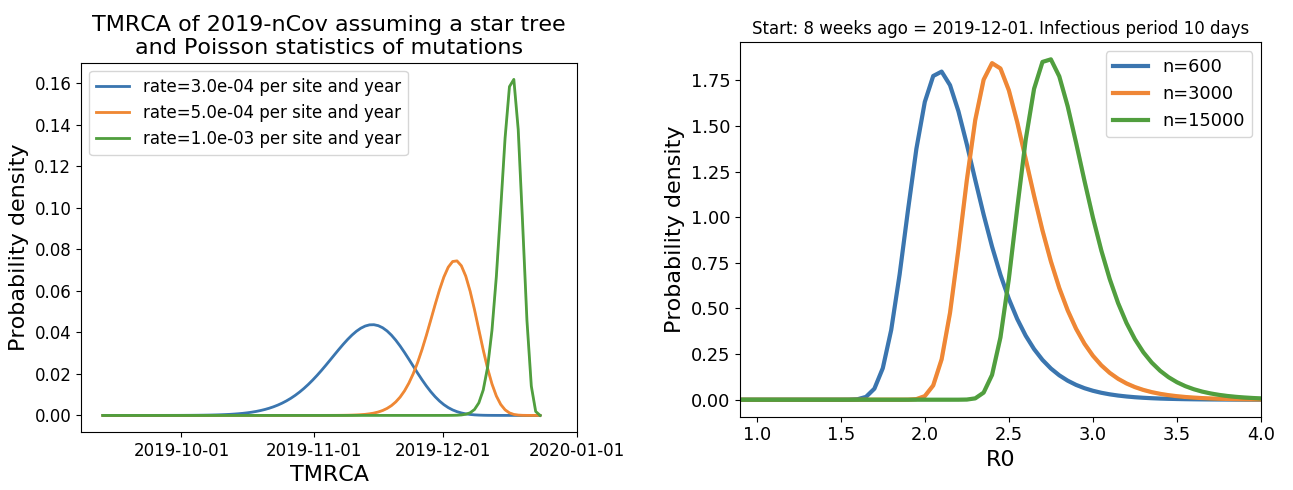
Single introduction into the human population between Nov 15 and Dec 15 and subsequent rapid human-to-human spread
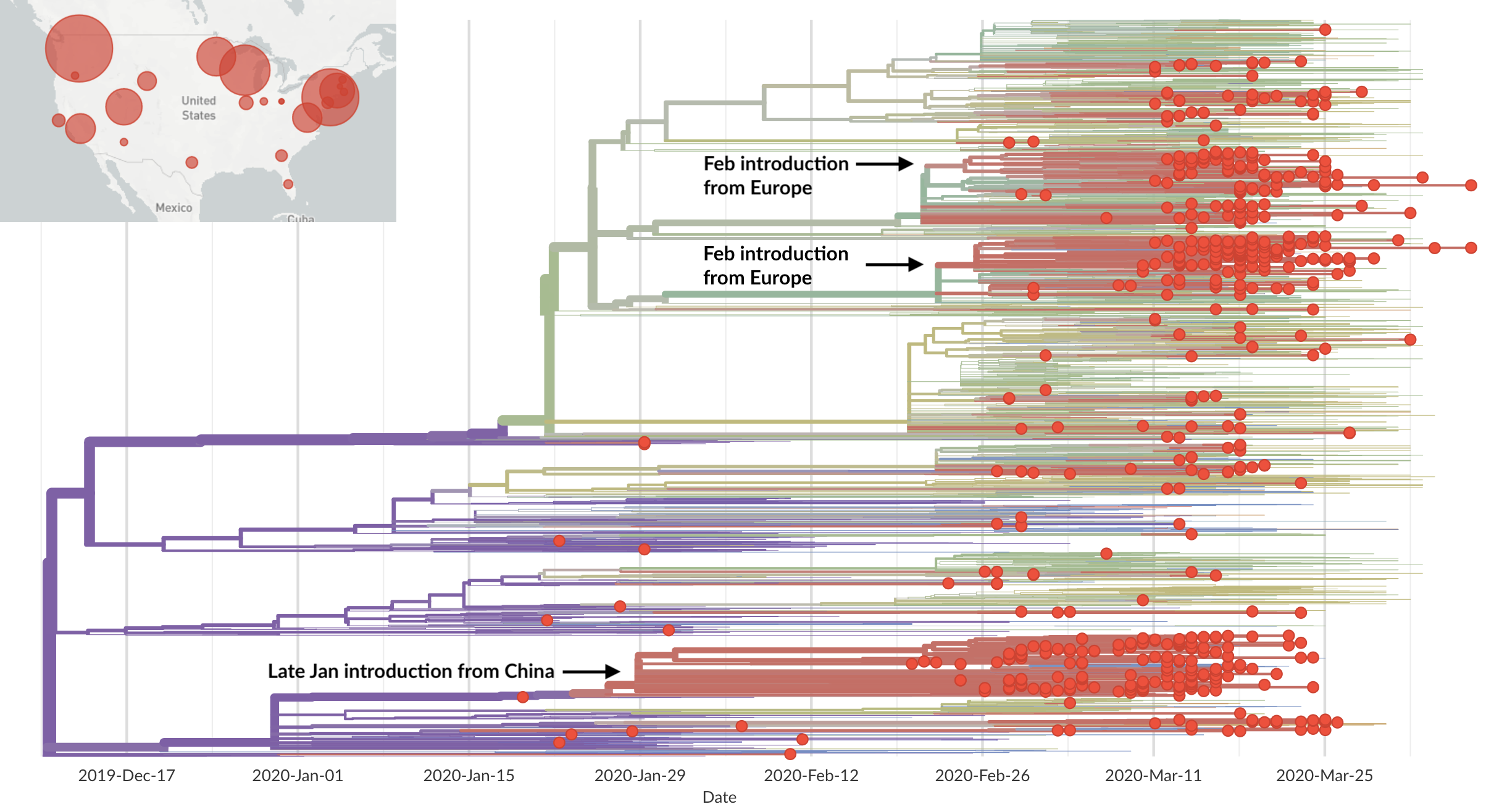
Rapid global epidemic spread from China
Epidemic in the USA was introduced from China in late Jan and from Europe during Feb
Seattle Flu Study detects local circulation and charts early epidemic in Washington State
