Global circulation and evolution of influenza virus
Trevor Bedford (@trvrb)
5 Mar 2018
NIHE/OUCRU Workshop on Influenza Epidemiology and Evolution in Vietnam
Population turnover (in H3N2) is extremely rapid
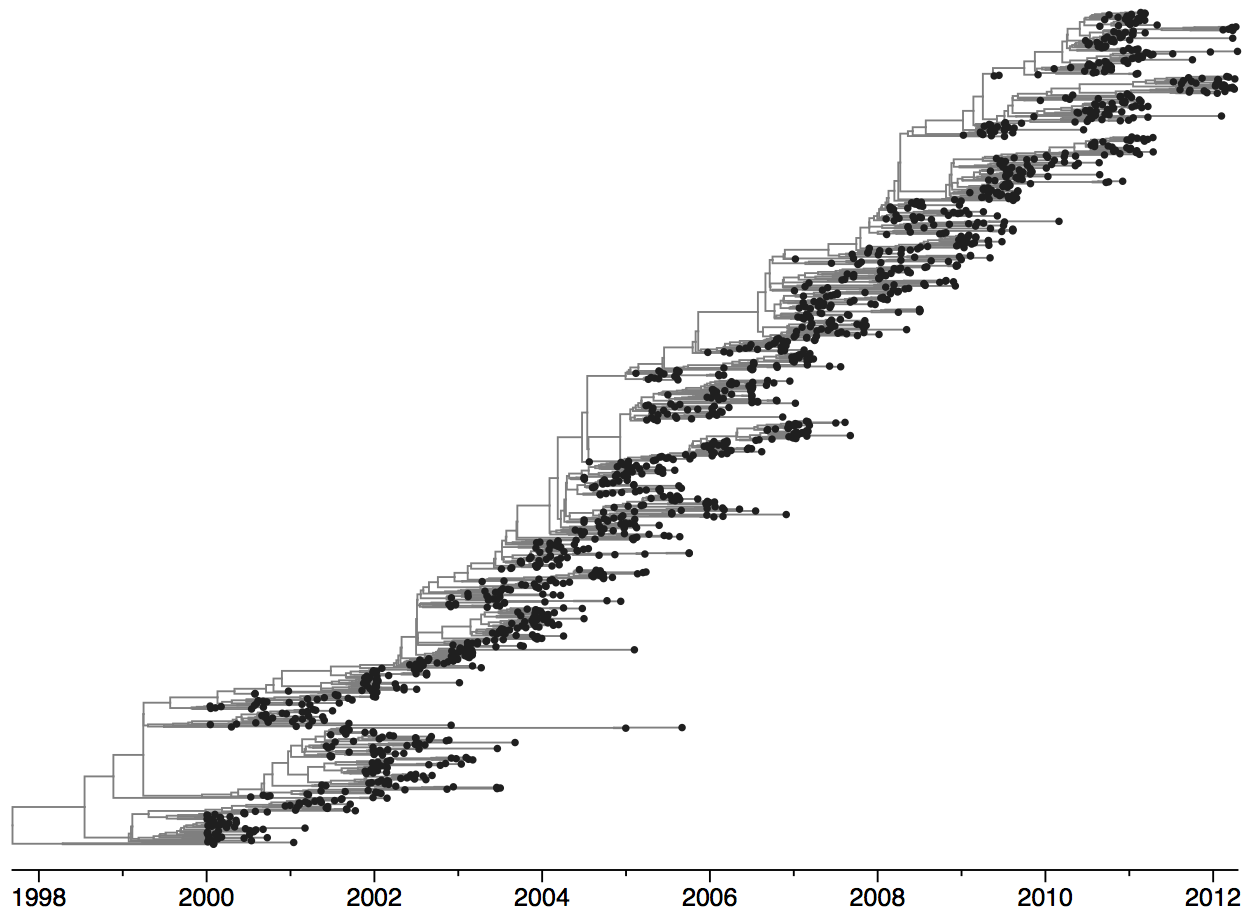
Clades emerge, die out and take over
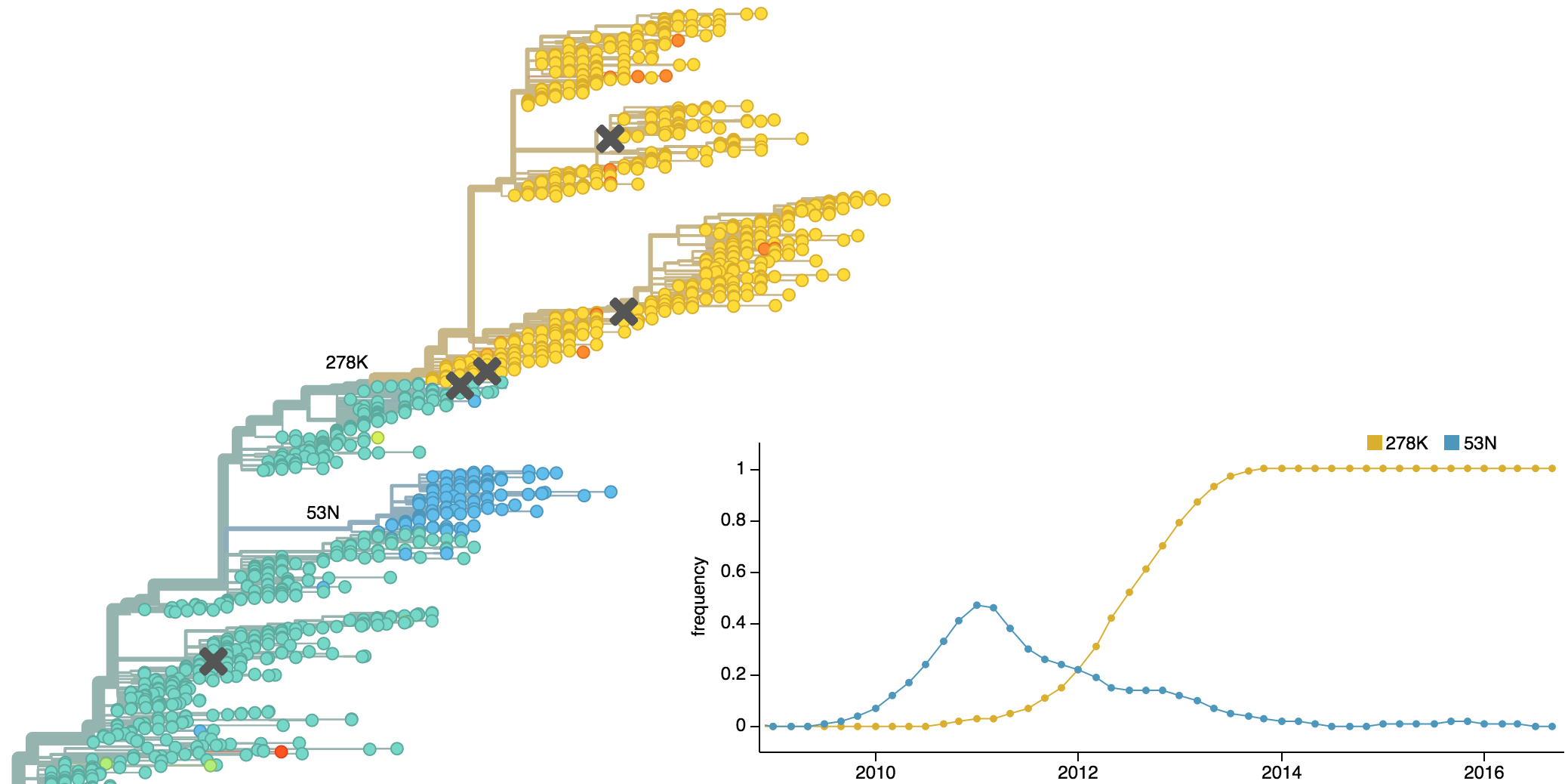
Clades show rapid turnover

Dynamics driven by antigenic drift
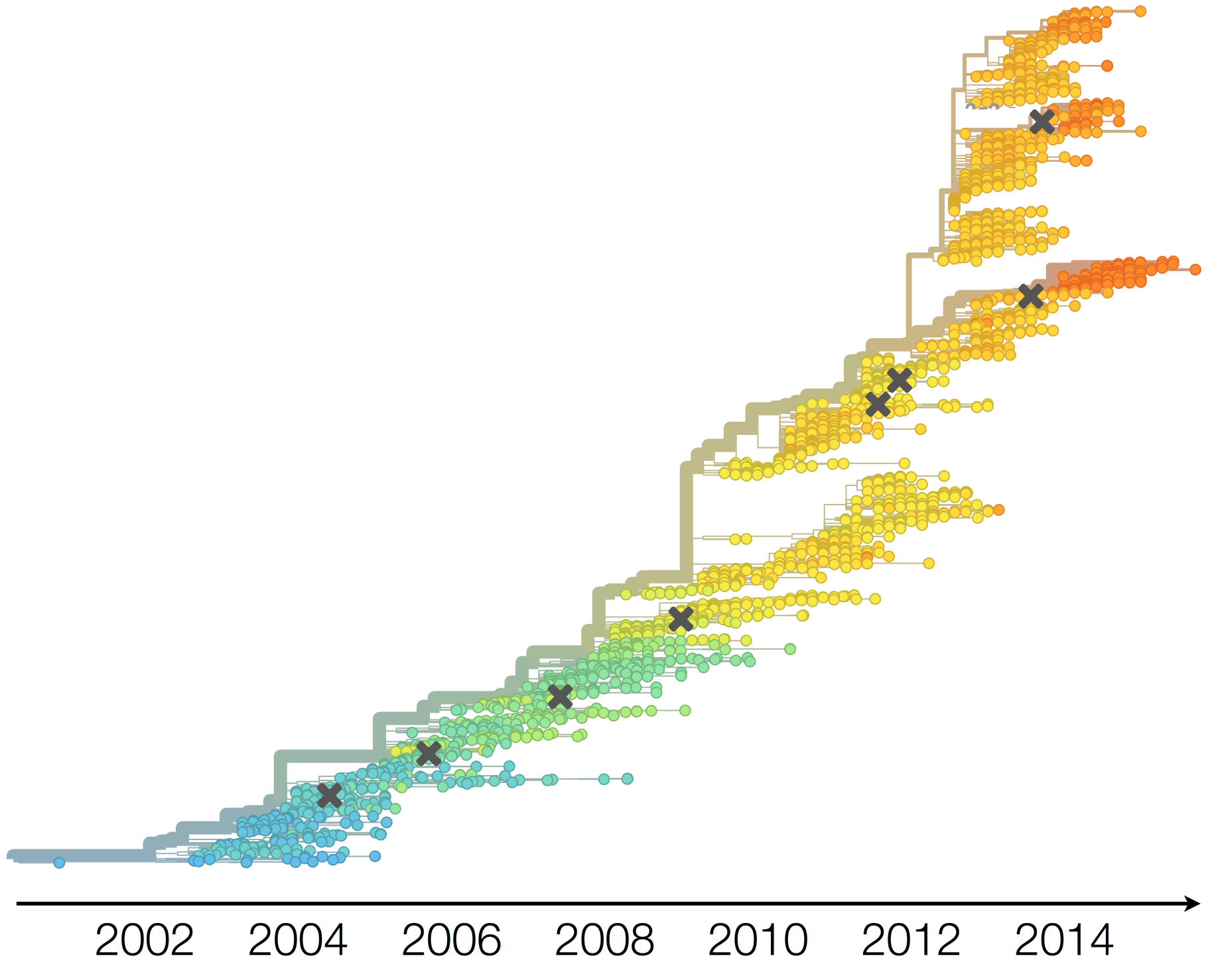
Drift necessitates vaccine updates
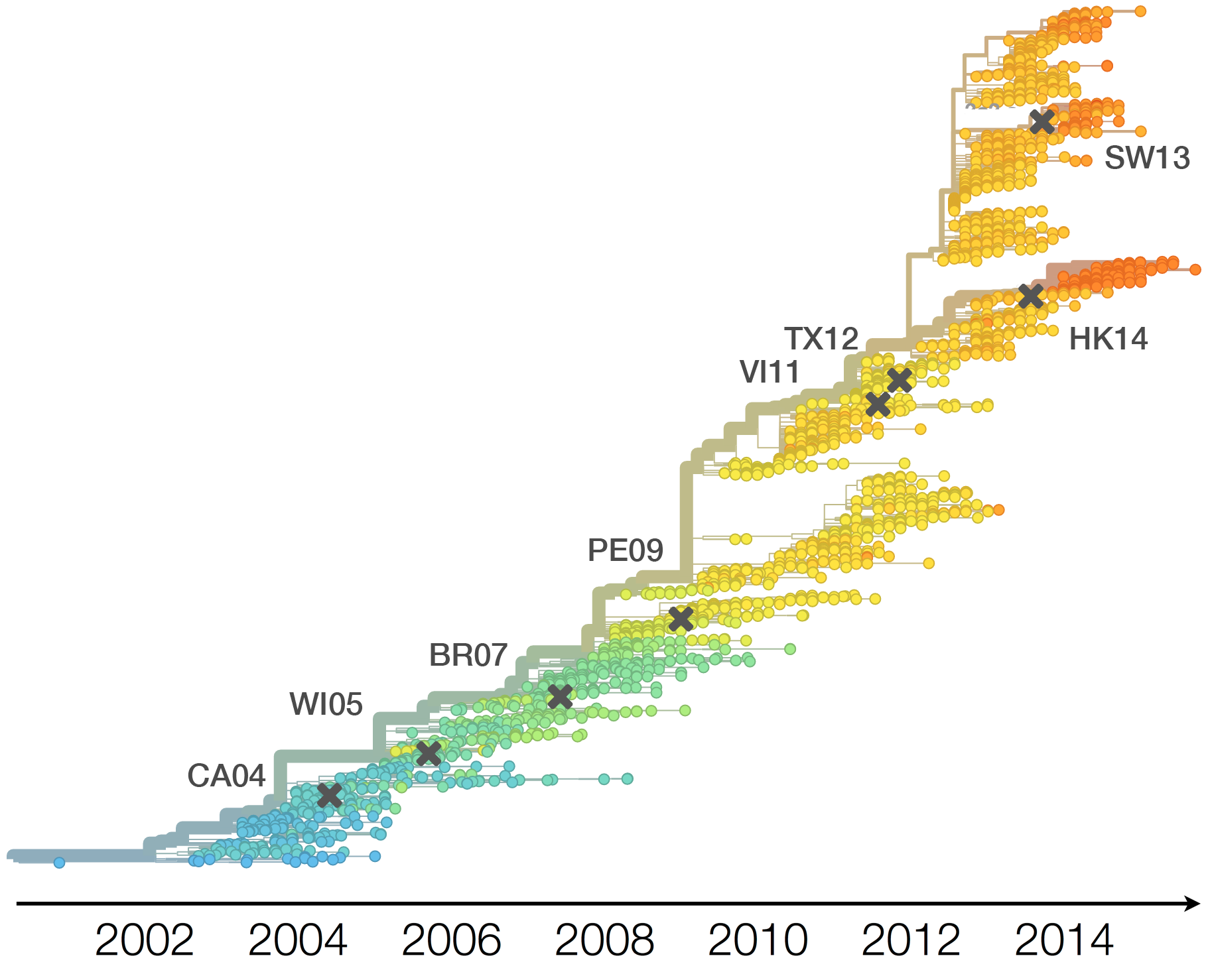
H3N2 vaccine updates occur every ~2 years
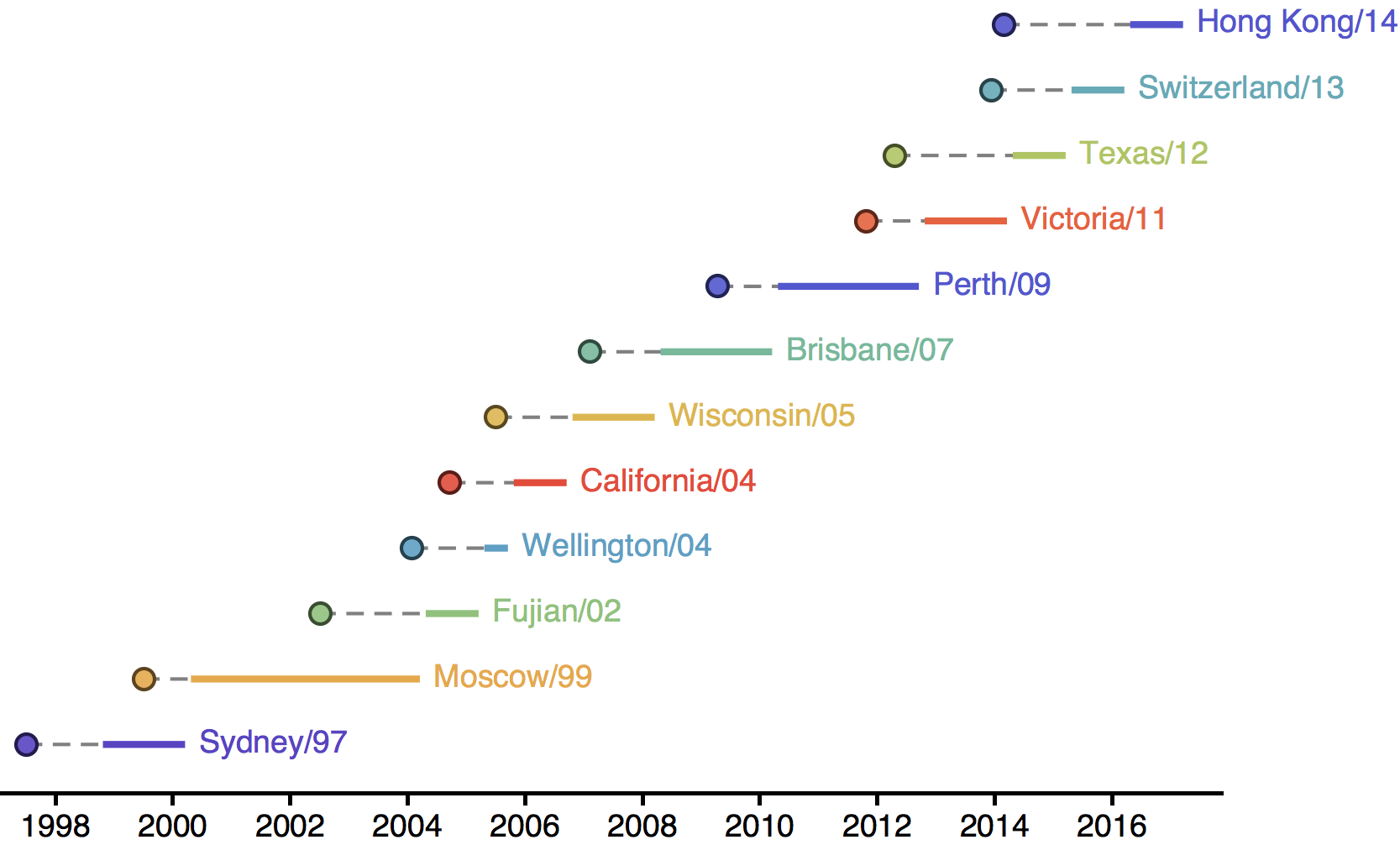
Timely surveillance and rapid analysis essential to vaccine strain selection
nextflu