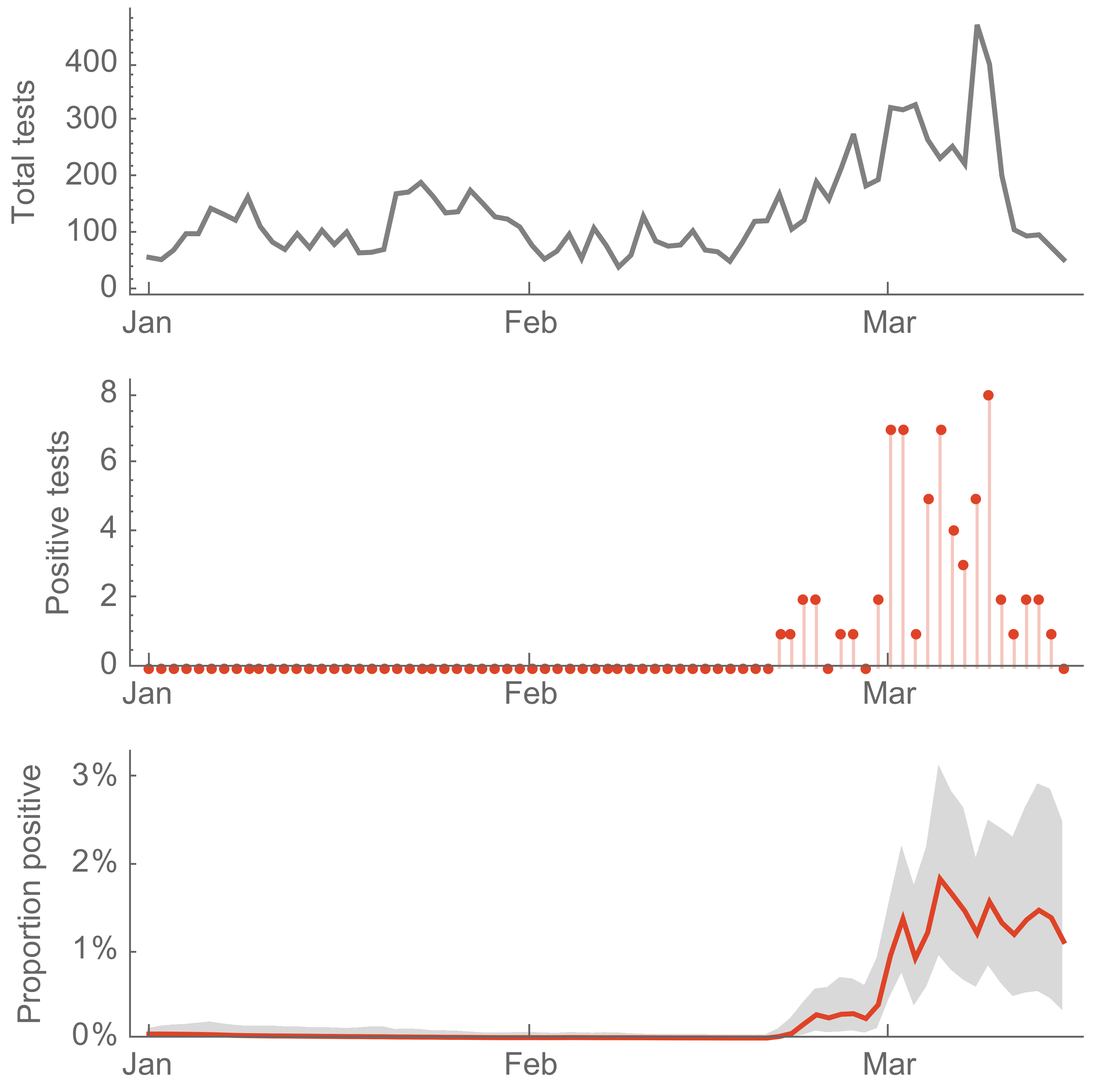
Direct introduction from China ~Feb 1 responsible for the majority of the epidemic
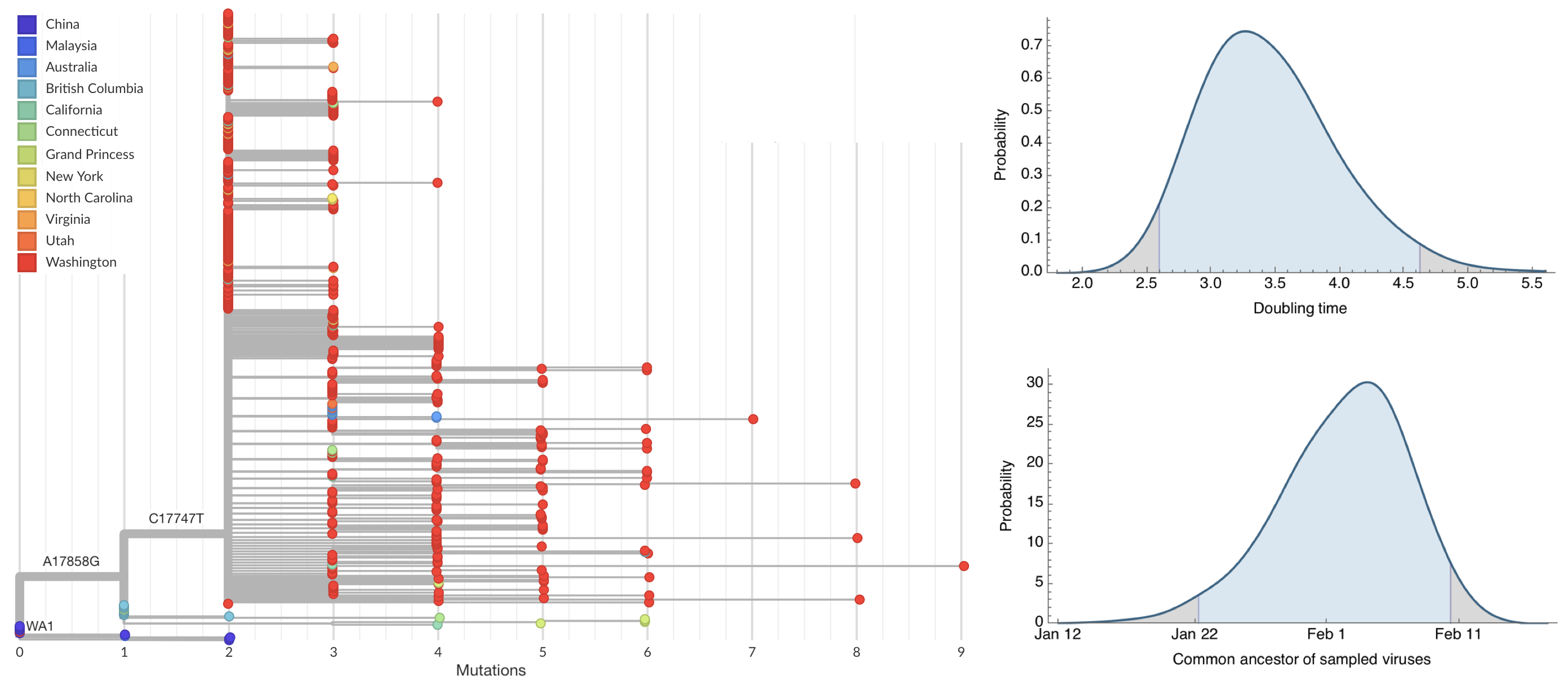
MPXV emergence
Monkeypox virus genome
Generally expect DNA viruses to have much slower evolutionary rate of roughly 1 mutation per year compared to RNA viruses like SARS-CoV-2 which mutate once every 2 weeks