Project to provide a real-time view of the evolving influenza population
nextflu pipeline
- Download all recent HA sequences from GISAID
- Filter to remove outliers
- Subsample across time and space
- Align sequences
- Build tree
- Estimate clade frequencies
- Infer antigenic phenotypes
- Export for visualization
Up-to-date analysis publicly available at:
nextflu.org
Competing clades central to influenza evolutionary dynamics
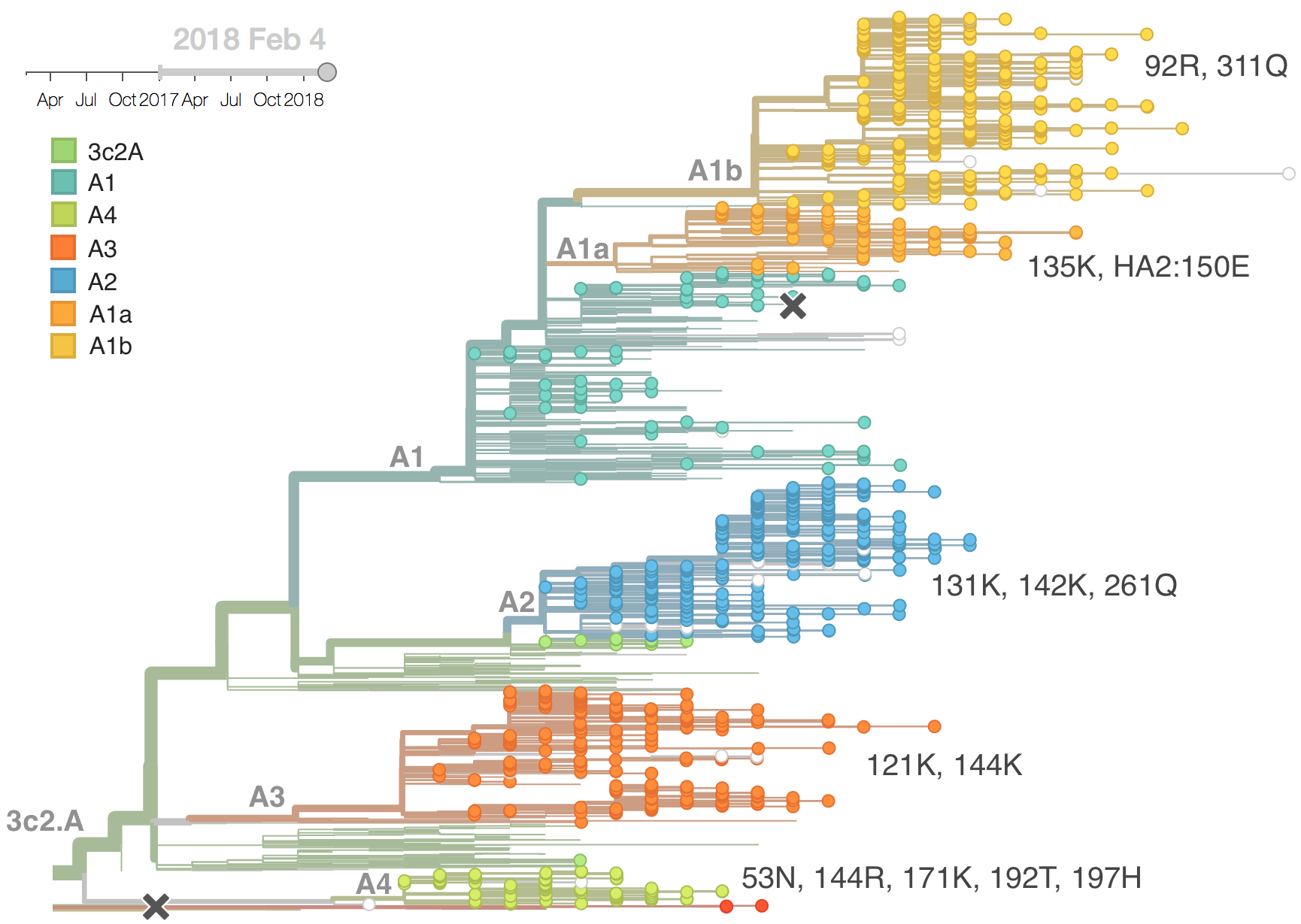
Clades grow and decline through time

Fitness models can project clade frequencies
Clade frequencies $X$ derive from the fitnesses $f$ and frequencies $x$ of constituent viruses, such that
$$\hat{X}_v(t+\Delta t) = \sum_{i:v} x_i(t) \, \mathrm{exp}(f_i \, \Delta t)$$
This captures clonal interference between competing lineages
Construct a fitness model based on:
- Amino acid changes at epitope sites
- Antigenic novelty based on HI
- Rapid phylogenetic growth
Clade growth rate is well predicted (ρ = 0.66)
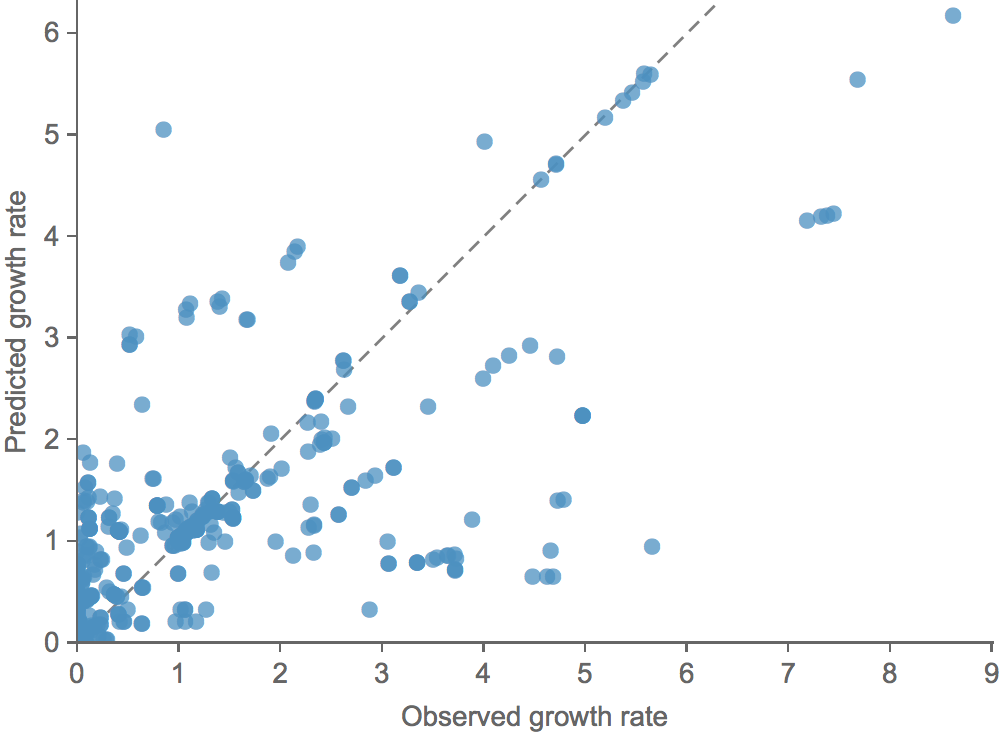