Monkeypox viral diversity and typical epidemiology
Previously we observed stuttering transmission chains in humans after spillover from reservoir, similar to viruses like MERS-CoV and Nipa

Outbreak viruses are closely related as descendants of clade 3
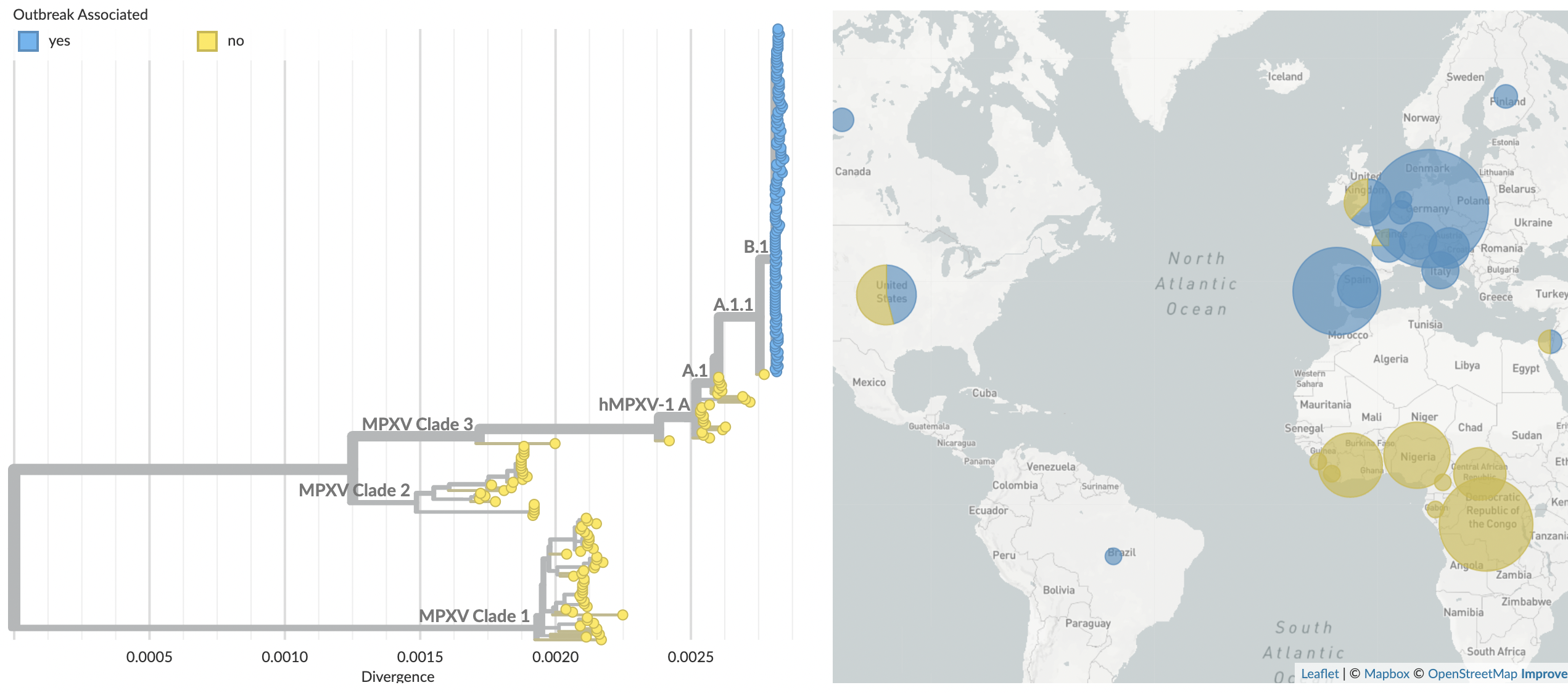
Outbreak viruses differ by 47 SNPs compared to 2018