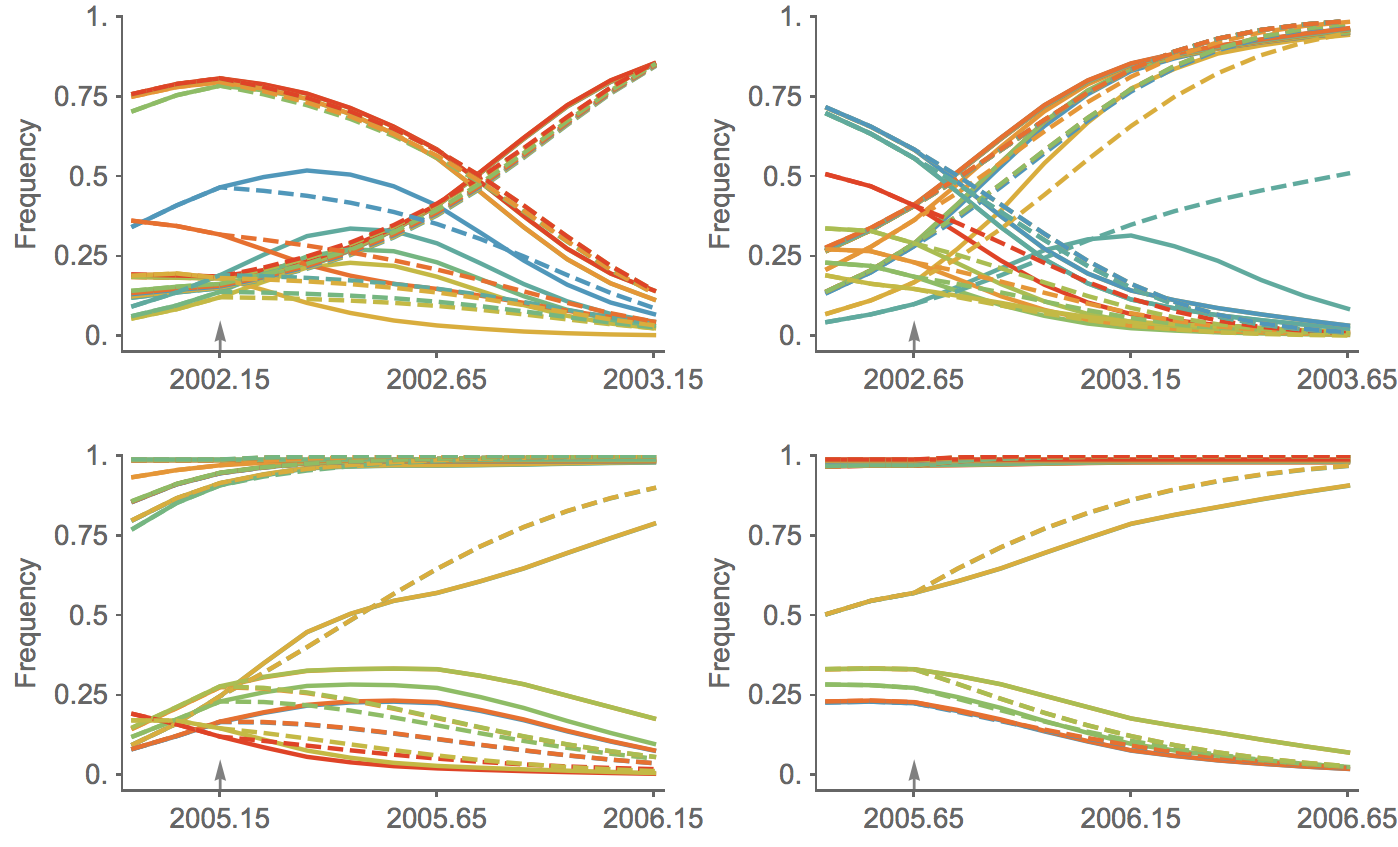
Trajectories show more detailed congruence
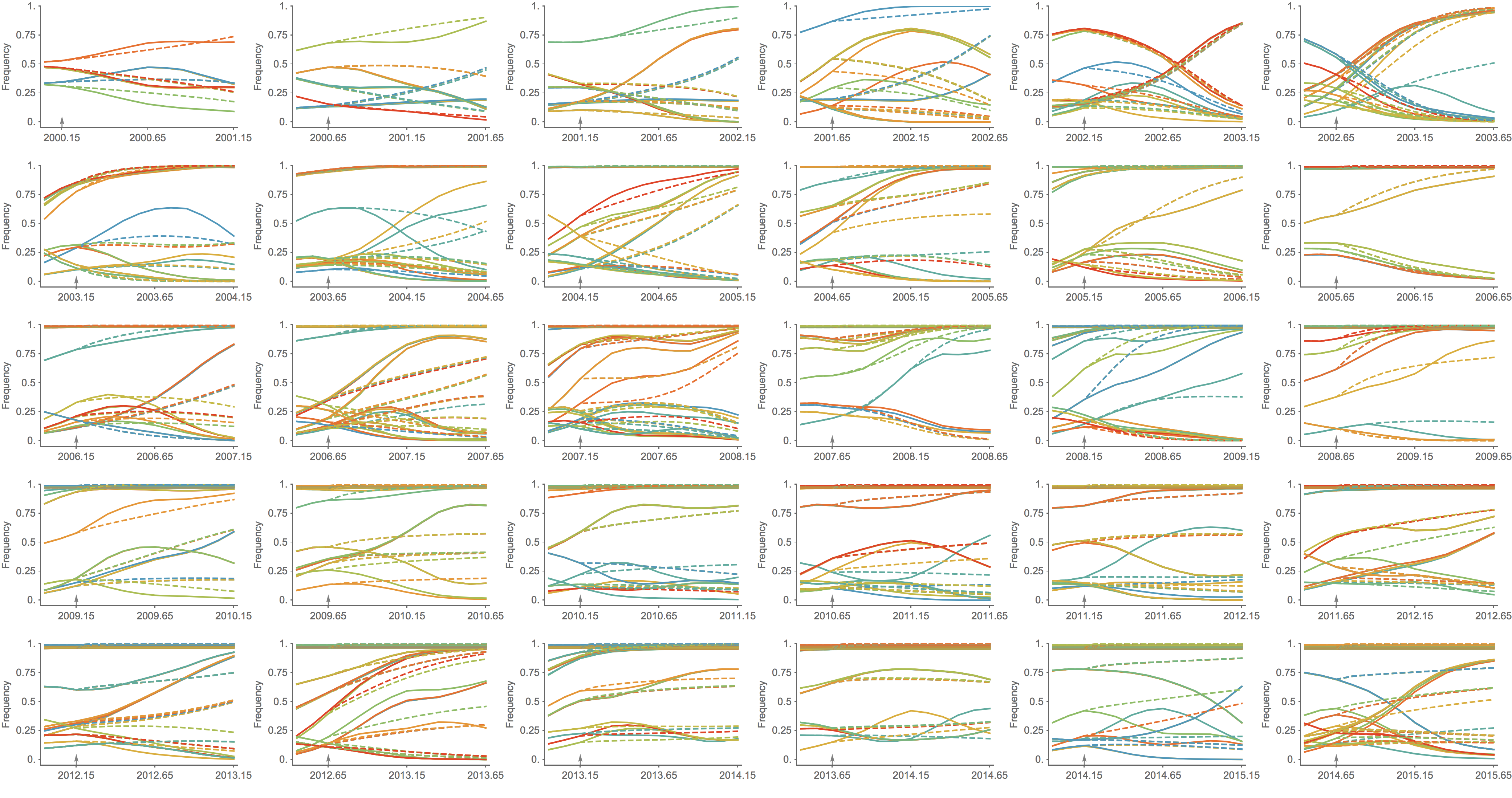
Trajectories show more detailed congruence
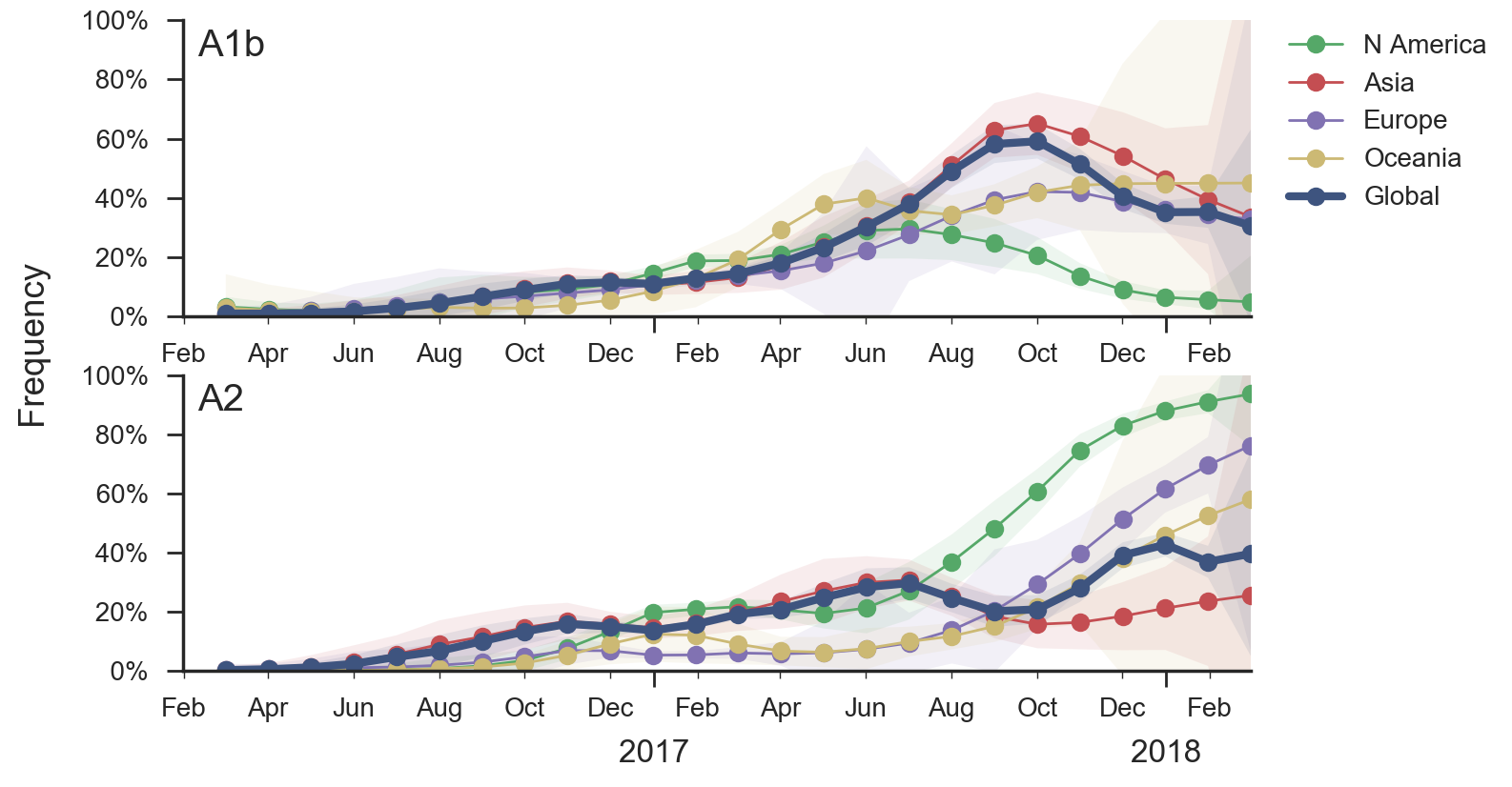
Geographic circulation drives evolutionary outcomes
Clades commonly show region-specific circulation patterns
Phylogeny of H3 with geographic history
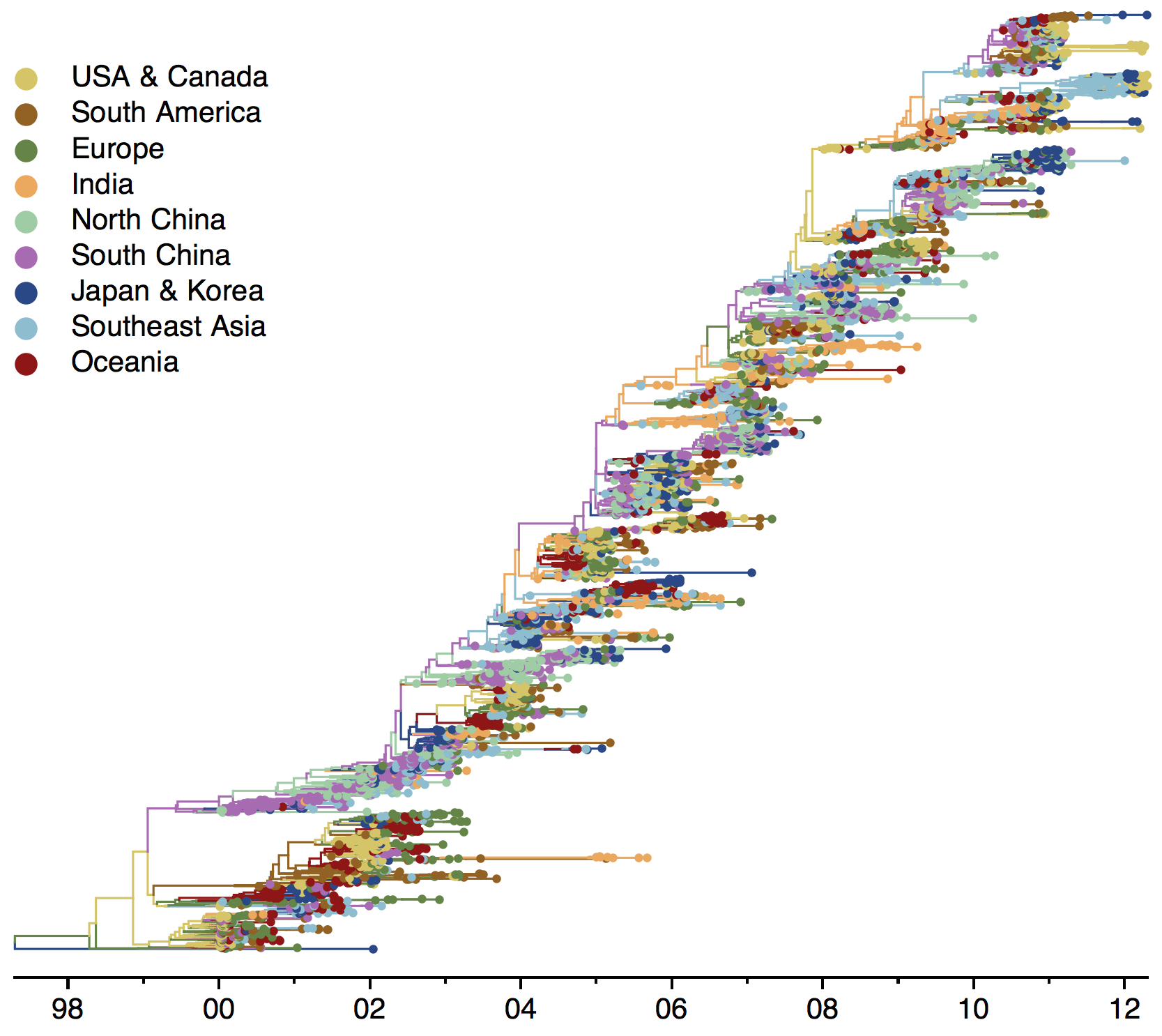
Infer geographic transition matrix
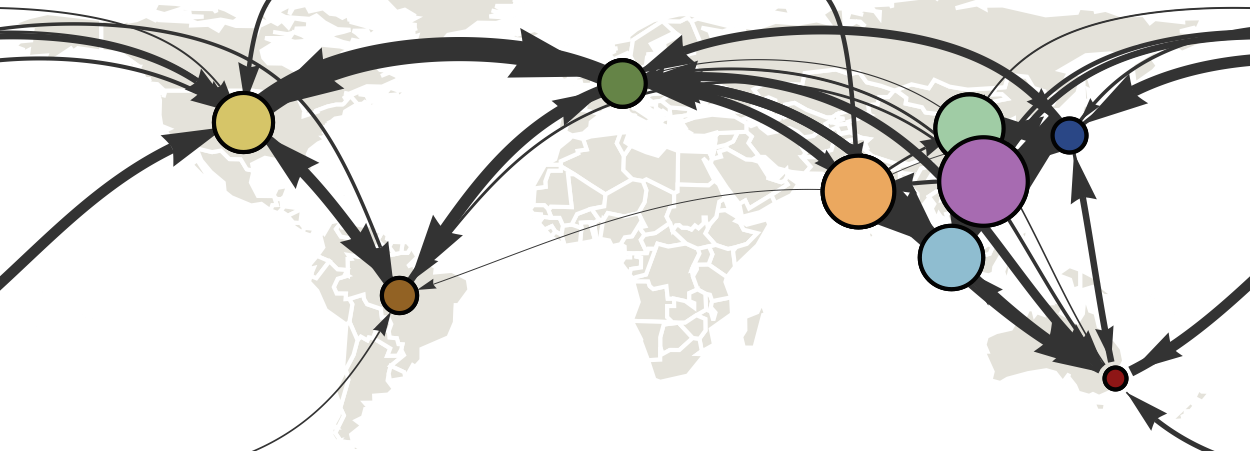
Geographic location of phylogeny trunk
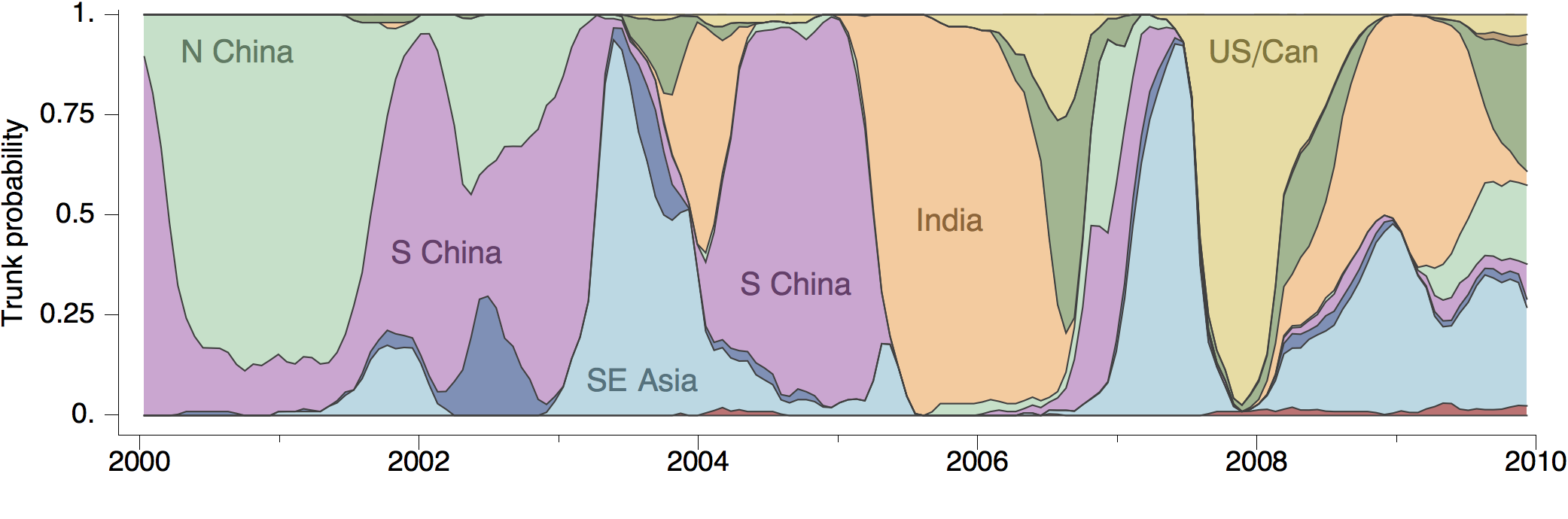
Region-specific ancestry
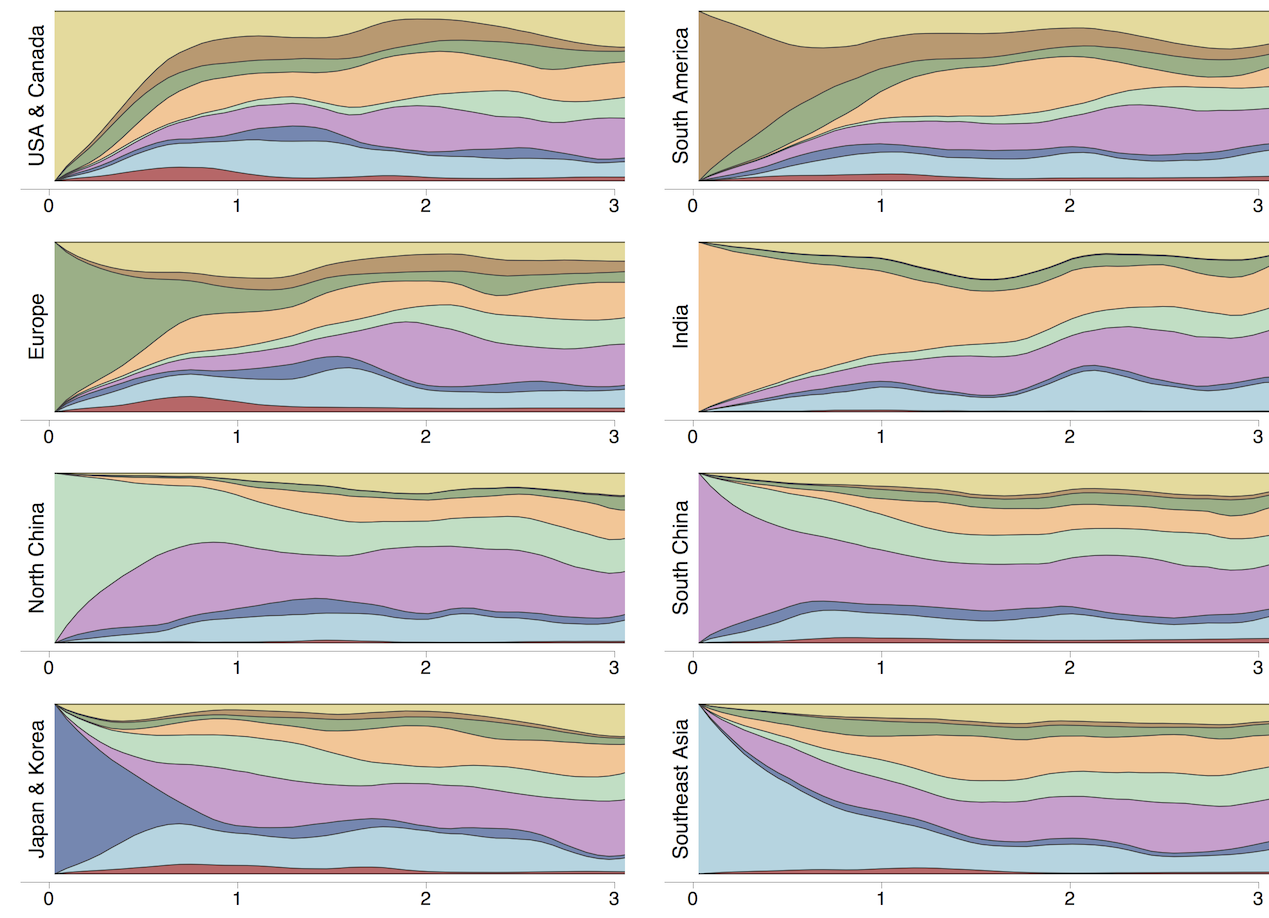
Which regions are best predictive of future outcomes?