Observe ~1 mutation per month instead of the DNA virus expectation of 1 mutation per year

Unusual mutation spectrum consistent with APOBEC editing

Mutations consistent with APOBEC editing begin in 2018 and continue through current outbreak

Conclude human-to-human transmission has been occuring since late 2017
SARS-CoV-2 continuing evolution
Global SARS-CoV-2 evolution over the course of the pandemic

Rapid accumulation of spike mutations driving displacement

S1 evolution remarkably fast relative to seasonal influenza

Initial variants characterized by repeated evolution at spike 484K and 501Y
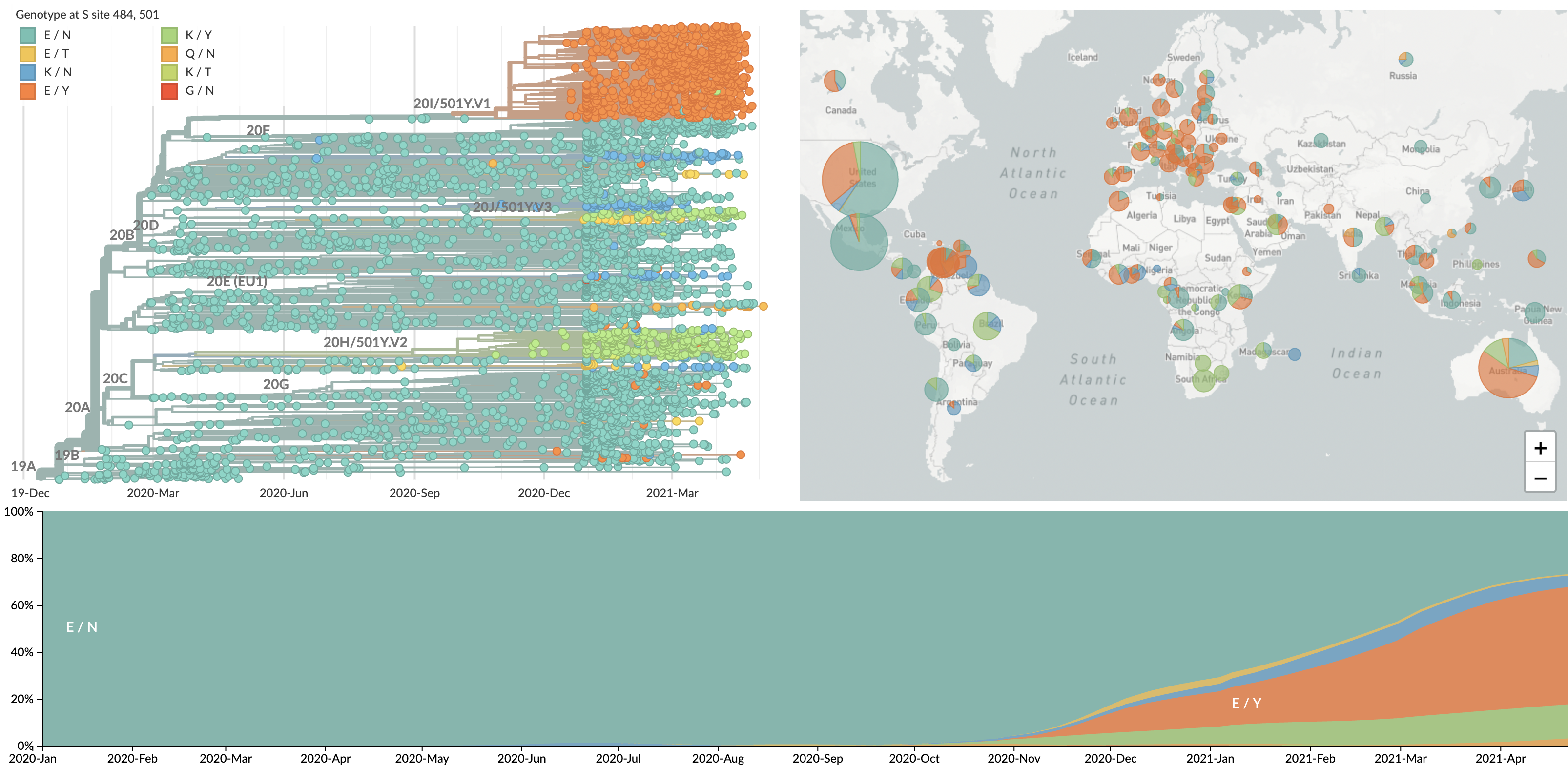
We're now seeing repeated evolution at spike 452

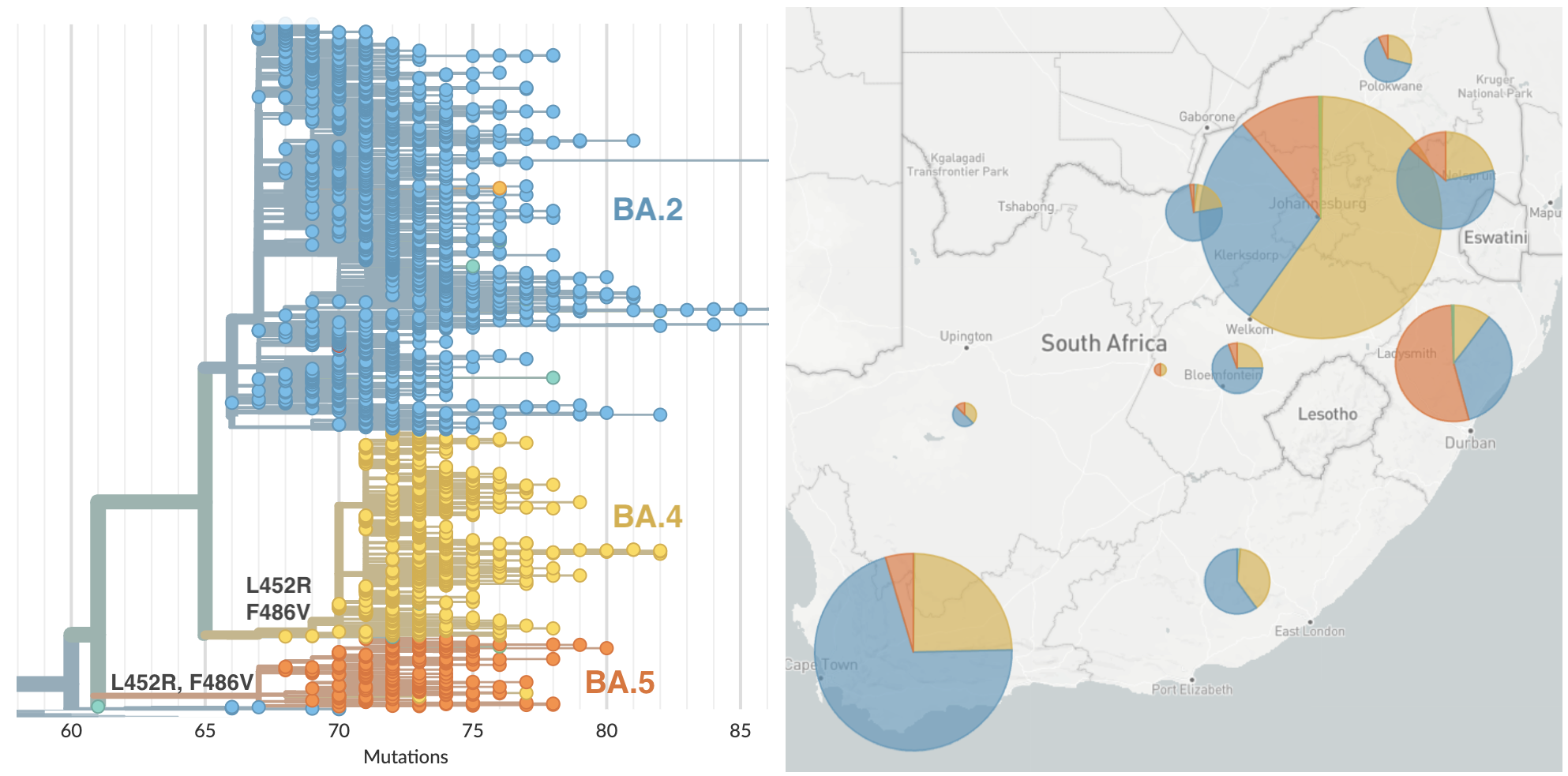
BA.4 and BA.5 emerging from South Africa with spike mutations 452R and 486V