Correlation between clade frequencies
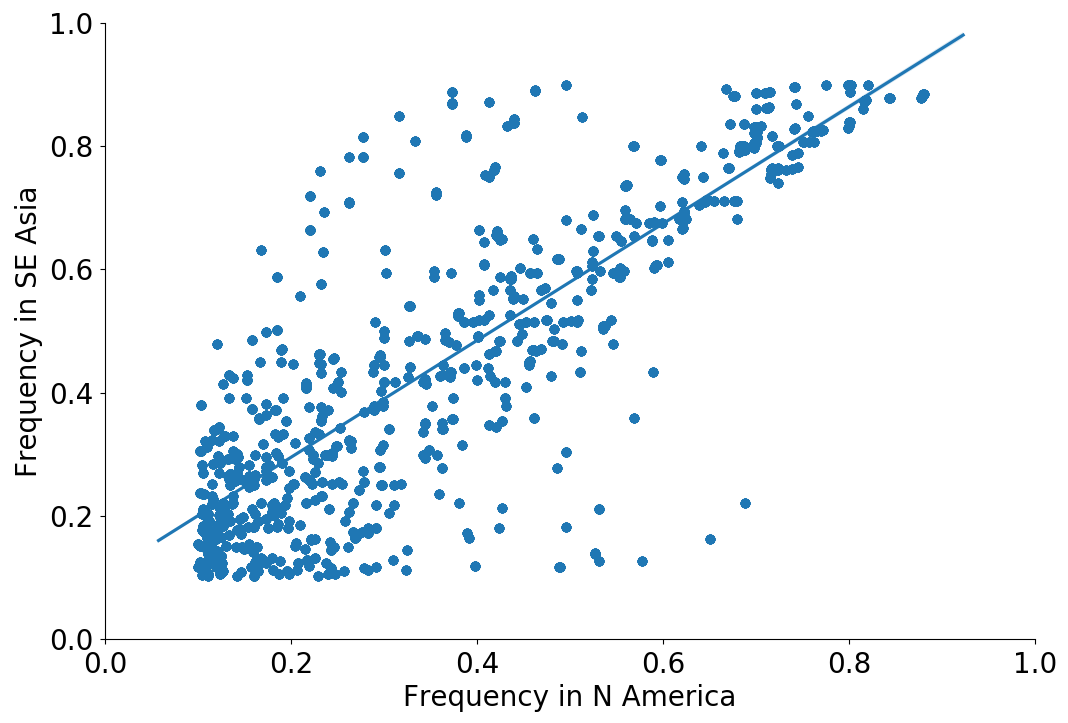
Correlations across regions
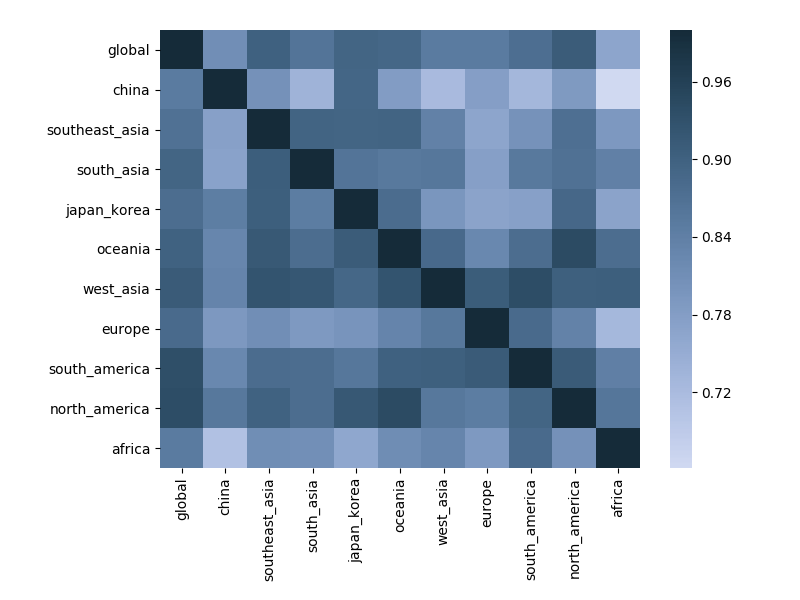
Correlations across regions with 6-month lag
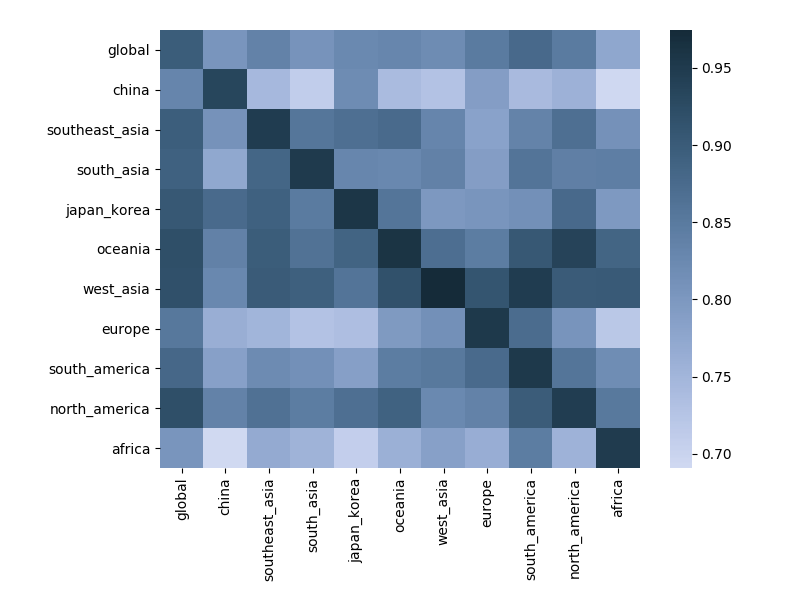
Correlations across regions with 12-month lag
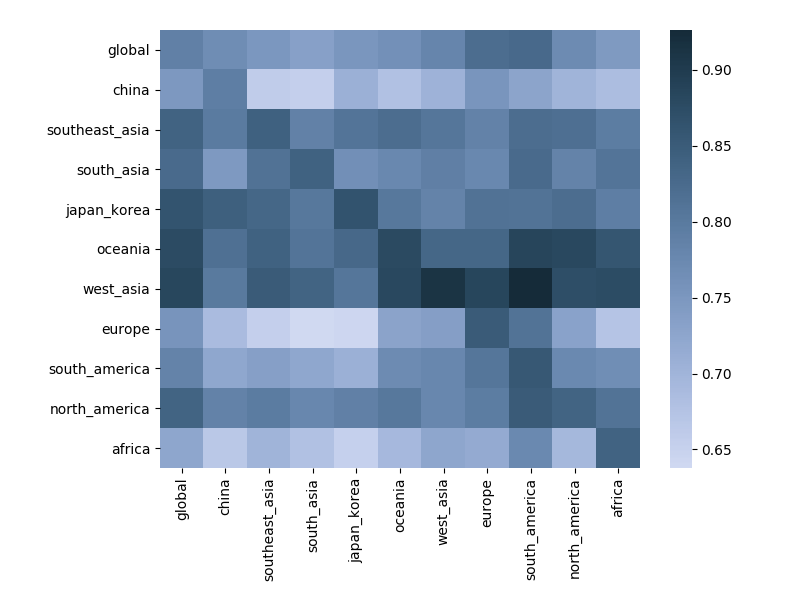
Goal is to include migration forcings in predictive fitness model
Acknowledgements
Bedford Lab:
![]() Alli Black,
Alli Black,
![]() Sidney Bell,
Sidney Bell,
![]() Gytis Dudas,
Gytis Dudas,
![]() John Huddleston,
John Huddleston,
![]() Barney Potter,
Barney Potter,
![]() James Hadfield,
James Hadfield,
![]() Louise Moncla
Louise Moncla
Influenza: WHO Global Influenza Surveillance Network, GISAID, Richard Neher, Colin Russell, Andrew Rambaut, Gytis Dudas, Dave Wentworth, Becky Garten, Marta Łuksza, Michael Lässig
Nextflu/Nextstrain: Richard Neher, James Hadfield, Colin Megill, Sidney Bell, Barney Potter, John Huddleston, Charlton Callender, Emma Hodcroft