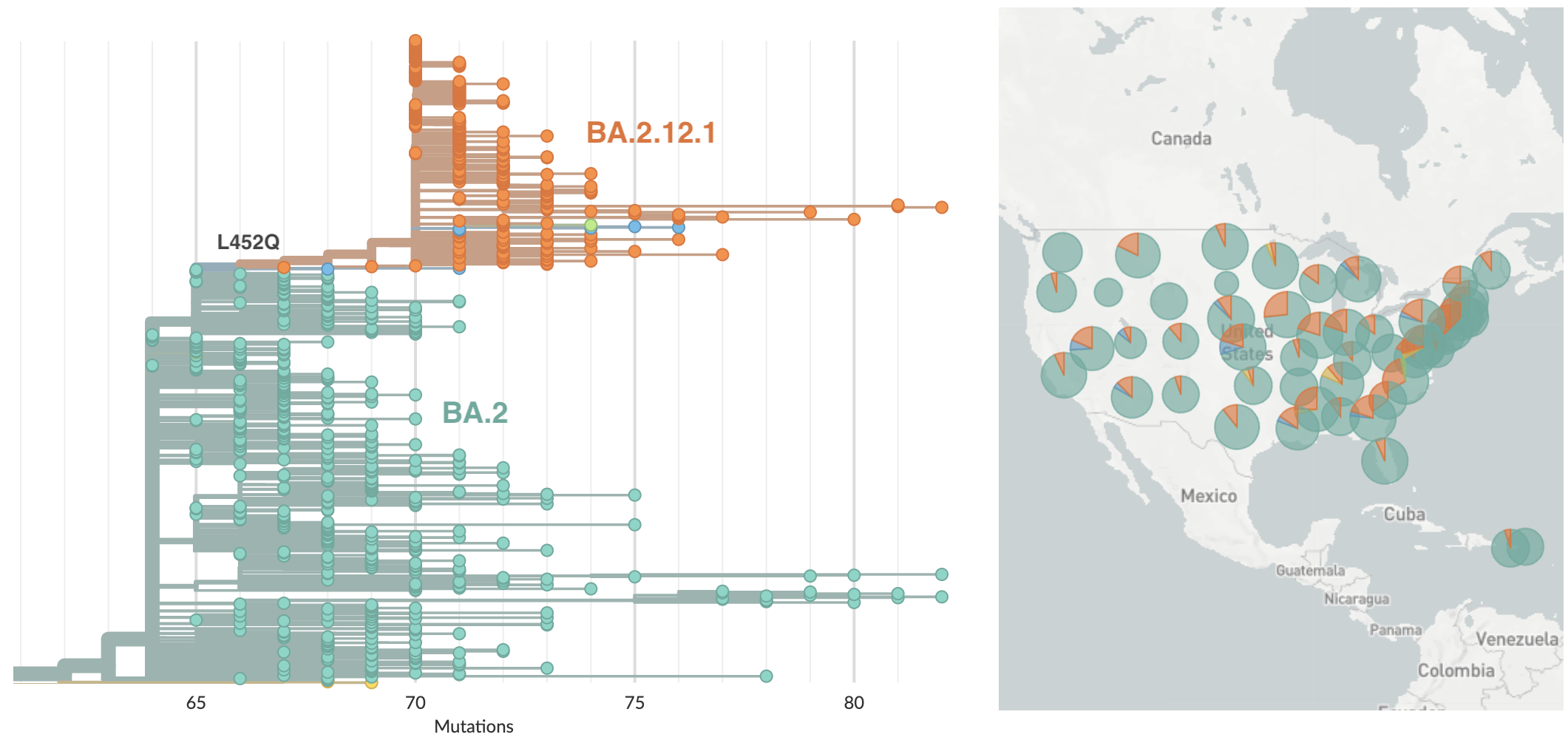
BA.2.12.1 emerging from Northeastern US bearing spike mutation 452Q
Variant-specific epidemics shown by partitioning case counts with variant frequency

BA.4, BA.5, BA.2.12.1 driving cases across countries

Rt for BA.2.12.1 greater than BA.2 and Rt for BA.4/5 greater still

More broadly, I expect continued genomic surveillance to respond to ongoing evolution of SARS-CoV-2, but that capacity generated for pandemic response will pivot to emerging and endemic pathogens according to threat / burden
Acknowledgements
SARS-CoV-2 and MPXV genomic epi: Data producers from all over the world, GISAID and the Nextstrain team
Bedford Lab:
![]() John Huddleston,
John Huddleston,
![]() James Hadfield,
James Hadfield,
![]() Katie Kistler,
Katie Kistler,
![]() Louise Moncla,
Louise Moncla,
![]() Maya Lewinsohn,
Maya Lewinsohn,
![]() Thomas Sibley,
Thomas Sibley,
![]() Jover Lee,
Jover Lee,
![]() Cassia Wagner,
Cassia Wagner,
![]() Miguel Paredes,
Miguel Paredes,
![]() Nicola Müller,
Nicola Müller,
![]() Marlin Figgins,
Marlin Figgins,
![]() Denisse Sequeira,
Denisse Sequeira,
![]() Victor Lin,
Victor Lin,
![]() Jennifer Chang
Jennifer Chang






